Abstract
Next-generation sequencing technologies provide unprecedented power to explore the repertoire of genes and their alternative splice variants, collectively defining the transcriptome of a species in great detail. However, assembling the short reads into full-length gene and transcript models presents significant computational challenges. We review current algorithms for assembling transcripts and genes from next-generation sequencing reads aligned to a reference genome, and lay out areas for future improvements.
Index Terms: Algorithms, biology and genetics, computer applications, medicine and science
1 Introduction
Identifying the genes being expressed by a cell is a critical step in studying a wide range of biological questions, ranging from what defines a cell’s type to what makes it turn cancerous. Despite tremendous progress in the dozen years since the human genome was first published [1], [2], our knowledge of transcriptomes remains incomplete, even for humans, owing to the high cost of generating the cDNA resources needed to characterize all genes and their possible isoforms in all cell types. Next-generation sequencing (NGS) technologies can now produce billions of short reads in a matter of days at a cost of pennies per megabase. In particular, RNA-seq, a technique in which RNA from a collection of cells is sampled and then sequenced, has become ubiquitous as a means to survey the cellular transcriptome [3]. This technology has dramatically expanded our view of the gene landscape, revealing thousands of novel splice variants and a wealth of previously unknown long, noncoding RNA genes. Analysis of RNA-seq data requires that the reads themselves be assembled together to reconstruct the transcripts from which they came, and this requirement has driven the development of sophisticated new computational methods.
Assembling reads into transcripts involves identifying overlaps between the reads, establishing their precise order, and orientation, and then connecting all reads so as to satisfy these relationships, some of which may be ambiguous. When a high-quality reference genome sequence is available, as it is for human, mouse, fly, and many other model organisms, assembly methods that first map the reads to the genome and then use that mapping to infer transcript structure [4], [5], [6], [7] are by far the most accurate. If the genome has not been sequenced or is available only as a highly fragmented draft assembly, de novo assembly methods that detect overlaps based on local similarities among read sequences must be employed instead. De novo transcriptome assemblers include Trinity [8], Oases [9], and TransABySS [10]; see Martin and Wang [11] for a review. In this perspective, we focus on the algorithmic challenges of genome-guided methods.
Perhaps the most fundamental challenge for reconstructing transcripts from NGS data is fragmentation, an unavoidable consequence of the fact that the reads are much shorter than the transcripts. A typical RNA-sequencing run today produces 50–200 million short read (50–250 bp) from a single sample, usually in pairs where the reads come from opposite ends of a short fragment. These reads must be aligned to the genome, and then pieced together to create gene and transcript models. Due to repetitive sequences in the genome, many reads cannot be placed unambiguously, as happens for example in a multigene family where two or more copies of the gene are nearly identical. Further complicating matters, even when a read can be aligned precisely, it may originate from any of several transcripts at the same gene locus. More than 90 percent of human genes have multiple isoforms due to the use of alternative splicing, alternative transcription initiation, or alternative transcription termination sites [12], [13], and high levels of alternative splicing have similarly been reported for many other eukaryotes. The number of transcripts per gene varies widely, with some genes potentially expressing thousands of isoforms [14]. And while the number of annotations in gene databases grows each year, there is no agreed-upon estimate yet for the true number of transcripts encoded in the human genome, let alone a complete catalog of these variants.
Furthermore, unlike whole-genome sequencing data where read coverage levels along the genome are relatively stable and statistically well characterized by a Poisson distribution, in RNA-seq data each gene may have a different expression level, with some genes represented by thousands of reads and others by just a few. Thus, every gene essentially poses a different transcript assembly problem, where the goal is to assemble all expressed isoforms, and then count the reads deriving from each isoform. Read coverage tends to be highly nonuniform even within a gene, due to biases introduced by the various steps during the library preparation, sequencing, and mapping procedures [15], [16]. Artifacts introduced by the alignment algorithm can confound the assembly by creating spurious splice junctions and exon boundaries, which in turn will lead to additional candidate gene variants. Transcript assembly programs must also be computationally efficient to process the vast amounts of data in an RNA-seq experiment. Several genome-guided transcript assembly algorithms have emerged over the past few years that address all of these challenges, albeit in different ways. Below, we give an overview of the basic design considerations and algorithmic techniques they employ, and lay out areas for further improvement.
