Abstract
Growth restriction by antibiotics is a common feature that pathogenic bacteria must overcome for survival. The struggle of bacteria to escape from growth restriction eventually results in development of antibiotic-resistance through the expression of a set of genes. Here we found that some physiologically important transcriptional regulators of Pseudomonas aeruginosa including QscR, a quorum sensing (QS) receptor, SoxR, a superoxide sensor-regulator, and AntR, a regulator of anthranilate-related secondary metabolism, are activated by various growth-restricted conditions. We generated the growth-restricted conditions by various methods, such as overexpression of PA2537 and treatment with antibiotics or disinfectants. The overexpression of PA2537, encoding an acyltransferase homologue, tightly restricted the growth of P. aeruginosa and significantly activated QscR during the growth restriction. Similarly, treatments with gentamycin, tetracycline, and ethanol also activated QscR near their minimal inhibitory concentrations (MICs). Some non-QS regulators, such as AntR and SoxR, were also activated near the MICs in the same conditions. However, LasR and PqsR, other QS receptors of P. aeruginosa, were not activated, suggesting that only a specific set of transcriptional regulators is activated by growth restriction. Since paraquat, a super-oxide generator, significantly activated QscR and AntR, we suggest that the oxidative stress generated by growth restriction may be partly involved in this phenomenon.
Keywords: antibiotic resistance, AntR, growth restriction, oxidative stress, Pseudomonas aeruginosa, QscR, quorum sensing, SoxR
INTRODUCTION
Growth restriction is a common condition that bacteria have to overcome for survival. In particular, pathogenic bacteria during infection must escape the growth restriction enforced by antibiotic medication and host immunity. The struggle against this challenge leads bacterial cells to express a set of genes and eventually enables them to develop resistance against antibiotics. Therefore, understanding how bacteria overcome the growth restriction caused by various environmental challenges is very important for understanding bacterial virulence and antibiotic resistance.
Pseudomonas aeruginosa is a Gram-negative and ubiquitous bacterium that is widely found in many diverse environments, such as soils, sea, lakes, mineral water, and in association with plants (Hardalo and Edberg, 1997). It is a serious opportunistic pathogen to human, continuously provoking nosocomial infections, and is responsible for severe morbidity and mortality in patients with burn wounds, cystic fibrosis, pneumonia, urinary tract infections, skin infections, cancer, acquired immunodeficiency syndrome (AIDS), and ocular diseases (Engel et al., 1998; Hancock and Speert, 2000). However, the challenge in treating the Pseudomonas infection is that P. aeruginosa has strong intrinsic resistance to many antibiotics including most penicillins, kanamycin, cephalothin, and moxifloxacin, and it very frequently develops acquired resistance to the antibiotics commonly used in the treatment of its infections, such as ceftazidime, piperacillin, quinolone, imipenem, and so forth, and even to recently developed antibiotics (Hancock and Speert, 2000; Lambert, 2002; Livermore, 2002).
Like many other pathogenic bacteria, P. aeruginosa expresses its virulence and antibiotic resistance via a cell density-dependent regulation mechanism, quorum sensing (QS). This mechanism regulates a large number of genes related to the production of virulence factors, motility, and the formation of biofilm, a very resistant mode of life; therefore, the activation of QS is generally considered very important in the physiology of P. aeruginosa (Fuqua and Greenberg, 2002; Hastings and Greenberg, 1999). For QS signaling, P. aeruginosa uses acyl homoserine lactones (acyl-HSLs) as signal molecules and multiple signal receptors (Lee et al., 2006; Schuster and Greenberg, 2006). Two major acyl-HSLs produced by P. aeruginosa are N-3-oxododecanoyl-L-homoserine lactone (3OC12-HSL) and N-butyryl-L-homoserine lactone (C4-HSL), which are synthesized by LasI and RhlI and received by LasR and RhlR, respectively (Fuqua and Greenberg, 2002). The signal-receptor complexes activate the transcription of their target genes, so called QS regulon. 3OC12-HSL has another receptor protein, QscR. QscR was originally characterized as a QS repressor that negatively regulates many genes induced by LasR and RhlR (Chugani et al., 2001). Recently, it was found that QscR directly regulates its own regulon genes including PA1897 and qscR, its own gene (Ha et al., 2012; Lee et al., 2006; Park et al., 2013). QscR was suggested to have a role in interspecies communication through its versatile signal binding ability (Ha et al., 2012; Lee et al., 2006; Oinuma and Greenberg, 2011). The 3-dimensional crystal structure of QscR was recently reported (Lintz et al., 2011).
So far, the activation of the QS system has been known to be provoked as cells grow to high population density. However, in this study, we found that QscR can be also activated by growth restriction. This finding was unexpected but interesting because it implied that QS may be modulated by a non-canonical mechanism and that growth restriction may generate some signals to trigger the expression of a set of genes. Based on the results of our study about this phenomenon, we suggest that growth restriction can modulate QS regulation, and oxidative stress generated by growth restriction may partially be involved in this phenomenon.
