Abstract
Inflammation-induced cholestasis (IIC) is a frequently occurring phenomenon. A central role in its pathogenesis is played by nuclear receptors (NRs). These ligand-activated transcription factors not only regulate basal expression of hepatobiliary transport systems, but also mediate adaptive responses and possess anti-inflammatory characteristics. The latter two functions may be exploited in the search for new treatments for IIC and likely for cholestasis in general as well. Current knowledge of the pathogenesis of IIC and the dual role NRs in this process are reviewed. Special interest is given to the use of NRs as potential targets for intervention.
2. INTRODUCTION
Inflammation-induced cholestasis (IIC) is a frequently occurring, well-recognized clinical entity. Molecular mechanisms underlying IIC have been partially unravelled over the past decades, facilitated by the increasing knowledge of mechanisms of bile formation and regulation of the transport systems involved, identification of nuclear factors controlling transporter gene expression and a broader understanding of molecular aspects of the inflammatory response. It is now evident that several nuclear receptors (NRs), i.e., ligand-activated transcription factors, play key roles in the regulation of bile formation and the pathogenesis of IIC. These receptors are not only important under physiological conditions but their involvement expands to pathophysiological situations, both as mediators, i.e., in a disease-promoting role, and as important modulators of adaptive responses. Recently, NRs have also been proposed as targets for intervention in IIC. This review focuses on the various roles of NRs in processes that lead to cholestasis during inflammation and on the ways in which NRs can be exploited for design of treatment options.
2.1. Clinical aspects of inflammation-induced cholestasis (IIC)
The link between inflammation and cholestasis has been recognized for centuries, with jaundice as the primary symptom of impaired bile formation (Byers (1), Bernstein (2), Hamilton (3), Crawford (4)). The underlying mechanisms of this association, however, have gone unexplained for a long time (Lester (5)).
Cholestasis associated with sepsis is generally regarded as the prototypical example of IIC, but bile formation is also affected in other conditions associated with an inflammatory state and cholestasis may thus be considered as a consequence of the so-called acute phase response (APR). The APR consists of a set of rapid, well-coordinated responses initiated by infection or tissue damage leading to the production of various soluble mediators (e.g., proteases, clotting factors, cytokines, etc.) aimed at restoration of homeostasis (Baumann (6), Moshage (7)). The APR also includes a broad suppression of many core intermediary metabolic functions within the liver – notably albumin synthesis and the metabolism/flux of carbohydrates, fats, and bile acids, the latter of which leads to cholestasis. Conditions besides sepsis that are associated with cholestasis include extrahepatic infections such as bacterial pneumonia and urinary tract infections (Birch (8), Bernstein (2), Byers (1), Eley (9), Hamilton (3), Garcia (10)), but this group can likely be expanded with conditions involving a systemic inflammatory response syndrome following burn injury, severe trauma and major surgery (Trauner (11)). The importance of circulating pro-inflammatory mediators in the pathogenesis of IIC was illustrated by the side-effects of therapeutical administration of these mediators to humans. Treatment of cancer patients with recombinant cytokines (TNFalpha or IL-2) in phase I/II clinical trials was shown to lead to hyperbilirubinemia and cholestasis (Sherman (12), Fisher (13)). Cholestasis seen in certain non-metastatic paraneoplastic syndromes, such as Stauffer’s syndrome, appear to be caused by secreted cytokines as well (Blay (14), Prommer (15)). Considering the plethora of conditions associated with IIC, it is not surprising that jaundice is frequently observed in intensive care units for children, most notably neonates, and adults. The importance of sepsis as an underlying cause of clinical cholestasis has often been overlooked (Whitehead (16)).
The presence and severity of cholestasis appears to be associated with poor prognosis of septicaemia (te Boekhorst (17)). This obviously does not imply that cholestasis itself is the causative factor of poor outcome: cholestasis is more likely an indicator of the severity of sepsis. Therefore, current treatment modalities are mainly aimed at treating sepsis with antibiotics and further supportive care and not at restoration of hepatic secretory function. Yet, it is easily appreciated that cholestasis per se will have immediate repercussions for the metabolism and elimination of drugs and toxins. Moreover, intestinal function will be impaired with reduced bile flow, with subsequent complications of malabsorption as well as bacterial overgrowth and translocation, further worsening the cholestatic state. The long-term effects of sepsis-associated cholestasis are largely unknown.
2.2. Experimental models of IIC
The pathogenesis of IIC has been studied using a variety of in vivo, ex vivo and in vitro models. These models generally involve the induction of a hepatic APR. A frequently used in vivo model involves administration of endotoxin, i.e., lipopolysaccharide (LPS), to rodents (Moseley (18)). LPS, a component of the outer membrane of Gram negative bacteria, is a ligand for two different pattern-recognition receptors, i.e., Toll-like receptor (TLR)-4 (Poltorak (19)) and CD14 (Wright (20)). LPS signaling is dependent on a complex arrangement that includes binding to soluble proteins (LBP) and both CD14 and TLR4. Mice deficient in either TLR4-signaling (mutant strain C3H/HeJ or null-mice C57BL/1-ScCr) or CD14 mice are resistant to LPS (Poltorak (19), Haziot (21)). These receptors are present at the surface of several cell types within the liver, including Kupffer cells (KC), and LPS binding elicits an immune response in these cells (Su (22)). KC are the resident liver macrophages and central mediators of the inflammatory cascade leading to IIC (see below). Mice of the C3H/HeJ strain are often used as LPS-resistant control mice (Miyake (23), Li (24), Wagner (25), Ghose (26)).
IIC has also been studied using different activators of innate immunity, such as zymosan (Kim (27)) or lipoteichoic (Quale (28)), or individual pro-inflammatory cytokines (Whiting (29), Green (30), Geier (31)). Other models include administration of chemical agents to rodents, e.g., turpentine (Piquette-Miller (32), Hartmann (33), Siewert (34)) or surgical procedures to induce polymicrobial sepsis (e.g., cecal-ligation and puncture (CLP) (Von Allmen (35), Reynolds (36), Kim (37), Deutschman (38) Chen (39))).
Isolated perfused rodent livers (IPRL) (Utili (40), Roelofsen (41), Kubitz (42), Mühlfeld (43)) allow for well-controlled experiments with regard to perfusate composition, use of tracers, etc. Precision-cut slices from both human and rodent liver have also been used to study the effects of LPS on cytokine expression and transporter expression (Luster (44), Olinga (45), Elferink (46)).
In addition to using intact animals/organs, IIC has been examined at the (sub)cellular level using primary hepatocytes or hepatoma cell-lines (Li (47), Hisaeda (48)). Treatment with (individual) cytokines or medium obtained from activated Kupffer cells or macrophages mimics the in vivo response at the hepatocellular level down-stream of KC activation (Sturm (49)).
3. NUCLEAR RECEPTOR (NR) BIOLOGY
NRs are ligand-activated transcription factors that play important roles in many aspects of metazoan life, including embryonic development, cell differentiation and maintenance of cellular homeostasis (Mangelsdorf (50)). NRs are expressed differentially amongst tissue and throughout day-night cycles (Bookout (51), Yang (52)). NRs are assumed to have arisen from constitutively active transcription factors (Gronemeyer (53)) having acquired the ability to be activated by hormones (e.g., glucocorticoid receptor (GR), estrogen receptor (ER)) or to sense local environmental and nutritional cues (e.g., liver X receptor (LXR), farnesoid X receptor (FXR)). This allows not only for concerted gene responses throughout the organism, initiating hormone-appropriate responses, such as the stress-response after glucocorticoid release, but also for cell-specific responses to altered local environmental conditions, as exemplified by induction of cholesterol efflux transporters during cellular sterol overload by LXR stimulation.
3.1. Classes and structures of NRs
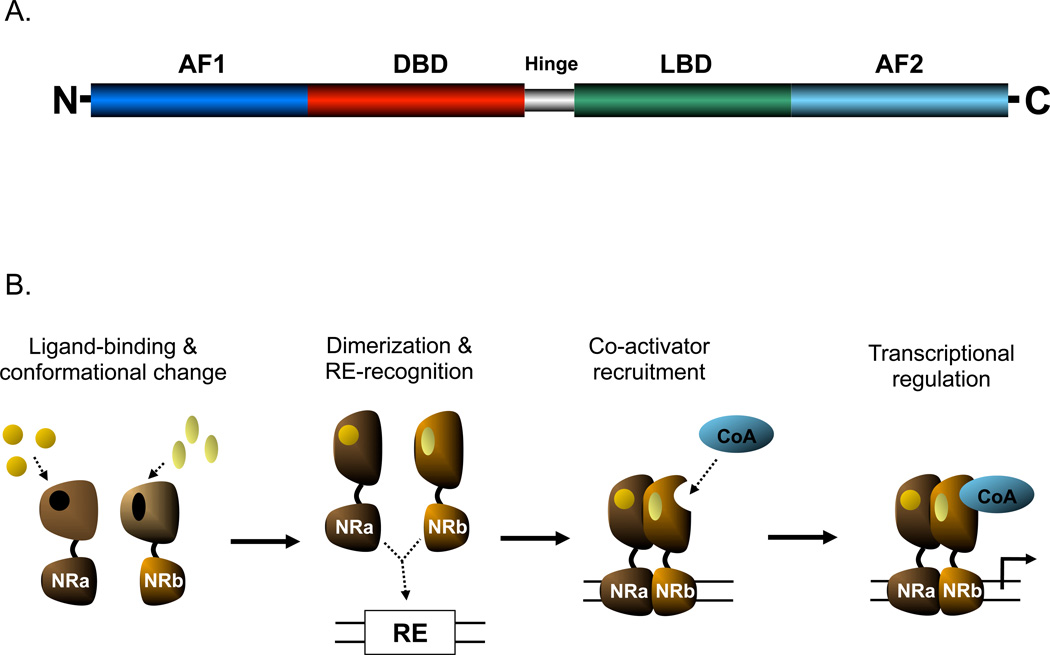
The various NRs share several structural similarities: a DNA-binding domain (DBD), a ligand-binding domain (LBD) and activator function domains (AF-1 and AF-2) (Figure 1A). A DBD contains two well-conserved zinc-finger domains, which determine the affinity for specific DNA sequences known as response elements (cis-acting elements) (Mangelsdorf (50)). The LBD determines the ligand-specificity of NRs and will thus differ most significantly between NR family members (Karpen (54)). Ligand binding will lead to conformational changes in the NR molecule, resulting in altered transcriptional activities through re-organization of the transcription complex at the promoter, generally involving removal of corepressors and recruitment of coactivators. The AF-1 (situated at the N-terminus) and AF-2 (actually contained within the LBD at the C-terminus) mediate interactions with coregulators.
Figure 1.
A) Schematic of NR structure (adapted from Karpen {5}) B) General mechanism of NR action (ligand-binding and conformational change not necessarily prior to DNA-binding; release of co-repressors shown)
The NR superfamily has been subdivided according to different classifications. Traditionally, NRs have been divided into three functional groups (Mangelsdorf (50)): 1) classic NRs such as glucocorticoid receptor (GR), estrogen receptor (ER), etc. These NRs generally form homodimers and are activated at high affinity by steroid hormones in an endocrine fashion. 2) heterodimers with RXR and partners such as LXR, FXR, RAR, PPARs, that are activated at lower affinity by metabolites or nutrients such as fatty acids and oxysterols, and 3) orphan NRs, referring to transcription factors expected to be NRs based on gene/protein structure, for which no specific ligands have been identified yet, or appear to lack a functional LBD based on structural analysis. Once members of this class of NRs have been assigned (specific) ligands, they become adopted, as occurred recently with the identification of heme as ligand of Rev-erb-alpha and Rev-erb-beta (Yin (55))
More recently, NRs nomenclature has been revised in a way analogous to that for cytochrome p450 enzyme systems using a coded numbering system. In this system, NRs are classified into 6 distinct groups based on molecular phylogeny (57). This system allows for classification of NRs from different species as well as NRs identified by genetic analyses without clarified functional and/or biological characteristics (53).
Inherent to their function as transcriptional regulators, NRs selectively recognize and bind to short DNA sequences located in gene-regulatory elements, either in close proximity to the transcriptional start sites (promoters) or in more distant elements (enhancers). These “cis-acting” response elements (REs) share several characteristics. REs that mediate transcriptional activity of RXR-heterodimers generally consist of two hexamers that are separated by zero to eight nucleotides and are direct repeats (DR), everted repeats (ER) or inverted repeats (IR). However, some of the NRs that form heterodimers with RXR may also regulate transcriptional activity by binding as a monomer (e.g., FXR (Claudel (58), Barbier (59)).
3.2. Mechanisms of genomic actions
A key characteristic of NRs that sets them apart from other transcription factors is altered function upon the binding of specific ligands. Although in reality much more intricate due to the involvement of many proteins (Gronemeyer (53)), the general mechanism of NR-regulated gene expression is depicted in Figure 1b. It involves a conformational change of the NR upon binding of its ligand. Some NRs will then relocate to the nucleus, bind to their corresponding response elements and recruit coactivator molecules. Other NRs are already bound to their response elements while being unliganded and associated with corepressor proteins. Upon ligand binding, these NRs undergo a conformational change (see below), allowing for the release of corepressors and recruitment of coactivators, a process that ultimately engages multitude of proteins with specific spatial and temporal characteristics. The conformational changes that occur upon ligand-binding invokes a closure of the ligand-binding pocket by helix 12 rendering the surface of the NR more available for binding of coactivators, while corepressors become less able to bind, and are released. Since NR-binding to promoters is a cyclic process, i.e., a continuous binding and removal of NRs from the response elements, increased stability of the coactivator complexes will shift the balance from inhibition to stimulation of transcription. One of the important molecular actions of these multi-functional coactivators is chromatin relaxation through histone acetyltransferase activity or mediating the recruitment of other proteins with such function. This will render target genes more accessible for the transcriptional machinery. Corepressors such as the nuclear receptor corepressors (NCoR) or silencing mediator of retinoid and thyroid hormone receptor (SMRT) either have histone deacetylating properties or stimulate the recruitment of other co-repressors with such enzymatic activity, the so-called HDACs, and thus reverse chromatin relaxation and inhibit gene transcription. In addition to (de)acetylating modification of histones, coregulators can also modify histones via (de)methylation and (de)phosphorylation mechanisms (Perissi (60)). There are also non-histone-mediated actions by which coregulators affect gene transcription, including ATP-dependent remodelling of chromatin and the recruitment of both basal transcription factors and co-regulators (Smith (61), Perissi (60)). In essence, NRs act as the “traffic cop” at the promoter, directing the flux and binding of regulatory molecules that ultimately drive gene expression. Thus, co-regulators are regarded as the actual determinants of NR-mediated transcriptional regulation and their tissue-specific expression patterns are responsible for the specific effects of NRs and their ligands in different tissues (Jordan (62)).
3.3. Non-genomic actions of NRs
Besides the “classical” NR mode of action, some of the NRs exert effects on gene transcription without DNA-binding, i.e., “non-genomically” by protein-protein interactions (Gronemeyer (53)). The small heterodimer partner (SHP) is one example of a NR that acts non-genomically as it lacks a DBD. SHP is often seen as a transcriptional repressor that acts by binding and interfering with the action of some NRs and transcriptional activators. SHP, however, is not the only NR that has non-genomic actions. There is substantial evidence that many NRs that possess a DBD regulate gene expression through other domains in the protein and without DNA-binding. This group includes GR, ER, PPARgamma and LXR, which are known to regulate gene expression via traditional REs, but have also been shown to suppress inflammatory signaling via non-genomic interactions.
3.4. Clinical relevance of NR ligands
The aspect of ligand-induced modification of NR activity and their generally well-matched sets of transcriptional targets has led to the concept that NRs represent attractive targets for pharmacological intervention in a wide range of pathophysiological processes. An estimated 20% of all US prescriptions exert their effects via NRs (Baxter (63)). Interestingly, some of these were already used clinically without knowledge of their primary target or molecular mode of action, such as hypolipidemic fibrates (e.g., clofibrate) and antidiabetic thiazolidinediones (e.g., rosiglitazone) which were later shown to be PPARalpha and PPARgamma ligands respectively (Issemann (64), Green (65), Lehmann (66)). For several drugs, their identification as ligands of NRs, more specifically of PXR and CAR, provided an explanation for their known interference with metabolism of other drugs. Examples of the latter group include phenobarbital, a CAR activator (Honkakoski (67), Sueyoshi (68), Tzameli (69)), and PXR ligands, rifampicin and nifedipine (Bertilsson (70))). These drugs regulate drug metabolism through activation of CAR and PXR and subsequent changes in the expression of cytochrome P450 (CYP) and other drug regulation genes.
3.5. NRs of specific relevance to IIC
A subset of NRs is of particular interest in relation to IIC. This subset includes RXRalpha for its central role as obligate heterodimerization partner for other class II NRs, RARalpha for its role in control of basal hepatocellular gene expression, FXR as bile acid sensor, PXR and CAR as xenobiotic sensors involved in detoxification pathways, LXR and PPARgamma for their recently identified anti-inflammatory activities and the orphan receptors LRH-1, HNF4alpha and SHP, involved in the transcriptional regulation of various genes involved in bile formation and hepatobiliary transport. Table 1 summarizes some of the characteristics of these NRs. In the paragraphs reviewing anti-inflammatory properties of NRs involved in IIC, GR will also be discussed as the prototypical example of NRs with anti-inflammatory properties. Some other NRs, e.g., ER or PR, are described to further illustrate general principles regarding NR biology.
Table 1.
NRs involved in bile-formation and IIC, their natural and synthetic ligands and examples of IIC-related target genes.
| NR | Official name | Ligands | Examples of target genes | |
|---|---|---|---|---|
| Natural | Synthetic | |||
| RXRalpha | NR2B1 | 9cRA | LG268, LG1069 | (obligate heterodimer partner) |
| RARalpha | NR1B1 | ATRA | TTNPB | Ntcp, Mrp2, Mrp3 |
| FXR | NR1H4 | CDCA, CA | GW4064 | Bsep, Mdr2, Shp, Ostalpha/beta |
| LXRalpha/beta | NR1H3/NR1H2 | Oxysterols | T0901317, GW3965 | Abcg5/8, (rodent) Cyp7a1 |
| PPARgamma | NR1C3 | Fatty acids, PGs | TZDs | |
| PXR | NR1I2 | 5beta-pregnane- 3,20-dione |
pregnenolone-16alpha- carbonitrile, rifampicin |
Mrp2, Mdr1b, Cyp-family |
| CAR | NR1I3 | Androstenol | TCBOPOP.… | Mrp2, Cyp-family, Ugt1a1 |
| Orphans | ||||
| HNF4alpha | NR2A1 | - would check here | - | ….,…, Abcg5/8, Cyp8b1 |
| LRH-1* | NR5A2 | -would check here | - | Cyp7a1, (human) ABCG5/8, |
| SHP | NR0B2 | - | - | (No DBD) |
Also known as CPF, FTF, (Adapted from Karpen (54))
4. MECHANISMS UNDERLYING IIC
It is generally accepted that IIC results from impairment of normal hepatobiliary transport functions through the effects of inflammatory mediators (Crawford (4), Moseley (18)). These mediators can either reach the liver via the systemic circulation or be produced locally in response to a variety of stimulants, including endotoxin (reviewed by Trauner (11), Moseley (18)). Inflammatory mediators are able to affect hepatobiliary transport functions via multiple signal transduction pathways, targeting events at the membrane, cytosol and the nucleus.