2 Overview of The RNA-seq Data Analysis Process
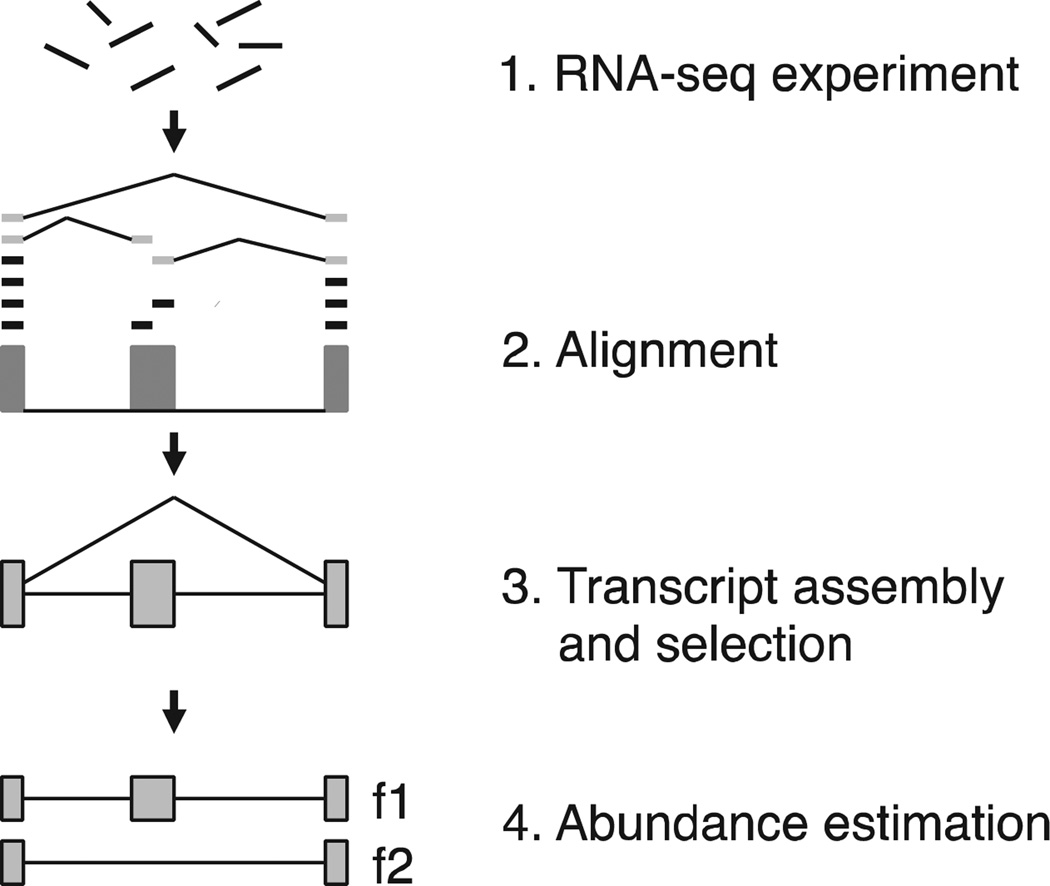
A typical transcriptome analysis consists of several stages (see Fig. 1). In an RNA-seq experiment, the RNA population in the cell is sampled, then fragmented and converted into a library of cDNAs, which are (sometimes) amplified and then sequenced at one or both ends. A single RNA-seq experiment produces tens to hundreds of millions of such reads, either single-end or paired-end. In stage 2, the reads are mapped to the genome using a fast alignment program that allows for introns and sequencing errors. Short read mapping programs such as Tophat [17], [18], MapSplice [19], and STAR [20] use compressed representations of the genome to quickly locate exact matches, then extend these matches to longer gapped alignments. More recently, tools such as TrueSight [21] and OLego [22] have incorporated splice site scoring schemes in an effort to increase the accuracy of some alignments. In stage 3, read alignments are assembled into gene and transcript models; note that this type of assembly is algorithmically much simpler than de novo assembly. In the fourth and last stage, reads are assigned to individual transcripts and genes to quantify their expression levels, which can later be compared between different samples to identify genes that are differentially expressed. Reads with multiple matches on the genome or reads that can be attributed to different isoforms of a same gene, significantly complicate the quantitation problem. Some transcript assembly programs generate the transcripts and quantify their abundance in one step, while others separate the two stages. Other approaches use statistical methods and models of fragment distributions to estimate transcript abundance levels, which can be done separately from the assembly itself; these methods are not reviewed here. A useful current listing of computational tools for RNA-seq data analysis can be found at http://www.rna-seqblog.com, and many of these tools can be run directly using the Galaxy system [23].
Fig. 1.
Overview of RNA-seq analysis. Reads produced by an RNA-seq experiment are aligned to the genome, then clustered into a graph structure that is traversed to recover all possible isoforms at one locus. Lastly, a subset of transcripts is selected and their abundance quantified from the input reads.
