Abstract
Allosterism is one of nature's principal methods for regulating protein function. Allosterism utilizes ligand binding at one site to regulate the function of the protein by modulating the structure and dynamics of a distant binding site. In this review, we first survey solution NMR techniques and how they may be applied to the study of allostery. Subsequently, we describe several examples of application of NMR to protein allostery and highlight the unique insight provided by this experimental technique.
Keywords: Solution NMR, Dynamics, Allostery, Conformational Changes, Relaxation Dispersion, Silent Allostery
Introduction
Allostery is the phenomenon in which allosteric ligand (A) binding alters the macromolecule's binding interaction (K-type allostery) with a second ligand or alters the turnover kinetics or Vmax (V-type allostery) in the case of an enzyme catalyzed reaction. The ability to utilize one ligand to tune affinity or reactivity at a distant site confers enormous adaptability to the cell or organism. [1-5]. Traditionally, allostery has been mechanistically described either by a concerted [6] or sequential [7] model in which the protein exists in either the relaxed (R) or tense (T) state. A key feature of all proteins is their inherent flexibility. Consequently, more recent notions of allostery have incorporated ensemble models in which allosteric effector binding causes a redistribution of the ensemble of protein structures with subsequent alteration in its functional properties [8, 9]. Regardless of the model, allostery is due to a thermodynamic linkage between the allosteric effector binding site and a distant biding site, which are in communication. The resulting thermodynamic coupling between the allosteric effector (A) binding site and ligand or substrate (S) binding site can be described by the following equilibrium model:
| (1) |
in which the magnitude of the interaction between binding sites is defined by a coupling free energy,
| (2) |
In equation (2), the terms on the right side represent the free energy difference between the enzyme complex indicated in the subscript and that of the apo (unligated) enzyme. Thus the coupling free energy, ΔGAS is the difference in free energy upon binding A and S simultaneously versus the sum of their individual affinities. If ΔGAS is negative (positive) then the allosteric effector is an activator (inhibitor). Furthermore, because ΔG = ΔH – T ΔS, this indicates that the allosteric effect can arise from changes that are purely enthalpic or entropic or a combination of both. Solution nuclear magnetic resonance (NMR) has the ability to monitor structural and dynamical changes, with atomic resolution, and is therefore ideally suited for the study of protein or enzyme allostery.
The remainder of this review will focus on NMR studies of protein allostery. First we discuss some general features of NMR. Next we survey some literature examples that utilize NMR chemical shifts, spin-relaxation methods, and residual dipolar couplings (RDC) to provide a spatial and temporal description of the allosteric phenomenon.
Investigation of allostery from NMR chemical shifts
The resonance frequencies observed in an NMR experiment vary due to the local magnetic environment of the nuclei. The frequency of these resonances, relative to a standard frequency, is called the chemical shift. The chemical shift can be measured in solution to a very high precision, typically to a few tenths of a Hz. In addition, the chemical shift is very sensitive to the local environment and therefore small changes in structure and dynamics of a particular nucleus is readily detectable by monitoring the change in chemical shift. In a simple, two-site case, say, of an allosteric protein converting between the R and T states, the observed chemical shift (δobs) represents the population-weighted average of the protein conformations.
| (3) |
in which pR and pT are the equilibrium populations for conformations R and T and δR/T are the chemical shifts of the respective conformations (Figure 1). This simple relationship was exploited for study of allostery in the protein NtrC[10]. Equation (3) is valid if the protein is in fast/intermediate exchange between the two conformations, meaning that the kinetics of the interconversion, kex ≥ Δω (|δR – δT|). In this case one observes a single resonance located at the population-weighted chemical shift. In the slow exchange case (kex < Δω), two resonances can be observed, each at their respective chemical shifts, and the equilibrium populations are related to the NMR peak volumes. Therefore, regardless of the allosteric model, the conversion of apo enzyme to the allosterically activated or inhibited enzyme can be followed by monitoring NMR chemical shifts (Fig. 1).
Figure 1.
General depiction of the shift of a two-dimensional NMR resonance as the protein is progressively saturated with ligand. The resonance shifts from red (apo) to purple (fully saturated).
NMR dynamics as a probe of allostery
Solution NMR is an extremely powerful technique for addressing the role of conformational motion and dynamics with atomic resolution on time scales ranging from picoseconds (ps) to seconds [11-15].
Fast (ps – ns) motions by NMR
Motions on these timescales, which are faster than overall rotational diffusion (typically tens of nanosecond) of the macromolecule, modulate the chemical shift anisotropy and dipolar interactions. These molecular fluctuations act to return the spin system to its Boltzmann equilibrium value. The rate constant for the return to equilibrium depends on the nuclei involved and importantly the frequencies of the molecular motions [16-29]. Below, the discussion is restricted to a heteronuclear two-spin-1/2 (I-S) system, such as an amide 1H-15N spin pair. Within this limit, longitudinal (Sz) and transverse (Sx,y) magnetization in the absence of slower motions, respectively, relax to their Boltzmann equilibrium value according to [16]
| (4) |
| (5) |
where is the dipolar coupling constant and is the permeability of free space, ħ is Planck's constant divided by 2π, γI(S) are the gyromagnetic ratios of the I and S nuclei, <rIS> is the average internuclear I-S bond length, ωI(S) is the Larmor frequency of the I(S) nuclei, Δσ is the chemical shift anisotropy (CSA) of the S nucleus. J(ω) is the spectral density function expressed as the cosine transform of the autocorrelation function (C(t)) of the I-S bond vector [16],
| (6) |
If intramolecular motion is faster and uncorrelated with overall rotational diffusion, the autocorrelation function can be written as the product of autocorrelation functions for overall (CO) and internal (CI) motions [30],
| (7) |
In the simplest case of a spherical macromolecule, which would experience isotropic rotation, CO decays in single exponential manner,
| (8) |
in which τc, is the rotational correlation time of the macromolecule. The corresponding spectral density function is
| (9) |
When combined with the correlation function for stochastic internal motions, the spectral density function becomes
| (10) |
where in which τe is the effective correlation time for internal motions and S2 is the generalized order parameter and ranges from 0 to 1, with lower values indicating increased flexibility. The S2 value provides insight into the configurational entropy of individual bond vectors [31-33]. In addition to these fast time scale motions, motions that are commonly referred to as chemical (conformational) exchange occur on the μs – ms time scale. Motions on this slower timescale are characterized as follows.
μs - ms motions by NMR
Motions on the μs - ms timescale that transfer a spin-1/2 nucleus in an I-S spin system between two chemically distinct sites (A and B, , or equivalently, R and T in allosteric terminology) results in an increase the transverse relaxation rate constant, R2 by an amount Rex.
| (11) |
is the base or ‘slow motion-free’ transverse relaxation rate as in equation (5) and in the fast limit Rex = pApB Δω2 / kex in which Δω is the difference in chemical shifts between the two interconverting sites and kex = k1+k2 is the microscopic exchange rate constant of the motional process. For these motional time scales, characterization of an equilibrium conformational exchange process, is performed using the relaxation-compensated Carr-Purcell-Meiboom-Gill (rcCPMG) experiment [34, 35] or the off-resonance rotating frame relaxation experiment (R1ρ) [36, 37] in what is termed relaxation dispersion analysis. In these experiments, Rex additionally depends on τcp, the variation of the repetition rate of the CPMG 180° pulse spacing [38-40] or the spin-locking field strength, ωe in the R1ρ experiment [37].
