Abstract
Engineering antibodies to utilize non-canonical amino acids (NCAA) should greatly expand the utility of an already important biological reagent. In particular, introducing crosslinking reagents into antibody complementarity determining regions (CDRs) should provide a means to covalently crosslink residues at the antibody–antigen interface. Unfortunately, finding the optimum position for crosslinking two proteins is often a matter of iterative guessing, even when the interface is known in atomic detail. Computer-aided antibody design can potentially greatly restrict the number of variants that must be explored in order to identify successful crosslinking sites. We have therefore used Rosetta to guide the introduction of an oxidizable crosslinking NCAA, l-3,4-dihydroxyphenylalanine (l-DOPA), into the CDRs of the anti-protective antigen scFv antibody M18, and have measured crosslinking to its cognate antigen, domain 4 of the anthrax protective antigen. Computed crosslinking distance, solvent accessibility, and interface energetics were three factors considered that could impact the efficiency of l-DOPA-mediated crosslinking. In the end, 10 variants were synthesized, and crosslinking efficiencies were generally 10% or higher, with the best variant crosslinking to 52% of the available antigen. The results suggest that computational analysis can be used in a pipeline for engineering crosslinking antibodies. The rules learned from l-DOPA crosslinking of antibodies may also be generalizable to the formation of other crosslinked interfaces and complexes.
Keywords: Computer-aided design, Structure-based design, Rosetta, Antibody, Non-canonical amino acid, Crosslinking, Binding affinity, l-DOPA
1. Introduction
Antibodies are key components of the immune system with broad diversity to recognize a variety of antigens. Antibody-based therapeutic, diagnostic, and industrial applications frequently require antibodies having high stability and strong binding affinity. With the development of computational techniques and a number of successful experiences in protein modeling and design (Lippow and Tidor, 2007; Mandell and Kortemme, 2009), computational antibody design has begun to play an important role in predicting improvements to antibody function. Computational design of antibodies has been used to enhance binding affinity (Barderas et al., 2008; Clark et al., 2006; Lippow et al., 2007), to improve stability by improvement of thermal/aggregation resistance (Chennamsetty et al., 2009; Miklos et al., 2012), and to alter binding specificity (Farady et al., 2009), and others (Caravella et al., 2010; Kuroda et al., 2012; Midelfort et al., 2004; Pantazes and Maranas, 2010).
To date, though, most computational design methods have focused on manipulating the twenty natural proteogenic amino acids to modify molecular forces such as electrostatics (Lippow et al., 2007), hydrophobic interactions (Chennamsetty et al., 2009), hydrogen bonds (Clark et al., 2006), and salt bridges (Miklos et al., 2012). However, recent advances in engineering the translation system have now allowed for the site-specific insertion of non-canonical amino acids (NCAAs) with a variety of functionalities into proteins with good efficiency (Wang et al., 2006; Xie and Schultz, 2006). Such NCAAs can be used to improve the stability and pharmacokinetics of therapeutic proteins (Cho et al., 2011), to augment binding (Liu et al., 2009), and to provide a myriad of chemical handles to study protein structure and function (Jones et al., 2010; Tsao et al., 2006; Zhang et al., 2002).
The generation of protein–protein crosslinks by inserting NCAAs into proteins could prove useful for a variety of applications. To this end, a number of crosslinking-capable NCAAs have been incorporated into proteins in a site-specific manner utilizing an array of functionalized amino acids. These crosslinking functionalities include photo-crosslinkable aryl-azides (Chin et al., 2002b), benzophenones (Chin et al., 2002a) and diazirines (Ai et al., 2011) as well as the oxidizable crosslinker, l-DOPA (Alfonta et al., 2003). While any of the crosslinkers might benefit from a quantitative placement methodology, we chose l-DOPA because the periodate induced oxidation allowed for more control over the crosslinking conditions relative to photo-inducible crosslinkers that have been found to spuriously crosslink during sample handling (Chin et al., 2002b). In addition, the nucleophile-driven cross-linking mechanism of l-DOPA has been extensively characterized with a variety of proteinaceous nucleophiles (Liu et al., 2006).
l-DOPA has previously been used to successfully crosslink the monomeric domains of a dimeric sortase A for structural studies (Umeda et al., 2009), to enhance the affinity of low-affinity peptide probes for a kinase SH3 bioassay (Umeda et al., 2010), and to site-specifically label proteins with polysaccharides (Ayyadurai et al., 2011). While previously reported uses of l-DOPA as a site-specific crosslinker have yielded examples of effective crosslinking (as shown by SDS–PAGE or Western blot analyses), the actual efficiencies of crosslinking have never been reported (Burdine et al., 2004; Umeda et al., 2009, 2010). These previous reports indicated that it was possible to place l-DOPA by intuition, but did not provide more quantitative assessments of what parameters impacted crosslinking efficiency (Umeda et al., 2009, 2010).
In this paper, we explore how the Rosetta suite of computational protein design tools might be used to predict the site-specific, functional incorporation of l-DOPA into an antibody, allowing it to crosslink to its cognate antigen. A better understanding of where and how to insert crosslinking moieties into an antibody combining site could lead to the development of tools for validating antibody–antigen structural models (Pimenova et al., 2008) and to reagents capable of binding analytes with extremely high affinities and specificities Kim and Yoon (2010).
As a proof-of principle demonstration, we chose a complex with a known structure, the anti-anthrax antibody M18 bound to anthrax protective antigen (PA) (Leysath et al., 2009). PA is a component of the tripartite toxin secreted by Bacillus anthracis which binds to cellular receptors, and assists host cellular targeting and transport of the lethal factor (LF) and edema factor (EF) into cytoplasm (Moayeri and Leppla, 2004; Young and Collier, 2007). M18 is a neutralizing antibody (Leysath et al., 2009) derived by directed evolution from monoclonal antibody 14B7 (Harvey et al., 2004; Little et al., 1988), which binds to the fourth domain of PA (PAD4), and effectively blocks PA binding to cellular receptors such as CMG2 to mitigate anthrax toxicity.
2. Methodology
2.1. Computational methods
2.1.1. Creation of models of l-DOPA antibody mutants in complex with antigen
Models for various mutants of the antibody–antigen complex were created using Rosetta (Leaver-Fay et al., 2011) with l-DOPA placed in various positions within the antibody paratope. Coordinates for the wild-type M18-PAD4 complex were downloaded from the Protein Data Bank (Berman et al., 2000) (PDB ID 3ETB). To remove crystal packing effects and obtain a Rosetta-minimized reference structure, fixed-backbone side-chain packing and minimization (1000 decoys) on the entire protein complex was performed using Rosetta's score12. The lowest-scoring structure was used for the calculations for the predictive introduction of l-DOPA into the CDRs of M18. The Rosetta 3.4 (revision 51671, available at www.rosettacommons.org) command line used to run the “fix_bb” protocol was:
fixbb.linuxgccrelease -s Crystal.pdb -nstruct 1000 -use_input_sc -minimize_sidechains -run:multiple_processes_writing_to_one_directory -packing:repack_only -ex1 -ex2aro
For each interface Lys on the antigen, neighboring antibody residues within 10 Å (Cβ–Cβ distance) were selected as potential mutation sites. Each antibody residue within the cutoff distance was substituted to l-DOPA individually, followed by fixed-backbone side-chain packing (20 decoys) of the nearby residues (<10 Å) to accommodate the local changes. For these and any further calculations where l-DOPA is present, Rosetta uses the molecular mechanics-based scoring function (mm_std) and associated NCAA rotamer library (Renfrew et al., 2012).