4.1. Physiology of bile formation
Bile formation represents an osmotic process, driven by active secretion of cholephils by hepatocytes into the minute bile canaliculi (reviewed by Koopen (71)). These canaliculi are separated from the circulation by tight junctions connecting adjacent hepatocytes. The active secretion of osmotically active solutes, most notably bile salts, leads to the passive transport of water and electrolytes into the canaliculi, thus generating bile flow (71). Total hepatic bile flow is considered to be the sum of bile salt-dependent flow (BSDF) and bile salt-independent flow (BSIDF). The latter is mainly driven by the secretion of substances such as glutathione (Ballatori (72), Ballatori (73)) and bicarbonate (Hardison (74)). The canaliculi join to form bile ductules that are lined with cholangiocytes and eventually converge into the major bile ducts that drain into the duodenum. Although bile ducts were earlier primarily regarded as a drainage system, it is now clear that the bile duct epithelium plays an active role in the generation of bile flow and regulation of bile composition (Prall (75)). Approximately 10–13% and 40% of total bile flow in rats and humans, respectively, is driven by secretin-stimulated secretion of chloride and bicarbonate by cholangiocytes ((Alpini (76), Boyer (77)). Not surprisingly, substantial compositional differences exist between canalicular and ductular bile. In humans and mice, but not in rats, bile is stored and further modified by concentration in the gallbladder.
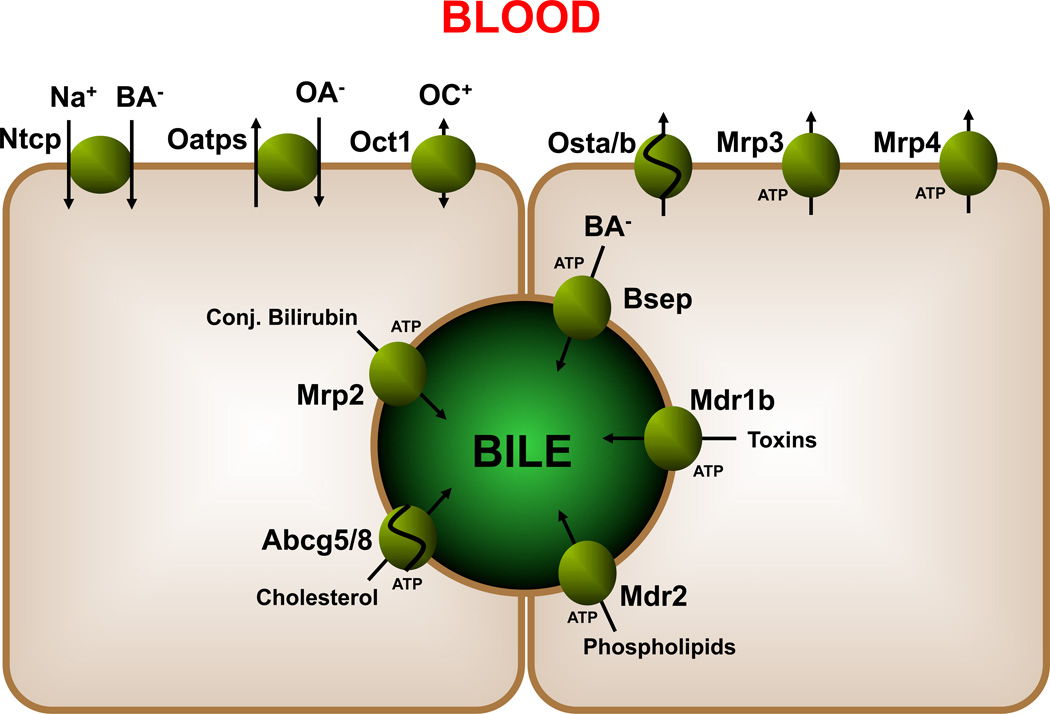
Over the past decades, knowledge of the molecular basis of bile formation has greatly expanded by the identification of several essential transporters that contribute to the process (Trauner (78)). Figure 2 shows the most important hepatocellular transporters involved in bile formation as well as in protection of hepatocytes against bile salt overload. At the basolateral side of hepatocytes, Ntcp, Oatps and Oct1 are responsible for sodium-dependent and sodium-independent uptake of bile salts, organic anions and cations. At this side of the cell, members of the multi drug resistance-related proteins, i.e., Mrp3 and Mrp4, are present too. These transporters exert hepatocyte-protecting effects during (extrahepatic) cholestasis most likely by facilitating the basolateral efflux of retained and potentially harmful substances including bile salts (Bohan (79), Mennone (80)). Recently, an additional bile salt transporter complex has been identified, i.e., the Ost-alpha/beta heterodimer (Wang (81), Seward (82)), which appeared to be the elusive bile salt efflux transporter at the basolateral side of enterocytes (Dawson (83), Rao (84)). Ost-alpha/beta heterodimers are expressed in the liver, both in cholangiocytes and hepatocytes (Ballatori (85)), differentially between species. In cholangiocytes, it may very well play a role in “cholehepatic shunting”of bile salts, while in hepatocytes it appears to function as an overflow efflux transporter, as its expression is markedly induced under cholestatic conditions in an FXR-dependent manner (Zollner (86), Boyer (87)).
Figure 2.
Hepatobiliary transporters: adjacent hepatocytes are shown and a bile canaliculus.
At the canalicular membrane of hepatocytes, several transporters are localized that are responsible for the biliary secretion of various biliary components. Many of these transporters belong to the ATP-binding cassette transporter family (ABC transporters) and actively transport their substrates against concentration gradients into the canalicular lumen. The bile salt export pump (Bsep or Abcb11) mediates biliary secretion of monovalent bile salts. Mrp2 (Abcc2) is responsible for efflux of divalent anions including conjugated bilirubin and sulfated bile salts. Mdr2/MDR3 (Abcb4) is involved in phospholipid secretion into the canalicular lumen by functioning as a floppase (Oude Elferink (88)). Abcg5 and -8 are two halftransporters that facilitate sterol export and Mdr1b/MDR1 (Abcb1) is involved in the excretion of many organic cations (endobiotics, xenobiotics). The importance of the various individual transporters in the process of bile formation is demonstrated in various human syndromes as well as various animal knock-out / mutant models (reviewed by Oude Elferink (88)).
Several other (non-ABC) transporter proteins are also present on the canalicular membrane including FIC1 and NPC1L1. The importance of FIC1 (ATP8b1) is evident since, when mutated, leads to reduced bile salt secretion and is recognized as the genetic defect underlying progressive familial intrahepatic cholestasis type 1 (PFIC1, hence its name FIC1), also known as Byler’s disease. In milder mutated forms, this can also lead to benign recurrent intrahepatic cholestasis type 1 (BRIC1). Although it is known to be a member of sub-family of ATP-transporters that act as aminophospholipid flippases, its exact modes of action under physiological conditions and in the development of PFIC1 and BRIC1 remain to be established (Oude Elferink (88)). NPC1L1 has been identified as the transporter responsible for cholesterol uptake in enterocytes and as the target of the cholesterol-lowering drug ezetimibe (Altmann (89)). NPC1L1 is highly expressed in human (but not mouse) liver ((89)), where it might mediate re-uptake of cholesterol from the canalicular lumen (Temel (90)). The physiological relevance of this process is, however, unknown.
4.2. Impaired bile formation during inflammation
LPS-treatment of perfused rat livers reduces bile flow and bilirubin/dye transport (Utili (40), Utili (91), Utili (92), Roelofsen (41), Whiting (29), Bolder (93), Mühlfeld (43)). Although LPS-treatment was initially thought to primarily affect BSIDF (Utili (92), Roelofsen (41), Trauner (94)), BSDF was also shown to be affected (Rustgi (95), Whiting (29), Bolder (93), Mühlfeld (43)), indicating that both components of bile flow are impaired upon inflammation. Similar results were obtained in another sepsis model, i.e., CLP (Reynolds (36)).
LPS interferes with normal bile flow generation via several mechanisms, which all ultimately lead to reduced activity of the transporters and enzymes involved. With regard to BSIDF, LPS reduces Na+-K+-ATPase activity (Utili (92), Green (30)) and also impairs glutathione secretion (Trauner (94)). The latter has been linked to reduced mRNA and protein expression of Mrp2 (see table 2), but this transporter has also been shown to undergo intracellular re-localization upon LPS-treatment (Kubitz (42), Dombrowski (96), Zinchuk (97)).
Table 2.
In vivo effects of LPS on hepatobiliary transporters in rodent models (mouse/rat)
| Transporter | Synonym | mRNA | References | Protein | References |
|---|---|---|---|---|---|
| Basolateral | |||||
| Slc10a1 | Ntcp | ↓ | Green (30), Moseley (275), Trauner (276), Lee (277), Zollner (193), Geier (109), Mesotten (278), Cherrington (124), Siewert (34), Ghose (131), Sturm (107), Lickteig (110), Zollner (139), Donner (279), Teng (150) | ↓ | Green (30), Moseley (275), Trauner (276), Sturm (107), Mesotten (278), Donner (279) |
| ↔ | Zollner (193) | ||||
| Slco1a1 | Oatp1 | ↓ | Geier (109), Cherrington (124), Mesotten (278), Hartmann (108), Siewert (34), Lickteig (110), Donner (279) | ↓ | Mesotten (278), Lund (280), Donner (279) |
| ↔ | Lund (280) | ||||
| Slco1a4 | Oatp2 | ↓ | Geier (109), Cherrington (124), Mesotten (278), Hartmann (108), Donner (279), Teng (150) | ↓ | Mesotten (278), Donner (279) |
| ↔ | Lickteig (110) | ||||
| Slco1b2 | Oatp4 | ↓ | Cherrington (124), Mesotten (278), Li (24), Lickteig (110), Donner (279), Li (281) | ↓ | Donner (279) |
| Slc22a1 | Oct1 | ↓ | Cherrington (124) | ||
| Abcc3 | Mrp3 | ↓ | Hartmann (108), Siewert (34), Teng (150) | ||
| ↔ | Lickteig (110), Donner (282) | ||||
| ↑ | Cherrington (124) | ↑ | Donner (282) | ||
| Abcc4 | Mrp4 | ↔ | Lickteig (110), Donner (279), Donner (282) | ||
| Apical | |||||
| Abcb11 | Bsep, Spgp | ↓ | Green (283), Lee (277), Mesotten (278), Hartmann (33), Hartmann (108), Siewert (34), Ghose (131), Hartmann (122), Lickteig (110), Donner (279), Cherrington (124), Teng (150) | ↓ | Lee (277), Mühlfeld (43) |
| ↔ | Donner (279) | ||||
| Abcc2 | Mrp2 | ↓ | Trauner (284), Vos (123), Kubitz (42), Nakamura (105), Lee (277), Geier (109), Mesotten (278), Teng (150), Mühlfeld (43), Hartmann (108), Siewert (34), Ghose (131), Cherrington (124), Donner (282) | ↓ | Trauner (284), Vos (123), Lee (277), Teng (150), Mühlfeld (43), Donner (282), Donner (282) |
| ↔ | Lickteig (110) | ||||
| Abcb4 | Mdr2 | ↓ | Hartmann (33), Hartmann (122), Teng (150) | ||
| ↔ | Vos (123) | ||||
| Abcb1b | Mdr1b, Pgp | ↓ | Hartmann (33), Hartmann (122) | ||
| ↔ | Lickteig (110), Mesotten (278), Teng (150) | ↔ | Mesotten (278) | ||
| ↑ | Vos (123), Cherrington (124) | ||||
| Abcg5/8 | Sterolin1/2 | ↓ | Khovidhunkit (159), Ghose (131) |
It is generally accepted that canalicular transport is rate-controlling in overall hepatobiliary transport (Reichen (98), (93)). Hence, impaired biliary secretion is likely to lead to accumulation of potentially toxic substances in hepatocytes. Simultaneous effects on basolateral transporters, whether there be a reduction of influx (Ntcp, Oatps) or an increase in efflux (Mrp3, Mrp4, Ost-alpha/beta), will therefore strongly influence the degree of bile salt retention and potential cellular damage.
Due to their important contribution to bile formation, effects of inflammatory signaling on the cholangiocytes also contribute significantly to the pathogenesis of IIC. Spirli et al. (99) analyzed the effects of inflammatory mediators on biliary fluid secretion using isolated bile duct units. These authors found that a mixture of cytokines, but not individual cytokines, reduced cAMP-dependent fluid secretion in isolated bile duct units (99). At the same time, they observed an impaired biliary epithelial barrier function, which most probably contributes to the emergence of cholestasis (99). These data suggest that effects on biliary epithelium indeed have a role in IIC, but further studies are necessary to gain insight into the quantitative and qualitative importance of the effects of inflammation on the biliary epithelium.
Important for bile flow generation, and bile salt metabolism in general, is the synthesis of bile salts by the hepatocytes. This process is tightly regulated and involves the actions of several NRs, e.g., FXR, SHP, LRH-1, HNF4alpha and LXR (Russell (100)). Due to the highly efficient reabsorption of bile salts by the epithelia of biliary system and in the distal ileum, leading to cycling of bile salts via the cholehepatic shunt and in the enterohepatic circulation, respectively, the loss of bile salts per cycle is small. Therefore, the quantitative contribution of newly synthesized bile salts to the bile formation process is limited. Inflammation-induced suppression of the expression and activity of Cyp7a1 and Cyp27 (Feingold (101), Memon (102)), which catalyze the first step of respectively the classic/neutral and the acidic pathway of bile salt synthesis from cholesterol (Russell (100)), will only become relevant after prolonged sepsis/inflammation. On the other hand, effects of inflammation on intestinal and gallbladder motility, leading to impaired enterohepatic cycling, may contribute to impaired bile formation.
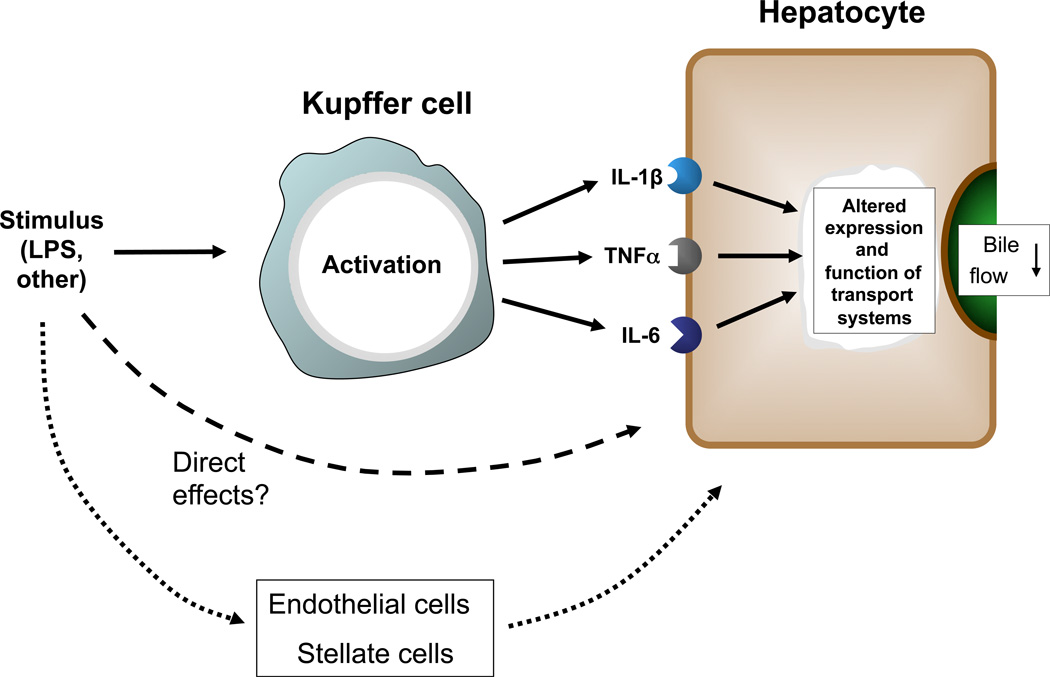
4.3. Inflammatory cascade and Kupffer cells
IIC can be elicited by various inflammatory mediators, which are either reaching the liver from the circulation or are produced locally (figure 3). The Kupffer cells (KC), resident liver macrophages, play a central role in local production (Chensue (103)). KCs form the primary line of defense against intestine-derived toxins that enter the liver via the portal circulation, as is illustrated by the nearly complete clearance of endotoxin from portal blood by KCs (Fox (104)). Activation of KC by LPS occurs via several different signal transduction pathways (Su (22)). The importance of KCs in the pathogenesis of IIC has been demonstrated in various in vivo studies in which suppression of transporter expression by LPS-administration was found to be reduced when KC had been inactivated by gadolinium chloride (Nakamura (105), Xu (106)) or selectively removed using liposomal clodronate (Sturm (107)).
Figure 3.
Linking inflammatory signals to hepatocellular effects
Mediators affecting bile formation in hepatocytes include KC-secreted pro-inflammatory cytokines such as IL-1beta, TNFalpha and IL-6. The importance of these cytokines was supported by experimental findings showing that individually administered cytokines elicited similar responses in vivo as LPS did (Green (30), Hartmann (108), Geier (31), Siewert (34)). In addition, cytokine-inactivation through the administration of anti-TNFalpha and anti-IL-1beta antibodies blunted the cholestatic response to endotoxin-treatment both in vitro and in vivo (Whiting (29), Sturm (49), Geier (109)), despite the fact that there appeared to be some degree of redundancy in the cytokine-signaling (Lickteig (110)).
Although the involvement of KCs in the pathogenesis of IIC is evident, it has also become clear that KC-independent mechanisms contribute. Suppression of Ntcp expression after LPS administration persisted in KC-depleted livers, albeit that the degree of suppression was lower than in control livers (Sturm (107)). It is plausible that either direct effects of LPS on hepatocytes or LPS activation of other non-parenchymal liver cells can be involved in the pathogenesis of IIC, e.g., endothelial cells and stellate cells. Endothelial cells, for instance, are known to express TLR4 and to respond to inflammatory stimuli (Dunzendorfer (111), Nakamura (112)). The importance of endothelial cells is further illustrated by the findings of Laschke et al. ((113)) using a combination of LPS and D-galactosamine (LPS/Gal) in mice. Pre-treatment of mice with antibodies against the adhesion molecule P-selectin, which is present on endothelial cells upon LPS-treatment (Essani (114)), reduced the recruitment of leukocytes and concomitantly diminished the effects of LPS/Gal on bile flow, transporter gene expression profile and liver morphology, while levels of inflammatory cytokines were not affected ((113)). These results indicate that processes such as leukocyte recruitment are also of critical importance in the pathophysiology of sepsis-associated cholestasis. The mechanistic link is between reduced leukocyte recruitment and inhibition of cytokine activity remains to be determined.
Finally, activated KCs not only secrete cytokines but also inflammatory mediators such as NO and prostaglandins (Bilzer (115)). Although NO had been shown to stimulate bile flow (Trauner (116)), LPS-induced NO-production did not have a significant effect on LPS-induced suppression of bile flow (Trauner (94)). Prostaglandins have also been shown to reduce bile flow and bile salt secretion (Beckh (117), Weidenbach (118)).
4.4. Inflammatory signaling in hepatocytes
Pro-inflammatory cytokines and other inflammatory mediators affect hepatocellular function via various, sometimes parallel, pathways (Moshage (7), Ramadori (119)). These include, but are not limited to, NF-kappaB, and MAPK routes. Examples of how these pathways participate in the development of IIC include NF-kappaB-mediated induction of Mdr1b expression (Ros (120)), JNK-mediated nuclear export of RXRalpha in vitro (Li (47), Zimmerman (121)) and ERK-mediated inhibition of IRF3 nuclear import (Hiseada (48)). Of note, the specific roles of these pathways have often been examined in vitro using hepatocytes or hepatoma cells treated with (individual) cytokines. Although this is an elegant approach to elucidate underlying mechanisms, one has to bear in mind that in the in vivo situation multiple, parallel pathways may be active with a certain degree of redundancy, while LPS signaling affects hepatocytes directly to some extent as well.