For the single quantum CPMG experiment [38-40]:
| (12) |
| (13) |
| (14) |
| (15) |
in which pA and pB are the equilibrium populations of the two sites, Δω is the difference in chemical shifts between the two sites, and and are the intrinsic transverse relaxation rates of the two sites in the absence of chemical exchange. In the off-resonance R1ρ experiment [41]:
| (16) |
in which are the effective fields for sites A/B and . Here ωA, ωB and ωiso are the frequency offsets of the A, B, and population averaged NMR resonances from the radio-frequency (RF) carrier, respectively and ω1 is the RF field strength with tilt angle, θ = arctan(ω1/ ωiso). R1 and R2 are measured separately [27]. Equations (12) and (16) are general equations that are used when the conformational motion exists in slow to intermediate exchange (kex ≤ Δω). Under fast exchange conditions (kex > Δω) equations (12) and (16) simplify to those given in (17) - (19) [39, 42]. For the R1ρ experiments in fast exchange
| (17) |
in which Rex is:
| (18) |
and for the rcCPMG experiment
| (19) |
in which ϕex = pApBΔω2. Full characterization requires measurements at two static magnetic fields [43-45]. More complex, three-site conformational exchange processes can also be characterized [46, 47]; and these experiments can also be performed on multiple quantum coherences [48, 49], which can provide additional chemical shift information. Motions slower than ca. 100 ms can also be examined using ZZ-exchange type measurements [50-52]. Lastly, for proteins with molecular weight > 30 kDa, TROSY enhancement [53] can be incorporated into any of the aforementioned experiments to provide increased signal-to-noise and resolution, extending the size limit more than 100 kDa.
Solution NMR relaxation experiments can provide detailed, atomic resolution characterization of molecular motions occurring over a wide range of time scales for highly skewed equilibrium populations (99.5%:0.5%) under physiological-like conditions.
Structural information from residual dipolar couplings (RDC)
Magnetic dipole interactions between nuclei contain an abundance of structural information. However, in isotropic solution these interactions are averaged to zero as a result of the rotational tumbling of the macromolecule. These dipolar couplings can be reintroduced at high static magnetic field by partial alignment of the macromolecule in solution with dilute liquid crystalline phase [54]. This partial alignment can be brought about by the introduction of bicelles [55, 56] or filamentous phage [57, 58] in the NMR tube. Alternatively, paramagnetic tags attached to the protein or self-alignment, for proteins with large intrinsic magnetic susceptibilities, can also provide the necessary degree of alignment for measurement of RDC [59, 60]. The measured RDC contains the bond vector orientational information that provides important structural constraints as shown in equation (20) [61].
| (20) |
Here is the measured RDC between nuclei i and j, with γ being their gyromagnetic ratios, μ0 and h are defined from equation (4) and (5). The effective internuclear distance between i and j is given by r. P2(cos(θ)) is the second rank Legendre polynomial with θ being the angle between the static magnetic field direction and the vector connecting nuclei i and j, in which the angle brackets indicate the time-average. Therefore measurement of for many spin-pairs (such as an amide H-N group) in a protein allows determination of their orientation with respect to B0 and thus their orientation with respect to each other. is typically measured as a contribution to the scalar coupling constant, is measured in isotropic solution in which , and again in the presence of alignment media where . These experiments are sensitive to small changes in protein conformation and provide valuable insight into structural changes that occur upon ligand binding [62].
In the remainder of this review we analyze recent representative research in which solution NMR was used to provide insight into protein allostery.
Applications: Chemical Shift Analysis
NMR chemical shifts can provide valuable information in allosteric studies as they are sensitive reporters of protein structural and dynamical changes. In this section we survey several studies that make primary use of NMR chemical shifts in the investigation of allostery.
To determine the allosteric mechanism of aminoglycoside N-(6’)-acetyltransferase-Ii (AAC(6’)-Ii) with its substrate, acetyl coenzyme A (AcCoA) a combination of isothermal titration calorimetry (ITC), circular dichroism (CD), and NMR was used to characterize the thermodynamics, structure, and dynamics of AcCoA binding [63]. AAC(6’)-Ii is a homodimeric enzyme that is known to bind AcCoA with positive cooperativity [64, 65]. Interestingly, ITC shows that the cooperativity is temperature dependent. From 10 °C to 37 °C, the second AcCoA binds more tightly than the first, but at temperatures greater than 37 °C this relationship is reversed with the first AcCoA binding more tightly than the second. This temperature dependent reversal in binding affinity indicates a shift from positive (low temperature) to negative cooperativity at the higher temperatures. ITC data showed that the enthalpy of binding for the second AcCoA molecule undergoes a dramatic decrease at temperatures over 30 °C while the binding enthalpy of the first molecule experiences only a slight increase over the same temperature range. It is noted that this behavior can be rationalized by a population of the active form (F state) in equilibrium with the partly or fully unfolded form (U state). At higher temperatures the U state becomes more populated requiring an additional folding step for substrate binding to occur. The CD results are consistent with the presences of both states in solution. ITC and CD results were able to provide a global view of events that occurred when AcCoA bound to AAC(6’)-Ii, but these experiments lack resolution at the atomic level. To provide additional insight, two-dimensional NMR 1H-15N heteronuclear single quantum coherence (HSQC) spectra were used to compare the differences in chemical shifts between apo and AcCoA saturated AAC(6’)-Ii. 137 of the 172 expected amide resonances were observed for apo AAC(6’)-Ii, while the AcCoA bound AAC(6’)-Ii spectrum contained 166. The difference in the number of observed resonances can be attributed to apo AAC(6’)-Ii being partially disordered or highly mobile leading to peak broadening or disappearance. By determining the minimal chemical shift difference between unbound and bound, the largest chemical shift differences were identified at the interface of the homodimer. Because the largest difference occurred at the interface the authors conclude that AcCoA altered the subunit interactions of the homodimer. This change may be an important aspect of allosteric communication of AAC(6’)-Ii. Titration experiments with AcCoA added to AAC(6’)-Ii were also performed and monitored by 1H-15N HSQC experiments. Over the course of the titration there are three possible states of AAC(6’)-Ii, apo, singly-bound, and/or doubly occupied AcCoA states. It is noted that the singly-bound state does not show distinct signals in the spectra, but is either grouped with the apo form or doubly bound states. To address the lack of a unique NMR signature for the singly-bound form, the fraction of AAC(6’)-Ii for apo, singly, and doubly occupied states determined previously by ITC were used in a joint analysis with the NMR peak intensity data. The method employed here allowed for the extraction of the contribution of the singly-bound form from the predominately apo and doubly-bound spectra. The singly-bound state monomer was found to resemble the doubly-bound state. Whereas the unbound monomer of the singly-bound homodimer showed some resemblance to the apo state indicating that the unbound monomer of the singly-bound homodimer has some properties similar to those of the apo conformation. These studies are limited to only noting the difference between the unbound domain of the 1-bound state and the unbound domains of the apo state due to experimental limitations, but it nonetheless provides valuable insight into the allosteric model of AAC(6’)-Ii [63] that being the temperature dependent switch between positive and negative cooperativity as well as the possibility of an unfolded state being crucial for the allosteric phenomenon.