3 Algorithmic Techniques in Transcript Assembly
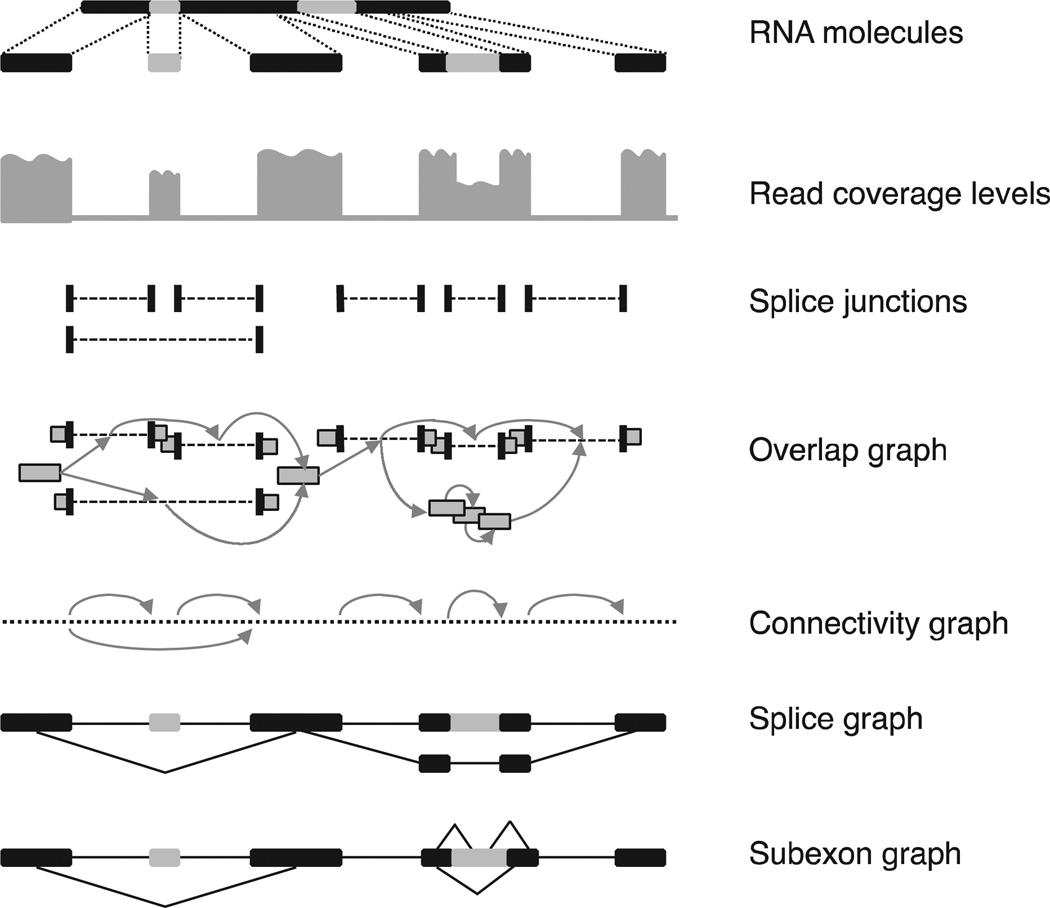
All current approaches for genome-based transcript assembly start by clustering overlapping reads from each locus, then building a graph that captures all possible isoforms, and traversing the graph to resolve individual isoforms (see Fig. 2). Since many such graphs encode a very large number of theoretically possible splice variants, many of them are artificial combinations of exons and introns, a crucial step is selecting a subset of transcripts that are the most likely to be represented in the RNA-seq sample. Next, we describe some of the representative data structures and transcript assembly and selection techniques.
Fig. 2.
Overview of data structures employed by reference-based transcript assembly. (Top) RNA-seq molecules are sequenced and reads are aligned to the reference genome. Light-colored segments are alternatively spliced. Spliced reads define the introns of a gene, whereas exons are derived from both unspliced and spliced reads. Transcripts are assembled from the reads using various algorithmic techniques. An overlap graph has a node for each read, and two reads are connected by an edge iff they are compatible (i.e., they have the same splicing patterns along the overlap segment). A connectivity graph connects any two consecutive bases on the chromosome, as well as the endpoints of introns. For simplicity, only spliced edges are shown, with arrows. A splice graph has exons as nodes, connected by introns (edges); splice variants can then be read from the graph as maximal paths. As a variation, a subexon graph connects gene segments if they are adjacent on the genome as part of the same exon, or are connected via a spliced read.