4.5. Hepatobiliary transporters
Inflammatory signaling has been shown to differentially affect hepatocellular transport function at both transcriptional and post-transcriptional level. Table 2 summarizes the effects of LPS/endotoxin on the expression of several physiologically relevant transporters in rats and mice. This list, however, is not complete and ought to be seen as an indicator of general effects. Figure 2 shows the localization of the various transporters. Most extensively studied transporters in this regard are the sodium-dependent taurocholate transporting polypeptide (Ntcp, Slc10a1), the bile salt export pump (Bsep, Abcb11), the multidrug resistance associated protein (Mrp)-2 (Abcc2). These three transporters are generally suppressed, both at mRNA and protein level. To our knowledge, no information is available on the effects of LPS on the expression of the canalicular transporters Fic1 and Npc1l1 or on that of the basolateral Ost-alpha/beta transporting complex.
Most of the transporters are suppressed under inflammatory conditions with rodent Mdr1b (Abcb1b) being the most consistent exception. Although some demonstrated suppressive effects of LPS on Mdr1b expression (Hartmann (33), Hartmann (122)), other groups have observed an induction of Mdr1b in vivo (Vos (123), Cherrington (124). Mdr1b, as a member of the multidrug resistance protein family serves as an inducible efflux transporter for organic cations, xenobiotics and toxins (Trauner (78)) and is directly regulated by NF-kappaB signaling (Ros (120), Zhou (125)). This role of inducible efflux transporter appears to be in contrast to that of its closely related family member Mdr1a, which is more consistently expressed (Vos (123)).
Water channels, the so-called aquaporins, have so far received relatively little attention in IIC research, despite the expression of at least 7 family members in the hepatobiliary system (Masyuk (126)). A recent report by Lehmann et al. showed that LPS-treatment of rats led to down-regulation of AQP8 protein expression, while AQP9 expression was not affected ((127)). The suppression of AQP8 expression was TNF-alpha-dependent and post-transcriptionally mediated through both lysosomal and proteasomal degradation (127). This led to a reduced osmotic water permeability of the canalicular membrane, which was suggested to contribute to the pathogenesis of inflammation-induced cholestasis (127).
4.6. NR expression and function during inflammation
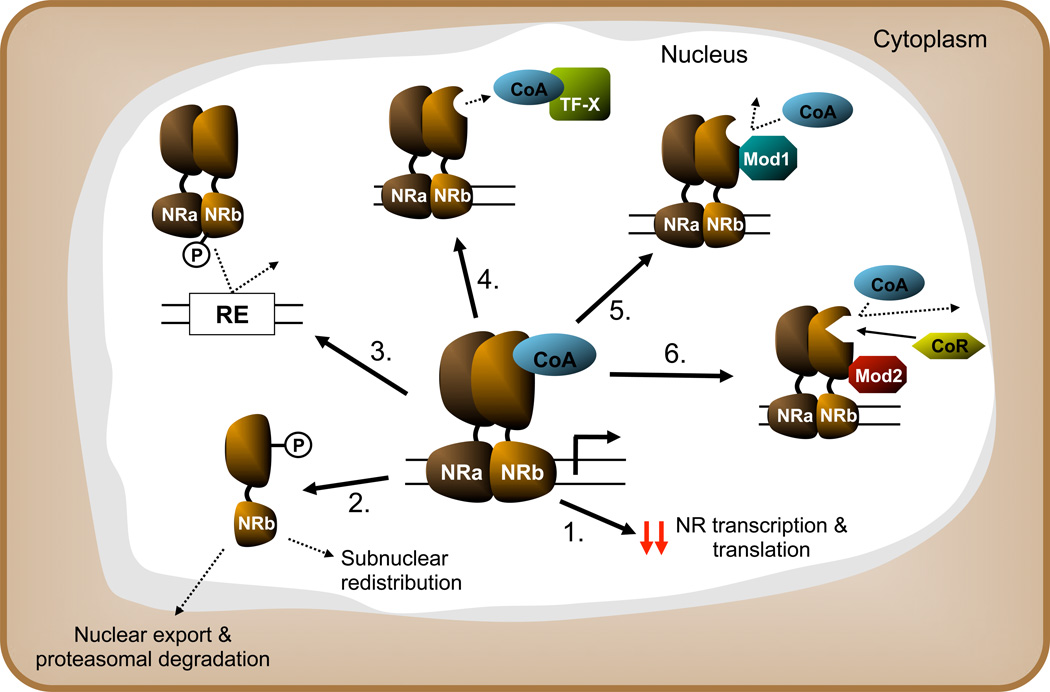
The observed reduction in hormone sensitivity during inflammatory conditions, for instance growth hormone (GH) resistance during chronic inflammation (Denson (128)), illustrates that there is a direct link between inflammation and altered NR function. This link has been addressed experimentally in many studies. Although some NRs are differentially regulated, e.g., induction of Nur77 in LPS-stimulated macrophages (Pei (129)), inflammation suppresses expression and function of most NRs studied at multiple levels (Figure 4). This general response is considered to be a crucial step in the negative hepatic APR (Beigneux (130), Ghose (131), Wang (132)).
Figure 4.
Shown effects of inflammatory signaling on NR expression and function in general (simplified/schematic): 1) suppression of NR transcription or translation 2) signal dependent relocalization and degradation 3) reduced DNA binding 4) competition for coactivator (CoA) 5) inhibition of CoA recruitment (hindrance/conformational change) 6) increased affinity for corepressor binding
4.6.1. RXRalpha
In view of its importance as obligate heterodimer partner of class II NRs, the effects of inflammatory signaling on RXRalpha expression and function have been studied extensively. Beigneux et al. (130) showed that both mRNA and protein expression of RXRalpha in livers of Syrian hamsters were strongly reduced upon treatment with either LPS or cytokines, which led to reduced binding activity of RXR-homodimers. This effect was shown to be mediated, at least partially, by reduced RXRalpha transcription (130). Similar effects of LPS on hepatic RXRalpha gene expression were obtained in mice (Kim (133)) and rats (Fang (134)) as well as in non-hepatic tissues, i.e., adipose tissue (Lu (135)) and heart (Feingold (136)).
At least two other mechanisms have been identified by which inflammatory signaling alters RXRalpha transcriptional activity. Firstly, as part of the protein complex originally known as Footprint B binding protein, RXRalpha was shown to be a key regulator of the rat Ntcp gene expression (Karpen (137), Denson (138)). Follow-up studies revealed that the nuclear abundance of this protein is rapidly reduced upon LPS-treatment of mice with a simultaneous, transient appearance in the cytosolic compartment, suggesting active nuclear export followed by degradation (Ghose (131)). At the same time, c-Jun N-terminal kinase (JNK) activity, important in the regulation of Ntcp promoter activity by RXRalpha during inflammation (Li (47)), was increased (131). This led to a reduced mRNA expression of NR-target genes, despite preserved expression of most heterodimer partners (131). Subsequent in vitro studies provided a potential mechanism for the nuclear export of RXRalpha (Zimmerman (121)). JNK-dependent phosphorylation of a serine residue at position 260 of RXRalpha was shown to induce nuclear export and subsequent proteasomal degradation (121). Reduced nuclear protein levels of RXRalpha have also been reported by others (Zollner (139)). Secondly, Gu et al. (140) presented another mechanism involved in altered RXR function during inflammation. Activated NF-kappaB suppressed DNA-binding of the PXR:RXR heterodimer to the promoter of the Cyp3a4 gene and this effect was in part mediated by direct interaction between the p65 subunit of NF-kappaB and RXR (140). Although this has not been shown to occur with other heterodimers, it may represent a more generally occurring mechanism and thus not only pertain to the PXR:RXR heterodimer.
These results combined suggest that inflammatory signaling suppresses RXRalpha expression and function in multiple ways. Considering its important role in the function of class II NRs, the regulation of RXRalpha alone could already be a central mediator of the negative hepatic APR.
4.6.2. RARalpha
In contrast to RXRalpha, limited information is available on the effects of inflammatory signaling specifically on RARalpha expression. TNFalpha treatment of mice led to a transient reduction in nuclear protein levels of RARalpha in liver, which corresponded well with transiently reduced mRNA expression (Geier (31)). Interestingly, treatment with IL-1beta also led to reduced nuclear protein levels of RARalpha, but at a later timepoint, while RARalpha mRNA expression was not altered (31). This indicates that the reduction in nuclear RARalpha by inflammatory cytokines is not only regulated at the transcriptional level.
Results obtained with individual cytokines were different from those with LPS-treatment. Ghose et al. ((131)) showed that RARalpha mRNA expression and nuclear protein levels remained unaffected in mice after LPS-treatment, while nuclear RXRalpha protein levels were rapidly reduced. The cause of these differential effects of individual cytokines and LPS remains unclear, but may be related to the dose of inflammatory mediators used, as RARalpha nuclear protein levels were strongly reduced after very high dose of LPS in mice (Zollner (139)).
4.6.3. FXR
Since its identification as an intracellular bile salt sensor (Makishima (141), Parks (142)), FXR has been shown to play a central role in control of expression of transporter genes such as Bsep (Abcb11) as well as in the adaptive response to a bile salt challenge (Sinal (143)). Studies with FXR-null mice revealed an impaired hepatocellular protection against bile salt overload ((143)). The expression of FXR at both mRNA and protein level is reduced upon LPS- or cytokine treatment of rodents or cytokine-treatment of hepatoma cells (Kim (144), Fang (134), Geier (31)). In the CLP-model, expression and binding activity of FXR:RXR were also reduced (Chen (39)). These results not only illustrate that FXR is similarly affected by inflammatory signaling as other NRs and thus may contribute to the pathogenesis of IIC, but also indicate that impaired FXR function will most likely prevent a proper adaptive response during IIC as well.
4.6.4. CAR and PXR
CAR and PXR play critical roles in xenobiotic metabolism and detoxification (Willson (145)). These NRs are distinct NR family members and show important differences in their LBD, with PXR being more promiscuous ((145)). Nevertheless, they are often grouped together due to several shared characteristics, including expression profile, with highest expression in liver and intestine, cross-over of some target genes as well as a subset of ligands. CAR and PXR have been studied extensively in relation to drug metabolism, which is altered by inflammation (Song (146), Farrell (147), Morgan (148)). CAR and PXR mRNA expression levels were found to be suppressed in LPS-treated rodents (Beigneux (149), Fang (134), Ghose (131), Teng (150), Xu (106)). Suppressed PXR-function was shown by Kim et al. ((151)), who demonstrated that pregnenolone 16alpha-carbonitrile (PCN)-mediated induction of dehydroepiandrosterone-sulfotransferase (Sult2a1) expression was suppressed after LPS administration in mice. This effect was most likely mediated by cytokines (TNF, IL-1, but not IL-6 as these induced similar effects in hepatoma cells (151). Interestingly, Sult2a1 expression was suppressed at much lower LPS doses than expression of PXR and CAR (151). It is unclear whether the transactivation capacity of PXR and CAR per se is affected, since both NRs were shown to retain their transactivational activity, albeit when overexpressed, in IL-6-treated human hepatocytes (Pascussi (152)).
Mechanisms underlying suppression of CAR expression were analyzed by Assenat et al. (153), who showed that inflammatory signaling interfered with GR-regulated CAR-expression via NF-kappaB signaling. Activation of the latter led to decreased histone acetylation of the proximal CAR promoter (153). Of note, NF-kappaB signaling has also been shown to affect PXR function (Gu (140), Zhou (154)).
Apart from their roles in xenobiotic/drug metabolism, CAR and PXR are also involved in the protection against cholestatic liver injury in general (Stedman (155)) and, more specifically, against various “cholestatic compounds” such as hepatotoxic bile salts, notably lithocholate (Xie (156)), bilirubin (Saini (157)) and cholesterol metabolites (Sonoda (158)). It can therefore be anticipated that inflammation will not only affect drug metabolism, but will also render the liver less capable to deal with the ensuing cholestatic insult.
4.6.5. LXR
LPS-treatment of both hamsters and mice led to reduced LXR DNA-binding in the liver (Beigneux (130), Ghose (131)). This occurred simultaneously with reduced LXRalpha mRNA expression in hamster (130), while hepatic LXRalpha mRNA expression was actually slightly increased in LPS-treated mice (131). Reduced DNA-binding in mouse liver, associated with suppressed expression of the LXR target gene Abcg5, was ascribed to reduced nuclear RXRalpha levels (131). Treatment of mice with either TNFalpha or IL-1 was found to slightly reduce liver LXRalpha mRNA level (Kim (133)). In vitro experiments with Hep3B cells confirmed suppression of LXRalpha mRNA expression, reduced transactivating activity and suppressed target gene expression (SREBP1c) ((133)). In contrast to LPS-treated livers tissue and cytokine-treated hepatoma cells, LPS treatment of macrophages did not affect LXR DNA-binding (Khovidhunkit (159)), indicating cell-specific effects. Expression of the LXRbeta isoform was not significantly reduced upon LPS-treatment in mouse liver (Beigneux (130)).
4.6.6. PPARgamma
Hepatic PPARgamma mRNA expression was suppressed by LPS treatment of Syrian hamsters (Beigneux (130)). Similar results were obtained in mice, where LPS-induced suppression of PPARgamma mRNA and protein expression was shown to be dependent on TNFalpha release (Zhou (160)). In vitro, TNFalpha and IL-1beta were also shown to suppress PPARgamma expression in Hep3B cells (Kim (133)).
The mechanism underlying the reduced gene expression of PPARgamma by inflammatory signalling may involve NF-kappaB as was shown for LPS-induced suppression of PPARgamma in macrophages (Necela (161)).
4.6.7. HNF4alpha
HNF4alpha is a transcriptional regulator of many genes involved in hepatic lipid and bile salt metabolism (Hayhurst (162)). HNF4alpha appears to be constitutively active, supposedly due to a permanent association of lipids with its LBD (Wisely (163)). However, HNF4alpha is still considered an orphan NR, since no specific ligand has been identified. Considering the broad impact of HNF4alpha on hepatocellular gene expression profiles (Odom (164)), it has been postulated that effects of inflammatory signaling on HNF4alpha function might provide a central mechanism for initiation of the hepatic APR (Wang (132)).
Inflammatory signalling affects HNF4alpha function via multiple, often simultaneously acting, mechanisms. These mechanisms include reduction of mRNA expression, acceleration of proteasomal degradation, reduction of DNA-binding through post-transcriptional modifications and inhibition of coactivator recruitment (Wang (165), De Fabiani (166), Li (167), Cheng (168), Jahan (169), Zollner (139), Li(170), Nikolaidou-Neokosmidou(171), Wang (132), Krajewski (172)). Since these data suggest that inflammatory signalling indiscriminately leads to reduced HNF4alpha activity, it is important to emphasize that HNF4alpha activity can also be regulated in an opposite manner by other inflammation-related cues. Kuo and colleagues showed that in a different in vitro model of sepsis/shock, i.e., combined treatment of hepatocytes with cytokines and hydrogen peroxide, alteration of the specific phosphorylation pattern of HNF4alpha was critical for interaction with a specific coactivator and thus for enhancement of cytokine-induced iNOS expression by oxidative stress (Guo (173), Guo (174), Guo (175)). This indicates that post-transcriptional modification of HNF4alpha in the context of inflammatory processes has promoter-specific effects.
Finally, HNF4alpha was also shown to undergo tyrosine-phosphorylation signal-dependent intranuclear redistribution (Ktistaki (176)). Whether this also occurs in the setting of inflammation is unclear. It may, however, provide a new level of complexity to the regulation of NR function.
4.6.8. LRH-1
Gerbod-Giannone et al. (177) identified another mechanism by which NR function can be inhibited during inflammation. Inflammation-induced production of alpha1-antitrypsin leads to increased production of the alpha1-antitrypsin-derived peptide C-36. This peptide was shown to specifically reduce DNA-binding of LRH-1 and to inhibit LRH-1 regulated gene transcription (including Cyp7a1, alpha-fetoprotein). C-36 physically interacts with LRH-1, but did not bind to its DBD suggesting that C-36 induced conformational changes in LRH-1 (177). This interaction between bio-active peptides and a NR suggests that there may be more of these unanticipated interactions.
4.6.9. SHP
The expression of the atypical NR SHP is regulated by several other NR superfamily members, including FXR, ER and HNF4alpha, but also via an AP-1 binding site (Lu (178), Sinal (143), Goodwin (179), Lai (180)). The latter is most likely regulated by inflammatory signaling. LPS-treatment of mice leads to strongly reduced hepatic mRNA expression of SHP (Ghose (131), Zollner (139)). However, the exact mechanism underlying this effect and whether reduced mRNA levels are translated into reduced protein expression and activity, remains to be elucidated. Considering the general repressive effect of SHP on other NRs, it is interesting to speculate on the functional consequences of reduced SHP expression. The net outcome will depend on whether reduced repression by SHP or reduced expression of the other NRs prevails.
4.7. Effects of inflammation on co-factor expression and function
The importance of coregulators in NR action on gene transcription implies that interactions between inflammatory signaling and these coregulators will directly affect NR-regulated gene transcription. Effects of inflammation on co-factor expression and function can be categorized into three different general categories, i. reduction of coregulator expression/abundances, ii. competition between transcription factors for critical cofactors and iii. post-transcriptional modification of cofactors (Figure 5).
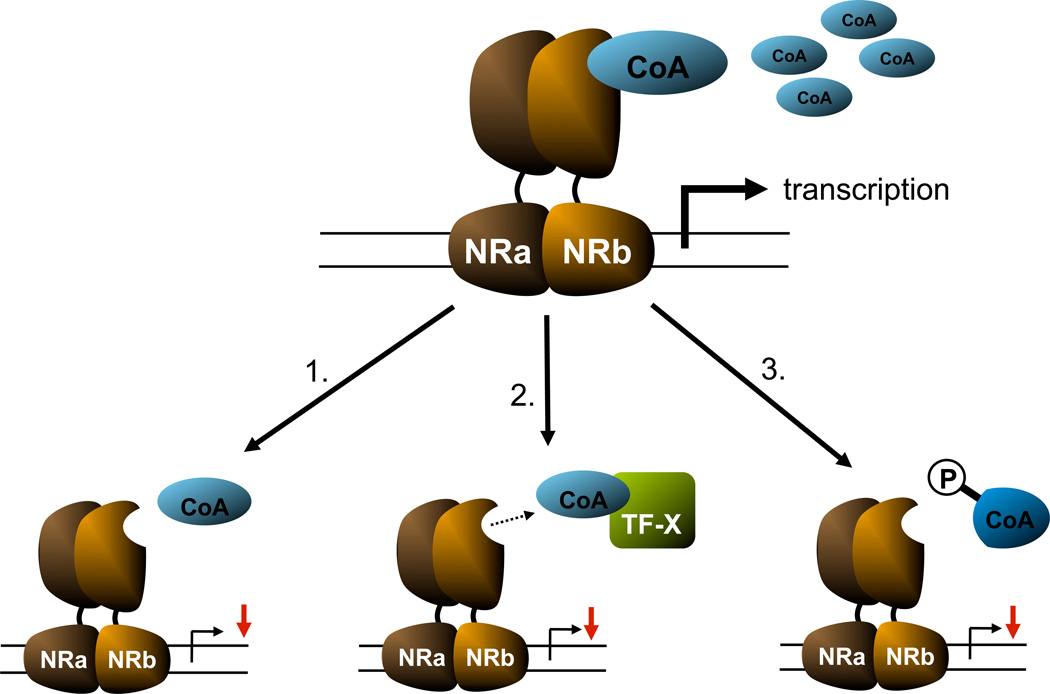
Figure 5.