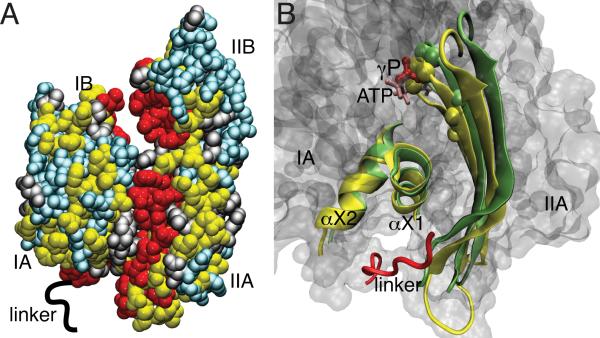
In another multidomain enzyme, NMR chemical shift titration studies were used to examine allostery in the nucleotide-binding domain (NBD) of the chaperone, DnaK [66]. DnaK is the Heat Shock Protein (Hsp70) protein of E. coli. Hsp70 consists of two domains, the aforementioned NBD, which binds ATP or ADP and the substrate-binding domain (SBD), which are linked by the conserved hydrophobic interdomain linker sequence, 389VLLL392 [67, 68]. ATP-bound NBD has a low affinity for substrate at the SBD binding site, while ADP-bound NBD has a high affinity for substrate at the SBD. It is also known that the ATPase activity of the NBD is stimulated by substrate binding at the SBD [69]. This activity is also observed for the NBD in the presence of the 389VLLL392 linker. Analysis of 1HN, 13C, 13Cα, 13Cβ, and 15N chemical shifts for a total of six different states of two NBD constructs, NBD388 (truncated at the VLLL linker) and NBD392 (with linker), was undertaken to investigate the role of the VLLL linker in the allosteric pathway in DnaK. The NBD with and without VLLL was investigated for the apo, ADP-bound, and ATP-bound states by study of NMR chemical shifts. In the ATP-bound state the interdomain linker has a large effect on the conformation of the NBD. Large chemical shift perturbations are seen in several sections of the NBD when ATP and the VLLL interdomain linker are present compared to ATP-bound to NBD388 indicating a reorganization in the NBD interfaces. 13Cα and 13Cβ chemical shifts correlate with protein secondary structure and were similar between NBD388 and NBD392, which indicated that there are minimal secondary structure differences between the two constructs. In contrast, roughly 60% of 1HN, 15N, 13C’ resonances were significantly different in the presence of nucleotide and/or the linker region. When these residues are mapped onto the DnaK NBD crystal structure the large and significant shifts are at or near the NBD subdomain interface and connect to the nucleotide binding site (Figure 2). These ‘hotspots’ suggest a network of residues that are important for relaying the allosteric signal. The combination of maintained secondary structure and changes in the subdomain interfaces implies that ligand binding and the interdomain linker change the orientation of the subdomains with respect to each other. This chemical shift analysis is in agreement previous RDC studies that also show significant subdomain reorganization in solution [70]. The spectral overlays show a nonlinear relationship for the chemical shifts of many resonances in their varying ligand states, with many having a unique resonance for each of the 12 states examined. This difference in chemical shift between liganded states suggests that the NBD can exist in more than two conformational states.
Figure 2.
Mechanism of intramolecular allostery in the Hsp70 NBD. (A) Mapping of allosteric hot spots onto the structure of the DnaK NBD: Residues with large and significant chemical shift differences are shown in red and yellow, respectively. Cyan and gray indicate significant changes and residues with no data, respectively. (B) Structural model for two way coupling pathway between the nucleotide-binding site and the interdomain linker: superposition of the IIA β-sheet and the crossing α-helices for the ATP- (yellow) and ADP- (green) bound conformations of the NBD. The ATP γ-phosphate and the linker are in red, and residues involved in nucleotide binding are shown as spheres.
A slightly more detailed analysis of chemical shifts by Melacini and coworkers was employed to investigate allostery [71] in the signaling protein EPAC, which is a guanine exchange factor that is modulated by cAMP [72]. Here, these investigators sought to understand the pathway by which the cAMP binding site signal is propagated to the adjacent α6 hinge helix to the distally located ionic latch region. Because much of this allosteric signal is relayed by changes in protein dynamics, monitoring cAMP induced structural changes was not sufficient to identify this pathway. In this work, these authors note that a set of amino acid residues belonging to the same allosteric network should have correlated chemical shift changes in response to allosteric perturbations. Melacini and coworkers monitor the covariance in chemical shifts as a result of binding five different allosteric ligands, each with different activating properties [71]. In this way they are able to map the protein regions that bridge the cAMP binding site and the ionic latch.
The use of multiple ligands has also proven fruitful in studying allosteric protein evidenced by the work of Lipchock and Loria [73]. Here, these investigators make use of the result of equation (2), that is the non-additivity of the allosteric phenomenon. The system under study was the heterodimeric enzyme imidazole glycerol phosphate synthase (IGPS), which catalyzes the hydrolysis of glutamine and subsequent production of imidazole glycerol phosphate and 5-aminoimidazole-4-carboxamide ribotide (AICAR) from N’-[(5’-phosphoribulosyl)formimino]-5-aminoimidazole-4-carboxamide ribonucleotide (PRFAR) and the NH3 generated from Gln hydrolysis [74, 75]. These two catalytic sites are ca. 30Å apart [76]. Furthermore, Gln hydrolysis is accelerated 5000-fold in the presence of PRFAR [77, 78]. These authors compared the changes in methyl chemical shifts, from the apo enzyme, for the two binary complexes, PRFAR bound (ΔδPRFAR) and acivicin (a Gln analog) bound (ΔδAcivicin) as well as chemical shift changes between apo and ternary complex in which both PRFAR and acivicin were bound (ΔδTernary). Methyl sites in which ΔΔδ ≠ 0 where,
| (21) |
are indicative of synergistic effects of ternary complex formation. Using this analysis, Lipchock and Loria identified ten residues with non-additive chemical shift changes [73]. The importance of these residues in propagating the allosteric signal is bolstered by mutagenesis and NMR dynamics measurements.
Applications: Atomic motions and their role in protein allostery
NMR measurement of changes in protein dynamics can provide additional information about how the allosteric signal is transmitted through a dense protein matrix. The NMR experiments can provide crucial, additional information particularly in allosteric proteins that exhibit minimal or no structural changes upon effector binding.
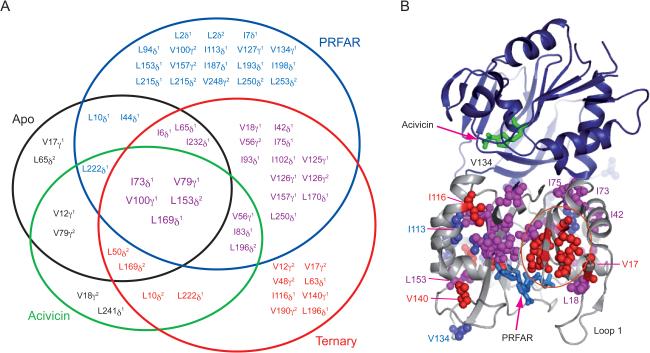
The application of Carr-Purcell-Meiboom-Gill (CPMG) NMR relaxation dispersion techniques to the V-type allosteric enzyme imidazole glycerol phosphate synthase was used assess the role of millisecond motions in this enzyme. IGPS, as noted in the previous section is a V-type allosteric enzyme consisting of a glutaminase domain (HisH) and a cyclase domain (HisF). X-ray structures of IGPS in the apo and PRFAR bound states show very little structural difference between the states, which suggest that the allostery of IGPS may have a significant dynamic component [79]. Methyl, multiple quantum CPMG relaxation dispersion [48] curves were measured for Isoleucine, Leucine, and Valine residues in the apo, acivicin (substrate mimic), PRFAR (effector ligand), and ternary enzyme complexes. While the apo and acivicin-bound complexes have a small subset of residues that display millisecond motions, the addition of PRFAR more than doubles the number of residues in the HisF domain that undergo millisecond motion and additionally results in exchange broadening of nearly 70 backbone amide resonances. The increase in millisecond motion upon PRFAR binding is domain wide in HisF. Interestingly the relaxation dispersion studies of the ternary complex, reveal eight resides that undergo millisecond motion that were not observed in the apo, acivicin, or PRFAR complexes (Figure 3) [73]. These residues are found to connect the PRFAR binding site to the HisF-HisH interface providing a path of residues that undergo millisecond motions and connect the effector binding site to the substrate binding site. These results suggest that the increased ms motions due to ternary complex formation, allow IGPS to populate an active conformation that is not present to an appreciable extent in the enzyme in the absence of bound allosteric effector.
Figure 3.
Summary of ms motions. (A) Venn-type diagram illustrating the relation between flexible residues from ILV relaxation dispersion experiments and the enzyme complex in which they occur for apo (black), acivicin (green), PRFAR (blue), and ternary (red). Residues common to all four are shown in a larger font size. (B) Residues exhibiting ILV methyl 13C1H3 MQ dispersion in the PRFAR bound and ternary states are shown mapped onto IGPS. Residues with dispersion in only the PRFAR bound state are shown in blue spheres, while those with dispersion in only the ternary complex are shown in red. Residues with dispersion in both states are shown in magenta.