To carry out these calculations, the position of the l-DOPA mutation and the positions of the neighboring residues were specified in a “resfile”, and the same “fixbb” protocol read the “resfile” and substituted the target residue to l-DOPA, followed by side chain repacking including all the neighboring residues. A Rosetta 3.4 (revision 51671) command line example is:
fixbb.linuxgccrelease -s Best_Prepacked.pdb -nstruct 20 -use_input_sc -resfile 315_LYS_J_679–139_SER_H_31.resfile -score:weights mm_std -minimize_sidechains -ex1 –ex2
2.1.2. Model relaxation with crosslink constraint
Some measures are performed on a structure where the l-DOPA is artificially constrained to be proximal to the target lysine residue. For these calculations, we employed an empirically-determined linear constraint potential,
where dXL is the distance in Ångstroms between Cγ atom on the l-DOPA ring (the atom bound to the Cβ atom) and the Lys Nε and Econstr is the constraint energy in Rosetta Energy Units (REU). The constraint weights were chosen to bring the l-DOPA and Lys into proximity, in order to evaluate interface compatibility. This constraint energy was not included in the final calculated interface score. All the neighboring residues within 10 Å (Cβ–Cβ distance) of the l-DOPA/Lys pair were repacked to accommodate the constraint. The constrained conformation was generated using a custom PyRosetta script with PyRosetta 2.012, revision 51671 (PyRosetta available at www.rosettacommons.org, script available upon request).
2.1.3. Crosslinking distance
After selecting the l-DOPA position and the target Lys, all the distances of potential crosslinking atom pairs (lysine Nε atom and l-DOPA atoms C2, C5, and C6) were evaluated, and the one with the minimum value represented the crosslinking distance (dXL).
2.1.4. Solvent-accessibility measures
As a proxy for solvent accessibility, the number of atoms within 3 Å of the l-DOPA side-chain atoms was computed (Simons et al., 1999). All non-l-DOPA atoms were considered, both from the antibody and antigen, and including hydrogens (as placed by Rosetta). A second solvent accessibility measure is the solvent accessible surface area (SASA) (Le Grand and Merz, 1993), calculated using an embedded function in PyRosetta 2.012 using a probe radius of 1.4 Å.
2.1.5. Rosetta energy calculations
The interface score ISC is defined as
where Sbound and Sunbound represent the total scores of the antibody-antigen complex in bound and unbound form, respectively. The unbound form was scored after separating the antibody and antigen to a very large distance without additional side chain repacking. Scores are given in Rosetta Energy Units (REU), which approximate kcal/mol.
After repacking side chains under a constraint on the l-DOPA/Lys pair, the new pose of the M18/PAD4 complex was saved in a separate file. The constrained interface score, , was then obtained by rescoring the new conformations both as a complex and as unbound components without retaining the constraint potential,
where the superscript indicates that the constrained conformations are used.
2.2. Experimental methods
2.2.1. Vector construction
The pMB1 origin of replication in the dual aminoacyl tRNA synthetase/tRNA expression vector, pRST.11B (Hughes and Ellington, 2010), was replaced by the p15 origin of replication from vector pACYC184 by amplification of the p15 origin from pACYC184 using primers p15AA.1, and p15A0.2 (Supplementary Material). The pRST.11B vector was amplified with primers VSP.2 and VSP.3 to generate a ∼6kbp fragment that lacks the original origin of replication and is flanked by the unique SpeI and XmaI restriction endonuclease sites. The 1.2kbp PCR product containing the p15A orgin and the 6kbp vector fragment was digested with SpeI and XmaI and ligated together to yield vector pRST.11C. The Nap1 mutant Methanococcus jannaschii tyrosyl amber suppressor tRNA (Guo et al., 2009) was assembled via PCR from the four overlapping oligonucleotides Nap.1, Nap.2, Nap.3 and Nap.4 (Supplemental Material). The assembled tRNA gene was digested with KpnI and BsrGI restriction enzymes and ligated into a similarly digested pRST.11C vector to yield vector pRST.11C-Nap1. A redundant XbaI site in pRST.11C-NapI was removed by quick change mutagenesis using primers, Qcxbaprstc.1 and Qcxbaprstc.2. The gene for the evolved l-DOPA utilizing aminoacyl tRNA synthetase (Alfonta et al., 2003) was assembled from overlapping oligonucleotides in-house using automated protein fabrication gene assembly process (Cox et al., 2007). The assembled gene was digested with XbaI and XhoI and ligated into a similarly digested pRST.11C vector to yield the l-DOPA incorporating tRNA synthetase/tRNA vector, pDopa.
The pAK400-M18 scFv antibody and pAK400-pAD4 expression vectors (Leysath et al., 2009) were obtained from George Georgiou's group at the University of Texas at Austin. The pAK400 terminal codon (TAG) was changed to the ochre stop codon (TAA) using Gibson Assembly PCR. Amber (TAG) codons were introduced into the coding sequence of the M18 antibody via quick change mutagenesis or Gibson Assembly PCR.
2.2.2. Expression and purification of M18 variants with NCAAs
The M18 antibody variants were expressed using a condensed culture labeling method (Liu et al., 2010) in the presence (or absence) of supplemented l-DOPA. Briefly, the M18 antibody and variants were expressed by inoculating 900 mLs of 2xYT media containing 35 lg/mL chloramphenicol and 100 lg/mL ampicillin with 1 mL of a saturated overnight culture. Expression cultures were grown at 37 °C to OD600 ∼ 0.8. Cultures were centrifuged at 3500g for 10 min, and resuspended in 100 mL 2xYT containing 5 mM DTT, 1.5 mM l-DOPA, and 1.5 mM IPTG. Condensed cultures were grown at 26 °C for 12 h. The PAD4 antigen was grown in Terrific Broth, induced at OD600 = 1.0 with 1 mM IPTG, and allowed to grow at 30 °C for 12 h.