MATERIALS AND METHODS
Bacterial strains and culture conditions
Both P. aeruginosa and Escherichia coli strains used in this study (Table 1) were cultured in Luria-Bertani medium (LB; 5 g/L yeast extract, 10 g/L bactotryptone, 5 g/L NaCl) at 37°C with vigorous shaking at 170 rpm. Cell growth was measured by optical density at 600 nm (OD600). For the selection of plasmid-carrying cells, carbenicillin or/and gentamicin were added at 150 μg/ml and 50 μg/ml, respectively. To induce the growth-restricted condition, L-arabinose or growth-inhibiting agents were added at various concentrations as indicated. Gentamicin was used in two ways: to select the cells carrying pJN105 and pDY101 at fixed concentration (50 μg/ml), and to restrict the growth of non-resistant cells at various concentrations. To generate oxidative stress, paraquat (1,1′-dimethyl-4,4′-bipyridinium dichloride) was added to media at a concentration of 1 mM, which did not affect cell growth.
Table 1.
Bacterial strains and plasmids used in this study
| Names | Genotype | References |
|---|---|---|
| Bacterial strains | ||
| PA01 | Wild type P. aeruginosa strain | Pearson et al. (1997) |
| DH5α | Escherichia coli strain, supE44, ϕ lacU169 (ϕ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Lab collection |
| Plasmids | ||
| pQF50 | Broad-host-range lacZ transcriptional fusion vector, Ori1600, pMB1, CbR | Farinha and Kropinski (1990) |
| pSC11 | lasI-lacZ fusion in pQF50, CbR | Chugani et al. (2001) |
| pJL101 | PA1897-lacZ fusion in pQF50, CbR | Lee et al. (2006) |
| pJL201 | antA-lacZ fusion in pQF50, CbR | Choi et al. (2011) |
| pJL301 | pqsA-lacZ fusion in pQF50, CbR | Choi et al. (2011) |
| pSJ101 | PA2274-lacZ fusion in pQF50, CbR | This study |
| pJN105 | araC-PBAD cassette cloned in pBBR1MCS-5, GmR | Newman and Fuqua (1999) |
| pDY101 | PA2537 in pJN105, GmR | Yeom et al. (2013) |
Plasmids and preparation of reporter strains
The plasmids used in this study are listed in Table 1. For the measurement of the activities of various sensor-regulator proteins, such as QscR, LasR, PqsR, and AntR, their target promoter-lacZ fusion plasmids (pJL101, pSC11, pJL301, and pJL201, respectively) were used as reporters in PA01 cells as described elsewhere (Choi et al., 2011; Ha et al., 2012; Kim et al., 2012; Lee et al., 2006). For the measurement of SoxR activity, PA2274 promoter-lacZ fusion plasmid, pSJ101, was constructed as a reporter, in which a 314-bp PCR fragment from −289 to +25 relative to the PA2274 translation start codon was ligated into BamHI-HindIII-digested pQF50. These fusion plasmids were introduced into PA01 cells by transformation and the transformed strains were used as the reporter strains to monitor the regulator activity.
Growth restriction and measurement of regulator activities
The growth restriction was induced by overexpressing PA2537 gene or by adding growth-inhibiting agents. PA2537 gene was previously reported to inhibit the growth of P. aeruginosa when overexpressed (Yeom et al., 2013). To overexpress PA2537, pDY101, a pJN105-based plasmid in which the PA2537 gene is under the control of the arabinose-inducible pBAD promoter, was introduced into P. aeruginosa cells harboring each reporter plasmid by transformation. The transformed cells were grown overnight in 5 ml of LB with vigorous shaking at 37°C and diluted to OD600 = 0.02 in fresh LB medium. The growth restriction was induced by adding L-arabinose into the medium at various concentrations. Growth inhibition was also induced by adding growth-inhibiting agents such as gentamicin, tetracycline, and ethanol into P. aeruginosa cells harboring reporter plasmids. The reporter cells were prepared in the same manner and the growth inhibitors were added at various concentrations instead of arabinose. After the addition of arabinose or growth inhibitors, the cells were incubated with vigorous shaking at 37°C and sampled at 4 h (for LasR and PqsR activities) or 7 h (for QscR, AntR, and SoxR activities). The cells were taken from the culture to measure the cell density (OD600) and the β-galactosidase activity to monitor the growth restriction and regulator activities, respectively. The whole procedure is schematically illustrated in Supplementary Fig. S2. With growth restriction, the final OD600 remained near the initial OD600 of 0.02 (C, the growth restriction range in Supplementary Fig. S2) and, with growth, the final OD600 was significantly higher than 0.02 (A, the normal growth range in Supplementary Fig. S2). The extent of growth was plotted with the concentration of the growth inhibitors or arabinose as illustrated in Supplementary Fig. S2. The β-galactosidase activity was measured with Tropix-Plus™ kit (Applied Biosystems, USA) according to the manufacturer’s instructions, as described elsewhere (Lee et al., 2006). The level of β-galactosidase is proportional to the activity of the respective regulators and plotted with the concentration of the growth inhibitors or arabinose as in Supplementary Fig. S2. To measure the activities of SoxR, QscR, and AntR in response to oxidative stress, P. aeruginosa cells harboring pSJ101, pJL201, and pJL101 were cultivated in LB medium for 2 h (for pSJ101), 4 h (for pJL201), and 6 h (for pJL101), treated with 1 mM paraquat for 30 min, and then β-galactosidase activity was measured with Tropix-Plus™ kit. The β-galactosidase activity is presented in arbitrary units of luminescence per OD600.