3.1 Transcript Representation and Enumeration
3.1.1 Overlap graph
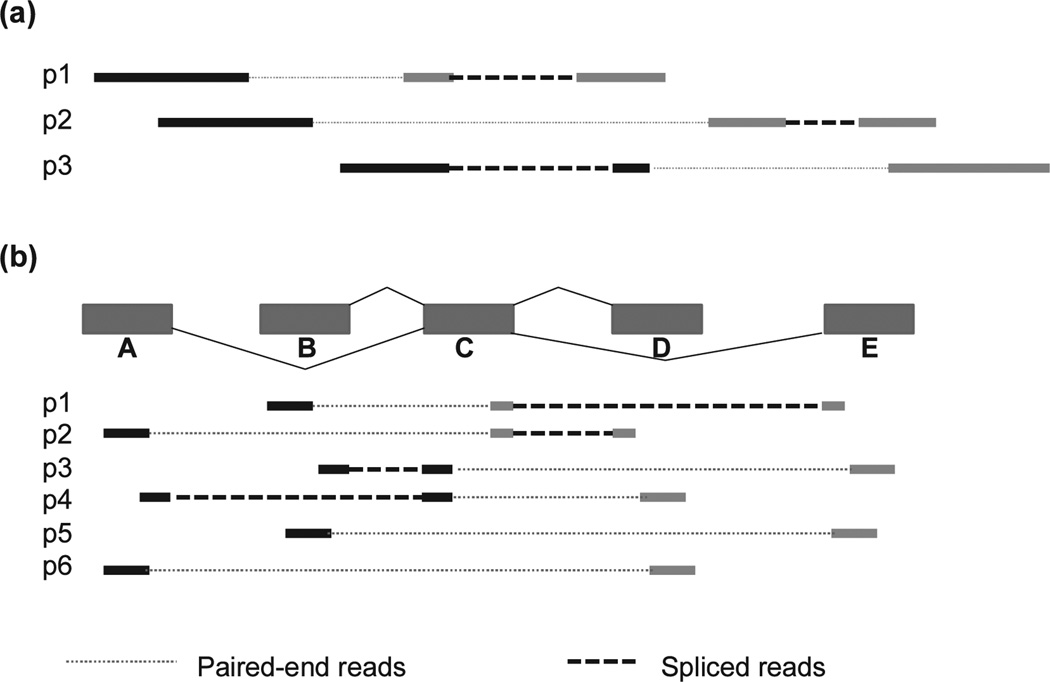
An overlap graph compactly represents the order of reads along putative transcripts based on each read location and its alignment pattern. Each read is a node in the graph, and two nodes are connected by an edge if the two reads overlap and have compatible alignments, meaning that they share the same splicing patterns along the overlap segment. For single-end reads, this condition forms a partial order. However, for paired-end reads the relationship is not always transitive, leading to reads whose placements are “uncertain” (see Fig. 3a). Consequently, overlap graph-based programs tend to ignore “uncertain” reads. To simplify the problem, reads contained within others are also ignored. The overlap graph has a directed acyclic (DAG) structure that can be traversed to produce paths that cover all qualifying reads, each representing a putative transcript. The representative program for this class of methods is Cufflinks [5], which was also the first widely used transcriptome assembler. Cufflinks uses an efficient polynomial time partitioning algorithm to select the minimum number of transcripts (isoforms) that can explain all the reads in the graph. It infers both the exons and the full transcripts in one sweep. Although the minimum number of isoforms is intuitively appealing as an explanation for the data, the true set of isoforms may sometimes contain more than the minimum number, in which cases Cufflinks will miss some variants.
Fig. 3.
Design choices implications for transcript assembly: (a) filtering out “uncertain” reads and (b) “unfeasible” transcripts. (Adapted from Li et al. [7] with permission from Mary Ann Liebert, Inc.) In (a), removing any of the three read pairs will cause some introns and splice variants to be missed. Read pairs (p1 and p2) and (p2 and p3) are compatible, but p1 and p3 could not have come from the same transcript. In (b), four possible combinations of segments are encoded in the graph: ACD, ACE, BCD, and BCE. Of these, ACE and BCD are unfeasible, i.e., cannot be assembled from the mapped paired-end reads.
3.1.2 Connectivity Graph
A connectivity graph connects any two bases that are next to each other on the genome or are connected via a spliced read. This is also a directed acyclic graph, and it too can be traversed to build a set of transcripts. Several transcript assembly programs use the connectivity graph, but they differ in how they extract exons and transcripts from the connectivity graph. In Scripture [6], for instance, a connectivity graph contains all bases in a single chromosome, with edges between any two consecutive positions in the genome and between the two endpoints of an intron. Scripture then uses a segmentation approach that takes into account the read coverage levels along the genome to determine significant paths. More specifically, it scans paths in the connectivity graph within fixed-size windows and compares the read coverage levels to those derived from a genome-wide Poisson distribution, retaining those in the top 5 percent significance interval. Significant paths are then merged into a splice graph (described below), from which transcripts can be enumerated. Scripture retains all transcripts enumerated from the graph, and hence it reconstructs the full set of true transcripts it encodes, but it may also produce many false variants, as well as unfeasible combinations that cannot be fully accounted for by the input reads (see Fig. 3b). A complementary approach is taken by IsoLasso [7]. IsoLasso builds a connectivity graph that contains only those genome bases that are covered by read alignments. It then traverses the graph to build all possible transcripts for a gene, which are later reduced using a quadratic program, described below.