Expression cultures were centrifuged at 3500g for 15 min, and resuspended in PBS with 1 mg/mL lysozyme and 0.25U/mL benzonase, and incubated on ice for 30 min. Cells were then sonicated for 4 min to further lyse the cells. Lysates were centrifuged at 35,000 g for 45 min, after which the liquid fraction was poured over a 1.5 mL Ni–NTA agarose column. The resin was washed with 45 mL wash buffer (60 mM imidazole, 200 mM NaCl, 50 mM Phosphate at pH 8), and the protein was eluted with elution buffer (400 mM imidazole, 200 mM NaCl, 50 mM phosphate at pH 8). The Proteins were concentrated using an Amiconcellulose based centrifugal concentrators. Concentrations of the proteins were determined using Abs280 measurements.
2.2.3. Crosslinking assay
81 pmol (2.7 μM) PAD4 and 81 pmol (2.7 lM) of the M18 scFv variant were mixed in assay buffer (200 mM NaCl, 50 mM Phosphate at pH 8.5). Crosslinking was accomplished by adding sodium periodate to 3.3 mM. Samples were incubated at room temperature for 30 min. Periodate was quenched with addition of DTT to 100 mM and 5× SDS loading dye. Samples were denatured by heating to 98 °C for ten minutes.
Samples were then run on Novex 4–12% Bis-tris SDS gels using MES-SDS running buffer at 200 V for 35 min. Proteins were transferred to nitrocellulose at 25 V for 1 h, using Invitrogen XCell II Blot Module. Nitrocellulose was blocked at room temperature for 30 min using Superblock in PBS (Thermo) (for α-His antibody) or Superblock in TBS (Thermo) (for α-Flag antibody). Nitrocellulose was rinsed three times with PBS or TBS. Nitrocellulose was immersed in 50 mL of PBS or TBS with 10 μL of α-His or α-Flag for 1 h at room temperature. Nitrocellulose was rinsed with PBS or TBS three times, and resolved with Promega Western Blue Stabilized AP substrate for α-His-AP conjugate, or with Promega ECL western blotting substrate for luminescent detection using the α-Flag-HRP conjugate.
M18 variants were resolved at ∼30 kDa, PAD4 resolved at 14 kDa. Crosslinked product appeared at ∼45 kDa. Luminescent western bands were quantified using ImageJ.
3. Results
We wished to test whether we could use computational design for the precise placement of crosslinks between antibodies and antigens. Our general plan was to use simple criteria and the Rosetta suite of software (Kaufmann et al., 2010; Leaver-Fay et al., 2011) to identify where we might place the NCAA l-DOPA, which upon activation to form the ortho-quinone in the presence of periodate (Burdine et al., 2004; Liu et al., 2006) can crosslink to a variety of residues, in particular the histidine imidazole cysteine thiol, and the ∈-amine of lysine. Thus, we first used Rosetta to build structural models of the antibody–antigen complex with candidate l-DOPA mutations, using energy parameters and rotamer libraries recently developed for NCAAs (Renfrew et al., 2012). For each l-DOPA position on the antibody, we evaluated various biophysical measurements that might affect crosslinking to an adjacent histidine or lysine on the bound antigen. The incorporation of l-DOPA into proteins was accomplished using an evolved tRNA synthetase from Methanacoccus jannaschii that when expressed in E. coli can specifically charge a suppressor tRNA with l-DOPA (Alfonta et al., 2003). In the following sections we detail each of these steps.
4. Biophysical criteria for rational design
4.1. Initial choice of residues for l-DOPA substitution
The X-ray structure of the M18-PAD4 complex, solved at 3.8 Å resolution, reveals a binding interface of ∼1700 Å2, with roughly equal contributions from the M18 light and heavy chains. All the antibody CDR loops except L3 contact the antigen, and CDR loops H3 and L2 appear to have the strongest interaction with PAD4 (Leysath et al., 2009). His and Lys residues are the only nucleophiles present on the antigen surface, however only Lys residues are near the epitope (Fig. 1). To identify candidate positions, we selected all pairs of antigen nucleophilic residues and antibody residues that have heavy atoms within 10 Å. Thus we considered three nucleophilic lysines (PAD4 residues 684, 679 and 653) as crosslinking targets for sixteen possible l-DOPA locations on the antibody (thirteen on the heavy chain and three on the light chain).
Fig.1.
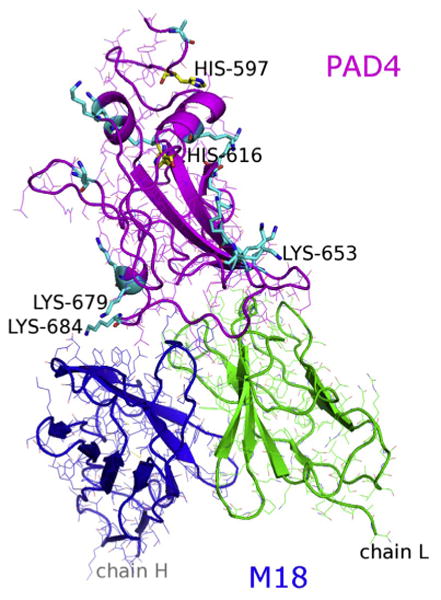
The M18/PAD4 antibody–antigen complex structure from PDB 3ETB highlighting the antigen's candidate crosslinking residues, Lys (cyan sticks) and His (yellow sticks).
Several biophysical parameters can be suggested as criteria for the evaluation and prediction of sites for l-DOPA insertion. First, the distance between the l-DOPA mutant and the reactive antigen nucleophilic residue should be close enough for them to react. Second, l-DOPA should be introduced to the antibody position where it is exposed to the solvent, so that periodate anions can access the reaction pair and trigger the reaction. Third, the original binding of M18/PAD4 should not be affected by the introduction of l-DOPA, meaning the interface must be compatible with the l-DOPA insertion, or the change of interface energy should be moderate. Each of these criteria will be examined in turn as the basis for design.
4.2. Crosslinking distance between l-DOPA and nucleophile
The most obvious structural criterion for l-DOPA crosslinking is that the residue must be close enough to a nucleophile to crosslink. To assess this criterion, we took those l-DOPA atoms that were already within 10 Å of a lysine, as measured from Cβ to Cβ, and further calculated the distance between C2, C5, and C6 on the ring and the lysine Nε atom. In preliminary calculations, Rosetta tended to extend both l-DOPA and lysine residues into solution, and thus the calculated distances would exclude the possibility of crosslinking. To bring the reactive atoms closer, we optimized side-chain conformations using an energy function augmented with a constraint potential (see Methods). The crosslinking distance thus obtained, dXL, varied from 2.9 to 9.8 Å (Table 1). For comparison, we also analyzed one l-DOPA substitution at a position far from the interface (L_D17, dXL = 32.1 Å).
Table 1.