Homeodomains are small DNA binding modules found in transcription factors. Lowly populated states detected by NMR relaxation dispersion of PBX1 homeodomain (PBX-HD) lead to the determination of the role of a transient conformation in the allosteric mechanism of PBXHD. While DNA binding causes a conformational change, the bound DNA molecule never comes in structural contact with the residues that undergo secondary structure rearrangement [80], particularly the C-terminal α-helix 4. Nonetheless, removal of the 15 amino acid residues that make up this fourth helix is known to reduce the affinity of PBX-HD for DNA complex 5-fold [81]. Thus, the dynamic characteristics of this C-terminal region were examined by 15N CPMG relaxation dispersion experiments of apo PBX-HD over a 15° temperature range [82]. Five of these 15 residues were amenable to NMR study by relaxation dispersion experiments. At each temperature (10, 15, 20, 25 °C), the dispersion curves of the C-terminus residues were globally fit to extract kex, pa and pb, and Δω. kex was found to increase with temperature, whereas pb decreased at higher temperature. It was determined that the residues behaved as a unit, exchanging between two conformations with an identical rate constant. Additional van't Hoff analysis, by examining the temperature dependence of ΔG, indicated that the minor state (B) is more ordered than the ground state (A) with an enthalpy difference ΔHAB = −9.5 kcal mol−1 and an entropy change of −38 cal mol−1 K−1. These thermodynamics for the A to B transition are consistent with the folding of an unstructured region into an α-helix. The hypothesis formed is that PBX-HD exists with the C-terminal residues in an unordered state with a population of 90-95% that is in equilibrium with an α-helical structure at about 5-10%. Correlations between the |Δω| derived from the CPMG experiments and |Δδ| calculated from the difference in apo-PBXHD and PBX-HD-DNA chemical shifts show that the structure of the transient alpha helix 4 is similar to the alpha helix 4 formed by DNA binding. These data collectively suggest a conformational selection model for allostery with regard to α-helix 4 because even in the absence of DNA the C-terminus assumes a measurable amount of helix structure. The perturbation of the homeodomain by the transient formation of the fourth alpha helix is also proposed to be a key aspect of the allosteric mechanism. The folding of the C-terminal residues alters the ensemble to favor ligand binding and as the changes in chemical shifts show, has an effect on the DNA binding site. [82], Based on the thermodynamics and NMR studies, the authors propose that the formation of the fourth alpha helix increases the affinity for DNA and pays for the energetic cost of the helix formation Catabolite activator protein (CAP) is allosterically activated for DNA binding by cyclic AMP (cAMP), switching the protein from an inactive conformation, which weakly and non-specifically interacts with DNA to an active conformation that increases the binding affinity and specificity for the DNA substrate [83] with sub-micromolar affinity. NMR studies of a constitutively active CAP mutant (S62F) indicated that it is in the inactive (DNA-binding incompetent) state in the apo form and furthermore that cAMP binding resulted in little change to the DNA binding domain (DBD) of CAP. This mutant seems to have significantly altered allosteric properties compared to the WT CAP. Nonetheless both cAMP activated proteins bind DNA with equivalent affinity. However, ITC experiments showed that these binding affinities are obtained through different thermodynamics. WT DNA binding is enthalpically favored (ΔH = – 23.2 kcal/mol; –TΔS = 14.3 kcal/mol), whereas S62F DNA binding is dominated by favorable entropy (–TΔS = –13.2 kcal/mol).
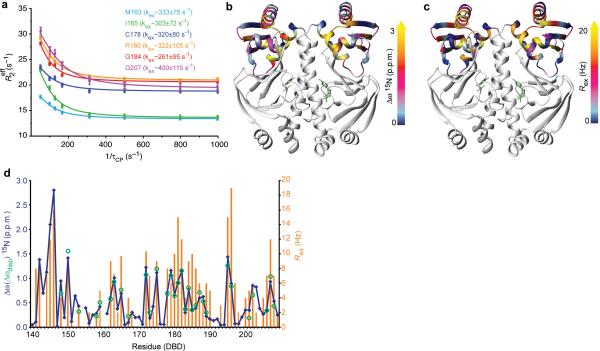
NMR relaxation dispersion experiments demonstrated that the DBD of cAMP-bound CAP-S62F exists in equilibrium on the millisecond time scale with a 2% populated conformation that resembles the DNA bound conformation – a conformational selection model (Figure 4) [84]. In addition, N-H bond order parameters (S2) for WT-CAP-cAMP2-DNA and CAP-S62F-cAMP2-DNA were used to estimate the conformational entropy of the ligand bound states. On binding of DNA the S2 for WT-CAP-cAMP2 increase throughout the protein indicating a protein-wide rigidification, which is consistent with the ITC results. In contrast, DNA binding to CAP-S62F-cAMP2 resulted in widespread decreases in S2 indicating the protein became more flexible upon binding its target DNA. Thus, NMR relaxation was used to demonstrate that a mutant of CAP that adopts the inactive structure when the allosteric effector is bound is still able to bind the DNA substrate by increasing the conformational entropy to compensate for lack of favorable enthalpy, which is observed for the WT protein [84].
Figure 4.
CAP-S62F-cAMP2 visits an active, low-populated conformational state. (A) Representative relaxation dispersion data of 15N backbone amides of CAP-S62F-cAMP2 DBD residues. (B) Backbone 15N chemical shift differences of the DBD residues between the apo and cAMP2-bound WT-CAP mapped on the structure. (C) Conformational exchange dynamics of CAP-S62F-cAMP2 on the μs – ms timescale as indicated by Rex, are mapped on the structure. (D) Plot of 15N static Δδ values (blue), Rex values (orange), and dynamic Δδ (green).
PDZ domains are found as parts of larger proteins where the main function of these domains is to bind to C-terminal tails of target proteins and play important roles in signaling [85]. The third PDZ domain (PDZ3) of PSD-95/SAP90 adheres to the conserved fold of the PDZ family but possesses an additional C-terminal alpha helix. This alpha helix is distal to the peptide binding site of the domain. Nonetheless, removal of helix 3 results in a 21-fold decrease in ligand affinity [86]. In addition, ITC experiments indicate that the source of this lowered affinity in the truncated protein (Δ7ct) is purely entropic in nature. To investigate the mechanism of the allosteric effect of helix 3, Lee and coworkers undertook a solution NMR comparison of PDZ3 and the Δ7ct variant protein [86]. Two-dimensional 1H-15N HSQC spectra of the two constructs showed that Δ7ct maintains a native PDZ structure similar to WT-PDZ3. However, quantitation of side-chain 2H-methyl order parameters (often referred to as S2axis) indicated that while both peptide bound PDZ3 and Δ7ct proteins displayed similar levels of ps – ns dynamics, the apo forms were significantly different with the Δ7ct for being more flexible. Thus in the absence of helix 3, the PDZ domain has a greater entropic penalty to pay for binding its target peptide, which leads to a reduction in affinity, in agreement with ITC data. Lee and coworkers therefore conclude that helix 3 plays an important biological function as a distal modulator of peptide binding affinity [86].
The catalytic subunit of protein kinase A (PKA-C) progresses through an open, intermediate, and closed conformation during its catalytic cycle [87]. Because of its crucial role in biology, the activity of PKA-C must be tightly controlled in vivo. PKA-C is inhibited by the protein kinase inhibitor (PKI), which can be mimicked by the inhibitory region of this protein (PKI5-24). Nucleotide binding allosterically modulates inhibitor and substrate (PLN1-20) binding to PKA-C. To investigate the details of conversion between conformational states of PKA-C, Masterson et al. used solution NMR and demonstrated that 1H-15N chemical shifts in PKA-C experienced a linear progression between the apo, binary, ternary, and fully inhibited ternary complexes suggesting that ligand binding shifts the populations of the major conformational states [88]. To gain further insight these investigators measured changes in ps – ns and μs – ms time scale motions using solution NMR spectroscopy. They observe quenched motions on both timescale ranges for the fully inhibited PKA-C (60 mM Mg2+/nucleotide/PKI5-24) (Figure 5) [88]. In contrast, the substrate bound form (10 mM Mg2+/nucleotide/PLN1-20), which they call “dynamically committed”, experiences enhanced flexibility on both the ps – ns and μs – ms time scales. Importantly, these data are consistent with calorimetry measurements and suggest mechanisms for modulation of PKA-C activity.