3.1.3 Splice Graph and Subexon Graph
A splice graph is a DAG in which nodes represent exons, edges are introns connecting the exons, and transcripts can be enumerated as maximal paths. These graphs offer an intuitive representation of a gene [24], [25], and have been previously used for transcript reconstruction from conventional (Sanger) cDNA data. One common variation is the subexon graph (see Fig. 2), where each exon segment between consecutive splice sites is a node, and two nodes are connected by edges if they are adjoined as part of the same exon or are connected by a spliced read. Because RNA-seq reads are typically too short to cover an entire exon, exons in the graph need to be first assembled from reads. Scripture, for instance, uses the segmentation procedure described earlier to determine enriched paths representing groups of exons or exon fragments. Other programs, such as SLIDE and SpliceGrapher [4], [26], use an existing set of gene annotations to build a subexon graph, augmenting it with new splicing events from the RNA-seq data. Both splice graphs and subexon graphs may encode thousands if not millions of potential splice variants, most of which are spurious combinations of exons and introns. Therefore, once the graph is constructed, a critical next step is to select a subset of likely transcripts. A variety of transcript selection strategies have been devised, including greedy methods, dynamic programming, linear programming, and LASSO or expectation maximization (EM) algorithms [4], [7], [27], some of which are described below.
3.2 Transcript Selection via Numerical Optimization
Intuitively, read coverage levels along the genome offer clues into the structure of genes, with exons represented by peaks and introns represented by valleys, and therefore can guide algorithms on how to reconstruct the set of transcript and quantify their abundance from reads. One class of algorithmic approaches has focused on simultaneously selecting a subset of candidate transcripts and estimating their abundance, using quadratic programming (QP) or expectation maximization techniques.
3.2.1 Quadratic Programming
Given a set of transcripts generated using one of the methods described previously, the quadratic programming approach assigns each possible transcript an (unknown) abundance level, typically expressed as average reads per base, and finds the combination of transcripts that best explains the observed read coverage levels, by minimizing the total estimation error. More specifically, starting with a collection of T transcripts, each with abundance xj, j = 1 … T, the problem of finding a subset of transcripts that most accurately explain the observed coverage levels along the gene can be formulated as a quadratic program:
where ri, li are the number of reads aligning to subexon i and the length of i; and aji is an indicator variable that, in the simplest case, has value 1 iff segment i is present in transcript j, or otherwise can incorporate more complex information about the probability of reads in segment i to be sampled from isoform j [4]. The accuracy of the solution depends critically on the completeness of the initial set of transcripts, hence the set has to be large enough to encompass nearly all of the true transcripts. However, too large a set of initial candidates may lead to solutions with a large number of transcripts with values close to 0, which is biologically implausible. Therefore, a penalty term (λ ∑j xj) is typically added to the objective function to minimize the number of isoforms in the solution. A representative program for this class is IsoLasso [7], which simultaneously selects a set of transcripts from among those enumerated from the connectivity graph, and estimates their abundance levels. A similar approach is taken by SLIDE [4], which however starts by enumerating transcripts from the subexon graph constructed from a known set of gene annotations.
3.2.2 Expectation Maximization
The abundance of each isoform can be determined in principle from the reads assigned to it. With short RNA-seq reads, however, their assignments can be ambiguous because they might be mapped to multiple genes at different genomic loci, or to multiple isoforms of a same gene. For this reason, each read is assigned a probability of belonging to any of the isoforms, a value that depends on the current abundance estimates for isoforms and, in some formulations, on the read alignment scores [27]. The EM algorithm, as used by iReckon and Cufflinks, then attempts to determine the maximum-likelihood expression levels for all isoforms, by iteratively assigning reads to isoforms according to the probabilities above, and re-estimating the abundances and the fitness of the solution. This iterative process can start by assigning reads in equal numbers to all isoforms of a given gene, or by distributing reads proportionally to the number of uniquely mapped reads for each isoform, or by other heuristics. The process evaluates the likelihood of the total set of transcripts and their expression levels at each iteration, and stops when the change in likelihood is smaller than a predefined cutoff. As with quadratic programming methods, EM methods may suffer from overfitting. Therefore, programs incorporate additional penalties and adopt criteria to discourage low-expressed isoforms and to simplify the system, for instance by splitting the isoforms into smaller independent groups to speed up calculations, as implemented in iReckon [27]. Unlike with simultaneous methods, the set of transcripts in Cufflinks is predetermined, and the EM procedure is used only to estimate the expression levels of the transcripts previously selected by the minimum partition procedure.
Lastly, all of these approaches can take advantage of the paired-end read information such as distance and orientation, to connect disparate portions of the genes and reduce the number of transcripts by eliminating unlikely or inconsistent isoforms. More specifically, all programs use paired reads in the graph construction stage, and overlap graph-based programs such as Cufflinks use the paired-read alignments to determine read pair compatibilities and to filter out “uncertain” pairs (see Fig. 3a). Furthermore, to allocate paired reads to isoforms during expression level estimations, programs may take into account their compatibility of both the exon-intron structure and the underlying fragment length distribution (e.g., reads placed too far apart can be penalized). Lastly, Scripture uses paired-end reads to connect splice graphs representing portions of the genes, resulting in more contiguous gene structures.