Candidate crosslink residue pairs, calculated biophysical properties, and experimental crosslinking of the M18/PAD4 interface with an l-DOPA mutation. Antigen residue nucleophile residues paired with antibody positions for placement of l-DOPA. The calculated values of factors (accessible distance, number of neighbors, interface score) after l-DOPA mutation that can affect l-DOPA-Lys crosslinking. Mutants are sorted by accessible distance. Grey shading indicates poor conditions that may reduce or prevent crosslinking (dXL > 7 Å, over ten neighbors, SASA < 90 Å2, Isc > 2.5 REU, or > 10REU). For the two cases where antibody positions have multiple candidate crosslink targets, only one is shown in the table (Position H_G53's second target is J_K684 at dXL = 4.1 Å with Isc = 1.0 REU and 11.5REU, and position H_S31's second target is J_K684 at dXL = 8.6 Å with Isc = 0.7 REU and 4.7REU.).
| Antigen Residue | Antibody Position | Crosslinking Distance, dxL(Å) | Number of Neighbors | SASA (Å2) | Interface Score, Isc (REU) | Interface Score (REU) | Experimental Crosslinking (%) |
|---|---|---|---|---|---|---|---|
| J_K684 | H_D54 | 2.9 | 4 | 124.9 | 0.8 | 52.0 | 30.4 (15.8) |
| J_K684 | H_D56 | 3.0 | 2 | 120.3 | 1.1 | 28.9 | 36.2 (5.0) |
| J_K684 | H_Y52 | 3.0 | 14 | 25.1 | 1.3 | 15.6 | 19.7 (4.4) |
| J_K684 | H_W33 | 3.4 | 9 | 14.1 | 4.8 | 8.4 | 13.9 (8.4) |
| J_K684 | H_N58 | 5.5 | 6 | 111.3 | 3.7 | 6.1 | 24.4 (3.3) |
| J_K679 | H_S31 | 5.7 | 1 | 132.3 | 0.7 | -0.6 | 51.9(11.5) |
| J_K679 | H_G53 | 6.1 | 8 | 124.8 | 1.0 | 0.2 | 40.3 (13.8) |
| J_K684 | H_R99 | 6.6 | 8 | 28.2 | 0.2 | 151.1 | Not tested |
| J_K684 | H_G55 | 8.3 | 1 | 161.6 | 2.8 | 1.8 | 5.1 (1.3) |
| J_K653 | L_N31 | 8.6 | 9 | 75.3 | 1.2 | -1.1 | Not tested |
| J_K684 | H_S57 | 8.8 | 2 | 122.7 | 2.7 | 12.7 | Not tested |
| J_K684 | H_S32 | 8.9 | 18 | 42.9 | 1.3 | 12.2 | Not tested |
| J_K653 | L_Y50 | 9.0 | 11 | 10.5 | 2.8 | 0.1 | Not tested |
| J_K684 | H_G96 | 9.3 | 11 | 56.8 | -1.4 | 11.5 | Not tested |
| J_K684 | H_I51 | 9.8 | 22 | 0.5 | 2.8 | 13.4 | 2.7 (0.14) |
| J_K653 | L_D17 | 32.1 | 4 | 139.9 | 2.7 | 0.2 | 1.5 (0.49) |
4.3. Solvent accessibility of l-DOPA
A second hypothesis is that a good crosslinking site needs access to the solvent so that the l-DOPA can be easily oxidized by periodate anions. In the presence of periodate, the hydroxyl groups on the phenolic ring of l-DOPA on M18 are oxidized to form the quinone intermediate, which activates the ring for potential nucleophilic attack by a neighboring lysine nucleophile on PAD4. We chose two methods to quantify the degree of solvent exposure. First, a simple metric was to count of the number of neighboring antibody and antigen atoms within 3 Å of the l-DOPA side chain. Mutants predicted to have fewer neighboring atoms were expected to have better chances for effective crosslinking. Second, the solvent-accessible surface area was calculated using a 1.4 Å probe. Both calculations were performed on the structure of antibody– antigen complex in which the l-DOPA and neighboring residue side chains were optimized by packing based on a rotamer library and the standard Rosetta “fixbb” protocol (see Computational Methods) to find an energetically favorable conformation. We ultimately sought to test whether there was a minimal solvent accessibility or whether crosslinking would improve as solvent accessibility increased (Table 1).
4.4. Mutant interface compatibility
While it is difficult to computationally determine the impact of l-DOPA substitution on antigen binding, we tested whether Rosetta's estimated energetics of the substituted interface would be helpful. Two energy measures were tested. First, we hypothesized that a successful crosslink would require that the introduced l-DOPA would not disrupt the favorable energy of interaction between the antigen and antibody. We therefore calculated the interface score, Isc, a measure used in docking calculations to approximate a binding energy (see Section 2) (Chaudhury and Gray, 2008; Chaudhury et al., 2011). Data from docking 116 complexes (Chaudhury et al., 2011) indicate that most protein–protein complexes have negative interface scores in the range of −4 to −12 REU, and comparisons to experimental alanine mutants suggest that changes in interface scores above +1 REU are likely to characterize mutations that will significantly reduce binding affinity (Kortemme and Baker, 2002). One complication is that NCAAs necessitate use of a Rosetta force field variation. In the case of the M18-PAD4 complex, the predicted interface score was -21.6 with the Rosetta's (standard) score12 and +2.7 with the NCAA score function, mm_std. The mm_std interface scores of the l-DOPA substituted variants ranged from -1.4 to 4.8 REU, with most interface scores being slightly positive, indicating that the l-DOPA mutated M18-PAD4 complex may be slightly disfavored.
A second energetic hypothesis is that an activated conformation with proximity between l-DOPA and the target lysine should be energetically accessible. Thus, we evaluated the interface score for the structure we generated with the crosslinking constraint that brought the reacting atoms together ; see Methods). Table 1 shows that these values vary from −0.6 to over 150 kcal/mol, showing that some cases exhibit significant energetic perturbations (e.g. H_D56) and even atomic clashes (H_R99 or H_D54) when the l-DOPA/Lys pair is constrained.
4.5. Crosslinking studies
We chose to test candidate mutations based on their predicted placement within the protein–protein interface. We anticipated that the distance between the l-DOPA and a lysine nucleophile was likely to be the most critical parameter for successful cross-linking. Therefore, we made the closest eight substitutions within M18 (Table 1) (crosslinking distance of between 2.9 and 8.3 Å), excluding only H_R99 because the high constrained interface energy suggested that it would bind antigen poorly. As controls for the crosslinking distance, two additional mutants were made by placing l-DOPA at two locations that were predicted to be unlikely to crosslink: H_I51 (9.8 Å, solvent excluded) and L_D17 (32.1 Å, solvent exposed).