Figure 5.
Backbone dynamics of PKA-C in different ternary complexes. Mapping of (A) fast and (B) slow backbone dynamics show that upon inhibition with PKI5-24 and with high Mg2+, a decrease of picosecond to millisecond dynamics occurs throughout the backbone. For the comparison, the dynamics of PKA-C with the substrate PLN1-20 is shown.
Applications: Residual dipolar couplings and protein allostery
Because of their sensitivity to structural changes, RDC have been used in the study of allosterism as a means to detect differences in conformational states due to ligand binding. Zuiderweg and coworkers [89] used 1H-15N RDC to study allostery in Hsp70. Hsp70 is a chaperone protein that couples nucleotide binding with the unfolding of misfolded proteins. These chaperones contain two domains, a 44 kDa nucleotide binding domain, termed the NBD and a 25 kDa substrate binding domain (SBD), which binds the protein substrate. Whereas, the NBD is responsible for binding ATP or ADP. It is believed that ATP binding to the NBD alters the conformation of the NBD such that productive interactions with SBD can take place. These favorable interactions are then propagated to the SBD substrate binding site, which leads to the release of the substrate. Thus, the ADP and ATP bound NBD differentially interact with the SBD. However there is little structural evidence for such different nucleotide bound NBDs. By measuring RDC of the T. thermophilus Hsp70 homolog, DnaK in the ADP and AMPPNP (a nonhydrolyzable ATP analogue) Zuiderweg and coworkers showed that a subdomain of the NBD, (subdomain IIb) assumes an orientation with 20° difference between the ADP and AMPPNP forms. Additional changes are also observed in subdomain IA. The authors suggest that these changes are also realized in the full-length protein and that their proximity to the linker binding between the SBD and NBD facilitate propagation to the substrate binding site.
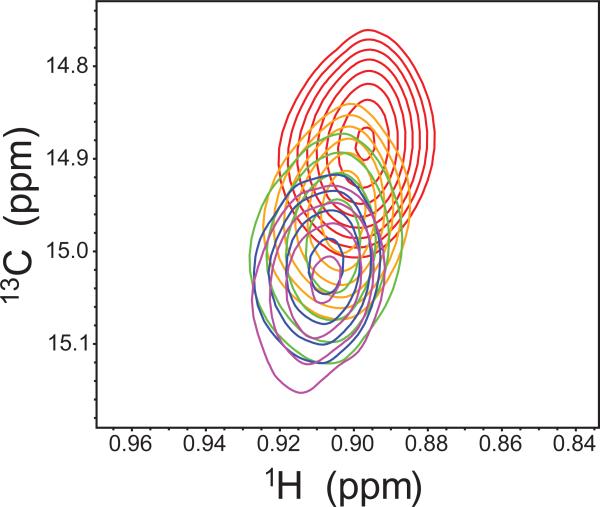
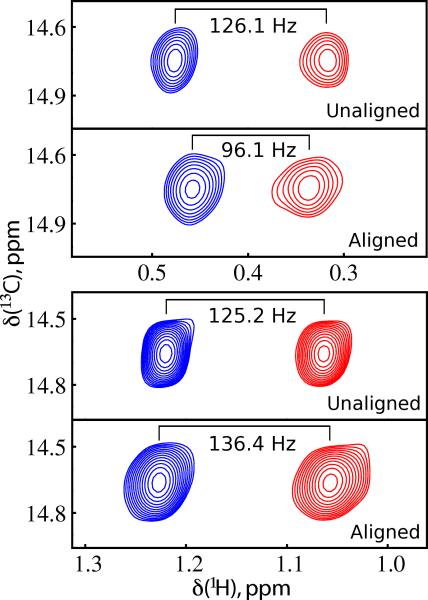
One of the most studied allosteric enzymes is aspartate transcarbamoylase (ATCase), which is a 300 kDa enzyme that catalyzes the first step in biosynthesis of pyrimidines. ATCase is an oligomer composed of six regulatory and six catalytic chains [90]. The catalytic chains promote the reaction between carbamoylphosphate and aspartate to generate carbamoylaspartate and inorganic phosphate and furthermore exhibit homotropic cooperativity with respect to the substrate aspartate. In addition, the regulatory chains exert a heterotropic allosteric effect on enzyme activity with activation by ATP binding and inhibition via CTP binding [91]. Numerous studies support a Monod-Wyman-Changeaux (MWC) model with respect to the substrate molecules [92-96]. However, there is not uniform agreement with regard to the effect of nucleotide binding, particularly as it pertains to the catalytic chains. Kay and Schachman [97] exploit the sensitivity and resolution of the methyl transverse relaxation optimized spectroscopy (TROSY) [98] to study the effect of ATP and CTP on the structure of the catalytic chains. To do this they specifically isotopically label the methyl groups of isoleucine, leucine, and valine (Ileδ1-[13CH3], Leu, Val-[13CH3, 12CD3]) in a perdeuterated catalytic chain and combine this with unlabeled regulatory chains to form ATCase. They spectroscopically observe 81% of the expected methyl correlations (109 of 135) and quantitate RDC for 46 methyl positions in the absence of nucleotide and in the presence of CTP and ATP (Figure 6). For each they observe no significant difference in RDC values between these complexes, which strongly argues for a lack of structural change in the catalytic chain due to nucleotide binding, in agreement with the MWC model. In support of this conclusion, these authors also demonstrate no significant changes in methyl chemical shifts due to ATP or CTP binding.
Figure 6.
Measurement of methyl dipolar (Dmethyl) coupling values. Top (bottom) panels of each pair are spectra obtained in the absence (presence) of alignment from which the values of Jmethyl (Jmethyl+Dmethyl) are measured.
Conclusion
Recent innovations in NMR methods and isotope labeling strategies now allow for relatively routine NMR study of very large proteins. Solution NMR is able to provide an atomic resolution view of structure and dynamics, which has led to unique insight into the allosteric mechanisms of many proteins. It is anticipated that future studies on even larger and more complex systems will continue to refine our understanding of the fundamentals of allosteric communication.
Highlights.
Solution NMR provides unique structural and dynamical detail of allosteric proteins Relaxation dispersion experiments provide evidence for lowly populated conformers NMR can provide insight into kinetics and thermodynamics of allostery
Acknowledgements
Greg Manley acknowledges support from NIH Biophysical training grant (T32GM008283). JPL acknowledges support from the National Science Foundation (MCB-0744161). There are many more quality examples of protein allostery than can possibly be covered in the scope of this review and we apologize to those researchers whose work we were not able to cover.