General effects of inflammation on coregulators: 1) reduction of abundance 2) competition 3) post-transcriptional modification (simplified version: hypothetical ligand-activated NR-heterodimer (NRa and NRb); binding by a positive coregulator (CoA), transcription factor X (TF-X) competing for CoA, P = phosphorylation of CoA)
Kim et al. showed that inflammation leads to a reduced expression of a subset of co-factors in vitro and in vivo (133). Cytokine treatment (IL-1 and TNF, but not IL-6) of hepatoma cells or cytokine and LPS-treatment of mice led to a specific reduction in mRNA expression of the coactivators PGC-1alpha, PGC-1beta and SRC-1, while the expression of other co-activators, including CBP and TRAP, as well as corepressors NCoR and SMRT was not affected (133). This selective effect in combination with reduced NR expression led to reduced NR-mediated transcription using reporter constructs (541}. This supports the concept that a specific reduction in co-activating complexes shifts the balance between transactivating and transrepressing complexes towards the latter, ultimately suppressing gene transcription. Reduced expression of specific subsets of co-activators upon inflammatory stimuli was not only seen in liver, but also in other organs and tissues, including heart (Feingold (136)), adipose tissue (Lu (135)) and uterine smooth muscle cells (Leite (181)).
Leite et al. ((181)) provided evidence that there is a critical window of coactivator abundance, as the activity of the progesterone receptor (PR) is reduced during inflammation despite the fact that protein levels are not altered. This suggests that the concomitantly reduced levels of co-activators SRC-1 and SRC-2 are responsible for reduced PR function (181). This is further supported by the fact that exogenous SRC-1 or SRC-2 can at least partially reverse the effects of TNFalpha on PR function (181).
The concept of critical co-activator levels within the nucleus for proper NR function also provides the basis for another mechanism by which inflammation may interfere with NR function, namely competition between transcription factors. Despite large functional and structural differences, the individual NRs often recruit the same co-factors as other types of transcription factors (Perissi (60)). Therefore, initiation of inflammatory signaling within cells and the subsequent activation of transcription factors such as NF-kappaB and AP-1 will lead to the recruitment of coactivators. If coactivators are not redundantly present within cells, competition for coactivators can lead to insufficient coactivator-NR interaction and thus to reduced NR function. This principle of competition has been shown to occur between GR and NF-kappaB, as these transcription factors were shown to both be dependent on coactivators SRC-1 and CBP for maximal activity (Sheppard (182)). Again, exogenous supplementation of either coactivator was able to reverse this competition (182). Similar findings were reported for other NRs, including RXR (Na (183)).
A third mechanism by which inflammation can affect co-factor function is by post-transcriptional modification. It is known that, similarly to NRs, co-factors can be subjected to (de)phosphorylation, (de)acetylation and (de)methylation and their activity can further be controlled by proteolytic processes and shuttling between nucleus and cytoplasm in response to various signals (Hermanson (184)). For example, TNF-alpha-induced, IKK-mediated phosphorylation of SRC-3 leads to preferential nuclear localization of this co-activator and enhanced NF-kappaB mediated gene transcription (Wu (185)). Although this is an example of increased co-activator activity, it is plausible that inflammatory signaling may lead to reduced activity of co-activators in other settings. The multiple sites in coregulators for post-transcriptional/translational modification provide a means to rapidly regulate coregulator function with a need to alter gene expression (McKenna (186)).
Thus, inflammatory stimuli can have multiple effects on co-factors that may directly translate into disturbed NR function. The overall effect, however, is most likely context-dependent, i.e., specific for type of stimulus and tissue involved, and not easily predicted. Moreover, a recent report by Lu et al. (187) adds another level of complexity of NR-cofactor function. These authors showed that the coactivator SRC-3 is specifically involved in suppressing the innate immune response. SRC-3 knock-out mice were shown to be highly sensitive to LPS-treatment. Surprisingly, SRC-3 was shown to act as a suppressor of mRNA translation. This unexpected interaction between inflammation and co-factor function warrants caution in predicting how co-factors will affect inflammatory processes.
5. PHARMACOLOGICAL/EXPERIMENTAL INTERVENTIONS IN MODELS OF IIC
Better insight into the pathogenesis of specifically IIC and the negative APR in general has provided the opportunity to explore different, intriguing approaches to intervene in these processes. These approaches have been aimed at different levels of the cascade leading to IIC (Figure 3).
Administration of high-density lipoprotein particles (Pajkrt (188), Thabut (189)), recombinant LPS-binding protein (Lamping (190)) or TLR4 antagonists, e.g., M62812 (Nakamura (112)) have been used to attenuate the initial step of KC activation. Liposomal siRNA against TNFalpha was used to specifically suppress production of this cytokine in KC, which rapidly take up liposomes (Jing (191)). Inhibition of signaling downstream of KCs has been achieved by using cytokine-inactivating antibodies, e.g. anti-TNFalpha, anti-IL-1beta (Whiting (29), Sturm (49), Geier (109)), or inhibitors of hepatocellular signaling, e.g., JNK-inhibitors as SP600125 (Zimmerman (121)).
6. DIFFERENT ROLES OF NRs IN IIC: MEDIATORS AND MODIFIERS
Thus far, this review has mainly dealt with effects of inflammatory signaling on NR function that contribute to the pathogenesis of IIC. Inflammatory signaling has multiple potential effects on NR function through reduction of NR mRNA and protein expression, subcellular localization, post-transcriptional modifications with subsequent reduced DNA-binding and/or coregulator recruitment and altered coregulator expression or activity (Figures 4 and 5). In this regard, NRs can be seen as mediators in the development of IIC. On the other hand, NRs can also play modifying roles, which can be divided into two modes of action. NRs can exert adaptive responses aimed at restoration of normal hepatocellular homeostasis and NRs have been shown to have anti-inflammatory properties.
6.1. Modifier function 1: adaptive responses
During cholestasis, in particular of extrahepatic origin, hepatocellular accumulation of bile salts and other potentially toxic compounds will lead to an adaptive cellular response (Trauner (192)). This means that, in theory, accumulation of bile salts could lead to an adapative response during IIC too. High intracellular bile salt concentrations will activate FXR and thus induce their export from the liver while reducing import and production. However, this adaptive response requires “sound” NR signaling. It is, however, unlikely that such an appropriate adaptive response can be fully engaged, since inflammation affects NR signaling in many different ways. Although the quantitative contribution of adaptive responses remains to be determined, it probably is limited. The latter idea is best illustrated by the reported findings of Zollner et al.: bile-duct ligation and bile salt treatment of mice leads to increased SHP mRNA expression, while LPS-treatment strongly suppresses SHP mRNA expression (Zollner (193), Zollner (139)). The latter suggests that in the context of LPS, direct suppressive effects of inflammatory signaling on SHP expression overrule the anticipated stimulating effect of bile salt accumulation.
In this regard, it is important to mention the concept of pharmacologic enhancement of normal adaptive responses (Boyer (194)) as a means to intervene in the pathogenesis of IIC. Although inflammation generally impairs NR function, boosting the remaining activity using pharmacological agents may provide an avenue to at least attenuate the effects of inflammation on hepatobiliary transporter expression.
6.2. Modifier function 2: anti-inflammatory actions
Several NRs have been shown to possess anti-inflammatory properties. Although most of the studies exploring these NR properties have dealt with other models of inflammatory diseases, most notably atherosclerosis, it implies that use of NR ligands to suppress inflammatory signaling may be a useful means to intervene in the pathogenesis of IIC as well. This approach has been explored already by several groups used by Miyake et al. (23), Ghose et al. (195), Wang et al. (196). The following sections will further focus on the mechanisms of anti-inflammatory effects of NRs (Figure 6).
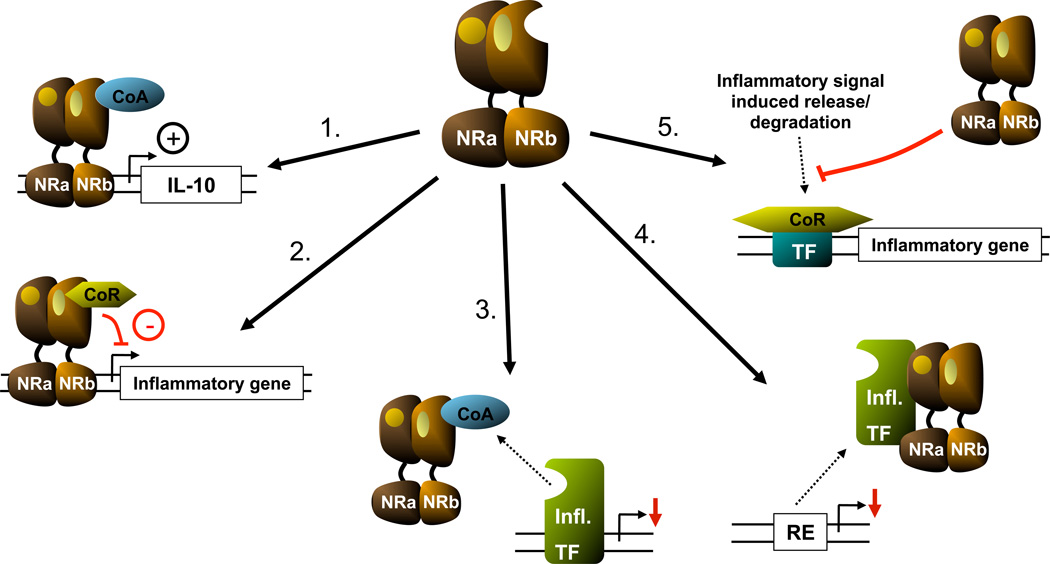
Figure 6.
Mechanisms behind anti-inflammatory actions of NRs: 1) induction of anti-inflammatory mediators through transactivation; 2) transrepression of (pro-)inflammatory genes; 3) competition for co-activators; 4) interaction with transcription factors induced by inflammatory signaling; 5) interference with release of corepressor complexes associated with inflammatory genes
6.2.1 Glucocorticoid receptor (GR)
The glucocorticoid receptor (GR) has been studied extensively for its anti-inflammatory properties (De Bosscher et al. (197)). Glucocorticoids remain the mainstay of therapeutic options for a wide variety of conditions involving a detrimental inflammatory response, including sepsis, asthma, and autoimmune diseases (Rhen (198)). GR, more in particular the GRalpha isoform, appears to have multiple effects on inflammatory signaling, both through activation of gene expression (transactivation) and through inhibition of gene expression (transrepression) (Smoak (199)). Examples of transactivation include the induction of the anti-inflammatory cytokine IL-10, the IL-1 receptor antagonist and the inhibitor of NF-kappaB. The transrepressive mechanisms appear to be more diverse and include, but are not limited to, inhibition of gene expression, competition for cofactors, the physical interaction between GR and other transcription factors, and effects on chromatin-remodelling (199). Glucocorticoids have been found to be effective in various experimental settings of inflammation-induced cholestasis, both in vitro and in vivo (Roelofsen (200), Kubitz (42), Chen (39), Cherrington (124)). However, despite these effects in experimental settings, this has not led to the widespread clinical use of glucocorticoids for IIC, although they are used in some other types of cholestatic liver disease (e.g., post-Kasai).
6.2.2 PPARgamma
PPARgamma was among the first to be attributed with anti-inflammatory properties (along with PPARalpha). Much of this initial work dealt with the development of atherosclerosis. A central role in the pathogenesis of atherosclerosis is played by peripheral macrophages. PPARgamma was found to exert anti-inflammatory effects (Ricote (201), Jiang (202)).
Considering the role of KCs in the pathogenesis of endotoxemic shock, Uchimura and colleagues (203) investigated the potential effect of the PPARgamma agonist pioglitazone and of the RXR agonist Ro47-5944, on LPS-stimulated rat Kupffer cells and found that both were able to suppress the production of TNFalpha and NO and the transactivation activity of NF-kappaB. This effect, however, was not mediated by reduced DNA-binding of the transcription factors AP-1/NF-kappaB (203). Since no PPARgamma/RXR response element was found in either the TNFalpha or iNOS promoter, these authors suggested activated PPARgamma/RXR to interfere non-genomically with the transcriptional activity of the pro-inflammatory transcription factors, perhaps through competition for important co-activators. This concept had been already been addressed by Li et al. (204), who showed that in macrophages PPARgamma transrepressed iNOS gene expression through interaction with co-activator CBP.
A different mechanism behind PPARgamma induced transrepression was elucidated by Pascual et al. (205) who showed that the transrepression of NF-kappaB signaling by ligand-activated PPARgamma involved prevention of the clearance of NCoR-HDAC3 from the basally repressed iNOS promoter, whose activation requires recruitment of NF-kB. This clearance is normally initiated upon a pro-inflammatory stimulus, but ligand-dependent SUMOylation of PPARgamma prevented this process. In retrospect, this mode of action appears to be in agreement with the anti-inflammatory effects of PPARgamma-RXR in rat Kupffer cells (Uchimura (203)), which might also involve prevention of co-repressor clearance.
The effect of PPARgamma-agonist treatment in the setting of IIC has been addressed by Ghose et al. (195), who showed that pre-treatment of mice with rosiglitazone partially preserved gene expression of critical hepatobiliary transporters involved in bile formation. This, however, did not seem to be mediated by Kupffer cells, as cytokine expression and production were not significantly altered by rosiglitazone-pretreatment, but most likely involves the preservation of nuclear RXRalpha levels in the hepatocytes (195). Miyake et al. (23) used rosiglitazone in a mouse model of hepatic inflammation induced by a bile-acid containing pro-atherogenic diet. Rosiglitazone was able to suppress the effect of diet-induced suppression of Cyp7a1, which was indirectly shown to be dependent on macrophage/KC inflammatory signaling (23).
Lastly, PPARgamma ligands also exert anti-inflammatory actions through gene transactivation. Similarly to the induction of IL-10 and the IL-1 receptor antagonist by glucocorticoids, PPARgamma ligands induce the soluble IL-1 receptor antagonist in the THP-1 macrophage cell-line (Meier (206)).
6.2.3 LXR
LXR is critical for maintenance of cellular cholesterol homeostasis (Janowski (207), Lehmann (208)). In peripheral macrophages, LXR activation leads to an increased expression of transporters involved in reverse cholesterol transport, including Abca1 (Venkateswaran (209)) and Abcg1 (Venkateswaran (210), Sabol (211)) and functionally to a reduction of atherosclerosis in relevant mouse models (Joseph (212), Tangirala (213), Terasaka (214)). LXR activation in hepatocytes induces expression of the canalicular half-transporters Abcg5 and Abcg8 and stimulates biliary cholesterol excretion (Repa (215), Plösch (216)). Joseph et al. (217) reported that LXR activation inhibited the macrophage inflammatory gene response both in vitro and in vivo. This effect could be exerted by either isoform of LXR and appeared to involve inhibition of NF-kappaB signaling (217). Follow-up studies by Castrillo et al. (218) demonstrated that ligand-activated LXR inhibited inflammatory signalling, leading to macrophage MMP-9 expression at least in part through interference with NF-kappaB signaling downstream of its binding to DNA. No evidence was found for interference with AP-1 signaling or for direct LXR:RXR binding to the 5’-flanking region (218). Since then, multiple groups have reported on this anti-inflammatory effect of LXR(-agonists) in macrophages (Terasaka (219), Ogawa (220), Birrell (221), Fontaine (222)) and other cell-types, including lymphocytes (Walcher (223)), keratinocytes (Fowler (224)), Kupffer cells (Wang (196)), hepatocytes (Blaschke (225), microglia and astrocytes (Zhang-Ghandi (226)) and polymorphonuclear neutrophils (Smoak (227)). Interestingly, Ogawa et al. ((220)) found LXR-agonists to inhibit inflammation-induced expression of osteopontin through interference with AP-1 signaling. It remains unclear what causes the discrepancy between this finding and the earlier observed lack of effect on AP-1 signaling (218;219). There may also be a difference between the anti-inflammatory actions in mice and humans as Fontaine et al. (222) showed that in human macrophages LXR(-agonists) increased the expression of TLR4 and the response to LPS-challenge via an LXRE in the TLR4 promoter, which is not conserved in mice.
Inspired by studies on anti-inflammatory actions of PPARgamma, Ghisletti et al. (228) sought to determine whether similar mechanisms might be involved in the anti-inflammatory actions of LXRs. Interestingly, similar mechanisms involving SUMOylation (albeit by SUMO2/3 rather than SUMO1) of ligand-bound LXR were found, which prevented the clearance of NCoR from the iNOS (or other inflammatory gene) promoter (228). Reduced clearance of NCoR by ligand activated LXR has also been shown to be the mechanism underlying the inhibition of hepatocellular C-reactive protein expression (Blaschke (225)).
Considering the anti-inflammatory effects of LXRs and their expression in KCs we recently examined the potential of LXR-agonists to interrupt the inflammatory cascade leading to LPS-induced suppression of hepatobiliary transporter expression. Although treatment of mice with T0901317 led to partial preservation of transporter gene expression and preserved Mrp2 protein expression, this appeared to be KC-independent, since cytokine expression was not altered. Wang et al. employed a different LXR-agonist in a more severe model of hepatic inflammation (LPS/Gal-treatment of rats) and found the LXR agonist to be protective and associated with a reduced KC-response (196).
Similarly to GR, LXR can also modify inflammatory response “genomically” through direct transactivation, e.g., through induction of ArgII, which can compete with iNOS for substrate (Marathe (229)) . Furthermore, it was recently shown that LXRalpha can directly repress gene expression through promoter specific recruitment of the corepressor RIP140 (Wang (230)). It is tempting to speculate that LXR may repress inflammatory genes via such a mechanism too.
6.2.4 RXRalpha and RARalpha
The anti-inflammatory effects of retinoids have been used clinically for several decades, especially in the treatment of various dermatological conditions (e.g., psoriasis, acne) (Orfanos (231)). Initial studies addressed the effects of both types of receptor on regulation of TGFbeta1 (Salbert (232)). Both receptors were shown to inhibit AP-1 signalling through physical interaction (232).
Pertaining to IIC, RXRalpha agonists were shown to inhibit TNFalpha release and nitrite production by primary Kupffer cell (Motomura (233)). These results were confirmed by Uchimura et al. ((203)), who went on to show that this effect was associated with reduced NF-kappaB transcriptional activity. RARalpha-selective ligands, on the other hand, were not effective in vitro (Motomura (233)). The latter was surprising as all-trans retinoic acid was shown to blunt LPS and Propionibacterium acnes induced liver injury in vivo (Motomura (234)), indicating that RARalpha agonists can initiate anti-inflammatory mechanisms at least in vivo.
Ligand-bound RXRalpha was able to inhibit NF-kappaB transcriptional activity (203) and appeared to do so similarly to PPARgamma:RXRalpha heterodimer, suggesting that similar mechanisms as described for PPARgamma and LXR might be active (i.e., inhibition of co-repressor clearance). Na et al. showed that RXR-ligands reduced LPS-induced cytokine expression in mouse macrophages via at least two mechanisms, i.e., either through physical interactions with NF-kappaB subunits p50 and p65, but also through co-activator competition ((183)). Anti-inflammatory effects of retinoids have also been shown in other cell-types, e.g., microglia and astrocytes (Zhang-Ghandi (226). Finally, the observation of increased liver cytokine gene expression in hepatocyte-specific RXRalpha-deficient mice after alcohol-treatment suggests that RXRalpha may very well have anti-inflammatory effects under basal conditions (Gyamfi (235))
6.2.5 Anti-inflammatory effects of other NRs
Besides GR, PPARgamma and LXRa, other NRs involved in bile formation, lipid homeostasis and bile salt metabolism have also been attributed anti-inflammatory properties.