Statistical analysis
In order to ensure the significance of the results in the virulence analyses, the data were statistically analyzed using t-test (two-sample assuming equal variances) in MS office Excel (Microsoft, USA). If the P-value was lower than 0.05, it was considered significant.
RESULTS
Growth restriction causes the activation of QscR, a QS regulator of P. aeruginosa
In our previous study, we found that the PA2537 gene encoding a probable acyltransferase severely restricted the growth of P. aeruginosa when overexpressed at a higher level than a certain threshold, while the mild expression of PA2537 below that threshold did not cause growth restriction (Yeom et al., 2013). Interestingly, the mild expression of PA2537 had differential effect on each of two 3OC12-HSL receptors, LasR and QscR, such that it alleviated the LasR activity but had no effect on QscR (Yeom et al., 2013). In this study we found that the over-expression of PA2537 over the threshold dramatically activated QscR with growth restriction (Supplementary Fig. S1). The QscR activity was measured using the PA1897-lacZ fusion and so the β-galactosidase activity reflects the QscR activity, because PA1897 is specifically induced by QscR (Lee et al., 2006). This result was interesting but it was unclear which of the acyltransferase function or growth restriction modulated the QscR activity. Since the growth restriction by PA2537 overexpression could be finely controlled by adjusting the arabinose concentration, we measured the QscR activity and growth under a wide range of arabinose concentrations covering the threshold for growth restriction and plotted them versus the concentration of arabinose as illustrated in Supplementary Fig. S2 and experimental procedures. The result showed that the QscR activity did not increase in the normal growth concentration range (A region in Fig. 1), but increased in the transition range where growth restriction began to occur (B region) and reached a plateau in the growth restriction range (C region). This result strongly suggests that QscR activation might be triggered by growth restriction, not by the function of PA2537, because the activation of QscR did not seem to correlate to the expression level of PA2537 itself.
Fig. 1.
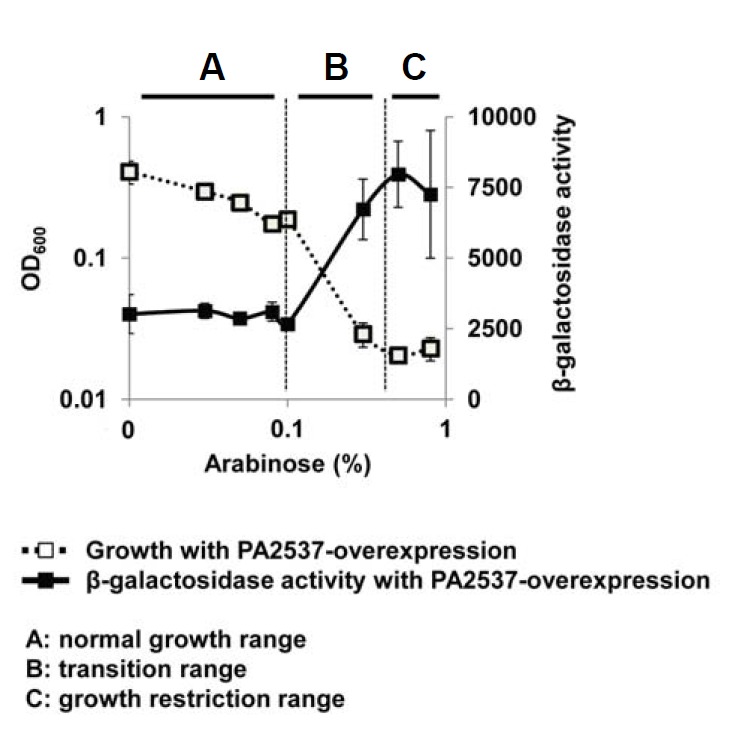
QscR activation by the growth restriction. QscR activity was measured by the PA1897-lacZ fusion and the β-galactosidase activity reflects the QscR activity, since PA1897 is specifically induced by QscR (Lee et al., 2006). Two compatible plasmids, pJL101 (PA1897-lacZ fusion plasmid) and pDY101 (PA2537-overexpressing plasmid) were co-introduced into PA01 cells and the transformed cells were cultivated for 7 h. The β-galactosidase activity and growth were measured at various concentrations of arabinose and plotted as illustrated in Supplementary Fig. S2.
If this is the case, QscR should be activated in other growth-inhibiting conditions, such as with antibiotic treatment. To prove this, we restricted the growth of P. aeruginosa by treatment with two structurally different antibiotics, gentamicin or tetracycline, and measured the QscR activity and growth to plot them versus the concentration of growth inhibitors just as in Fig. 1. Interestingly, QscR was significantly activated by these antibiotic treatments (Figs. 2A and 2B). A growth-inhibiting disinfectant, ethanol, also activated QscR (Fig. 2C). In all cases, the activation of QscR was triggered at the concentration of the transition range where the growth inhibition started to occur (Figs. 2A–2C). Therefore, we concluded that QscR activation was mediated by growth restriction.