The recombinant M18 antibodies were expressed in the periplasm of E. coli as a 28.9 kDa single-chain variable fragment (scFv) with a N-terminal FLAG epitope tag and a C-terminal hexa-His tag to enable purification via immobilized metal affinity chromatography (IMAC). This tag also allows ready detection of the antibody via Western blot analysis. Similarly, the PAD4 antigen was expressed as a 17.5 kDa protein with a C-terminal His tag to enable purification of the antigen via IMAC. l-DOPA was introduced site-specifically into the chosen sites in the CDRs of M18 by changing the codon for the naturally occurring amino acid at these positions to an Amber (TAG) stop codon. The M18 amber variants were then co-expressed with an orthogonal tRNA suppressor and tRNA synthetase pair that had previously been evolved to be specific for l-DOPA (Alfonta et al., 2003). Each M18 variant was incubated with PAD4, and crosslinking was catalyzed by the addition of sodium periodate. Crosslinking between predicted l-DOPA-containing M18 variants and PAD4 was probed and quantitated using a Western blot assay to the FLAG epitope tag (Fig. 2). A covalent crosslink between antibody and antigen was apparent by a shift in the molecular mass of the antibody. The efficiency of crosslinking was determined by the percentage of antibody that underwent a mass shift. Each variant was tested a minimum of two times. The distal crosslinking controls (H_I51 and L_D17) showed no appreciable crosslinking (<3%), and eight of the substitutions exhibited crosslinking values ranging from 5% to 52% (Table 1).
Fig.2.

A western blot of M18 and variants treated with periodate in the absence and presence of antigen PAD4. α-Flag-HRP was used to detect a Flag-tag on the M18 variants. The shift in mass in the presence of PAD4 corresponds to the mass of PAD4 antigen (∼14 kDa), signaling covalent crosslinking between antibody and antigen. Bands were quantified using ImageJ to determine crosslinking efficiencies, which were listed in Table 1. D17L is the only tested position on M18 light chain. The rest of variants were all on heavy chain.
4.6. Correlations between Rosetta criteria and crosslinking
We can inspect the biophysical criteria and determine whether and how they impact crosslinking efficiency. Fig. 3 shows the plots of each of the five biophysical criteria described above versus the experimentally observed crosslinking extent. Crosslinking efficiency dropped off appreciably (<3%) at distances greater than 7 Å [e.g. variants H_G55 (8.3 Å), H_I51 (9.8 Å), and L_D17 (32.1 Å)] relative to l-DOPA substitutions that were predicted to lie closer to lysines (Fig. 3a). There was no correlation between solvent accessibility measures and crosslinking efficiency when all data are considered (Fig. 3b and c). However, when the points representing mutants with long-distance crosslinks are excluded (black in Fig. 3b and c), both the number of residues that surround an l-DOPA substitution and the solvent accessible surface area (SASA) show relationships to crosslinking efficiency (Fig. 3c). In particular, variants H_Y52 and H_W33, with SASA of only 25.1 and 14.1 Å, respectively, have the lowest crosslinking efficiencies (19.7% and 13.9%, respectively) amongst variants with dXL under 7 Å.
Fig.3.
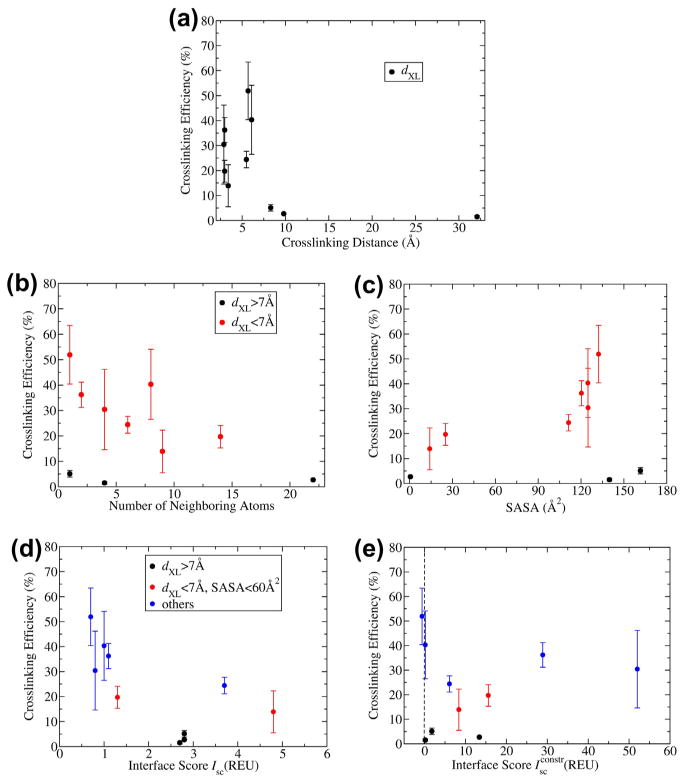
Plots relating the biophysical criteria to experimental crosslinking efficiency. Error bars display the standard deviations obtained from multiple trials in experiments. (a) Effect of crosslinking distance. (b and c) Effect of solvent accessibility by (b) l-DOPA neighboring atoms or (c) solvent-accessible surface area. Black circles represent positions with dXL over 7 Å (H_G55, H_I51, and L_D17). (d and e). Effect of interface energetics by (d) interface energy or (e) crosslink fluctuation energy. Again black circles represent positions with dXL over 7 Å, and additionally red circles represent positions with solvent-accessible surface area under 60 Å (H_Y52 and H_W33).
The energetic parameters do not provide clear trends for the analysis (or prediction) of crosslinking efficiency even after filtering for distance and solvent accessibility (Fig. 3d and e). However, it is notable that the two most efficient crosslinking sites (H_S31 and H_G53) have low energies (Isc and ) of 1 REU or less in both cases). At best, we can say that by also choosing positions that pass the energy filter, we may identify variants that exhibit efficient crosslinking. Overall, these plots emphasize the key importance of distance, followed by solvent accessibility, in identifying positions that may be capable of significant crosslinking.
4.7. Interface structure of a successful crosslinking case
Fig. 4 shows a detail of the structural model of the best cross-linking case with the l-DOPA mutant at position H_S31 targeting the Lys at position 679. This mutant exhibits 52% crosslinking (standard deviation 12%). The crosslinking distance is a moderate 5.7 Å, and the SASA of 132 Å2 shows good solvent accessibility. The calculated interface energies of Isc = 0.7 REU and REU suggest that both the unconstrained and constrained conformations are energetically favorable in the context of the bound antibody–antigen complex. As seen in the figure, the aromatic carbon atoms C2, C5, and C6 are all positioned for nucleophilic attack by the amine.
Fig.4.