Although FXR has been shown to modify disease processes involving inflammation (including atherosclerosis (Hanniman (236)) and cholesterol gallstone disease (237)), limited information is available on direct effects of FXR on inflammatory signaling in liver cells. Hepatic cytokine gene expression after LPS treatment was not different between in FXR+/+ and FXR −/− mice (Zollner (139)). However, one has to bear in mind that, in this study, FXR-agonists were not used to analyze the effect of pharmacological activation of FXR on LPS-induced cytokine expression (139). In CBDL mice, FXR-activation by GW4064 led to a decreased expression of the pro-fibrogenic cytokine TGFbeta (Liu (238), although no underlying mechanism was provided. It remains to be determined whether this reduced expression is a concurrent effect of disease process modification rather than a direct effect on TGFbeta expression. Interestingly, it was recently shown in models of diabetic nephropathy, which is associated with increased fibrogenesis/inflammation, that FXR agonists such as cholate and GW4064 reduced IL-6 and TGFbeta mRNA expression (Jiang (239)). This suggests that this effect of FXR is a more general phenomenon, because it is not limited to liver cells and because different disease processes that lead to fibrogenesis in different cell types respond similarly. Li et al. (240) showed that FXR-ligands were able to inhibit inflammatory response in rat/human vascular smooth muscle cells (VSMCs) in a FXR- and SHP-dependent manner, with concurrent reduction in NF-kappaB activation.
In contrast with a potential anti-inflammatory role, Qin et al. (241) showed that FXR mediates the bile salt-induced expression of intracellular adhesion molecule-1 (ICAM-1), which is involved in the recruitment of neutrophils to the site of inflammation. By inducing ICAM-1, FXR will actually enhance the inflammatory process. Furthermore, FXR activation led to a SHP-mediated derepression of MMP-9 in endothelial cells (242). MMP-9, involved in tissue remodelling including atherosclerosis, was shown to be negatively regulated by LXR (218), suggesting opposite effects of these two nuclear receptors.
It clearly remains to be further investigated how FXR modulates inflammatory processes overall and whether some the effects described above are concurrent effects of FXR agonists on disease progression or distinct anti-inflammatory effects.
PXR is another NR that is able to interfere with NF-kappaB signaling (Zhou (154)). Different PXR ligands were able to suppress basal or induced expression of NF-kappaB target genes and this suppression was dependent on PXR expression. Furthermore, primary hepatocytes derived from PXR-KO mice showed increased expression of NF-kappaB target genes, indicating a general anti-inflammatory effect. A mechanistic explanation, however, was not provided for these observations. Similar observations of the anti-inflammatory characteristics of PXR were done in a mouse model of inflammatory-bowel disease (Shah (243)).
LRH-1 was recently also shown to inhibit the hepatic inflammatory response via at least two different mechanisms (Venteclef (244), Venteclef (245)). First, LRH-1 was shown to specifically interfere with C/EBPbeta transcriptional activity induced by different cytokines, while it had no effect on simultaneous induction of NF-kappaB and AP-1 activity (244). Subsequent studies revealed that LRH-1 also induces expression of secreted IL-1RA by hepatocytes under inflammatory conditions (245). The latter indicates that LRH-1 not only directly interferes with inflammatory signaling (C/EBPbeta), but also indirectly through induction of an anti-inflammatory mediator (IL-1RA). Both mechanisms appear to act synergistically. Of interest, Mueller et al. reported that LRH-1 mediates the induction of extra-adrenal glucocorticoid synthesis, i.e. by the intestine (246)), which has been linked to reduced inflammatory responses in the setting of inflammatory bowel disease (Coste (247)). It would be relevant to examine whether such mechanisms might also be at work in the pathogenesis of IIC.
Although generally regarded as a transcriptional suppressor, only limited information is available on a potential role of SHP in modulation of inflammatory signaling. Some of the mechanisms by which SHP exerts its repressive effects on gene transcription, e.g., competition for binding with coactivators through physical interaction with NRs/TFs (Lee (248), Yamagata (249)) are very similar to those by which other NRs interfere with inflammatory signaling. It seems therefore plausible that SHP interferes with inflammatory cascades. This has already been shown for TGFbeta1 signaling (Suh (250)). TGFbeta1, a cytokine involved liver regeneration and fibrosis, activate, amongst other pathways, SMAD-signaling (Gressner (251)). SHP was shown to inhibit TGFbeta1-induced gene responses through physical interaction with SMAD3, preventing binding of the coactivator p300 ((250)). As mentioned above, Li (240) showed that FXR-mediated induction of SHP expression was able to inhibit the inflammatory response in rat and human VSMCs by interfering with NF-kappaB activation. The exact molecular mechanism of action, however, remains to be revealed.
6.2.6 General considerations regarding the anti-inflammatory effects of NRs
Although some NRs seem to act clearly in an anti-inflammatory fashion (e.g., GR), the effects of others, i.e., FXR and LXR, appear to be rather unpredictable and are gene-specific. The inflammatory response is an extremely complex process which balances effects aimed at protection of the organism with potential detrimental effects. This balance, in turn, involves multiple delicately organized processes in different cell-types in response to multiple stimuli which change over time with various feedback systems. It might therefore be an oversimplification to expect that one particular NR will either be pro-inflammatory, neutral or anti-inflammatory as this may be cell type-, species- and context-dependent. In its overall complexity, it appears that inflammatory signaling involves several different, often redundant pathways. Considering the different modes of anti-inflammatory actions of the NRs, one could propose that targeting multiple NRs simultaneously may lead to synergistic anti-inflammatory effects. The latter has indeed been shown for combinations of GR- and PPARgamma ligands (Ogawa (252)) as well as LXR- and PPARgamma ligands (Piraino (253)). This concept is especially interesting with regard to clinical application of NR ligands in inflammatory therapeutic regimes as combining ligands may reduce side-effects associated with the use of single ligands (Glass (254)).
7. DESIGN OF OPTIMAL NR LIGANDS FOR INTERVENTION IN IIC
In the previous sections, the involvement of NRs in the pathophysiology of IIC as well as their potential role as modifiers of this condition have been discussed. The latter concept makes use of the key characteristic of NRs that separates them from other TFs, namely their ligand-binding properties. Although natural ligands have been identified for several of the NRs discussed above and synthetic ligands have become available, there remains a clear necessity for greater understanding, and a broadening of exploration of the efficacy of different ligands as anti-inflammatory agents. Besides optimizing the general pharmacokinetic characteristics, i.e., absorption, distribution, elimination and metabolism of NR-ligands, and perhaps the application of advanced drug targeting strategies, the concept of selective NR modulators (SNuRMs) may provide an alternative approach in the quest for ideal NR ligands.
NRs function as molecular links between genomic information (genes/promoters), environmental/hormonal cues and the effector machinery for active gene transcription. However, NRs should not be regarded as rigid mediators that serve as “on/off switches”, whose interactions with ligands resemble key-lock interactions. The overall outcome of ligand-binding to a NR is determined at multiple levels. First of all, although not all NRs are as promiscuous with regard to their ligand-binding as for instance CAR and PXR, NRs can generally be bound by different, more or less related ligands. Differential ligand binding will lead to different conformational changes, which will affect co-factor recruitment. Secondly, it has also been shown that very subtle differences in cis-acting response-elements can have an influence of cofactor recruitment by NRs as well as other TFs. The latter was illustrated by Leung et al. who showed that a single-nucleotide difference in a kappaB-site affected cofactor recruitment to NF-kappaB dimers. This illustrates the extent to which NR/TF actions are fine-tuned and that DNA can be considered as an allosteric modifier of TF function (Rosenfeld (255)) Thirdly, cofactors can be regarded as the effector molecules of NR function. Since this large group of molecules has its own (spatial and temporal) expression profile, binding of a single ligand to a specific NR may have transcriptional effects that are context-dependent (organ, tissue, cell-type, promoter). These aspects all contribute to the complexity of DNA-NR-cofactor interaction. This feature of NR biology makes it near to impossible to accurately predict the outcome of NR-binding by a single ligand, yet, it simultaneously provides an opportunity to search for compounds that acts as SNuRMs. Kremoser et al. reviewed a series of approaches to search for effective SNuRMs ((256)).
A good example of a clinically used SNuRMs are the selective modulators of the estrogen receptor (SERMs), tamoxifen and raloxifene (Jordan (62)). Tamoxifen is widely used as an adjuvant treatment in breast cancer care, while raloxifene is used as anti-osteoporosis agent. These drugs act as antagonists of the estrogen receptor in some tissues, but as agonists of the same receptor in other tissues. The underlying mechanism of this tissue specificity primarily involves differences in coregulator expression and activity (Jordan (62)). SERMs are actually mixed agonists/antagonists and their binding induces conformational changes, which lead to changes co-activator and co-regulator recruitment. The exact pattern of recruitment depends on coregulator expression and post-transcriptional modification. The development/identification of such selective modulators for other NRs would potentially be a great stride forward, especially for those NRs whose general modulation/activation appears to be of benefit in certain settings, but whose further development is hampered by undesired activation of other subsets of genes.
Tissue- and gene-specific activation of LXR is probably one of the greatest challenges in the NR field. Although the anti-inflammatory (Joseph (217)) and plaque-reducing effects (Joseph (212), Tangirala (213), Terasaka (214))) are obvious from mouse studies, current LXR agonists have also been shown to induce de novo lipogenesis in hepatocytes with increased plasma triglycerides and hepatic steatotosis as detrimental results. The metabolic consequences of these particular agonists currently preclude their clinical use. In the search for other LXR ligands with great therapeutic potential, one can take different approaches. First of all, ligands which are preferentially taken up by macrophages or specifically targeted to KCs, rather than by hepatocytes, should in theory limit the detrimental hepatocellular LXR-activation. Organ selectivity has been described for one of the current LXR-agonists, i.e., GW3965 (Brunham (257)). LXRbeta selective ligands would also be appealing alternatives, since LXRbeta expression is lower than that of LXRalpha in hepatocytes, while expression levels of both are more comparable in macrophages. Although LXRbeta-selectivity may a priori not be easily achieved as the LBDs of the two LXR-isoforms are highly similar, several groups have reported on LXRbeta selective LXR-ligands (Molteni (258), Hu (259)). In addition to cell-, organ- and isoform-specific LXR-ligands, development of SLXRMs that are specifically active in non-hepatocyte cells or inactive on promoter of lipogenic genes would be a great therapeutic potential.
7.1. Other approaches
As described previously, coregulators are very important in regulation of gene transcription by NRs. Initially considered to be mere executors in the regulation of gene expression by NRs, this clearly underestimates the role of coregulators. This can be illustrated by the transcriptional regulation of phosphoenolpyruvate carboxykinase (PEPCK), an enzyme centrally involved in gluconeogenesis. The gluconeogenic response is dependent on proper HNF4alpha function, but binding of this NR is not altered during fasting, when PEPCK transcription is induced. This induction was regulated by increased expression of coactivator PGC1alpha (Yoon (260), Rhee (261)), illustrating both that coregulators are more than mere bystanders in the process of gene transcription and that methods to alter coregulator abundance/modification may provide another approach to modify NR-function. Although even more far-fetched, the concept of coactivator rescue, e.g., restoring NF-kappaB transactivation capacity by supplying an exogenous coactivator (Leung (262)) is also intriguing. However, due to the complexity of the transcriptional complex, it is difficult to predict what the implications of alterations of single coregulators may be in vivo. So far, this concept has not been pursued in the context of IIC.
8. SUMMARY AND PERSPECTIVE
IIC is a frequently occurring phenomenon and with substantial morbidity and mortality. NRs play a dual role in the pathogenesis of IIC. On one hand, NRs contribute to disease development, as they are targets of inflammatory signalling themselves. Whether it be through direct effects on their expression levels or functionality or through indirect effects on their coregulators, altered NR activity affects basal expression levels of important hepatobiliary transport systems. Hence, NRs can be considered to be important mediators of IIC. On the other hand, NRs also play important roles in adaptive responses, for instance during bile salt accumulation within hepatocytes through FXR activation. Several NRs have also been shown to possess anti-inflammatory effects. Therefore, NRs can also act as modifiers of IIC. The latter concept makes it interesting to determine whether NRs can be exploited as pharmacological targets to intervene in IIC, either through boosting adaptive responses or induce anti-inflammatory responses. Identification of new NR ligands or SNuRMs with characteristics as described in the previous section would be ideal.
Although sepsis-associated cholestasis is the most widely known example of IIC, cholestasis can also occur in the setting of many other inflammation-related conditions associated with an generalized APR, e.g., severe burn injury or trauma. Moreover, inflammatory signalling also appears to be involved in other conditions characterized by cholestasis. An example of the latter is biliary atresia (BA), the most common neonatal cholestatic disorder, which is characterized by progressive inflammation, fibrosis and, subsequently, obliteration of the bile ducts ultimately leading to bile retention and biliary cirrhosis (Mack (263)). This multifactorial pathophysiological process appears to include components of a persistent (auto)immune response after a perinatal, infectious insult or aberrant bile duct formation (263). Several cytokines, e.g., TNFalpha, IFN-gamma and osteopontin (Mack (264), Shivakumar (265), Whitington (266)), have been implicated as potentially important mediators and are in part secreted by recruited macrophages/monocytes (Mack (267)). This indicates that anti-inflammatory agents aimed at macrophages may have a role in treatment of BA. Perhaps PPARgamma or LXR agonists may be suitable agents to explore. Interestingly, glucocorticoids have often been used in the treatment of BA, but only after the primary surgical intervention, i.e., Kasai portoenterostomy, with the underlying thoughts that glucocorticoids will enhance bile formation and reduce inflammation, and thus increase the chances of establishing permanent post-Kasai bile drainage (Sokol (268)).
Total parenteral nutrition associated cholestasis (TPNAC) is another, rather frequently occurring condition that shows the close link between inflammation and cholestasis. Although the exact pathogenesis of TPNAC remains unclear, many factors have be implicated to play a causative role, including increased portal LPS-levels and individual components of TPN formulas (Carter (269)). With regard to the latter, it was recently shown that a the phytosterol, stigmasterol, can act as an FXR antagonist (Carter (270)), which is expected to impair hepatic adaptive responses to bile salt retention. Implicitly, this also suggests that FXR-agonists are potentially of benefit under such conditions by presumably enhancing adaptive responses. Furthermore, FXR also appears to have anti-inflammatory characteristics within the intestine (Inagaki (271)). This suggests that NR ligands are of potential use in TPNAC either as anti-inflammatory agents or as adaptation-boosting agents.
These two examples (BA and TPNAC) further illustrate the link between inflammation and cholestasis, which probably ought to be regarded as intertwined phenomena. Not only does inflammation lead to cholestasis (as in IIC), cholestasis per se leads to hepatic inflammation as well, as reduced intestinal delivery of bile may lead to bacterial overgrowth, translocation and an inflammatory response (Trauner (11)). Finally, hepatocellular damage, regardless of its cause, will activate inflammatory signaling and thus further impair liver function in part via inducing cholestasis. Studies in bile duct-ligated mice, a well known model of acute, extrahepatic cholestasis, have shown that there is an inflammatory response that further worsens liver injury (Gujral (272), Gujral (273)). The underlying mechanisms, however, remain unclear as BDL also leads to reduced expression of hepatic transporters independently of cytokine expression (Geier (274)) or degree of inflammatory response (Wagner (25)).
In conclusion, NRs play dual roles in the setting of IIC. Increased understanding of the pathogenesis of IIC has shown us how centrally involved NRs and their coregulators are in the regulation of hepatobiliary transport systems. Simultaneously, this has provided us with novel therapeutic strategies aimed at maintaining or even boosting NR function during inflammation, enabling proper/necessary adaptive responses. Furthermore, future application of new selective anti-inflammatory agents (such as SPARMs, SLXRMs) would expand the therapeutic arsenal importantly making us less dependent on traditional compounds as glucocorticoids.
ACKNOWLEDGEMENT
This work was supported by grants from ZonMW (AGIKO-stipend NR-920-03-287 to JM), KNAW Ter Meulen Fonds (to JM) and TI/Pharma (T2.110 to FK).