Fig. 2.
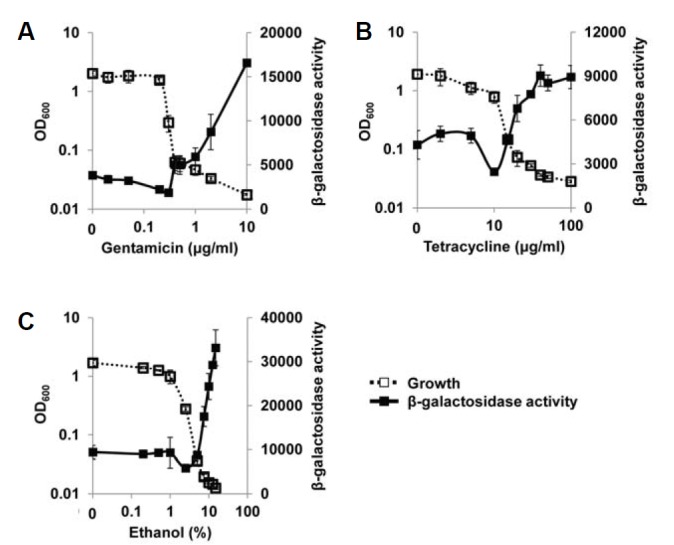
Change of QscR activity in growth-restricted conditions. The growth of P. aeruginosa cells harboring pJL101 was finely restricted by gentamicin (A), tetracycline (B), and ethanol (C), and the β-galactosidase activity and growth were measured and plotted as in Fig. 1. The β-galactosidase activity reflects the QscR activity.
Growth restriction affects only QscR, not the whole QS system
QscR is hierarchically downstream of LasR in the QS cascade, since it uses 3CC12-HSL synthesized by the LasR-I system for its ligand (Schuster and Greenberg, 2006). So, we investigated whether the activity of LasR could be affected by the same growth restriction. The LasR activity was measured using the lasI-lacZ fusion that is specifically induced by LasR (Lee et al., 2006). When LasR activity was finely measured with PA2537 overexpression and plotted along with growth and concentration of arabinose in the same manner, it was not significantly affected by growth restriction unlike QscR (Fig. 3A). Although a certain degree of oscillation was observed in the transition range, the normal growth and growth restriction ranges showed similar levels of LasR activity (Fig. 3A). The antibiotic-induced growth restriction by gentamicin and tetracycline also failed to change the LasR activity between normal growth and growth restriction ranges (Figs. 3B and 3C). These results strongly suggest that only QscR is specifically activated by the growth restriction, rather than the entire QS system.
Fig. 3.
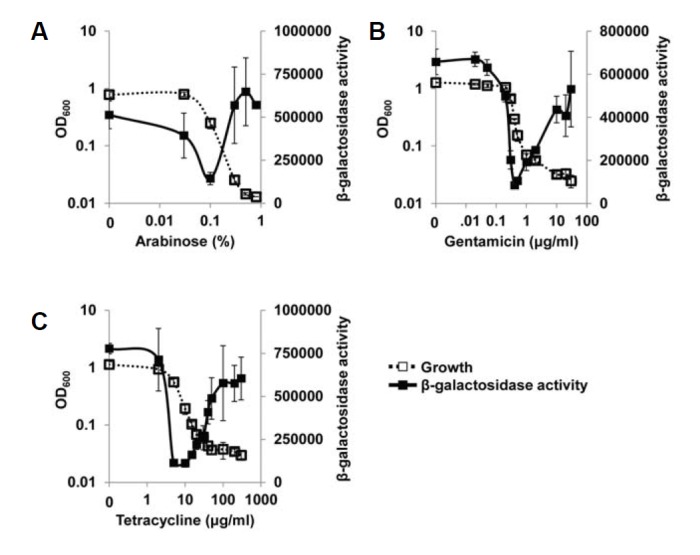
LasR activity with growth restriction. (A) Two plasmids, pSC11 (lasI-lacZ fusion plasmid) and pDY101 were co-introduced into PA01 cells, and the growth of the transformed cells was finely restricted by various concentrations of arabinose. The β-galactosidase activity and growth were measured and plotted. Since lasI gene is specifically induced by LasR (Lee et al., 2006), the β-galactosidase activity reflects the LasR activity. (B, C) The growth of P. aeruginosa cells harboring only pSC11 was restricted by gentamicin (B) and tetracycline (C). The β-galactosidase activity and growth were measured and plotted in the same manner.
To confirm this suggestion, we investigated the activity of PqsR, another QS receptor protein of PQS signaling system, with growth restriction. When the activity of PqsR was measured using the pqsA-lacZ fusion that is specifically induced by PqsR (Choi et al., 2011), it was not influenced by the growth restriction caused by gentamicin and tetracycline (Figs. 4A and 4B). Similar to LasR, there was some fluctuation of PqsR activity in the transition range, but there was no significant difference in the activity between normal growth and growth restriction ranges. This oscillation may be relatively magnified due to the small difference in the activity before and after growth restriction. Taken together, we suggest that only QscR, rather than the entire QS system, is influenced by growth restriction.
Fig. 4.