4 Algorithm Design Considerations
In addition to the choice of data structure and algorithmic technique, there are several other design considerations that programs must take into account (see Table 1). The first is condensing the very large set of input reads into a relatively small number of transcripts for each gene. Early approaches that have gained wide popularity, such as Cufflinks and later IsoLasso, take the parsimonious approach by seeking the minimum number of isoforms that can explain the input reads. This strategy based on Occam’s Razor is mathematically appealing, but it may miss some transcripts, for at least two reasons. First, it might be that the true set of isoforms is larger than the minimal set. And second, the minimal set is not unique; for example, if the minimal set has four splice variants at a given locus, there could be more than one set of four variants that explain the data. At the opposite end of the spectrum, Scripture reports all possible isoforms that can be formed by connecting gene segments, which can generate very large numbers of isoforms, many of them likely spurious. Other approaches use a “best fit” strategy, minimizing a mathematically defined cost function implemented as a quadratic program or expectation maximization system. Even for this class of methods, however, the large number of candidate transcripts for a gene can lead to large systems that are difficult to solve and are prone to overfitting. In the end, in practice most algorithms and annotation pipelines apply filters based on the abundance and structural properties of the predicted transcripts, such as length, number of exons and positioning in the context of other genes, to reduce the number of transcripts to a relative small subset, typically capturing the major isoforms of the gene. Overall, the wide variability in the numbers of transcripts produced by all these methods, and the lack of a true gold standard for human gene annotation, makes it very difficult to determine which program is more accurate for any given RNA-seq data set.
TABLE 1.
Design Characteristics of Transcript Assembly Programs
| Program | Condensing strategy |
Read com pleteness |
Transcript feasibility |
Transcript repre sentation |
Transcript selection |
Intronic reads con text |
Novel gene ends |
|---|---|---|---|---|---|---|---|
| Cufflinks | Parsimony | No | Yes | Overlap graph | Minimum partition | Exon | Partial |
| IsoLasso | Best fit | No | Yes | Connectivity graph | QP | Not available | Partial |
| Scripture | Exhaustive | Yes | No | Connectivity graph, splice graph | None | Genome | Partial |
| iReckon | Best fit | Yes | No | Splice graph | EM | Gene | No |
| SLIDE | Best fit | No | No | Subexon graoh | QP | Ignore | No |
| SpliceGrapher | None | Yes | No | Splice graph | None | Ignore | No |
QP - quadratic programming; EM - expectation maximization.
Read completeness, which refers to how many of the input reads are contained in the solution, is another important algorithm constraint. Scripture, iReckon, and SpliceGrapher produce assemblies that explain all the reads, but Cufflinks ignores “uncertain” reads, and as a result may miss some splice variations (see Fig. 3a). Similarly, transcript feasibility [7] refers to whether all transcripts produced by the method can be explained by the input reads. All the isoforms produced by Cufflinks and IsoLasso can be fully reconstructed from the input reads, whereas some combinations of exons reported by Scripture and SLIDE cannot be explained by the read data alone (see Fig. 3b). While no method can reconstruct the set of transcripts with 100 percent accuracy, methods that report only “feasible” transcripts generally produce a more conservative and more precise set of annotations, albeit at the expense of occasionally missing variants with low expression levels.
An important practical choice that programs must make is the treatment of intronic reads. A typical RNA-seq experiment will sample RNA that has not been completely processed and that therefore still contains introns [28]. Incompletely spliced RNA, which can be extremely difficult to remove completely in the lab, greatly complicates exon prediction and confounds the detection of true intron retention events. To distinguish the true mRNA signal from transcriptional “noise,” some methods largely ignore intronic reads when inferring exons. Others judge the validity of intronic reads by comparing them to a background distribution that they estimate either locally (at the intron or gene level) or genome wide (see Table 1). For instance, Cufflinks compares the level of intronic reads to the coverage of the flanking exons to determine whether the intronic reads represent true intron retention. iReckon explicitly considers the unprocessed primary transcript in its list of transcript candidates, and includes the intron with the strongest read support, if that intron is significantly enriched over the gene’s background. Scripture uses a segmentation approach to define exons, including those containing retained introns, by comparing the read coverage against the genome-wide distribution. Since the number and preponderance of intron retention events in the human genome and in individual samples are unknown, the performance of any of these approaches is difficult to assess without extensive experimental validation.
Apart from alternative splicing, alternative polyadenylation and alternative promoter usage also contribute to transcriptome diversity. More than 50 percent of human genes have multiple polyadenylation sites that create different 3’ ends of the gene [29]. The recent ENCODE project identified over 120,000 transcription start sites (for approximately 25,000 genes) and a similar number of 3’ transcription termination sites across 15 cell lines [30]. Most transcript assembly programs can identify events with distinct splicing patterns of the alternative last or first exons (see Table 1). However, detection of tandem polyadenylation events, where the gene uses different termination sites within the same terminal exon, or transcript ends that occur at internal exons, is particularly difficult and not yet systematically addressed by existing methods.