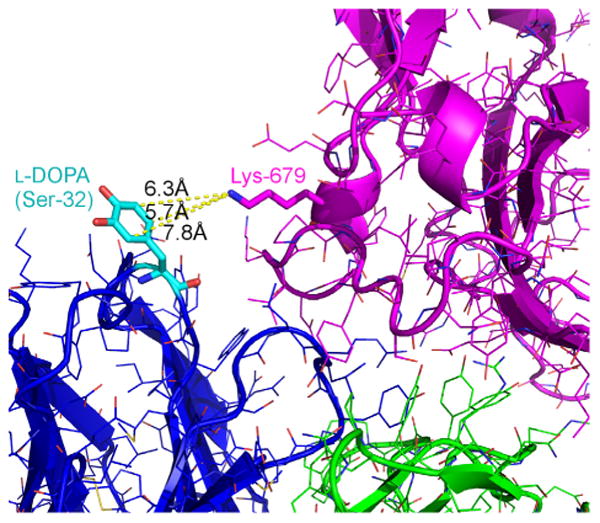
Structural detail of the successfully crosslinking l-DOPA mutant at position H_S31 with its targeted Lys at position 679. Crosslinking distance of 5.7,132 Å2 of solvent-accessible surface area, and calculated interface energies of Isc = 0.7 REU and . Aromatic carbon atoms C2, C5, and C6 are all positioned for nucleophilic attack by the amine. The crosslinking distances for these three sites are 7.8, 6.3, and 5.7 Å, respectively.
5. Discussion
The insertion of new, functional amino acids into proteins, even proteins of known structure, remains an enterprise that is fraught with uncertainty. While there have been several successful demonstrations of the rational placement of crosslinking amino acids into proteins (Alfonta et al., 2003; Forne et al., 2012; Sato et al., 2011; Umeda et al., 2009, 2010), to date there has not been a thorough analysis of how a detailed structural analysis might be used to guide the placement of amino acids that would crosslink efficiently. Currently, placement of NCAAs into proteins is based on intuition and no computational tools exist to guide the design of crosslinks. As a first step towards the development of tools for engineering crosslinks, we sought to develop a rational, rules-based approach for predicting the placement of a crosslinkng NCAA within a protein–protein complex. The placement of l-DOPA was correlated with its ability to crosslink, and the biophysical parameters that most impacted successful crosslinking were compared. Based on computational predictions, a number of high efficiency crosslinking sites were recovered. The best engineered antibody could crosslink with very high efficiency (52%) with little to no observable side reactions. Additional optimizations of crosslinking by varying buffer conditions and periodate concentrations may lead to further enhancements in crosslinking efficiency.
Our results are especially significant because typical crosslinking reactions for rationally placed variants have been reported to suffer from a lack of specificity, low yield, and uncharacterized side reactions (Fancy and Kodadek, 1999; Fancy et al., 1996; Sinz, 2006). Additionally, several commercially available ‘random’ crosslinkers have been reported to be inefficient and barely detectable by MS analysis (Dihazi and Sinz, 2003). In our study, we found that adjacency is an excellent primary criterion for success, and additionally, that greater solvent accessibility increases crosslinking efficiency for proximal residues. Although energetic calculations were noisy, the best two positions for crosslinking l-DOPA moieties showed low interface energies, both when free and when constrained to be near the nucleophile.
Direct comparison of the crosslinking efficiencies obtained herein to previously published results proved difficult because crosslinking efficiencies have generally not been evaluated quantitatively, but rather just examined qualitatively by gel mobility shifts (Burdine et al., 2004; Umeda et al., 2009, 2010). Our results therefore likely represent the first quantitative study of l-DOPA crosslinking.
That said, the utility of the computational methods employed can perhaps be discerned by comparing them with intuition. In the case of antibody M18 and PAD, positions H_D54, H_D56, or H_Y52 on the antibody would likely have been chosen as the best locations for l-DOPA, since they lie close to a lysine in the complex. While l-DOPA insertions at all three of these positions did show crosslinking (19.7–36.2%), the most efficient crosslinking (40.3% and 51.9%) instead occurred at the computer-chosen positions H_S31 and H_R99 that scored well by all three biophysical criteria: distance, solvent accessibility, and energetics.
As this study relied on a single antibody–antigen target with a known crystal structure of the complex, the conclusions should be generalized with caution. Nonetheless, a strategy for identifying high-probability antibody–antigen crosslinking positions might consist of a three-part filter that requires: (i) crosslinking distance under 7 Å; (ii) sufficient solvent-accessibility (SASA over 90 Å2 or fewer than ten neighboring atoms); and (iii) compatible interface energies (Isc under 2.5 REU and under 10 REU). Application of these filters in this case led to the quick and accurate prediction of a small set of substitutions from which generally excellent cross-linking antibodies were derived. This differs from previous attempts where iterations of insertion and crosslinking conditions had to be considered in order to identify functional crosslinks. Further studies will be needed to test the utility of these approaches with antibody homology models (e.g., (Marcatili et al., 2008; Pantazes and Maranas, 2010; Sivasubramanian et al., 2009)) or docked structures (e.g., (Sircar and Gray, 2010)).
Supplementary Material
Acknowledgments
This project was supported by the DARPA Antibody Technology Program (HR-0011-10-1-0052). The authors thank Dr. P. Douglas Renfrew for his help implementing non-canonical amino acids in Rosetta, and we thank George Georgiou, Brian Kuhlman, and Bryan Der for helpful discussions.
Footnotes
Appendix A. Supplementary data: Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jsb.2013.05.003.
Contributor Information
Andrew D. Ellington, Email: ellingtonlab@gmail.com.
Jeffrey J. Gray, Email: jgray@jhu.edu.