Effect of growth restriction on the activity of PqsR, another QS receptor. The growth of P. aeruginosa cells harboring pJL301 (pqsA-lacZ fusion plasmid) was inhibited by antibiotic treatments, gentamicin (A) and tetracycline (B), and the β-galactosidase activity was measured and plotted. Since pqsA gene is specifically regulated by PqsR (Choi et al., 2011), the β-galactosidase activity reflects the PqsR activity.
Some non-QS regulators were also activated in growth-restricted conditions
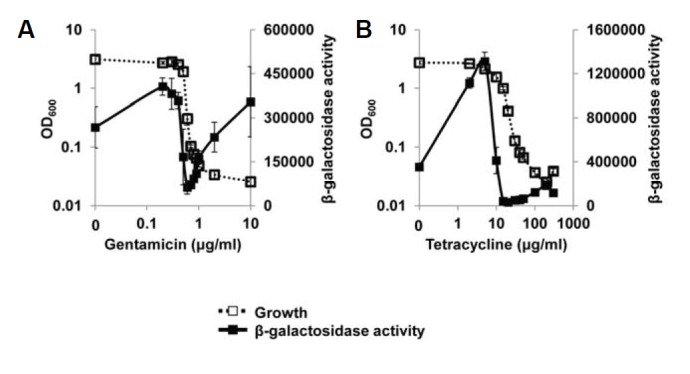
Several years ago, Kohanski et al. (2007) suggested a common mechanism of cellular death induced by antibiotics. They suggested that bactericidal antibiotics generate oxidative stress within bacterial cells and ultimately cause cell death. Although their suggestion excluded bacteriostatic antibiotics that only inhibit cell growth, the clinical definition of bactericidals and bacteriostatics is arbitrary and most antibacterials are better described as potentially having both bactericidal and bacteriostatic activities (Pankey and Sabath, 2004). So, we were curious whether the growth restriction used in this study could generate the oxidative stress that is sensible to P. aeruginosa cells. To know this, we investigated the influence of growth restriction on the activity of SoxR, a superoxide sensor of P. aeruginosa. When we measured the SoxR activity using PA2274-lacZ fusion that is specifically regulated by SoxR (Palma et al., 2005), it was significantly activated in the growth-restricted conditions caused by PA2537 overexpression and by antibiotics (Figs. 5A–5C). This result indicates that growth restriction most likely generates oxidative stress within cells, and also implies that the influence of growth restriction might be not limited to QscR, but might be pleiotropic.
Fig. 5.
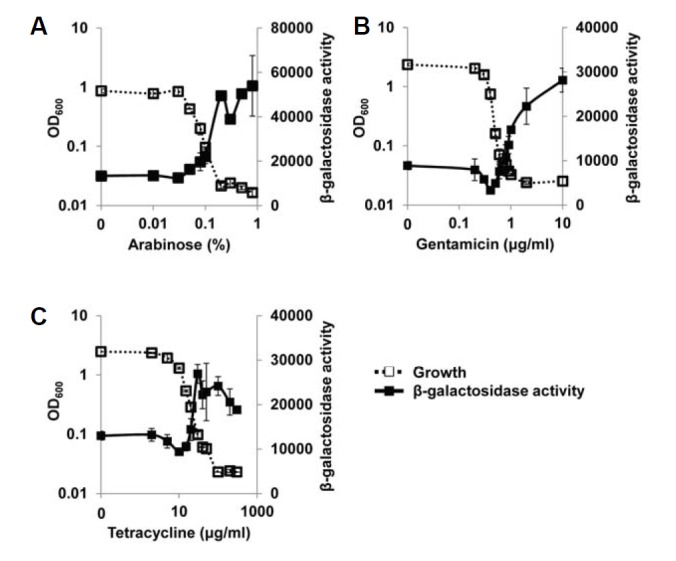
Effect of growth restriction on SoxR, a redox sensor protein. (A) Two plasmids, pSJ101 (PA2274-lacZ fusion plasmid) and pDY101 were co-introduced into PA01 cells and the growth of the transformed cells was restricted by various concentrations of arabinose. (B, C) The growth of P. aeruginosa cells harboring only pSJ101 was restricted by gentamicin (B) and tetracycline (C). The β-galactosidase activity and growth was measured and plotted in the same manner as in the previous figures. Since PA2274 is specifically induced by SoxR (Kobayashi and Tagawa, 2004), the β-galactosidase activity reflects the SoxR activity.
In general, secondary metabolites are produced when the cell growth is arrested in the stationary phase. We wondered whether secondary metabolism could be affected by growth-restricted conditions. We examined the activities of AntR, a regulator of anthranilate metabolism that is closely related with the TCA cycle, tryptophan biosynthesis and degradation, and PQS biosynthesis (Choi et al., 2011; Oglesby et al., 2008). Since AntR specifically regulates the transcription of the antA gene (Kim et al., 2012), we measured AntR activity using the antA-lacZ fusion. Interestingly, AntR was significantly activated by antibiotic-induced growth restriction (Figs. 6A and 6B).
Fig. 6.