5 CONCLUSIONS
Despite the fact that the gene annotation problem has been in computational biologists’ cross hairs for more than two decades, and despite the recent influx of RNA-seq data, we still have many challenges to overcome before we can precisely identify and quantify all the transcripts expressed in a cell. With new RNA-seq surveys emerging, and with different algorithms producing vastly different accounts of the number and structure of transcripts, gene annotations are still in flux, even for the intensely studied human genome. New and more accurate transcript assembly tools are needed to further refine the gene and transcript models, in view of the expanding biological knowledge about gene structure and variation and advances in sequencing technologies. Future algorithms may do a better job at distinguishing transcriptional “noise” from intronic retention, or at removing read mapping artifacts. Additional work too is needed on methods to more accurately assess the number of transcripts and the expression level of each one. Some help may come from sequencing technology: longer reads will make it easier to link exons together in the same isoform, and eventually the assembly problem may become entirely an alignment problem, if reads can capture entire transcripts. Over time, publicly available catalogs of carefully validated splicing variations across human cell types will provide a gold standard that can be used to measure the effectiveness of transcript assemblers, and at the same time bring us closer to the goal of fully characterizing the human transcriptome.
Acknowledgments
The authors would like to thank Stefan Canzar for his helpful comments on the manuscript. This work was supported in part by the US National Science Foundation (NSF) Award ABI-1159078 to LF and by NIH grants R01-HG006102 and R01-HG006677 to SLS.
Biographies

Liliana D. Florea received the PhD degree in computer science and engineering from the Pennsylvania State University in 2000, and the MSc degree in 1998. From 2000 to 2004, she was a senior scientist at Celera Genomics and Applied Biosystems. She subsequently held faculty positions at the George Washington University and the University of Maryland. As an assistant professor with the McKusick-Nathans Institute of Genetic Medicine at the Johns Hopkins University, she currently develops algorithms for analyzing next-generation sequencing data to determine genes and their alternative splicing variations.

Steven L. Salzberg received the PhD degree in computer science from Harvard University in 1989. From 1989 to 1997, he was a faculty member of computer science at Johns Hopkins University. In 1997, he joined The Institute for Genomic Research, where he was director of Bioinformatics from 1998 to 2005. From 2005 to 2011, he was the Horvitz professor of computer science and the director of the Center for Bioinformatics and Computational Biology at the University of Maryland, College Park. Since 2011, he has been a professor of medicine and biostatistics in the McKusick-Nathans Institute of Genetic Medicine at Johns Hopkins University, where he is the director of the Center for Computational Biology. He is among the most highly cited scientists in computational biology, with more than 200 scientific papers and an H-index of 102. He is a fellow of the American Association for the Advancement of Science and the International Society for Computational Biology.
Footnotes
⊳For more information on this or any other computing topic, please visit our Digital Library at www.computer.org/publications/dlib.
References
- 1.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The Sequence of the Human Genome. Science. 2001;291(5507):1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 2.The Int’l Human Genome Sequencing Consortium. Initial Sequencing and Analysis of the Human Genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Salzberg SL. Recent Advances in RNA Sequence Analysis. F1000 Biology Reports. 2010;2:64. doi: 10.3410/B2-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li JJ, Jiang CR, Brown JB, Huang H, Bickel PJ. Sparse Linear Modeling of Next-Generation mRNA Sequencing (RNASeq) Data for Isoform Discovery and Abundance Estimation. Proc. Nat’l Academy of Sciences USA. 2001;108(50):19867–19872. doi: 10.1073/pnas.1113972108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript Assembly and Quantification by RNA-Seq Reveals Unannotated Transcripts and Isoform Switching During Cell Differentiation. Nature Biotechnology. 2009;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, Rinn JL, Lander ES, Regev A. Ab Initio Reconstruction of Cell Type-Specific Transcriptomes in Mouse Reveals the Conserved Multi-Exonic Structure of LincRNAs. Nature Biotechnology. 2010;28(5):503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W, Feng J, Jiang T. IsoLasso: A LASSO Regression Approach to RNA-Seq Based Transcriptome Assembly. J. Computational Biology. 