References
- 1.Carpenter JF, Hand SC. Comparison of pH-dependent allostery and dissociation for phosphofructokinases from Artemia embryos and rabbit muscle: nature of the enzymes acylated with diethylpyrocarbonate. Arch Biochem Biophys. 1986;248:1–9. doi: 10.1016/0003-9861(86)90394-2. [DOI] [PubMed] [Google Scholar]
- 2.Liu L, Wales ME, Wild JR. Temperature effects on the allosteric responses of native and chimeric aspartate transcarbamoylases. Journal Of Molecular Biology. 1998;282:891–901. doi: 10.1006/jmbi.1998.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Onuchic JN, Kobayashi C, Miyashita O, Jennings P, Baldridge KK. Exploring biomolecular machines: energy landscape control of biological reactions. Philos Trans R Soc Lond B Biol Sci. 2006;361:1439–1443. doi: 10.1098/rstb.2006.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ackers GK, Doyle ML, Myers D, Daugherty MA. Molecular code for cooperativity in hemoglobin. Science. 1991;255:54–63. doi: 10.1126/science.1553532. [DOI] [PubMed] [Google Scholar]
- 5.Monod J, Changeux JP, Jacob F. Allosteric proteins and cellular control systems. Journal Of Molecular Biology. 1963;6:306–329. doi: 10.1016/s0022-2836(63)80091-1. [DOI] [PubMed] [Google Scholar]
- 6.Monod J, Wyman J, Changeux J-P. On the nature of allosteric transitions: A plausible model. J. Mol. Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 7.Koshland DE, Nemethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 8.Pan H, Lee JC, Hilser VJ. Binding sites in Escherichia coli dihydrofolate reductase communicate by modulating the conformational ensemble. Proc Natl Acad Sci U S A. 2000;97:12020–12025. doi: 10.1073/pnas.220240297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duke TA, Bray D. Heightened sensitivity of a lattice of membrane receptors. Proc Natl Acad Sci U S A. 1999;96:10104–10108. doi: 10.1073/pnas.96.18.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Volkman BF, Lipson D, Wemmer DE, Kern D. Two-state allosteric behavior in a single-domain signaling protein. Science. 2001;291:2429–2433. doi: 10.1126/science.291.5512.2429. [DOI] [PubMed] [Google Scholar]
- 11.Kempf JG, Loria JP. Protein dynamics from solution NMR: Theory and applications. Cell Biochem. & Biophys. 2002;39:187–212. doi: 10.1385/cbb:37:3:187. [DOI] [PubMed] [Google Scholar]
- 12.Kempf JG, Loria JP. In: Protein NMR Techniques. Downing AK, editor. Humana Press; Totowa: 2004. pp. 185–231. [Google Scholar]
- 13.Lipchock JM, Loria JP. Monitoring molecular interactions by NMR. Methods Mol Biol. 2009;490:115–134. doi: 10.1007/978-1-59745-367-7_5. [DOI] [PubMed] [Google Scholar]
- 14.Loria JP, Watt ED, Berlow RB. Characterization of enzyme motions by solution NMR relaxation dispersion. Acc Chem Res. 2007;41:214–221. doi: 10.1021/ar700132n. [DOI] [PubMed] [Google Scholar]
- 15.Palmer AG, Kroenke CD, Loria JP. Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Meth. Enzymol. 2001;339:204–238. doi: 10.1016/s0076-6879(01)39315-1. Part B. [DOI] [PubMed] [Google Scholar]
- 16.Abragam A. Principles of Nuclear Magnetism. Clarendon Press; Oxford: 1961. [Google Scholar]
- 17.Skelton NJ, Palmer AG, Akke M, Kördel J, Rance M, Chazin WJ. Practical aspects of two-dimensional proton-detected 15N spin relaxation measurements. J. Magn. Reson., Ser. B. 1993;102:253–264. [Google Scholar]
- 18.Mandel AM, Akke M, Palmer AG. Backbone dynamics of Escherichia coli ribonuclease HI: correlations with structure and function in an active enzyme. J. Mol. Biol. 1995;246:144–163. doi: 10.1006/jmbi.1994.0073. [DOI] [PubMed] [Google Scholar]
- 19.Farrow NA, Zhang O, Szabo A, Torchia DA, Kay LE. Spectral density function mapping using 15N relaxation data exclusively. J. Biomol. NMR. 1995;6:153–162. doi: 10.1007/BF00211779. [DOI] [PubMed] [Google Scholar]
- 20.Ishima R, Nagayama K. Protein backbone dynamics revealed by quasi spectral density function analysis of amide N-15 nuclei. Biochemistry. 1995;34:3162–3171. doi: 10.1021/bi00010a005. [DOI] [PubMed] [Google Scholar]
- 21.Peng JW, Wagner G. Mapping spectral density functions using heteronuclear NMR relaxation measurements. J. Magn. Reson. 1992;98:308–332. [Google Scholar]
- 22.d'Auvergne EJ, Gooley PR. The use of model selection in the model-free analysis of protein dynamics. J. Biomol. NMR. 2003;25:25–39. doi: 10.1023/a:1021902006114. [DOI] [PubMed] [Google Scholar]
- 23.Cole R, Loria JP. FAST-Modelfree: A program for rapid automated analysis of solution NMR spin-relaxation data. J. Biomol. NMR. 2003;26:203–213. doi: 10.1023/a:1023808801134. [DOI] [PubMed] [Google Scholar]
- 24.Clore GM, Szabo A, Bax A, Kay LE, Driscoll PC, Gronenborn AM. Deviations from the simple two-parameter model-free approach to the interpretation of nitrogen-15 nuclear magnetic relaxation of proteins. J. Am. Chem. Soc. 1990;112:4989–4991. [Google Scholar]
- 25.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J. Am. Chem. Soc. 1982;104:4546–4559. [Google Scholar]
- 26.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 2. Analysis of experimental results. J. Am. Chem. Soc. 1982;104:4559–4570. [Google Scholar]
- 27.Cavanagh J, Fairbrother WJ, Palmer AG, Rance M, Skelton NJ. Protein NMR Spectroscopy: Principles and Practice. Elsevier Academic Press; San Diego: 2007. [Google Scholar]
- 28.Palmer AG, 3rd, Massi F. Characterization of the dynamics of biomacromolecules using rotating-frame spin relaxation NMR spectroscopy. Chem Rev. 2006;106:1700–1719. doi: 10.1021/cr0404287. [DOI] [PubMed] [Google Scholar]
- 29.Palmer AG, Williams J, McDermott A. Nuclear magnetic resonance studies of biopolymer dynamics. J. Phys. Chem. 1996;100:13293–13310. [Google Scholar]
- 30.Wallach D. Effect of internal rotation on angular correlation functions. J. Chem. Phys. 1967;47:5258–5268. [Google Scholar]
- 31.Akke M, Brüschweiler R, Palmer AG. NMR order parameters and free energy: An analytic approach and application to cooperative Ca2+ binding by calbindin D9k. J. Am. Chem. Soc. 1993;115:9832–9833. [Google Scholar]
- 32.Li Z, Raychaudhuri S, Wand AJ. Insights into the local residual entropy of proteins provided by NMR relaxation. Protein Sci. 1996;5:2647–2650. doi: 10.1002/pro.5560051228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang D, Kay LE. Contributions to conformational entropy arising from bond vector fluctuations measured from NMR-derived order parameters: Application to protein folding. J. Mol. Biol. 1996;263:369–382. doi: 10.1006/jmbi.1996.0581. [DOI] [PubMed] [Google Scholar]
- 34.Loria JP, Rance M, Palmer AG. A Relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. J. Am. Chem. Soc. 1999;121:2331–2332. [Google Scholar]
- 35.Loria JP, Rance M, Palmer AG. A TROSY CPMG sequence for characterizing chemical exchange in large proteins. J. Biomol. NMR. 1999;15:151–155. doi: 10.1023/a:1008355631073. [DOI] [PubMed] [Google Scholar]
- 36.Akke M, Palmer AG. Monitoring macromolecular motions on microsecond-millisecond time scales by R1ρ-R1 constant-relaxation-time NMR spectroscopy. J. Am. Chem. Soc. 1996;118:911–912. [Google Scholar]
- 37.Deverell C, Morgan RE, Strange JH. Studies of chemical exchange by nuclear magnetization relaxation in the rotating frame. Mol. Phys. 1970;18:553–559. [Google Scholar]