REFERENCES
- 1.Byers JW. 1887 Clinical lecture on the jaundice of infants. [Google Scholar]
- 2.Bernstein J, Brown AK. Pediatrics. 1962;29:873–882. [PubMed] [Google Scholar]
- 3.Hamilton JR, Sass-Kortsak A. J. Pediatr. 1963;63:121–132. doi: 10.1016/s0022-3476(63)80310-8. [DOI] [PubMed] [Google Scholar]
- 4.Crawford JM, Boyer JL. Hepatology. 1998;28:253–260. doi: 10.1002/hep.510280133. [DOI] [PubMed] [Google Scholar]
- 5.Lester R. Am. J. Surg. 1968;116:342–345. doi: 10.1016/0002-9610(68)90221-3. [DOI] [PubMed] [Google Scholar]
- 6.Baumann H, Gauldie J. Immunol. Today. 1994;15:74–80. doi: 10.1016/0167-5699(94)90137-6. [DOI] [PubMed] [Google Scholar]
- 7.Moshage H. J. Pathol. 1997;181:257–266. doi: 10.1002/(SICI)1096-9896(199703)181:3<257::AID-PATH756>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 8.Birch CA. 1937 Jaundice complicating pneumonia. [Google Scholar]
- 9.Eley A, Hargreaves T, Lambert HP. Br. Med. J. 1965;2:75–77. doi: 10.1136/bmj.2.5453.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia FJ, Nager AL. Pediatrics. 2002;109:846–851. doi: 10.1542/peds.109.5.846. [DOI] [PubMed] [Google Scholar]
- 11.Trauner M, Fickert P, Stauber RE. J Gastroenterol Hepatol. 1999;14:946–959. doi: 10.1046/j.1440-1746.1999.01982.x. [DOI] [PubMed] [Google Scholar]
- 12.Sherman ML, Spriggs DR, Arthur KA, Imamura K, Frei E, III, Kufe DW. J. Clin. Oncol. 1988;6:344–350. doi: 10.1200/JCO.1988.6.2.344. [DOI] [PubMed] [Google Scholar]
- 13.Fisher B, Keenan AM, Garra BS, Steinberg SM, White DE, DiBisceglie AM, Hoofnagle JH, Yolles P, Rosenberg SA, Lotze MT. J. Clin. Oncol. 1989;7:1852–1862. doi: 10.1200/JCO.1989.7.12.1852. [DOI] [PubMed] [Google Scholar]
- 14.Blay JY, Rossi JF, Wijdenes J, Menetrier-Caux C, Schemann S, Negrier S, Philip T, Favrot M. Int. J. Cancer. 1997;72:424–430. doi: 10.1002/(sici)1097-0215(19970729)72:3<424::aid-ijc9>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 15.Prommer E. J. Pain Symptom. Manage. 2007;34:225–226. doi: 10.1016/j.jpainsymman.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 16.Whitehead MW, Hainsworth I, Kingham JG. Gut. 2001;48:409–413. doi: 10.1136/gut.48.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.te Boekhorst T, Urlus M, Doesburg W, Yap SH, Goris RJ. J Hepatol. 1988;7:111–117. doi: 10.1016/s0168-8278(88)80514-2. [DOI] [PubMed] [Google Scholar]
- 18.Moseley RH. Clin. Liver Dis. 2004;8:83–94. doi: 10.1016/S1089-3261(03)00134-X. [DOI] [PubMed] [Google Scholar]
- 19.Poltorak A, He X, Smirnova I, Liu MY, Van HC, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 20.Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 21.Haziot A, Ferrero E, Kontgen F, Hijiya N, Yamamoto S, Silver J, Stewart CL, Goyert SM. Immunity. 1996;4:407–414. doi: 10.1016/s1074-7613(00)80254-x. [DOI] [PubMed] [Google Scholar]
- 22.Su GL. Am. J. Physiol Gastrointest. Liver Physiol. 2002;283:G256–G265. doi: 10.1152/ajpgi.00550.2001. [DOI] [PubMed] [Google Scholar]
- 23.Miyake JH, Wang SL, Davis RA. J Biol Chem. 2000;275:21805–21808. doi: 10.1074/jbc.C000275200. [DOI] [PubMed] [Google Scholar]
- 24.Li N, Choudhuri S, Cherrington NJ, Klaassen CD. Drug Metab Dispos. 2004;32:1265–1271. doi: 10.1124/dmd.32.11.. [DOI] [PubMed] [Google Scholar]
- 25.Wagner M, Zollner G, Fickert P, Gumhold J, Silbert D, Fuchsbichler A, Gujral J, Zatloukal K, Denk H, Jaeschke H, Trauner M. Drug Metab Dispos. 2007 doi: 10.1124/dmd.107.015610. [DOI] [PubMed] [Google Scholar]
- 26.Ghose R, White D, Guo T, Vallejo J, Karpen SJ. Drug Metab Dispos. 2007 doi: 10.1124/dmd.107.018051. [DOI] [PubMed] [Google Scholar]
- 27.Kim MS, Shigenaga JK, Moser AH, Feingold KR, Grunfeld C. J. Lipid Res. 2005;46:2282–2288. doi: 10.1194/jlr.M500217-JLR200. [DOI] [PubMed] [Google Scholar]
- 28.Quale JM, Mandel LJ, Bergasa NV, Straus EW. Am. J. Med. 1988;85:615–618. doi: 10.1016/s0002-9343(88)80231-6. [DOI] [PubMed] [Google Scholar]
- 29.Whiting JF, Green RM, Rosenbluth AB, Gollan JL. Hepatology. 1995;22:1273–1278. doi: 10.1016/0270-9139(95)90639-8. [DOI] [PubMed] [Google Scholar]
- 30.Green RM, Beier D, Gollan JL. Gastroenterology. 1996;111:193–198. doi: 10.1053/gast.1996.v111.pm8698199. [DOI] [PubMed] [Google Scholar]
- 31.Geier A, Dietrich CG, Voigt S, Ananthanarayanan M, Lammert F, Schmitz A, Trauner M, Wasmuth HE, Boraschi D, Balasubramaniyan N, Suchy FJ, Matern S, Gartung C. Am. J. Physiol Gastrointest. Liver Physiol. 2005 doi: 10.1152/ajpgi.00307.2004. [DOI] [PubMed] [Google Scholar]
- 32.Piquette-Miller M, Pak A, Kim H, Anari R, Shahzamani A. Pharm Res. 1998;15:706–711. doi: 10.1023/a:1011962818051. [DOI] [PubMed] [Google Scholar]
- 33.Hartmann G, Kim H, Piquette-Miller M. Int Immunopharmacol. 2001;1:189–199. doi: 10.1016/s0162-3109(00)00271-x. [DOI] [PubMed] [Google Scholar]
- 34.Siewert E, Dietrich CG, Lammert F, Heinrich PC, Matern S, Gartung C, Geier A. Biochem. Biophys. Res. Commun. 2004;322:232–238. doi: 10.1016/j.bbrc.2004.07.102. [DOI] [PubMed] [Google Scholar]
- 35.von AD, Hasselgren PO, Higashiguchi T, Fischer JE. Metabolism. 1992;41:961–969. doi: 10.1016/0026-0495(92)90121-P. [DOI] [PubMed] [Google Scholar]
- 36.Reynolds MA, Siow Y, Collet D, Klar E, Richardson JD, Vitale GC. Am. Surg. 1995;61:469–474. [PubMed] [Google Scholar]
- 37.Kim PK, Chen J, Andrejko KM, Deutschman CS. Shock. 2000;14:176–181. doi: 10.1097/00024382-200014020-00017. [DOI] [PubMed] [Google Scholar]
- 38.Deutschman CS, Cereda M, Ochroch EA, Raj NR. Crit Care Med. 2006;34:2613–2620. doi: 10.1097/01.CCM.0000240229.98275.07. [DOI] [PubMed] [Google Scholar]
- 39.Chen YH, Hong IC, Kuo KK, Hsu HK, Hsu C. Shock. 2007;28:65–70. doi: 10.1097/shk.0b013e31802ec5d2. [DOI] [PubMed] [Google Scholar]
- 40.Utili R, Abernathy CO, Zimmerman HJ. Gastroenterology. 1976;70:248–253. [PubMed] [Google Scholar]
- 41.Roelofsen H, van der Veere CN, Ottenhoff R, Schoemaker B, Jansen PL, Oude Elferink RP. Gastroenterology. 1994;107:1075–1084. doi: 10.1016/0016-5085(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 42.Kubitz R, Wettstein M, Warskulat U, Haussinger D. Gastroenterology. 1999;116:401–410. doi: 10.1016/s0016-5085(99)70138-1. [DOI] [PubMed] [Google Scholar]
- 43.Muhlfeld A, Kubitz R, Dransfeld O, Haussinger D, Wettstein M. Arch. Biochem. Biophys. 2003;413:32–40. doi: 10.1016/s0003-9861(03)00098-5. [DOI] [PubMed] [Google Scholar]
- 44.Luster MI, Germolec DR, Yoshida T, Kayama F, Thompson M. Hepatology. 1994;19:480–488. [PubMed] [Google Scholar]
- 45.Olinga P, Merema MT, de Jager MH, Derks F, Melgert BN, Moshage H, Slooff MJ, Meijer DK, Poelstra K, Groothuis GM. J. Hepatol. 2001;35:187–194. doi: 10.1016/s0168-8278(01)00103-9. [DOI] [PubMed] [Google Scholar]
- 46.Elferink MG, Olinga P, Draaisma AL, Merema MT, Faber KN, Slooff MJ, Meijer DK, Groothuis GM. Am. J. Physiol Gastrointest. Liver Physiol. 2004;287:G1008–G1016. doi: 10.1152/ajpgi.00071.2004. [DOI] [PubMed] [Google Scholar]
- 47.Li D, Zimmerman TL, Thevananther S, Lee HY, Kurie JM, Karpen SJ. J Biol Chem. 2002;277:31416–31422. doi: 10.1074/jbc.M204818200. [DOI] [PubMed] [Google Scholar]
- 48.Hisaeda K, Inokuchi A, Nakamura T, Iwamoto Y, Kohno K, Kuwano M, Uchiumi T. Hepatology. 2004;39:1574–1582. doi: 10.1002/hep.20216. [DOI] [PubMed] [Google Scholar]
- 49.Sturm E, Zimmerman TL, Crawford AR, Svetlov SI, Sundaram P, Ferrara JL, Karpen SJ, Crawford JM. Hepatology. 2000;31:124–130. doi: 10.1002/hep.510310120. [DOI] [PubMed] [Google Scholar]
- 50.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Cell. 2006;126:789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 53.Gronemeyer H, Gustafsson JA, Laudet V. Nat. Rev. Drug Discov. 2004;3:950–964. doi: 10.1038/nrd1551. [DOI] [PubMed] [Google Scholar]
- 54.Karpen SJ. J Hepatol. 2002;36:832–850. doi: 10.1016/s0168-8278(02)00129-0. [DOI] [PubMed] [Google Scholar]
- 55.Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, Lazar MA. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- 56.Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, Rastinejad F. Nat. Struct. Mol. Biol. 2007;14:1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cell. 1999;97:161–163. doi: 10.1016/s0092-8674(00)80726-6. [DOI] [PubMed] [Google Scholar]
- 58.Claudel T, Sturm E, Duez H, Torra IP, Sirvent A, Kosykh V, Fruchart JC, Dallongeville J, Hum DW, Kuipers F, Staels B. J. Clin. Invest. 2002;109:961–971. doi: 10.1172/JCI14505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barbier O, Torra IP, Sirvent A, Claudel T, Blanquart C, Duran-Sandoval D, Kuipers F, Kosykh V, Fruchart JC, Staels B. Gastroenterology. 2003;124:1926–1940. doi: 10.1016/s0016-5085(03)00388-3. [DOI] [PubMed] [Google Scholar]
- 60.Perissi V, Rosenfeld MG. Nat. Rev. Mol. Cell Biol. 2005;6:542–554. doi: 10.1038/nrm1680. [DOI] [PubMed] [Google Scholar]
- 61.Smith CL, O’malley BW. Endocr. Rev. 2004;25:45–71. doi: 10.1210/er.2003-0023. [DOI] [PubMed] [Google Scholar]
- 62.Jordan VC, O’malley BW. J. Clin. Oncol. 2007 doi: 10.1200/JCO.2007.11.3886. [DOI] [PubMed] [Google Scholar]
- 63.Baxter JD, Webb P, Grover G, Scanlan TS. Trends Endocrinol. Metab. 2004;15:154–157. doi: 10.1016/j.tem.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 64.Issemann I, Green S. Nature. 1990;347:645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- 65.Green S, Wahli W. Mol. Cell Endocrinol. 1994;100:149–153. doi: 10.1016/0303-7207(94)90294-1. [DOI] [PubMed] [Google Scholar]
- 66.Lehmann JM, Moore LB, Smith-Oliver TA, Wilkison WO, Willson TM, Kliewer SA. J. Biol. Chem. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- 67.Honkakoski P, Zelko I, Sueyoshi T, Negishi M. Mol. Cell Biol. 1998;18:5652–5658. doi: 10.1128/mcb.18.10.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sueyoshi T, Kawamoto T, Zelko I, Honkakoski P, Negishi M. J. Biol. Chem. 1999;274:6043–6046. doi: 10.1074/jbc.274.10.6043. [DOI] [PubMed] [Google Scholar]
- 69.Tzameli I, Pissios P, Schuetz EG, Moore DD. Mol. Cell Biol. 2000;20:2951–2958. doi: 10.1128/mcb.20.9.2951-2958.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bertilsson G, Heidrich J, Svensson K, Asman M, Jendeberg L, Sydow-Backman M, Ohlsson R, Postlind H, Blomquist P, Berkenstam A. Proc. Natl. Acad. Sci. U. S. A. 1998;95:12208–12213. doi: 10.1073/pnas.95.21.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koopen NR, Muller M, Vonk RJ, Zimniak P, Kuipers F. Biochim. Biophys. Acta. 1998;1408:1–17. doi: 10.1016/s0925-4439(98)00053-2. [DOI] [PubMed] [Google Scholar]
- 72.Ballatori N, Truong AT. Am. J. Physiol. 1989;256:G22–G30. doi: 10.1152/ajpgi.1989.256.1.G22. [DOI] [PubMed] [Google Scholar]
- 73.Ballatori N, Truong AT. Am. J. Physiol. 1992;263:G617–G624. doi: 10.1152/ajpgi.1992.263.5.G617. [DOI] [PubMed] [Google Scholar]
- 74.Hardison WG, Wood CA. Am. J. Physiol. 1978;235:E158–E164. doi: 10.1152/ajpendo.1978.235.2.E158. [DOI] [PubMed] [Google Scholar]
- 75.Prall RT, LaRusso NF. Curr. Opin. Gastroenterol. 2000;16:432–436. doi: 10.1097/00001574-200009000-00008. [DOI] [PubMed] [Google Scholar]
- 76.Alpini G, Lenzi R, Zhai WR, Slott PA, Liu MH, Sarkozi L, Tavoloni N. Am. J. Physiol. 1989;257:G124–G133. doi: 10.1152/ajpgi.1989.257.1.G124. [DOI] [PubMed] [Google Scholar]
- 77.Boyer JL. Am. J. Physiol. 1996;270:G1–G5. doi: 10.1152/ajpgi.1996.270.1.G1. [DOI] [PubMed] [Google Scholar]
- 78.Trauner M, Boyer JL. Physiol Rev. 2003;83:633–671. doi: 10.1152/physrev.00027.2002. [DOI] [PubMed] [Google Scholar]
- 79.Bohan A, Chen WS, Denson LA, Held MA, Boyer JL. J Biol Chem. 2003;278:36688–36698. doi: 10.1074/jbc.M304011200. [DOI] [PubMed] [Google Scholar]
- 80.Mennone A, Soroka CJ, Cai SY, Harry K, Adachi M, Hagey L, Schuetz JD, Boyer JL. Hepatology. 2006;43:1013–1021. doi: 10.1002/hep.21158. [DOI] [PubMed] [Google Scholar]
- 81.Wang W, Seward DJ, Li L, Boyer JL, Ballatori N. Proc. Natl. Acad. Sci. U. S. A. 2001;98:9431–9436. doi: 10.1073/pnas.161099898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Seward DJ, Koh AS, Boyer JL, Ballatori N. J. Biol. Chem. 2003;278:27473–27482. doi: 10.1074/jbc.M301106200. [DOI] [PubMed] [Google Scholar]
- 83.Dawson PA, Hubbert M, Haywood J, Craddock AL, Zerangue N, Christian WV, Ballatori N. J. Biol. Chem. 2005;280:6960–6968. doi: 10.1074/jbc.M412752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rao A, Haywood J, Craddock AL, Belinsky MG, Kruh GD, Dawson PA. Proc. Natl. Acad. Sci. U. S. A. 2008;105:3891–3896. doi: 10.1073/pnas.0712328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ballatori N, Christian WV, Lee JY, Dawson PA, Soroka CJ, Boyer JL, Madejczyk MS, Li N. Hepatology. 2005;42:1270–1279. doi: 10.1002/hep.20961. [DOI] [PubMed] [Google Scholar]
- 86.Zollner G, Wagner M, Moustafa T, Fickert P, Silbert D, Gumhold J, Fuchsbichler A, Halilbasic E, Denk H, Marschall HU, Trauner M. Am. J. Physiol Gastrointest. Liver Physiol. 2006;290:G923–G932. doi: 10.1152/ajpgi.00490.2005. [DOI] [PubMed] [Google Scholar]
- 87.Boyer JL, Trauner M, Mennone A, Soroka CJ, Cai SY, Moustafa T, Zollner G, Lee JY, Ballatori N. Am. J. Physiol Gastrointest. Liver Physiol. 2006;290:G1124–G1130. doi: 10.1152/ajpgi.00539.2005. [DOI] [PubMed] [Google Scholar]
- 88.Oude Elferink RP, Paulusma CC, Groen AK. Gastroenterology. 2006;130:908–925. doi: 10.1053/j.gastro.2005.08.052. [DOI] [PubMed] [Google Scholar]
- 89.Altmann SW, Davis HR, Jr, Zhu LJ, Yao X, Hoos LM, Tetzloff G, Iyer SP, Maguire M, Golovko A, Zeng M, Wang L, Murgolo N, Graziano MP. Science. 2004;303:1201–1204. doi: 10.1126/science.1093131. [DOI] [PubMed] [Google Scholar]
- 90.Temel RE, Tang W, Ma Y, Rudel LL, Willingham MC, Ioannou YA, Davies JP, Nilsson LM, Yu L. J. Clin. Invest. 2007;117:1968–1978. doi: 10.1172/JCI30060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Utili R, Abernathy CO, Zimmerman HJ. J. Lab Clin. Med. 1977;89:471–482. [PubMed] [Google Scholar]
- 92.Utili R, Abernathy CO, Zimmerman HJ. J. Infect. Dis. 1977;136:583–587. doi: 10.1093/infdis/136.4.583. [DOI] [PubMed] [Google Scholar]
- 93.Bolder U, Ton-Nu HT, Schteingart CD, Frick E, Hofmann AF. Gastroenterology. 1997;112:214–225. doi: 10.1016/s0016-5085(97)70238-5. [DOI] [PubMed] [Google Scholar]
- 94.Trauner M, Nathanson MH, Rydberg SA, Koeppel TA, Gartung C, Sessa WC, Boyer JL. Hepatology. 1997;25:1184–1191. doi: 10.1002/hep.510250522. [DOI] [PubMed] [Google Scholar]
- 95.Rustgi VK, Jones DB, Dinarello CA, Hoofnagle JH. Liver. 1987;7:149–154. doi: 10.1111/j.1600-0676.1987.tb00335.x. [DOI] [PubMed] [Google Scholar]
- 96.Dombrowski F, Kubitz R, Chittattu A, Wettstein M, Saha N, Haussinger D. Biochem. J. 2000;348 Pt 1:183–188. [PMC free article] [PubMed] [Google Scholar]
- 97.Zinchuk V, Zinchuk O, Okada T. Microsc. Res. Tech. 2005;67:65–70. doi: 10.1002/jemt.20184. [DOI] [PubMed] [Google Scholar]
- 98.Reichen J, Paumgartner G. Am. J. Physiol. 1976;231:734–742. doi: 10.1152/ajplegacy.1976.231.3.734. [DOI] [PubMed] [Google Scholar]
- 99.Spirli C, Nathanson MH, Fiorotto R, Duner E, Denson LA, Sanz JM, Di Virgilio F, Okolicsanyi L, Casagrande F, Strazzabosco M. Gastroenterology. 2001;121:156–169. doi: 10.1053/gast.2001.25516. [DOI] [PubMed] [Google Scholar]
- 100.Russell DW. Annu. Rev. Biochem. 2003;72:137–174. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- 101.Feingold KR, Spady DK, Pollock AS, Moser AH, Grunfeld C. J Lipid Res. 1996;37:223–228. [PubMed] [Google Scholar]
- 102.Memon RA, Moser AH, Shigenaga JK, Grunfeld C, Feingold KR. J Biol Chem. 2001;276:30118–30126. doi: 10.1074/jbc.M102516200. [DOI] [PubMed] [Google Scholar]
- 103.Chensue SW, Terebuh PD, Remick DG, Scales WE, Kunkel SL. Am. J. Pathol. 1991;138:395–402. [PMC free article] [PubMed] [Google Scholar]