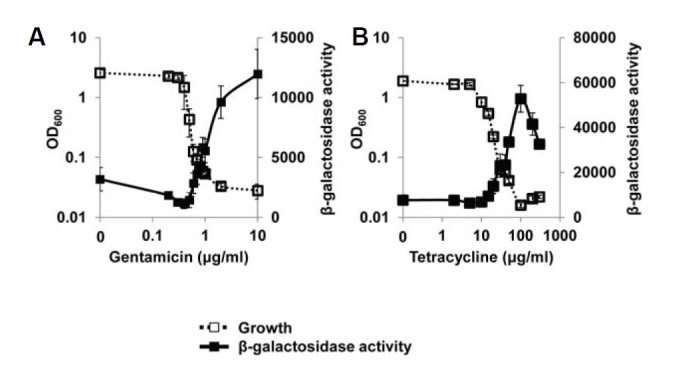
Effect of growth restriction on AntR, a regulator of anthranilate metabolism. The growth of P. aeruginosa cells harboring pJL201 (antA-lacZ fusion plasmid) was inhibited by gentamicin (A) and tetracycline (B), and the β-galactosidase activity was measured and plotted in the same manner. Since antA gene is specifically regulated by AntR (Kim et al., 2012), the β-galactosidase activity reflects the AntR activity.
Oxidative stress may be a partial cause of the activation of regulators in growth-restricted conditions
The activation of SoxR strongly suggested that intracellular oxidative stress might mediate the effects of growth restriction, regardless of its cause, as suggested by Kohanski et al. (2007). To address this possibility, we treated P. aeruginosa cells with paraquat, a well-known superoxide-generating agent, and measured the activities of the regulators that were activated by growth restriction. SoxR was strongly activated by paraquat treatment, as expected (Fig. 7A). QscR and AntR were also slightly but significantly activated (Figs. 7B and 7C). This means that oxidative stress may be a cause of the activation of the regulators in growth-restricted conditions. However, the extent of the activation in the paraquat treatment was small compared with that in the growth restriction. Growth restriction activated QscR by 2.6-fold (PA2537 overexpression), 4.4-fold (gentamicin), 2.1-fold (tetracycline), and 3.5-fold (ethanol), respectively. But paraquat activated QscR by 1.6-fold. In the case of SoxR, growth restriction activated it by 4.0-fold (PA2537 overexpression), 3.2-fold (gentamicin), and 2.1-fold (tetracycline), respectively, but paraquat activated it by 11-fold. Growth restriction activated AntR by 3.8-fold (gentamicin) and 6.9-fold (tetracycline), but paraquat activated AntR by 1.4-fold. This difference in the extent of the activation indicates that while oxidative stress may be involved in the activation of these regulators by growth restriction, it may not be the only cause of the growth restriction effect.
Fig. 7.
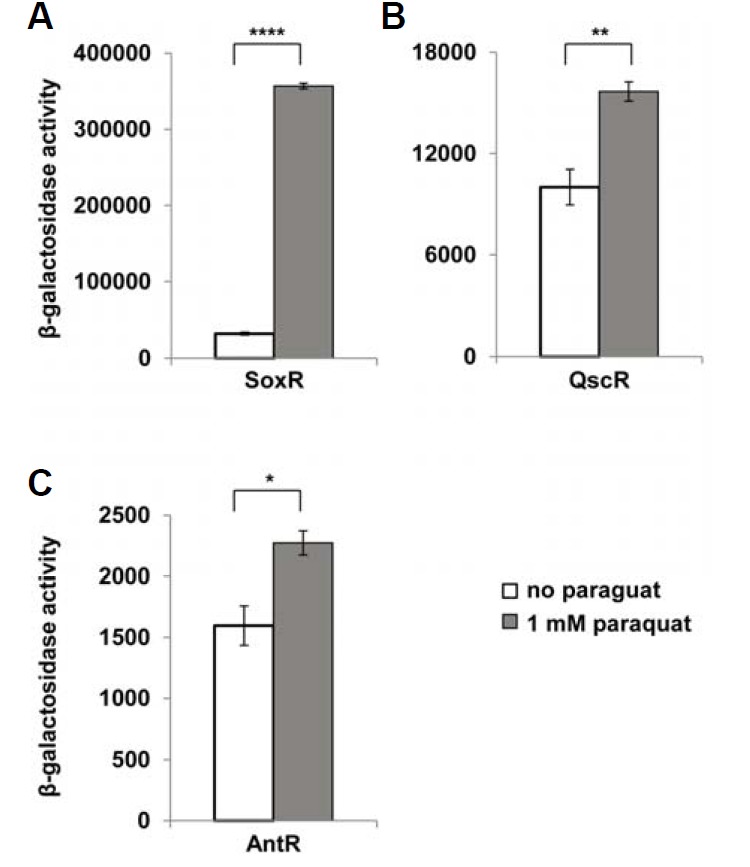
Activities of SoxR (A), QscR (B), and AntR (C) in response to the oxidative stress along growth. P. aeruginosa cells harboring pSJ101 (A), pJL101 (B) and pJL201 (C) were cultivated and treated with 1 mM paraquat for 30 min, and the β-galactosidase activity was measured. P-value: *< 0.05; **< 0.001; ****< 0.0000001.