- 38.Carver JP, Richards RE. A general two-site solution for the chemical exchange produced dependence of T2 upon the Carr-Purcell pulse separation. J. Magn. Res. 1972;6:89–105. [Google Scholar]
- 39.Davis DG, Perlman ME, London RE. Direct measurements of the dissociation-rate constant for inhibitor-enzyme complexes via the T1ρ and T2 (CPMG) methods. J. Magn. Reson., Ser B. 1994;104:266–275. doi: 10.1006/jmrb.1994.1084. [DOI] [PubMed] [Google Scholar]
- 40.Jen J. Chemical exchange and NMR T2 relaxation — the multisite case. J. Magn. Reson. 1978;30:111–128. [Google Scholar]
- 41.Trott O, Palmer AG., 3rd R1rho relaxation outside of the fast-exchange limit. J. Magn. Reson. 2002;154:157–160. doi: 10.1006/jmre.2001.2466. [DOI] [PubMed] [Google Scholar]
- 42.Luz Z, Meiboom S. Nuclear magnetic resonance study of the protolysis of trimethylammonium ion in aqueous solution—order of the reaction with respect to solvent. J. Chem. Phys. 1963;39:366–370. [Google Scholar]
- 43.Ishima R, Torchia DA. Estimating the time scale of chemical exchange of proteins from measurements of transverse relaxation rates in solution. J. Biomol. NMR. 1999;14:369–372. doi: 10.1023/a:1008324025406. [DOI] [PubMed] [Google Scholar]
- 44.Kovrigin EL, Kempf JG, Grey M, Loria JP. Faithful estimation of dynamics parameters from CPMG relaxation dispersion measurements. J. Magn. Reson. 2006;180:93–104. doi: 10.1016/j.jmr.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 45.Millet OM, Loria JP, Kroenke CD, Pons M, Palmer AG. The static magnetic field dependence of chemical exchange linebroadening defines the NMR chemical shift time scale. J. Am. Chem. Soc. 2000;122:2867–2877. [Google Scholar]
- 46.Grey MJ, Wang C, Palmer AG., 3rd Disulfide bond isomerization in basic pancreatic trypsin inhibitor: multisite chemical exchange quantified by CPMG relaxation dispersion and chemical shift modeling. J. Am. Chem. Soc. 2003;125:14324–14335. doi: 10.1021/ja0367389. [DOI] [PubMed] [Google Scholar]
- 47.Trott O, Palmer AG., 3rd Theoretical study of R(1rho) rotating-frame and R2 free-precession relaxation in the presence of n-site chemical exchange. J. Magn. Reson. 2004;170:104–112. doi: 10.1016/j.jmr.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 48.Korzhnev DM, Kloiber K, Kanelis V, Tugarinov V, Kay LE. Probing slow dynamics in high molecular weight proteins by methyl-TROSY NMR spectroscopy: application to a 723-residue enzyme. J. Am. Chem. Soc. 2004;126:3964–3973. doi: 10.1021/ja039587i. [DOI] [PubMed] [Google Scholar]
- 49.Korzhnev DM, Neudecker P, Mittermaier A, Orekhov VY, Kay LE. Multiple-site exchange in proteins studied with a suite of six NMR relaxation dispersion experiments: an application to the folding of a Fyn SH3 domain mutant. J. Am. Chem. Soc. 2005;127:15602–15611. doi: 10.1021/ja054550e. [DOI] [PubMed] [Google Scholar]
- 50.Led JJ, Gesmar H, Abildgaard F. Applicability of magnetization transfer nuclear magnetic resonance to study chemical exchange reactions. Meth. Enzymol. 1989;176:311–329. doi: 10.1016/0076-6879(89)76017-1. [DOI] [PubMed] [Google Scholar]
- 51.Montelione GT, Wagner G. 2D chemical exchange NMR spectroscopy by proton-detected heteronuclear correlation. J. Am. Chem. Soc. 1989;111:3096–3098. [Google Scholar]
- 52.Wider G, Neri D, Wuthrich K. Studies of slow conformational equilibria in macromolecules by exchange of heteronuclear longitudinal 2-spin-order in a 2D difference correlation experiment. J. Biomol. NMR. 1991;1:93–98. [Google Scholar]
- 53.Sahu D, Clore GM, Iwahara J. TROSY-Based z-Exchange Spectroscopy: Application to the Determination of the Activation Energy for Intermolecular Protein Translocation between Specific Sites on Different DNA Molecules. J. Am. Chem. Soc. 2007 doi: 10.1021/ja074604f. [DOI] [PubMed] [Google Scholar]
- 54.Saupe A, Englert G. High-resolution nuclear magnetic resonance spectra of oriented molecules. Phys. Rev. Lett. 1963;11:462–464. [Google Scholar]
- 55.Sanders CR, Prosser RS. Bicelles: a model membrane system for all seasons? Structure with Folding and Design. 1998;6:1227–1234. doi: 10.1016/s0969-2126(98)00123-3. [DOI] [PubMed] [Google Scholar]
- 56.Tjandra N, Bax A. Direct measurement of distances and angles in biomolecules by NMR in dilute liquid crystalline medium. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 57.Clore GM, Starich MR, Gronenborn AM. Measurement of residual dipolar couplings of macromolecules aligned in the nematic phase of a colloidal suspension of rod-shaped viruses. J Am Chem Soc. 1998;120:10571–10572. [Google Scholar]
- 58.Hansen MR, Mueller L, Pardi A. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat Struct Biol. 1998;5:1065–1074. doi: 10.1038/4176. [DOI] [PubMed] [Google Scholar]
- 59.Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH. Nuclear magnetic dipole interactions in field-oriented proteins: information for structure determination in solution. Proc Natl Acad Sci U S A. 1995;92:9279–9283. doi: 10.1073/pnas.92.20.9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beger RD, Marathias VM, Volkman BF, Bolton PH. Determination of internuclear angles of DNA using paramagnetic-assisted magnetic alignment. J Magn Reson. 1998;135:256–259. doi: 10.1006/jmre.1998.1527. [DOI] [PubMed] [Google Scholar]
- 61.Prestegard JH, al-Hashimi HM, Tolman JR. NMR structures of biomolecules using field oriented media and residual dipolar couplings. Q Rev Biophys. 2000;33:371–424. doi: 10.1017/s0033583500003656. [DOI] [PubMed] [Google Scholar]
- 62.Jain NU, Tjioe E, Savidor A, Boulie J. Redox-dependent structural differences in putidaredoxin derived from homologous structure refinement via residual dipolar couplings. Biochemistry. 2005;44:9067–9078. doi: 10.1021/bi050152c. [DOI] [PubMed] [Google Scholar]
- 63.Freiburger LA, Baettig OM, Sprules T, Berghuis AM, Auclair K, Mittermaier AK. Competing allosteric mechanisms modulate substrate binding in a dimeric enzyme. Nat Struct Mol Biol. 2011;18:288–294. doi: 10.1038/nsmb.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wright GD, Ladak P. Overexpression and characterization of the chromosomal aminoglycoside 6′-N-acetyltransferase from Enterococcus faecium. Antimicrob Agents Chemother. 1997;41:956–960. doi: 10.1128/aac.41.5.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Freiburger LA, Auclair K, Mittermaier AK. Elucidating protein binding mechanisms by variable-c ITC. Chembiochem. 2009;10:2871–2873. doi: 10.1002/cbic.200900614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhuravleva A, Gierasch LM. Allosteric signal transmission in the nucleotide-binding domain of 70-kDa heat shock protein (Hsp70) molecular chaperones. Proc Natl Acad Sci U S A. 2011;108:6987–6992. doi: 10.1073/pnas.1014448108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Swain JF, Dinler G, Sivendran R, Montgomery DL, Stotz M, Gierasch LM. Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol Cell. 2007;26:27–39. doi: 10.1016/j.molcel.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vogel M, Mayer MP, Bukau B. Allosteric regulation of Hsp70 chaperones involves a conserved interdomain linker. J Biol Chem. 2006;281:38705–38711. doi: 10.1074/jbc.M609020200. [DOI] [PubMed] [Google Scholar]