References
- Ai HW, Shen W, Sagi A, Chen PR, Schultz PG. Probing protein-protein interactions with a genetically encoded photo-crosslinking amino acid. Chembiochem. 2011;12:1854–1857. doi: 10.1002/cbic.201100194. [DOI] [PubMed] [Google Scholar]
- Alfonta L, Zhang Z, Uryu S, Loo JA, Schultz PG. Site-Specific Incorporation of a Redox-Active Amino Acid into Proteins. J Am Chem Soc. 2003;125:14662–14663. doi: 10.1021/ja038242x. [DOI] [PubMed] [Google Scholar]
- Ayyadurai N, Prabhu NS, Deepankumar K, Jang YJ, Chitrapriya N, Song E, Lee N, Kim SK, Kim BG, Soundrarajan N, Lee S, Cha HJ, Budisa N, Yun H. Bioconjugation of L-3,4-dihydroxyphenylalanine containing protein with a polysaccharide. Bioconjug Chem. 2011;22:551–555. doi: 10.1021/bc2000066. [DOI] [PubMed] [Google Scholar]
- Barderas R, Desmet J, Timmerman P, Meloen R, Casal JI. Affinity maturation of antibodies assisted by in silico modeling. Proc Natl Acad Sci USA. 2008;105:9029–9034. doi: 10.1073/pnas.0801221105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdine L, Gillette TG, Lin HJ, Kodadek T. Periodate-triggered cross-linking of DOPA-containing peptide-protein Complexes. J Am Chem Soc. 2004;126:11442–11443. doi: 10.1021/ja045982c. [DOI] [PubMed] [Google Scholar]
- Caravella AJ, Wang DM, Glaser S, Lugovskoy A. Structure-Guided Design of Antibodies. Curr Comput Aided Drug Des. 2010;6:128–138. [PubMed] [Google Scholar]
- Chaudhury S, Gray JJ. Conformer selection and induced fit in flexible backbone protein-protein docking using computational and NMR ensembles. J Mol Biol. 2008;381:1068–1087. doi: 10.1016/j.jmb.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury S, Berrondo M, Weitzner BD, Muthu P, Bergman H, Gray JJ. Benchmarking and Analysis of Protein Docking Performance in Rosetta v3.2. Plos One. 2011;6:e22477. doi: 10.1371/journal.pone.0022477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chennamsetty N, Voynov V, Kayser V, Helk B, Trout BL. Design of therapeutic proteins with enhanced stability. Proc Natl Acad Sci USA. 2009;106:11937–11942. doi: 10.1073/pnas.0904191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin JW, Martin AB, King DS, Wang L, Schultz PG. Addition of a photocrosslinking amino acid to the genetic code of Escherichiacoli. Proc Natl Acad Sci USA. 2002a;99:11020–11024. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin JW, Santoro SW, Martin AB, King DS, Wang L, Schultz PG. Addition of p-azido-l-phenylalanine to the genetic code of Escherichia coli. J Am Chem Soc. 2002b;124:9026–9027. doi: 10.1021/ja027007w. [DOI] [PubMed] [Google Scholar]
- Cho H, Daniel T, Buechler YJ, Litzinger DC, Maio Z, Putnam AMH, Kraynov VS, Sim BC, Bussell S, Javahishvili T, Kaphle S, Viramontes G, Ong M, Chu S, GC B, Lieu R, Knudsen N, Castiglioni P, Norman TC, Axelrod DW, Hoffman AR, Schultz PG, DiMarchi RD, Kimmel BE. Optimized clinical performance of growth hormone with an expanded genetic code. Proc Natl Acad Sci USA. 2011 doi: 10.1073/pnas.1100387108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark LA, Boriack-Sjodin PA, Eldredge J, Fitch C, Friedman B, Hanf KJM, Jarpe M, Liparoto SF, Li Y, Lugovskoy A, Miller S, Rushe M, Sherman W, Simon K, Van Vlijmen H. Affinity enhancement of an in vivo matured therapeutic antibody using structure-based computational design. Protein Sci. 2006;15:949–960. doi: 10.1110/ps.052030506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JC, Lape J, Sayed MA, Hellinga HW. Protein fabrication automation. Protein Sci. 2007;16:379–390. doi: 10.1110/ps.062591607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dihazi GH, Sinz A. Mapping low-resolution three-dimensional protein structures using chemical cross-linking and Fourier transform ion-cyclotron resonance mass spectrometry. Rapid Commun Mass Spectrom. 2003;17:2005–2014. doi: 10.1002/rcm.1144. [DOI] [PubMed] [Google Scholar]
- Fancy DA, Kodadek T. Chemistry for the analysis of protein-protein interactions: rapid and efficient cross-linking triggered by long wavelength light. Proc Natl Acad Sci U S A. 1999;96:6020–6024. doi: 10.1073/pnas.96.11.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fancy DA, Melcher K, Johnston SA, Kodadek T. New chemistry for the study of multiprotein complexes: the six-histidine tag as a receptor for a protein crosslinking reagent. Chem Biol. 1996;3:551–559. doi: 10.1016/s1074-5521(96)90146-5. [DOI] [PubMed] [Google Scholar]
- Farady CJ, Sellers BD, Jacobson MP, Craik CS. Improving the species cross-reactivity of an antibody using computational design. Bioorg Med Chem Lett. 2009;19:3744–3747. doi: 10.1016/j.bmcl.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forne I, Ludwigsen J, Imhof A, Becker PB, Mueller-Planitz F. Probing the conformation of the ISWI ATPase domain with genetically encoded photoreactive crosslinkers and mass spectrometry. Mol Cell Proteomics. 2012;11:M111 012088. doi: 10.1074/mcp.M111.012088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Melancon CE, 3rd, Lee HS, Groff D, Schultz PG. Evolution of amber suppressor tRNAs for efficient bacterial production of proteins containing nonnatural amino acids. Angew Chem Int Ed Engl. 2009;48:9148–9151. doi: 10.1002/anie.200904035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey BR, Georgiou G, Hayhurst A, Jeong KJ, Iverson BL, Rogers GK. Anchored periplasmic expression, a versatile technology for the isolation of high-affinity antibodies from Escherichia coli-expressed libraries. Proc Natl Acad Sci USA. 2004;101:9193–9198. doi: 10.1073/pnas.0400187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes RA, Ellington AD. Rational design of an orthogonal tryptophanyl nonsense suppressor tRNA. Nucl Acids Res. 2010;38:6813–6830. doi: 10.1093/nar/gkq521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DH, Cellitti SE, Hao X, Zhang Q, Jahnz M, Summerer D, Schultz PG, Uno T, Geierstanger BH. Site-specific labeling of proteins with NMR-active unnatural amino acids. J Biomol NMR. 2010;46:89–100. doi: 10.1007/s10858-009-9365-4. [DOI] [PubMed] [Google Scholar]
- Kaufmann KW, Lemmon GH, DeLuca SL, Sheehan JH, Meiler J. Practically useful: what the Rosetta protein modeling suite can do for you. Biochemistry. 2010;49:2987–2998. doi: 10.1021/bi902153g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Yoon MY. Recent advances in rapid and ultrasensitive biosensors for infectious agents: lesson from Bacillus anthracis diagnostic sensors. Analyst. 2010;135:1182–1190. doi: 10.1039/c0an00030b. [DOI] [PubMed] [Google Scholar]
- Kortemme T, Baker D. A simple physical model for binding energy hot spots in protein-protein complexes. Proc Natl Acad Sci USA. 2002;99:14116–14121. doi: 10.1073/pnas.202485799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda D, Shirai H, Jacobson MP, Nakamura H. Computer-aided antibody design. Protein Eng Des Sel. 2012;25:507–522. doi: 10.1093/protein/gzs024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Grand SM, Merz KM. Rapid approximation to molecular surface area via the use of Boolean logic and look-up tables. J Comp Chem. 1993;14:349–352. [Google Scholar]
- Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman KW, Renfrew PD, Smith CA, Sheffler W, Davis IW, Cooper S, Treuille A, Mandell DJ, Richter F, Ban YEA, Fleishman SJ, Corn JE, Kim DE, Lyskov S, Berrondo M, Mentzer S, Popović Z, Havranek JJ, Karanicolas J, Das R, Meiler J, Kortemme T, Gray JJ, Kuhlman B, Baker D, Bradley P. Chapter nineteen - Rosetta3: An Object-Oriented Software Suite for the Simulation and Design of Macromolecules. In: Michael LJ, Ludwig B, editors. Methods Enzymol. Academic Press; 2011. pp. 545–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leysath CE, Monzingo AF, Maynard JA, Barnett J, Georgiou G, Iverson BL, Robertus JD. Crystal structure of the engineered neutralizing antibody M18 complexed to domain 4 of the anthrax protective antigen. J Mol Bio. 2009;387:680–693. doi: 10.1016/j.jmb.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippow SM, Tidor B. Progress in computational protein design. Curr Opin Biotechnol. 2007;18:305–311. doi: 10.1016/j.copbio.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippow SM, Wittrup KD, Tidor B. Computational design of antibody-affinity improvement beyond in vivo maturation. Nat Biotech. 2007;25:1171–1176. doi: 10.1038/nbt1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little SF, Leppla SH, Cora E. Production and characterization of monoclonal antibodies to the protective antigen component of Bacillus anthracis toxin. Infec Immun. 1988;56:1807–1813. doi: 10.1128/iai.56.7.1807-1813.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Burdine L, Kodadek T. Chemistry of periodate-mediated cross-linking of 3,4-Dihydroxylphenylalanine-containing molecules to proteins. J Am Chem Soc. 2006;128:15228–15235. doi: 10.1021/ja065794h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CC, Choe H, Farzan M, Smider VV, Schultz PG. Mutagenesis and evolution of sulfated antibodies using an expanded genetic code. Biochemistry. 2009;48:8891–8898. doi: 10.1021/bi9011429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Castaneda CA, Wilkins BJ, Fushman D, Cropp TA. Condensed E. coli cultures for highly efficient production of proteins containing unnatural amino acids. Bioorg Med Chem Lett. 2010;20:5613–5616. doi: 10.1016/j.bmcl.2010.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell DJ, Kortemme T. Computer-aided design of functional protein interactions. Nat Chem Biol. 2009;5:797–807. doi: 10.1038/nchembio.251. [DOI] [PubMed] [Google Scholar]
- Marcatili P, Rosi A, Tramontano A. PIGS: automatic prediction of antibody structures. Bioinformatics. 2008;24:1953–1954. doi: 10.1093/bioinformatics/btn341. [DOI] [PubMed] [Google Scholar]
- Midelfort KS, Hernandez HH, Lippow SM, Tidor B, Drennan CL, Wittrup KD. Substantial energetic improvement with minimal structural perturbation in a high affinity mutant antibody. J Mol Biol. 2004;343:685–701. doi: 10.1016/j.jmb.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Miklos AE, Kluwe C, Der BS, Pai S, Sircar A, Hughes RA, Berrondo M, Xu J, Codrea V, Buckley PE, Calm AM, Welsh HS, Warner CR, Zacharko MA, Carney JP, Gray JJ, Georgiou G, Kuhlman B, Ellington AD. Structure-based design of supercharged, highly thermoresistant antibodies. Chem Biol. 2012;19:449–455. doi: 10.1016/j.chembiol.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moayeri M, Leppla SH. The roles of anthrax toxin in pathogenesis. Curr Opin Microbiol. 2004;7:19–24. doi: 10.1016/j.mib.2003.12.001. [DOI] [PubMed] [Google Scholar]
- Pantazes RJ, Maranas CD. OptCDR: a general computational method for the design of antibody complementarity determining regions for targeted epitope binding. Protein Eng Des Sel. 2010;23:849–858. doi: 10.1093/protein/gzq061. [DOI] [PubMed] [Google Scholar]
- Pimenova T, Nazabal A, Roschitzki B, Seebacher J, Rinner O, Zenobi R. Epitope mapping on bovine prion protein using chemical cross-linking and mass spectrometry. J Mass Spectrom. 2008;43:185–195. doi: 10.1002/jms.1280. [DOI] [PubMed] [Google Scholar]
- Renfrew PD, Choi EJ, Bonneau R, Kuhlman B. Incorporation of noncanonical amino acids into Rosetta and use in computational protein– peptide interface design. PLoS One. 2012;7:e32637. doi: 10.1371/journal.pone.0032637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Mimasu S, Sato A, Hino N, Sakamoto K, Umehara T, Yokoyama S. Crystallographic study of a site-specifically cross-linked protein complex with a genetically incorporated photoreactive amino acid. Biochemistry. 2011;50:250–257. doi: 10.1021/bi1016183. [DOI] [PubMed] [Google Scholar]
- Simons KT, Ruczinski I, Kooperberg C, Fox BA, Bystroff C, Baker D. Improved recognition of native-like protein structures using a combination of sequence-dependent and sequence-independent features of proteins. Proteins struct Funct Genet. 1999;34:82–95. doi: 10.1002/(sici)1097-0134(19990101)34:1<82::aid-prot7>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Sinz A. Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass Spectrom Rev. 2006;25:663–682. doi: 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- Sircar A, Gray JJ. SnugDock: paratope structural optimization during antibody–antigen docking compensates for errors in antibody homology models. Plos Comput Biol. 2010;6:e1000644. doi: 10.1371/journal.pcbi.1000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasubramanian A, Sircar A, Chaudhury S, Gray JJ. Toward high-resolution homology modeling of antibody Fv regions and application to antibody–antigen docking. Proteins: Structure Function, and Bioinformatics. 2009;74:497–514. doi: 10.1002/prot.22309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao ML, Summerer D, Ryu Y, Schultz PG. The genetic incorporation of a distance probe into proteins in Escherichia coli. J Am Chem Soc. 2006;128:4572–4573. doi: 10.1021/ja058262u. [DOI] [PubMed] [Google Scholar]
- Umeda A, Thibodeaux GN, Zhu J, Lee Y, Zhang ZJ. Site-specific protein cross-linking with genetically incorporated 3,4-dihydroxy-L-phenylalanine. Chembiochem. 2009;10:1302–1304. doi: 10.1002/cbic.200900127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda A, Thibodeaux GN, Moncivais K, Jiang F, Zhang ZJ. A versatile approach to transform low-affinity peptides into protein probes with cotranslationally expressed chemical cross-linker. Anal Biochem. 2010;405:82–88. doi: 10.1016/j.ab.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xie J, Schultz PG. Expanding the genetic code. Ann Rev Biophys Biomol Struct. 2006;35:225–249. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- Xie J, Schultz PG. A chemical toolkit for proteins - an expanded genetic code. Nat Rev Mol Cell Biol. 2006;7:775–782. doi: 10.1038/nrm2005. [DOI] [PubMed] [Google Scholar]
- Young JAT, Collier RJ. Anthrax Toxin: Receptor Binding, Internalization, Pore Formation, and Translocation. Ann Rev Biochem. 2007;76:243–265. doi: 10.1146/annurev.biochem.75.103004.142728. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Wang L, Brock A, Schultz PG. The selective incorporation of alkenes into proteins in Escherichia coli. Angew Chem Int Ed Engl. 2002;41:2840–2842. doi: 10.1002/1521-3773(20020802)41:15<2840::AID-ANIE2840>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