DISCUSSION
P. aeruginosa, a causative agent of many serious infections in human, is a very difficult pathogen to treat because of its resistance to antibiotics. It is a highly adaptable organism and a small change in susceptibility can frustrate the effectiveness of even antibiotics currently used in Pseudomonas infections (Hancock and Speert, 2000). Here we investigated the relationship between growth restriction and the activities of some physiologically important regulators of P. aeruginosa. Our results give a clue about how P. aeruginosa changes its gene expression profile when its growth is threatened by antibiotics. Although the mechanism of the growth restriction by PA2537 expression is not known yet, since it is most likely different from the mechanism by antibiotics, the mechanisms of growth restriction used for this study are considered all different. Nevertheless, these distinct causes of growth restriction resulted in the same consequence: the activation of QscR, SoxR, and AntR. This implies that a common cue may mediate the activation of these regulators. However, these regulators are activated by completely different molecules, 3OC12-HSL, superoxide, and anthranilate, respectively. Then, what can be the common cue?
Despite the activation of QscR, 3OC12-HSL is not the cue by growth restriction, because LasR is not activated in growth-restricted conditions (Fig. 3). While the signal to activate QscR is 3OC12-HSL in the canonical QS regulation (Lee et al., 2006), LasR is also activated by 3OC12-HSL more sensitively (Schuster et al., 2004). Therefore, QscR must use different signals in growth restriction. As we mentioned earlier, QS is a cell density-dependent phenomenon and is generally considered to be provoked as cells grow to high population density. So, growth restriction is the opposite of the canonical QS-triggering condition. This indicates that QscR may be activated by another cue brought about by growth restriction.
While we don’t fully understand what that cue is, one promising candidate was oxidative stress provoked during growth restriction because SoxR, the sensor of superoxide radicals, was activated in the same conditions (Fig. 5). The antibiotic treatment has been suggested to induce the generation of intracellular reactive oxygen species (Kohanski et al., 2007). In this hypothesis, bactericidal antibiotics kill bacteria in the following sequence, regardless of their drug-target interaction: 1) they stop the tricarboxylic acid cycle, 2) which depletes NADH in bacterial cells, 3) which in turn generates reactive oxygen species like hydroxyl radicals, 4) which damages intracellular macromolecules (Kohanski et al., 2007; Wright, 2007). The partial activation of QscR and AntR by paraquat may support this suggestion to some extent. However, this suggestion was originally limited to only bactericidal antibiotics, in that the bacteriostatic antibiotics do not induce the production of hydroxyl radicals or trigger cell death, and this suggestion is still controversial with the recent studies by independent groups (Keren et al., 2013; Liu and Imlay, 2013). Moreover, since the activation of QscR and AntR by paraquat was just limited, we think that oxidative stress can explain the growth restriction effects only partially and it is hard to say that oxidative stress is the only and major cue. The underlying mechanism remains to be unraveled.
We note that the classification of antibiotics as bactericidals and bacteriostatics is not always obvious practically, since a drug that is bactericidal for one strain may only inhibit the growth of another strain, and high concentrations of some bacteriostatic agents are also bactericidal, whereas low concentrations of some bactericidal agents are bacteriostatic (Pankey and Sabath, 2004). In this study, we used two antibiotics, gentamicin and tetracycline. Although both antibiotics work against protein synthesis, gentamicin is generally classified as bactericidal and tetracycline as bacteriostatic (Kohanski et al., 2007; Pankey and Sabath, 2004). However, our study showed no significant difference with two antibiotics. Since P. aeruginosa has no dormant form of life like an endospore, and the prolonged growth restriction would eventuate in the cell death in P. aeruginosa, we think that two antibiotics similarly affect cell physiology in our condition for growth restriction.