- 69.Jiang J, Maes EG, Taylor AB, Wang L, Hinck AP, Lafer EM, Sousa R. Structural basis of J cochaperone binding and regulation of Hsp70. Molecular cell. 2007;28:422–433. doi: 10.1016/j.molcel.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bhattacharya A, Kurochkin AV, Yip GNB, Zhang Y, Bertelsen EB, Zuiderweg ERP. Allostery in Hsp70 chaperones is transduced by subdomain rotations. Journal Of Molecular Biology. 2009;388:475–490. doi: 10.1016/j.jmb.2009.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Selvaratnam R, Chowdhury S, VanSchouwen B, Melacini G. Mapping allostery through the covariance analysis of NMR chemical shifts. Proc Natl Acad Sci U S A. 2011;108:6133–6138. doi: 10.1073/pnas.1017311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gloerich M, Bos JL. Epac: defining a new mechanism for cAMP action. Annu Rev Pharmacol Toxicol. 50:355–375. doi: 10.1146/annurev.pharmtox.010909.105714. [DOI] [PubMed] [Google Scholar]
- 73.Lipchock JM, Loria JP. Nanometer propagation of millisecond motions in V-type allostery. Structure. 2010;18:1596–1607. doi: 10.1016/j.str.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Klem TJ, Davisson VJ. Imidazole Glycerol Phosphate Synthase - the Glutamine Amidotransferase in Histidine Biosynthesis. Biochemistry. 1993;32:5177–5186. doi: 10.1021/bi00070a029. [DOI] [PubMed] [Google Scholar]
- 75.Klem TJ, Davisson VJ. Imidazole glycerol phosphate synthase: The glutamine amidotransferase in histidine biosynthesis. Biochemistry. 1993;32:5177–5186. doi: 10.1021/bi00070a029. [DOI] [PubMed] [Google Scholar]
- 76.Chaudhuri BN, Lange SC, Myers RS, Chittur SV, Davisson VJ, Smith JL. Crystal structure of imidazole glycerol phosphate synthase: a tunnel through a (beta/alpha)8 barrel joins two active sites. Structure. 2001;9:987–997. [PubMed] [Google Scholar]
- 77.Myers RS, Amaro RE, Luthey-Schulten ZA, Davisson VJ. Reaction coupling through interdomain contacts in imidazole glycerol phosphate synthase. Biochemistry. 2005;44:11974–11985. doi: 10.1021/bi050706b. [DOI] [PubMed] [Google Scholar]
- 78.Myers RS, Jensen JR, Deras IL, Smith JL, Davisson VJ. Substrate-induced changes in the ammonia channel for imidazole glycerol phosphate synthase. Biochemistry. 2003;42:7013–7022. doi: 10.1021/bi034314l. [DOI] [PubMed] [Google Scholar]
- 79.Douangamath A, Walker M, Beismann-Driemeyer S, Vega-Fernandez MC, Sterner R, Wilmanns M. Structural evidence for ammonia tunneling across the (beta alpha)(8) barrel of the imidazole glycerol phosphate synthase bienzyme complex. Structure. 2002;10:185–193. doi: 10.1016/s0969-2126(02)00702-5. [DOI] [PubMed] [Google Scholar]
- 80.Piper DE, Batchelor AH, Chang CP, Cleary ML, Wolberger C. Structure of a HoxB1-Pbx1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell. 1999;96:587–597. doi: 10.1016/s0092-8674(00)80662-5. [DOI] [PubMed] [Google Scholar]
- 81.Green NC, Rambaldi I, Teakles J, Featherstone MS. A conserved C-terminal domain in PBX increases DNA binding by the PBX homeodomain and is not a primary site of contact for the YPWM motif of HOXA1. The Journal of biological chemistry. 1998;273:13273–13279. doi: 10.1074/jbc.273.21.13273. [DOI] [PubMed] [Google Scholar]
- 82.Farber PJ, Mittermaier A. Concerted dynamics link allosteric sites in the PBX homeodomain. Journal Of Molecular Biology. 2010;405:819–830. doi: 10.1016/j.jmb.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 83.Passner J, Schultz S, Steitz T. Modeling the cAMP-induced Allosteric Transition Using the Crystal Structure of CAP-cAMP at 2.1 Å Resolution. Journal Of Molecular Biology. 2000 doi: 10.1006/jmbi.2000.4231. [DOI] [PubMed] [Google Scholar]
- 84.Tzeng SR, Kalodimos CG. Dynamic activation of an allosteric regulatory protein. Nature. 2009;462:368–372. doi: 10.1038/nature08560. [DOI] [PubMed] [Google Scholar]
- 85.Zhang M, Wang W. Organization of signaling complexes by PDZ-domain scaffold proteins. Accounts of Chemical Research. 2003;36:530–538. doi: 10.1021/ar020210b. [DOI] [PubMed] [Google Scholar]
- 86.Petit CM, Zhang J, Sapienza PJ, Fuentes EJ, Lee AL. Hidden dynamic allostery in a PDZ domain. Proc Natl Acad Sci U S A. 2009;106:18249–18254. doi: 10.1073/pnas.0904492106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Johnson DA, Akamine P, Radzio-Andzelm E, Madhusudan M, Taylor SS. Dynamics of cAMP-dependent protein kinase. Chem Rev. 2001;101:2243–2270. doi: 10.1021/cr000226k. [DOI] [PubMed] [Google Scholar]
- 88.Masterson LR, Shi L, Metcalfe E, Gao J, Taylor SS, Veglia G. Dynamically committed, uncommitted, and quenched states encoded in protein kinase A revealed by NMR spectroscopy. Proc Natl Acad Sci U S A. 108:6969–6974. doi: 10.1073/pnas.1102701108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bhattacharya A, Kurochkin AV, Yip GN, Zhang Y, Bertelsen EB, Zuiderweg ER. Allostery in Hsp70 chaperones is transduced by subdomain rotations. Journal Of Molecular Biology. 2009;388:475–490. doi: 10.1016/j.jmb.2009.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gerhart JC, Schachman HK. Distinct subunits for the regulation and catalytic activity of aspartate transcarbamylase. Biochemistry. 1965;4:1054–1062. doi: 10.1021/bi00882a012. [DOI] [PubMed] [Google Scholar]
- 91.Newell JO, Markby DW, Schachman HK. Cooperative binding of the bisubstrate analog N-(phosphonacetyl)-L-aspartate to aspartate transcarbamoylase and the heterotropic effects of ATP and CTP. J Biol Chem. 1989;264:2476–2481. [PubMed] [Google Scholar]
- 92.Fetler L, Tauc P, Herve G, Moody MF, Vachette P. X-ray scattering titration of the quaternary structure transition of aspartate transcarbamylase with a bisubstrate analogue: influence of nucleotide effectors. Journal Of Molecular Biology. 1995;251:243–255. doi: 10.1006/jmbi.1995.0432. [DOI] [PubMed] [Google Scholar]
- 93.Herve G, Moody MF, Tauc P, Vachette P, Jones PT. Quaternary structure changes in aspartate transcarbamylase studied by X-ray solution scattering. Signal transmission following effector binding. Journal Of Molecular Biology. 1985;185:189–199. doi: 10.1016/0022-2836(85)90190-1. [DOI] [PubMed] [Google Scholar]
- 94.Howlett GJ, Blackburn MN, Compton JG, Schachman HK. Allosteric regulation of aspartate transcarbamoylase. Analysis of the structural and functional behavior in terms of a two-state model. Biochemistry. 1977;16:5091–5100. doi: 10.1021/bi00642a023. [DOI] [PubMed] [Google Scholar]
- 95.Velyvis A, Yang YR, Schachman HK, Kay LE. A solution NMR study showing that active site ligands and nucleotides directly perturb the allosteric equilibrium in aspartate transcarbamoylase. Proc Natl Acad Sci U S A. 2007;104:8815–8820. doi: 10.1073/pnas.0703347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Werner WE, Schachman HK. Analysis of the ligand-promoted global conformational change in aspartate transcarbamoylase. Evidence for a two-state transition from boundary spreading in sedimentation velocity experiments. Journal Of Molecular Biology. 1989;206:221–230. doi: 10.1016/0022-2836(89)90535-4. [DOI] [PubMed] [Google Scholar]
- 97.Velyvis A, Schachman HK, Kay LE. Application of methyl-TROSY NMR to test allosteric models describing effects of nucleotide binding to aspartate transcarbamoylase. Journal Of Molecular Biology. 2009;387:540–547. doi: 10.1016/j.jmb.2009.01.066. [DOI] [PubMed] [Google Scholar]
- 98.Sprangers R, Kay LE. Probing supramolecular structure from measurement of methyl (1)H-(13)C residual dipolar couplings. J Am Chem Soc. 2007;129:12668–12669. doi: 10.1021/ja075846i. [DOI] [PubMed] [Google Scholar]