- 104.Fox ES, Broitman SA, Thomas P. Lab Invest. 1990;63:733–741. [PubMed] [Google Scholar]
- 105.Nakamura J, Nishida T, Hayashi K, Kawada N, Ueshima S, Sugiyama Y, Ito T, Sobue K, Matsuda H. Biochem. Biophys. Res. Commun. 1999;255:143–149. doi: 10.1006/bbrc.1999.0160. [DOI] [PubMed] [Google Scholar]
- 106.Xu DX, Wei W, Sun MF, Wu CY, Wang JP, Wei LZ, Zhou CF. Free Radic. Biol. Med. 2004;37:10–22. doi: 10.1016/j.freeradbiomed.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 107.Sturm E, Havinga R, Baller JF, Wolters H, van Rooijen N, Kamps JA, Verkade HJ, Karpen SJ, Kuipers F. J. Hepatol. 2005;42:102–109. doi: 10.1016/j.jhep.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 108.Hartmann G, Cheung AK, Piquette-Miller M. J Pharmacol Exp Ther. 2002;303:273–281. doi: 10.1124/jpet.102.039404. [DOI] [PubMed] [Google Scholar]
- 109.Geier A, Dietrich CG, Voigt S, Kim SK, Gerloff T, Kullak-Ublick GA, Lorenzen J, Matern S, Gartung C. Hepatology. 2003;38:345–354. doi: 10.1053/jhep.2003.50317. [DOI] [PubMed] [Google Scholar]
- 110.Lickteig AJ, Slitt AL, Arkan MC, Karin M, Cherrington NJ. Drug Metab Dispos. 2007;35:402–409. doi: 10.1124/dmd.106.012138. [DOI] [PubMed] [Google Scholar]
- 111.Dunzendorfer S, Lee HK, Soldau K, Tobias PS. J. Immunol. 2004;173:1166–1170. doi: 10.4049/jimmunol.173.2.1166. [DOI] [PubMed] [Google Scholar]
- 112.Nakamura M, Shimizu Y, Sato Y, Miyazaki Y, Satoh T, Mizuno M, Kato Y, Hosaka Y, Furusako S. Eur. J. Pharmacol. 2007;569:237–243. doi: 10.1016/j.ejphar.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 113.Laschke MW, Menger MD, Wang Y, Lindell G, Jeppsson B, Thorlacius H. Am. J. Physiol Gastrointest. Liver Physiol. 2007;292:G1396–G1402. doi: 10.1152/ajpgi.00539.2006. [DOI] [PubMed] [Google Scholar]
- 114.Essani NA, Fisher MA, Simmons CA, Hoover JL, Farhood A, Jaeschke H. J. Leukoc. Biol. 1998;63:288–296. doi: 10.1002/jlb.63.3.288. [DOI] [PubMed] [Google Scholar]
- 115.Bilzer M, Roggel F, Gerbes AL. Liver Int. 2006;26:1175–1186. doi: 10.1111/j.1478-3231.2006.01342.x. [DOI] [PubMed] [Google Scholar]
- 116.Trauner M, Nathanson MH, Mennone A, Rydberg SA, Boyer JL. Hepatology. 1997;25:263–269. doi: 10.1002/hep.510250202. [DOI] [PubMed] [Google Scholar]
- 117.Beckh K, Kneip S, Arnold R. Hepatology. 1994;19:1208–1213. [PubMed] [Google Scholar]
- 118.Weidenbach H, Scheibner J, Stange EF, Adler G, Beckh K. Life Sci. 1996;58:1531–1538. doi: 10.1016/0024-3205(96)00127-0. [DOI] [PubMed] [Google Scholar]
- 119.Ramadori G, Christ B. Semin. Liver Dis. 1999;19:141–155. doi: 10.1055/s-2007-1007106. [DOI] [PubMed] [Google Scholar]
- 120.Ros JE, Schuetz JD, Geuken M, Streetz K, Moshage H, Kuipers F, Manns MP, Jansen PL, Trautwein C, Muller M. Hepatology. 2001;33:1425–1431. doi: 10.1053/jhep.2001.24667. [DOI] [PubMed] [Google Scholar]
- 121.Zimmerman TL, Thevananther S, Ghose R, Burns AR, Karpen SJ. J. Biol. Chem. 2006;281:15434–15440. doi: 10.1074/jbc.M508277200. [DOI] [PubMed] [Google Scholar]
- 122.Hartmann G, Vassileva V, Piquette-Miller M. Drug Metab Dispos. 2005;33:820–828. doi: 10.1124/dmd.104.002568. [DOI] [PubMed] [Google Scholar]
- 123.Vos TA, Hooiveld GJ, Koning H, Childs S, Meijer DK, Moshage H, Jansen PL, Muller M. Hepatology. 1998;28:1637–1644. doi: 10.1002/hep.510280625. [DOI] [PubMed] [Google Scholar]
- 124.Cherrington NJ, Slitt AL, Li N, Klaassen CD. Drug Metab Dispos. 2004;32:734–741. doi: 10.1124/dmd.32.7.734. [DOI] [PubMed] [Google Scholar]
- 125.Zhou G, Kuo MT. J. Biol. Chem. 1997;272:15174–15183. doi: 10.1074/jbc.272.24.15174. [DOI] [PubMed] [Google Scholar]
- 126.Masyuk AI, LaRusso NF. Hepatology. 2006;43:S75–S81. doi: 10.1002/hep.20996. [DOI] [PubMed] [Google Scholar]
- 127.Lehmann GL, Carreras FI, Soria LR, Gradilone SA, Marinelli RA. Am. J. Physiol Gastrointest. Liver Physiol. 2008 doi: 10.1152/ajpgi.00232.2007. [DOI] [PubMed] [Google Scholar]
- 128.Denson LA, Menon RK, Shaufl A, Bajwa HS, Williams CR, Karpen SJ. J Clin Invest. 2001;107:1451–1458. doi: 10.1172/JCI10994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pei L, Castrillo A, Chen M, Hoffmann A, Tontonoz P. J. Biol. Chem. 2005;280:29256–29262. doi: 10.1074/jbc.M502606200. [DOI] [PubMed] [Google Scholar]
- 130.Beigneux AP, Moser AH, Shigenaga JK, Grunfeld C, Feingold KR. J Biol Chem. 2000;275:16390–16399. doi: 10.1074/jbc.M000953200. [DOI] [PubMed] [Google Scholar]
- 131.Ghose R, Zimmerman TL, Thevananther S, Karpen SJ. Nucl. Recept. 2004;2:4. doi: 10.1186/1478-1336-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang Z, Burke PA. J. Mol. Biol. 2007;371:323–335. doi: 10.1016/j.jmb.2007.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kim MS, Sweeney TR, Shigenaga JK, Chui LG, Moser A, Grunfeld C, Feingold KR. Metabolism. 2007;56:267–279. doi: 10.1016/j.metabol.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fang C, Yoon S, Tindberg N, Jarvelainen HA, Lindros KO, Ingelman-Sundberg M. Biochem. Pharmacol. 2004;67:1389–1397. doi: 10.1016/j.bcp.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 135.Lu B, Moser AH, Shigenaga JK, Feingold KR, Grunfeld C. J. Lipid Res. 2006;47:2179–2190. doi: 10.1194/jlr.M500540-JLR200. [DOI] [PubMed] [Google Scholar]
- 136.Feingold K, Kim MS, Shigenaga J, Moser A, Grunfeld C. Am J Physiol Endocrinol Metab. 2004;286:E201–E207. doi: 10.1152/ajpendo.00205.2003. [DOI] [PubMed] [Google Scholar]
- 137.Karpen SJ, Sun AQ, Kudish B, Hagenbuch B, Meier PJ, Ananthanarayanan M, Suchy FJ. J Biol Chem. 1996;271:15211–15221. doi: 10.1074/jbc.271.25.15211. [DOI] [PubMed] [Google Scholar]
- 138.Denson LA, Auld KL, Schiek DS, McClure MH, Mangelsdorf DJ, Karpen SJ. J Biol Chem. 2000;275:8835–8843. doi: 10.1074/jbc.275.12.8835. [DOI] [PubMed] [Google Scholar]
- 139.Zollner G, Wagner M, Fickert P, Geier A, Fuchsbichler A, Silbert D, Gumhold J, Zatloukal K, Kaser A, Tilg H, Denk H, Trauner M. Am. J. Physiol Gastrointest. Liver Physiol. 2005;289:G798–G805. doi: 10.1152/ajpgi.00319.2004. [DOI] [PubMed] [Google Scholar]
- 140.Gu X, Ke S, Liu D, Sheng T, Thomas PE, Rabson AB, Gallo MA, Xie W, Tian Y. J. Biol. Chem. 2006;281:17882–17889. doi: 10.1074/jbc.M601302200. [DOI] [PubMed] [Google Scholar]
- 141.Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, Luk A, Hull MV, Lustig KD, Mangelsdorf DJ, Shan B. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- 142.Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, Stimmel JB, Willson TM, Zavacki AM, Moore DD, Lehmann JM. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- 143.Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. Cell. 2000;102:731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 144.Kim MS, Shigenaga J, Moser A, Feingold K, Grunfeld C. J Biol Chem. 2003;278:8988–8995. doi: 10.1074/jbc.M212633200. [DOI] [PubMed] [Google Scholar]
- 145.Willson TM, Kliewer SA. Nat. Rev. Drug Discov. 2002;1:259–266. doi: 10.1038/nrd753. [DOI] [PubMed] [Google Scholar]
- 146.Song CS, Gelb NA, Wolff SM. J. Clin. Invest. 1972;51:2959–2966. doi: 10.1172/JCI107120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Farrell GC. Pharmacol. Ther. 1987;35:375–404. doi: 10.1016/0163-7258(87)90101-x. [DOI] [PubMed] [Google Scholar]
- 148.Morgan ET, Goralski KB, Piquette-Miller M, Renton KW, Robertson GR, Chaluvadi MR, Charles KA, Clarke SJ, Kacevska M, Liddle C, Richardson TA, Sharma R, Sinal CJ. Drug Metab Dispos. 2008;36:205–216. doi: 10.1124/dmd.107.018747. [DOI] [PubMed] [Google Scholar]
- 149.Beigneux AP, Moser AH, Shigenaga JK, Grunfeld C, Feingold KR. Biochem. Biophys. Res. Commun. 2002;293:145–149. doi: 10.1016/S0006-291X(02)00196-1. [DOI] [PubMed] [Google Scholar]
- 150.Teng S, Piquette-Miller M. J. Pharmacol. Exp. Ther. 2005;312:841–848. doi: 10.1124/jpet.104.076141. [DOI] [PubMed] [Google Scholar]
- 151.Kim MS, Shigenaga J, Moser A, Grunfeld C, Feingold KR. Am. J. Physiol Endocrinol. Metab. 2004;287:E731–E738. doi: 10.1152/ajpendo.00130.2004. [DOI] [PubMed] [Google Scholar]
- 152.Pascussi JM, Gerbal-Chaloin S, Pichard-Garcia L, Daujat M, Fabre JM, Maurel P, Vilarem MJ. Biochem. Biophys. Res. Commun. 2000;274:707–713. doi: 10.1006/bbrc.2000.3219. [DOI] [PubMed] [Google Scholar]
- 153.Assenat E, Gerbal-Chaloin S, Larrey D, Saric J, Fabre JM, Maurel P, Vilarem MJ, Pascussi JM. Hepatology. 2004;40:951–960. doi: 10.1002/hep.20387. [DOI] [PubMed] [Google Scholar]
- 154.Zhou C, Tabb MM, Nelson EL, Grun F, Verma S, Sadatrafiei A, Lin M, Mallick S, Forman BM, Thummel KE, Blumberg B. J. Clin. Invest. 2006;116:2280–2289. doi: 10.1172/JCI26283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Stedman CA, Liddle C, Coulter SA, Sonoda J, Alvarez JG, Moore DD, Evans RM, Downes M. Proc. Natl. Acad. Sci. U. S. A. 2005;102:2063–2068. doi: 10.1073/pnas.0409794102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Xie W, Radominska-Pandya A, Shi Y, Simon CM, Nelson MC, Ong ES, Waxman DJ, Evans RM. Proc. Natl. Acad. Sci. U. S. A. 2001;98:3375–3380. doi: 10.1073/pnas.051014398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Saini SP, Mu Y, Gong H, Toma D, Uppal H, Ren S, Li S, Poloyac SM, Xie W. Hepatology. 2005;41:497–505. doi: 10.1002/hep.20570. [DOI] [PubMed] [Google Scholar]
- 158.Sonoda J, Chong LW, Downes M, Barish GD, Coulter S, Liddle C, Lee CH, Evans RM. Proc. Natl. Acad. Sci. U. S. A. 2005;102:2198–2203. doi: 10.1073/pnas.0409481102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Khovidhunkit W, Moser AH, Shigenaga JK, Grunfeld C, Feingold KR. J. Lipid Res. 2003;44:1728–1736. doi: 10.1194/jlr.M300100-JLR200. [DOI] [PubMed] [Google Scholar]
- 160.Zhou M, Wu R, Dong W, Jacob A, Wang P. Am. J. Physiol Regul. Integr. Comp Physiol. 2008;294:R84–R92. doi: 10.1152/ajpregu.00340.2007. [DOI] [PubMed] [Google Scholar]
- 161.Necela BM, Su W, Thompson EA. Immunology. 2008 doi: 10.1111/j.1365-2567.2008.02849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Mol. Cell Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wisely GB, Miller AB, Davis RG, Thornquest AD, Jr, Johnson R, Spitzer T, Sefler A, Shearer B, Moore JT, Miller AB, Willson TM, Williams SP. Structure. 2002;10:1225–1234. doi: 10.1016/s0969-2126(02)00829-8. [DOI] [PubMed] [Google Scholar]
- 164.Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, Fraenkel E, Bell GI, Young RA. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wang B, Cai SR, Gao C, Sladek FM, Ponder KP. Hepatology. 2001;34:979–989. doi: 10.1053/jhep.2001.28885. [DOI] [PubMed] [Google Scholar]
- 166.De Fabiani E, Mitro N, Anzulovich AC, Pinelli A, Galli G, Crestani M. J Biol Chem. 2001;276:30708–30716. doi: 10.1074/jbc.M103270200. [DOI] [PubMed] [Google Scholar]
- 167.Li X, Salisbury-Rowswell J, Murdock AD, Forse RA, Burke PA. Biochem. J. 2002;368:203–211. doi: 10.1042/BJ20020233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Cheng PY, Wang M, Morgan ET. J Pharmacol Exp Ther. 2003;307:1205–1212. doi: 10.1124/jpet.103.057174. [DOI] [PubMed] [Google Scholar]
- 169.Jahan A, Chiang JY. Am. J. Physiol Gastrointest. Liver Physiol. 2005;288:G685–G695. doi: 10.1152/ajpgi.00207.2004. [DOI] [PubMed] [Google Scholar]
- 170.Li T, Jahan A, Chiang JY. Hepatology. 2006;43:1202–1210. doi: 10.1002/hep.21183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Nikolaidou-Neokosmidou V, Zannis VI, Kardassis D. Biochem. J. 2006;398:439–450. doi: 10.1042/BJ20060169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Krajewski J, Batmunkh C, Jelkmann W, Hellwig-Burgel T. Cell Mol. Life Sci. 2007;64:989–998. doi: 10.1007/s00018-007-6561-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Guo H, Wei J, Inoue Y, Gonzalez FJ, Kuo PC. Am. J. Physiol Cell Physiol. 2003;284:C1090–C1099. doi: 10.1152/ajpcell.00394.2002. [DOI] [PubMed] [Google Scholar]
- 174.Guo H, Gao C, Mi Z, Wai PY, Kuo PC. Biochem. J. 2006;394:379–387. doi: 10.1042/BJ20051730. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 175.Guo H, Gao C, Mi Z, Zhang J, Kuo PC. J. Biochem. 2007;141:635–640. doi: 10.1093/jb/mvm066. [DOI] [PubMed] [Google Scholar]
- 176.Ktistaki E, Ktistakis NT, Papadogeorgaki E, Talianidis I. Proc. Natl. Acad. Sci. U. S. A. 1995;92:9876–9880. doi: 10.1073/pnas.92.21.9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gerbod-Giannone MC, Del Castillo-Olivares A, Janciauskiene S, Gil G, Hylemon PB. J. Biol. Chem. 2002;277:42973–42980. doi: 10.1074/jbc.M205089200. [DOI] [PubMed] [Google Scholar]
- 178.Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, Mangelsdorf DJ. Mol. Cell. 2000;6:507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- 179.Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME, Maloney PR, Willson TM, Kliewer SA. Mol. Cell. 2000;6:517–526. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 180.Lai K, Harnish DC, Evans MJ. J. Biol. Chem. 2003;278:36418–36429. doi: 10.1074/jbc.M303913200. [DOI] [PubMed] [Google Scholar]
- 181.Leite RS, Brown AG, Strauss JF., III FASEB J. 2004;18:1418–1420. doi: 10.1096/fj.04-1684fje. [DOI] [PubMed] [Google Scholar]
- 182.Sheppard KA, Phelps KM, Williams AJ, Thanos D, Glass CK, Rosenfeld MG, Gerritsen ME, Collins T. J. Biol. Chem. 1998;273:29291–29294. doi: 10.1074/jbc.273.45.29291. [DOI] [PubMed] [Google Scholar]
- 183.Na SY, Kang BY, Chung SW, Han SJ, Ma X, Trinchieri G, Im SY, Lee JW, Kim TS. J. Biol. Chem. 1999;274:7674–7680. doi: 10.1074/jbc.274.12.7674. [DOI] [PubMed] [Google Scholar]
- 184.Hermanson O, Glass CK, Rosenfeld MG. Trends Endocrinol. Metab. 2002;13:55–60. doi: 10.1016/s1043-2760(01)00527-6. [DOI] [PubMed] [Google Scholar]
- 185.Wu RC, Qin J, Hashimoto Y, Wong J, Xu J, Tsai SY, Tsai MJ, O’malley BW. Mol. Cell Biol. 2002;22:3549–3561. doi: 10.1128/MCB.22.10.3549-3561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.McKenna NJ, O’malley BW. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 187.Yu C, York B, Wang S, Feng Q, Xu J, O’malley BW. Mol. Cell. 2007;25:765–778. doi: 10.1016/j.molcel.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Pajkrt D, Doran JE, Koster F, Lerch PG, Arnet B, van der PT, ten Cate JW, van Deventer SJ. J. Exp. Med. 1996;184:1601–1608. doi: 10.1084/jem.184.5.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Thabut D, Tazi KA, Bonnefont-Rousselot D, Aller M, Farges O, Guimont MC, Tellier Z, Guichard C, Ogier-Denis E, Poynard T, Moreau R, Lebrec D. Hepatology. 2007;46:1893–1906. doi: 10.1002/hep.21875. [DOI] [PubMed] [Google Scholar]
- 190.Lamping N, Dettmer R, Schroder NW, Pfeil D, Hallatschek W, Burger R, Schumann RR. J. Clin. Invest. 1998;101:2065–2071. doi: 10.1172/JCI2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Jing Y, Shishkov A, Ponnappa BC. Biochim. Biophys. Acta. 2008;1780:34–40. doi: 10.1016/j.bbagen.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Trauner M, Wagner M, Fickert P, Zollner G. J. Clin. Gastroenterol. 2005;39:S111–S124. doi: 10.1097/01.mcg.0000155551.37266.26. [DOI] [PubMed] [Google Scholar]
- 193.Zollner G, Fickert P, Silbert D, Fuchsbichler A, Stumptner C, Zatloukal K, Denk H, Trauner M. Am. J. Physiol Gastrointest. Liver Physiol. 2002;282:G184–G191. doi: 10.1152/ajpgi.00215.2001. [DOI] [PubMed] [Google Scholar]
- 194.Boyer JL. Gastroenterology. 2005;129:735–740. doi: 10.1016/j.gastro.2005.06.053. [DOI] [PubMed] [Google Scholar]
- 195.Ghose R, Mulder J, von Furstenberg RJ, Thevananther S, Kuipers F, Karpen SJ. J. Hepatol. 2007;46:115–123. doi: 10.1016/j.jhep.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wang YY, Dahle MK, Agren J, Myhre AE, Reinholt FP, Foster SJ, Collins JL, Thiemermann C, Aasen AO, Wang JE. Shock. 2006;25:141–146. doi: 10.1097/01.shk.0000191377.78144.d9. [DOI] [PubMed] [Google Scholar]