Supplemental data
Acknowledgments
This research was also supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2013R1A1A20122-20)
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Choi Y., Park H.Y., Park S.J., Kim S.K., Ha C., Im S.J., Lee J.H. Growth phase-differential quorum sensing regulation of anthranilate metabolism in Pseudomonas aeruginosa. Mol. Cells. 2011;32:57–65. doi: 10.1007/s10059-011-2322-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugani S.A., Whiteley M., Lee K.M., D’Argenio D., Manoil C., Greenberg E.P. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA. 2001;98:2752–2757. doi: 10.1073/pnas.051624298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel L.S., Hill J.M., Moreau J.M., Green L.C., Hobden J.A., O’Callaghan R.J. Pseudomonas aeruginosa protease IV produces corneal damage and contributes to bacterial virulence. Invest. Ophthalmol. Vis. Sci. 1998;39:662–665. [PubMed] [Google Scholar]
- Farinha M.A., Kropinski A.M. Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J. Bacteriol. 1990;172:3496–3499. doi: 10.1128/jb.172.6.3496-3499.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua C., Greenberg E.P. Listening in on bacteria: acyl-homoserine lactone signalling. Nat. Rev. Mol. Cell Biol. 2002;3:685–695. doi: 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- Ha C., Park S.J., Im S.J., Lee J.H. Interspecies signaling through QscR, a quorum receptor of Pseudomonas aeruginosa. Mol. Cells. 2012;33:53–59. doi: 10.1007/s10059-012-2208-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R.E., Speert D.P. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Resist. updat. 2000;3:247–255. doi: 10.1054/drup.2000.0152. [DOI] [PubMed] [Google Scholar]
- Hardalo C., Edberg S.C. Pseudomonas aeruginosa: assessment of risk from drinking water. Crit. Rev. Microbiol. 1997;23:47–75. doi: 10.3109/10408419709115130. [DOI] [PubMed] [Google Scholar]
- Hastings J.W., Greenberg E.P. Quorum sensing: the explanation of a curious phenomenon reveals a common characteristic of bacteria. J. Bacteriol. 1999;181:2667–2668. doi: 10.1128/jb.181.9.2667-2668.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren I., Wu Y., Inocencio J., Mulcahy L.R., Lewis K. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science. 2013;339:1213–1216. doi: 10.1126/science.1232688. [DOI] [PubMed] [Google Scholar]
- Kim S.K., Im S.J., Yeom D.H., Lee J.H. AntR-mediated bidirectional activation of antA and antR, anthranilate degradative genes in Pseudomonas aeruginosa. Gene. 2012;505:146–152. doi: 10.1016/j.gene.2012.05.004. [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Tagawa S. Activation of SoxR-dependent transcription in Pseudomonas aeruginosa. J. Biochem. 2004;136:607–615. doi: 10.1093/jb/mvh168. [DOI] [PubMed] [Google Scholar]
- Kohanski M.A., Dwyer D.J., Hayete B., Lawrence C.A., Collins J.J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- Lambert P.A. Mechanisms of antibiotic resistance in Pseudomonas aeruginosa. J. R. Soc. Med. 2002;95(Suppl 41):22–26. [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Lequette Y., Greenberg E.P. Activity of purified QscR, a Pseudomonas aeruginosa orphan quorum-sensing transcription factor. Mol. Microbiol. 2006;59:602–609. doi: 10.1111/j.1365-2958.2005.04960.x. [DOI] [PubMed] [Google Scholar]
- Lintz M.J., Oinuma K., Wysoczynski C.L., Greenberg E.P., Churchill M.E. Crystal structure of QscR, a Pseudomonas aeruginosa quorum sensing signal receptor. Proc. Natl. Acad. Sci. USA. 2011;108:15763–15768. doi: 10.1073/pnas.1112398108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore D.M. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin. Infect. Dis. 2002;34:634–640. doi: 10.1086/338782. [DOI] [PubMed] [Google Scholar]
- Newman J.R., Fuqua C. Broad-host-range expression vectors that carry the L-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene. 1999;227:197–203. doi: 10.1016/s0378-1119(98)00601-5. [DOI] [PubMed] [Google Scholar]
- Oglesby A.G., Farrow J.M., 3rd, Lee J.H., Tomaras A.P., Greenberg E.P., Pesci E.C., Vasil M.L. The influence of iron on Pseudomonas aeruginosa physiology: a regulatory link between iron and quorum sensing. J. Biol. Chem. 2008;283:15558–15567. doi: 10.1074/jbc.M707840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oinuma K., Greenberg E.P. Acyl-homoserine lactone binding to and stability of the orphan Pseudomonas aeruginosa quorum-sensing signal receptor. QscR. J. Bacteriol. 2011;193:421–428. doi: 10.1128/JB.01041-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma M., Zurita J., Ferreras J.A., Worgall S., Larone D.H., Shi L., Campagne F., Quadri L.E. Pseudomonas aeruginosa SoxR does not conform to the archetypal paradigm for SoxR-dependent regulation of the bacterial oxidative stress adaptive response. Infect. Immun. 2005;73:2958–2966. doi: 10.1128/IAI.73.5.2958-2966.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankey G.A., Sabath L.D. Clinical relevance of bacteriostatic versus bactericidal mechanisms of action in the treatment of Gram-positive bacterial infections. Clin. Infect. Dis. 2004;38:864–870. doi: 10.1086/381972. [DOI] [PubMed] [Google Scholar]
- Park S.J., Liu H.B., Park S., Lee J.H. Modulation of QscR, a quorum sensing receptor of Pseudomonas aeruginosa, by truncation of a signal binding domain. Res. Microbiol. 2013;164:375–381. doi: 10.1016/j.resmic.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Pearson J.P., Pesci E.C., Iglewski B.H. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M., Greenberg E.P. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int. J. Med. Microbiol. 2006;296:73–81. doi: 10.1016/j.ijmm.2006.01.036. [DOI] [PubMed] [Google Scholar]
- Schuster M., Urbanowski M.L., Greenberg E.P. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. Proc. Natl. Acad. Sci. USA. 2004;101:15833–15839. doi: 10.1073/pnas.0407229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright G.D. On the road to bacterial cell death. Cell. 2007;130:781–783. doi: 10.1016/j.cell.2007.08.023. [DOI] [PubMed] [Google Scholar]
- Yeom D.H., Kim S.K., Lee M.N., Lee J.H. Pleiotropic effects of acyltransferases on various virulence-related phenotypes of Pseudomonas aeruginosa. Genes Cells. 2013;18:682–693. doi: 10.1111/gtc.12076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