2011;18(11):1693–1707. doi: 10.1089/cmb.2011.0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. Full-Length Transcriptome Assembly from RNA-Seq Data without a Reference Genome. Nature Biotechnology. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schulz MH, Zerbino DR, Vingron M, Birney E. Oases: Robust De Novo RNA-Seq Assembly across the Dynamic Range of Expression Levels. Bioinformatics. 2012;28(8):1086–1092. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robertson G, Schein J, Chiu R, Corbett R, Field M, Jackman SD, Mungall K, Lee S, Okada HM, Qian JQ, Griffith M, Raymond A, Thiessen N, Cezard T, Butterfield YS, Newsome R, Chan SK, She R, Varhol R, Kamoh B, Prabhu AL, Tam A, Zhao YJ, Moore RA, Hirst M, Marra MA, Jones SJM, Hoodless PA, Birol I. De Novo Assembly and Analysis of RNA-Seq Data. Nature Methods. 2010;7(11):909–U962. doi: 10.1038/nmeth.1517. [DOI] [PubMed] [Google Scholar]
- 11.Martin JA, Wang Z. Next-Generation Transcriptome Assembly. Nature Rev. Genetics. 2011;12(10):671–682. doi: 10.1038/nrg3068. [DOI] [PubMed] [Google Scholar]
- 12.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative Isoform Regulation in Human Tissue Transcriptomes. Nature. 2008;456(7221):470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe, B.J BJ. Deep Surveying of Alternative Splicing Complexity in the Human Transcriptome by High-Throughput Sequencing. Nature Genetics. 2008;40(12):1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 14.Graveley BR. Alternative Splicing: Increasing Diversity in the Proteomic World. Trends in Genetics. 2001;17(2):100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 15.Hansen KD, Brenner SE, Dudoit S. Biases in Illumina Transcriptome Sequencing Caused by Random Hexamer Priming. Nucleic Acids Research. 2010;38(12):e131. doi: 10.1093/nar/gkq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z, Gerstein,M M, Snyder M. RNA-Seq: a Revolutionary Tool for Transcriptomics. Nature Rev. Genetics. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering Splice Junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: Accurate Alignment of Transcriptomes in the Presence of Insertions, Deletions and Gene Fusions. Genome Biology. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, He X, Mieczkowski P, Grimm SA, Perou CM, MacLeod JN, Chiang DY, Prins JF, Liu J. MapSplice: Accurate Mapping of RNA-Seq Reads for Splice Junction Discovery. Nucleic Acids Research. 2010;38(18):e178. doi: 10.1093/nar/gkq622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics. 2012;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Li-Byarlay H, Burns P, Borodovsky M, Robinson GE, Ma J. TrueSight: A New Algorithm for Splice Junction Detection Using RNA-Seq. Nucleic Acids Research. 2013;41(4):e51. doi: 10.1093/nar/gks1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu J, Anczukow O, Krainer AR, Zhang MQ, Zhang C. OLego: Fast and Sensitive Mapping of Spliced mRNA-Seq Reads using Small Seeds. Nucleic Acids Research. 2013;41(10):5149–5163. doi: 10.1093/nar/gkt216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goecks J, Nekrutenko A, Taylor J. Galaxy: A Comprehensive Approach for Supporting Accessible, Reproducible, and Transparent Computational Research in the Life Sciences. Genome Biology. 2010;11(8):R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heber S, Alekseyev M, Sze SH, Tang H, Pevzner PA. Splicing Graphs and the EST Assembly Problem. Bioinformatics. 2002;18(Suppl. 1):S181–S188. doi: 10.1093/bioinformatics/18.suppl_1.s181. [DOI] [PubMed] [Google Scholar]
- 25.Florea L, Di Francesco V, Miller J, Turner R, Yao A, Harris M, Walenz B, Mobarry C, Merkulov GV, Charlab R, Dew I, Deng Z, Istrail S, Li P, Sutton G. Gene and Alternative Splicing Annotation with AIR. Genome Research. 2005;15(1):54–66. doi: 10.1101/gr.2889405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogers MF, Thomas J, Reddy AS, Ben-Hur A. Splice-Grapher: Detecting Patterns of Alternative Splicing from RNA-Seq Data in the Context of Gene Models and EST Data. Genome Biology. 2012;13(1):R4. doi: 10.1186/gb-2012-13-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mezlini AM, Smith EJ, Fiume M, Buske O, Savich GL, Shah S, Aparicio S, Chiang DY, Goldenberg, A A, Brudno M. iReckon: Simultaneous Isoform Discovery and Abundance Estimation from RNA-Seq Data. Genome Research. 2013;23(3):519–529. doi: 10.1101/gr.142232.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ameur A, Zaghlool A, Halvardson J, Wetterbom A, Gyllensten U, Cavelier L, Feuk L. Total RNA Sequencing Reveals Nascent Transcription and Widespread Co-Transcriptional Splicing in the Human Brain. Nature Structural and Molecular Biology. 2011;18(12):1435–1440. doi: 10.1038/nsmb.2143. [DOI] [PubMed] [Google Scholar]
- 29.Tian B, Hu J, Zhang H, Lutz CS. A large-Scale Analysis of mRNA Polyadenylation of Human and Mouse Genes. Nucleic Acids Research. 2005;33(1):201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigo R, Gingeras TR. Landscape of Transcription in Human Cells. Nature. 2012;489(7414):101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]