- 197.De Bosscher K, Vanden BW, Haegeman G. Endocr. Rev. 2003;24:488–522. doi: 10.1210/er.2002-0006. [DOI] [PubMed] [Google Scholar]
- 198.Rhen T, Cidlowski JA. N. Engl. J. Med. 2005;353:1711–1723. doi: 10.1056/NEJMra050541. [DOI] [PubMed] [Google Scholar]
- 199.Smoak KA, Cidlowski JA. Mech. Ageing Dev. 2004;125:697–706. doi: 10.1016/j.mad.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 200.Roelofsen H, Schoemaker B, Bakker C, Ottenhoff R, Jansen PL, Elferink RP. Am. J. Physiol. 1995;269:G427–G434. doi: 10.1152/ajpgi.1995.269.3.G427. [DOI] [PubMed] [Google Scholar]
- 201.Ricote M, Li AC, Willson TM, Kelly CJ, Glass CK. Nature. 1998;391:79–82. doi: 10.1038/34178. [DOI] [PubMed] [Google Scholar]
- 202.Jiang C, Ting AT, Seed B. Nature. 1998;391:82–86. doi: 10.1038/34184. [DOI] [PubMed] [Google Scholar]
- 203.Uchimura K, Nakamuta M, Enjoji M, Irie T, Sugimoto R, Muta T, Iwamoto H, Nawata H. Hepatology. 2001;33:91–99. doi: 10.1053/jhep.2001.21145. [DOI] [PubMed] [Google Scholar]
- 204.Li M, Pascual G, Glass CK. Mol Cell Biol. 2000;20:4699–4707. doi: 10.1128/mcb.20.13.4699-4707.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Pascual G, Fong AL, Ogawa S, Gamliel A, Li AC, Perissi V, Rose DW, Willson TM, Rosenfeld MG, Glass CK. Nature. 2005;437:759–763. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Meier CA, Chicheportiche R, Juge-Aubry CE, Dreyer MG, Dayer JM. Cytokine. 2002;18:320–328. doi: 10.1006/cyto.2002.1945. [DOI] [PubMed] [Google Scholar]
- 207.Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ. Nature. 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- 208.Lehmann JM, Kliewer SA, Moore LB, Smith-Oliver TA, Oliver BB, Su JL, Sundseth SS, Winegar DA, Blanchard DE, Spencer TA, Willson TM. J. Biol. Chem. 1997;272:3137–3140. doi: 10.1074/jbc.272.6.3137. [DOI] [PubMed] [Google Scholar]
- 209.Venkateswaran A, Laffitte BA, Joseph SB, Mak PA, Wilpitz DC, Edwards PA, Tontonoz P. Proc. Natl. Acad. Sci. U. S. A. 2000;97:12097–12102. doi: 10.1073/pnas.200367697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Venkateswaran A, Repa JJ, Lobaccaro JM, Bronson A, Mangelsdorf DJ, Edwards PA. J. Biol. Chem. 2000;275:14700–14707. doi: 10.1074/jbc.275.19.14700. [DOI] [PubMed] [Google Scholar]
- 211.Sabol SL, Brewer HB, Jr, Santamarina-Fojo S. J. Lipid Res. 2005;46:2151–2167. doi: 10.1194/jlr.M500080-JLR200. [DOI] [PubMed] [Google Scholar]
- 212.Joseph SB, McKilligin E, Pei L, Watson MA, Collins AR, Laffitte BA, Chen M, Noh G, Goodman J, Hagger GN, Tran J, Tippin TK, Wang X, Lusis AJ, Hsueh WA, Law RE, Collins JL, Willson TM, Tontonoz P. Proc Natl Acad Sci U S A. 2002;99:7604–7609. doi: 10.1073/pnas.112059299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Tangirala RK, Bischoff ED, Joseph SB, Wagner BL, Walczak R, Laffitte BA, Daige CL, Thomas D, Heyman RA, Mangelsdorf DJ, Wang X, Lusis AJ, Tontonoz P, Schulman IG. Proc Natl Acad Sci U S A. 2002;99:11896–11901. doi: 10.1073/pnas.182199799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Terasaka N, Hiroshima A, Koieyama T, Ubukata N, Morikawa Y, Nakai D, Inaba T. FEBS Lett. 2003;536:6–11. doi: 10.1016/s0014-5793(02)03578-0. [DOI] [PubMed] [Google Scholar]
- 215.Repa JJ, Berge KE, Pomajzl C, Richardson JA, Hobbs H, Mangelsdorf DJ. J. Biol. Chem. 2002;277:18793–18800. doi: 10.1074/jbc.M109927200. [DOI] [PubMed] [Google Scholar]
- 216.Plosch T, Kok T, Bloks VW, Smit MJ, Havinga R, Chimini G, Groen AK, Kuipers F. J Biol Chem. 2002;277:33870–33877. doi: 10.1074/jbc.M206522200. [DOI] [PubMed] [Google Scholar]
- 217.Joseph SB, Castrillo A, Laffitte BA, Mangelsdorf DJ, Tontonoz P. Nat Med. 2003;9:213–219. doi: 10.1038/nm820. [DOI] [PubMed] [Google Scholar]
- 218.Castrillo A, Joseph SB, Marathe C, Mangelsdorf DJ, Tontonoz P. Journal of Biological Chemistry. 2003;278:10443–10449. doi: 10.1074/jbc.M213071200. [DOI] [PubMed] [Google Scholar]
- 219.Terasaka N, Hiroshima A, Ariga A, Honzumi S, Koieyama T, Inaba T, Fujiwara T. FEBS J. 2005;272:1546–1556. doi: 10.1111/j.1742-4658.2005.04599.x. [DOI] [PubMed] [Google Scholar]
- 220.Ogawa D, Stone JF, Takata Y, Blaschke F, Chu VH, Towler DA, Law RE, Hsueh WA, Bruemmer D. Circ. Res. 2005;96:e59–e67. doi: 10.1161/01.RES.0000163630.86796.17. [DOI] [PubMed] [Google Scholar]
- 221.Birrell MA, Catley MC, Hardaker E, Wong S, Willson TM, McCluskie K, Leonard T, Farrow SN, Collins JL, Haj-Yahia S, Belvisi MG. J. Biol. Chem. 2007 doi: 10.1074/jbc.M703278200. [DOI] [PubMed] [Google Scholar]
- 222.Fontaine C, Rigamonti E, Nohara A, Gervois P, Teissier E, Fruchart JC, Staels B, Chinetti-Gbaguidi G. Circ. Res. 2007;101:40–49. doi: 10.1161/CIRCRESAHA.106.135814. [DOI] [PubMed] [Google Scholar]
- 223.Walcher D, Kummel A, Kehrle B, Bach H, Grub M, Durst R, Hombach V, Marx N. Arterioscler. Thromb. Vasc. Biol. 2006;26:1022–1028. doi: 10.1161/01.ATV.0000210278.67076.8f. [DOI] [PubMed] [Google Scholar]
- 224.Fowler AJ, Sheu MY, Schmuth M, Kao J, Fluhr JW, Rhein L, Collins JL, Willson TM, Mangelsdorf DJ, Elias PM, Feingold KR. J Invest Dermatol. 2003;120:246–255. doi: 10.1046/j.1523-1747.2003.12033.x. [DOI] [PubMed] [Google Scholar]
- 225.Blaschke F, Takata Y, Caglayan E, Collins A, Tontonoz P, Hsueh WA, Tangirala RK. Circ. Res. 2006;99:e88–e99. doi: 10.1161/01.RES.0000252878.34269.06. [DOI] [PubMed] [Google Scholar]
- 226.Zhang-Gandhi CX, Drew PD. J. Neuroimmunol. 2007;183:50–59. doi: 10.1016/j.jneuroim.2006.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Smoak K, Madenspacher J, Jeyaseelan S, Williams B, Dixon D, Poch KR, Nick JA, Worthen GS, Fessler MB. J. Immunol. 2008;180:3305–3312. doi: 10.4049/jimmunol.180.5.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Ghisletti S, Huang W, Ogawa S, Pascual G, Lin ME, Willson TM, Rosenfeld MG, Glass CK. Mol. Cell. 2007;25:57–70. doi: 10.1016/j.molcel.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Marathe C, Bradley MN, Hong C, Lopez F, Ruiz de Galarreta CM, Tontonoz P, Castrillo A. J. Biol. Chem. 2006;281:32197–32206. doi: 10.1074/jbc.M605237200. [DOI] [PubMed] [Google Scholar]
- 230.Wang H, Zhang Y, Yehuda-Shnaidman E, Medvedev AV, Kumar N, Daniel KW, Robidoux J, Czech MP, Mangelsdorf DJ, Collins S. Mol. Cell Biol. 2008;28:2187–2200. doi: 10.1128/MCB.01479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Orfanos CE, Zouboulis CC, mond-Roesler B, Geilen CC. Drugs. 1997;53:358–388. doi: 10.2165/00003495-199753030-00003. [DOI] [PubMed] [Google Scholar]
- 232.Salbert G, Fanjul A, Piedrafita FJ, Lu XP, Kim SJ, Tran P, Pfahl M. Mol. Endocrinol. 1993;7:1347–1356. doi: 10.1210/mend.7.10.8264664. [DOI] [PubMed] [Google Scholar]
- 233.Motomura K, Sakai H, Isobe H, Nawata H. Cell Biochem. Funct. 1997;15:95–101. doi: 10.1002/(SICI)1099-0844(19970601)15:2<95::AID-CBF727>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 234.Motomura K, Sakai H, Isobe H, Nawata H. J. Gastroenterol. Hepatol. 1997;12:887–892. doi: 10.1111/j.1440-1746.1997.tb00388.x. [DOI] [PubMed] [Google Scholar]
- 235.Gyamfi MA, He L, French SW, Damjanov I, Wan YJ. J. Pharmacol. Exp. Ther. 2008;324:443–453. doi: 10.1124/jpet.107.132258. [DOI] [PubMed] [Google Scholar]
- 236.Hanniman EA, Lambert G, McCarthy TC, Sinal CJ. J. Lipid Res. 2005;46:2595–2604. doi: 10.1194/jlr.M500390-JLR200. [DOI] [PubMed] [Google Scholar]
- 237.Moschetta A, Bookout AL, Mangelsdorf DJ. Nat. Med. 2004;10:1352–1358. doi: 10.1038/nm1138. [DOI] [PubMed] [Google Scholar]
- 238.Liu Y, Binz J, Numerick MJ, Dennis S, Luo G, Desai B, MacKenzie KI, Mansfield TA, Kliewer SA, Goodwin B, Jones SA. J. Clin. Invest. 2003;112:1678–1687. doi: 10.1172/JCI18945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Jiang T, Wang XX, Scherzer P, Wilson P, Tallman J, Takahashi H, Li J, Iwahashi M, Sutherland E, Arend L, Levi M. Diabetes. 2007;56:2485–2493. doi: 10.2337/db06-1642. [DOI] [PubMed] [Google Scholar]
- 240.Li YT, Swales KE, Thomas GJ, Warner TD, Bishop-Bailey D. Arterioscler. Thromb. Vasc. Biol. 2007;27:2606–2611. doi: 10.1161/ATVBAHA.107.152694. [DOI] [PubMed] [Google Scholar]
- 241.Qin P, Borges-Marcucci LA, Evans MJ, Harnish DC. Am. J. Physiol Gastrointest. Liver Physiol. 2005;289:G267–G273. doi: 10.1152/ajpgi.00043.2005. [DOI] [PubMed] [Google Scholar]
- 242.Das A, Fernandez-Zapico ME, Cao S, Yao J, Fiorucci S, Hebbel RP, Urrutia R, Shah VH. J. Biol. Chem. 2006;281:39105–39113. doi: 10.1074/jbc.M607720200. [DOI] [PubMed] [Google Scholar]
- 243.Shah YM, Ma X, Morimura K, Kim I, Gonzalez FJ. Am. J. Physiol Gastrointest. Liver Physiol. 2007;292:G1114–G1122. doi: 10.1152/ajpgi.00528.2006. [DOI] [PubMed] [Google Scholar]
- 244.Venteclef N, Smith JC, Goodwin B, Delerive P. Mol. Cell Biol. 2006;26:6799–6807. doi: 10.1128/MCB.00579-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Venteclef N, Delerive P. J. Biol. Chem. 2007;282:4393–4399. doi: 10.1074/jbc.M608993200. [DOI] [PubMed] [Google Scholar]
- 246.Mueller M, Cima I, Noti M, Fuhrer A, Jakob S, Dubuquoy L, Schoonjans K, Brunner T. J. Exp. Med. 2006;203:2057–2062. doi: 10.1084/jem.20060357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Coste A, Dubuquoy L, Barnouin R, Annicotte JS, Magnier B, Notti M, Corazza N, Antal MC, Metzger D, Desreumaux P, Brunner T, Auwerx J, Schoonjans K. Proc. Natl. Acad. Sci. U. S. A. 2007;104:13098–13103. doi: 10.1073/pnas.0702440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Lee YK, Moore DD. J. Biol. Chem. 2002;277:2463–2467. doi: 10.1074/jbc.M105161200. [DOI] [PubMed] [Google Scholar]
- 249.Yamagata K, Daitoku H, Shimamoto Y, Matsuzaki H, Hirota K, Ishida J, Fukamizu A. J. Biol. Chem. 2004;279:23158–23165. doi: 10.1074/jbc.M314322200. [DOI] [PubMed] [Google Scholar]
- 250.Suh JH, Huang J, Park YY, Seong HA, Kim D, Shong M, Ha H, Lee IK, Lee K, Wang L, Choi HS. J. Biol. Chem. 2006;281:39169–39178. doi: 10.1074/jbc.M605947200. [DOI] [PubMed] [Google Scholar]
- 251.Gressner AM, Weiskirchen R. J. Cell Mol. Med. 2006;10:76–99. doi: 10.1111/j.1582-4934.2006.tb00292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Ogawa S, Lozach J, Benner C, Pascual G, Tangirala RK, Westin S, Hoffmann A, Subramaniam S, David M, Rosenfeld MG, Glass CK. Cell. 2005;122:707–721. doi: 10.1016/j.cell.2005.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Piraino G, Cook JA, O’Connor M, Hake PW, Burroughs TJ, Teti D, Zingarelli B. Shock. 2006;26:146–153. doi: 10.1097/01.shk.0000223121.03523.69. [DOI] [PubMed] [Google Scholar]
- 254.Glass CK, Ogawa S. Nat. Rev. Immunol. 2006;6:44–55. doi: 10.1038/nri1748. [DOI] [PubMed] [Google Scholar]
- 255.Rosenfeld MG, Lunyak VV, Glass CK. Genes Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 256.Kremoser C, Albers M, Burris TP, Deuschle U, Koegl M. Drug Discov. Today. 2007;12:860–869. doi: 10.1016/j.drudis.2007.07.025. [DOI] [PubMed] [Google Scholar]
- 257.Brunham LR, Kruit JK, Pape TD, Parks JS, Kuipers F, Hayden MR. Circ. Res. 2006;99:672–674. doi: 10.1161/01.RES.0000244014.19589.8e. [DOI] [PubMed] [Google Scholar]
- 258.Molteni V, Li X, Nabakka J, Liang F, Wityak J, Koder A, Vargas L, Romeo R, Mitro N, Mak PA, Seidel HM, Haslam JA, Chow D, Tuntland T, Spalding TA, Brock A, Bradley M, Castrillo A, Tontonoz P, Saez E. J. Med. Chem. 2007;50:4255–4259. doi: 10.1021/jm070453f. [DOI] [PubMed] [Google Scholar]
- 259.Hu B, Quinet E, Unwalla R, Collini M, Jetter J, Dooley R, Andraka D, Nogle L, Savio D, Halpern A, Goos-Nilsson A, Wilhelmsson A, Nambi P, Wrobel J. Bioorg. Med. Chem. Lett. 2007 doi: 10.1016/j.bmcl.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 260.Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, Newgard CB, Spiegelman BM. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 261.Rhee J, Inoue Y, Yoon JC, Puigserver P, Fan M, Gonzalez FJ, Spiegelman BM. Proc. Natl. Acad. Sci. U. S. A. 2003;100:4012–4017. doi: 10.1073/pnas.0730870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Leung TH, Hoffmann A, Baltimore D. Cell. 2004;118:453–464. doi: 10.1016/j.cell.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 263.Mack CL, Sokol RJ. Pediatr. Res. 2005;57:87R–94R. doi: 10.1203/01.PDR.0000159569.57354.47. [DOI] [PubMed] [Google Scholar]
- 264.Mack CL, Tucker RM, Sokol RJ, Karrer FM, Kotzin BL, Whitington PF, Miller SD. Pediatr. Res. 2004;56:79–87. doi: 10.1203/01.PDR.0000130480.51066.FB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Shivakumar P, Campbell KM, Sabla GE, Miethke A, Tiao G, McNeal MM, Ward RL, Bezerra JA. J. Clin. Invest. 2004;114:322–329. doi: 10.1172/JCI21153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Whitington PF, Malladi P, Melin-Aldana H, Azzam R, Mack CL, Sahai A. Pediatr. Res. 2005;57:837–844. doi: 10.1203/01.PDR.0000161414.99181.61. [DOI] [PubMed] [Google Scholar]
- 267.Mack CL, Tucker RM, Sokol RJ, Kotzin BL. Clin. Immunol. 2005;115:200–209. doi: 10.1016/j.clim.2005.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Sokol RJ. Hepatology. 2007;46:1675–1678. doi: 10.1002/hep.22001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Carter BA, Shulman RJ. Nat. Clin. Pract. Gastroenterol. Hepatol. 2007;4:277–287. doi: 10.1038/ncpgasthep0796. [DOI] [PubMed] [Google Scholar]
- 270.Carter BA, Taylor OA, Prendergast DR, Zimmerman TL, Von FR, Moore DD, Karpen SJ. Pediatr. Res. 2007;62:301–306. doi: 10.1203/PDR.0b013e3181256492. [DOI] [PubMed] [Google Scholar]
- 271.Inagaki T, Moschetta A, Lee YK, Peng L, Zhao G, Downes M, Yu RT, Shelton JM, Richardson JA, Repa JJ, Mangelsdorf DJ, Kliewer SA. Proc. Natl. Acad. Sci. U. S. A. 2006;103:3920–3925. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Gujral JS, Farhood A, Bajt ML, Jaeschke H. Hepatology. 2003;38:355–363. doi: 10.1053/jhep.2003.50341. [DOI] [PubMed] [Google Scholar]
- 273.Gujral JS, Liu J, Farhood A, Hinson JA, Jaeschke H. Am. J. Physiol Gastrointest. Liver Physiol. 2004;286:G499–G507. doi: 10.1152/ajpgi.00318.2003. [DOI] [PubMed] [Google Scholar]
- 274.Geier A, Zollner G, Dietrich CG, Wagner M, Fickert P, Denk H, van Rooijen N, Matern S, Gartung C, Trauner M. Hepatology. 2005;41:470–477. doi: 10.1002/hep.20594. [DOI] [PubMed] [Google Scholar]
- 275.Moseley RH, Wang W, Takeda H, Lown K, Shick L, Ananthanarayanan M, Suchy FJ. Am J Physiol. 1996;271:137–146. doi: 10.1152/ajpgi.1996.271.1.G137. [DOI] [PubMed] [Google Scholar]
- 276.Trauner M, Arrese M, Lee H, Boyer JL, Karpen SJ. J Clin Invest. 1998;101:2092–2100. doi: 10.1172/JCI1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Lee JM, Trauner M, Soroka CJ, Stieger B, Meier PJ, Boyer JL. Gastroenterology. 2000;118:163–172. doi: 10.1016/s0016-5085(00)70425-2. [DOI] [PubMed] [Google Scholar]
- 278.Mesotten D, Van den BG, Liddle C, Coulter S, McDougall F, Baxter RC, Delhanty PJ. Endocrinology. 2003;144:4008–4017. doi: 10.1210/en.2003-0139. [DOI] [PubMed] [Google Scholar]
- 279.Donner MG, Schumacher S, Warskulat U, Heinemann J, Haussinger D. Am. J. Physiol Gastrointest. Liver Physiol. 2007 doi: 10.1152/ajpgi.00079.2007. [DOI] [PubMed] [Google Scholar]
- 280.Lund M, Kang L, Tygstrup N, Wolkoff AW, Ott P. Am. J. Physiol. 1999;277:G91–G100. doi: 10.1152/ajpgi.1999.277.1.G91. [DOI] [PubMed] [Google Scholar]
- 281.Li N, Klaassen CD. Mol. Pharmacol. 2004;66:694–701. doi: 10.1124/mol.66.3.. [DOI] [PubMed] [Google Scholar]
- 282.Donner MG, Warskulat U, Saha N, Haussinger D. Biol. Chem. 2004;385:331–339. doi: 10.1515/BC.2004.029. [DOI] [PubMed] [Google Scholar]
- 283.Green RM, Hoda F, Ward KL. Gene. 2000;241:117–123. doi: 10.1016/s0378-1119(99)00460-6. [DOI] [PubMed] [Google Scholar]
- 284.Trauner M, Arrese M, Soroka CJ, Ananthanarayanan M, Koeppel TA, Schlosser SF, Suchy FJ, Keppler D, Boyer JL. Gastroenterology. 1997;113:255–264. doi: 10.1016/s0016-5085(97)70103-3. [DOI] [PubMed] [Google Scholar]