Abstract
Background
Recent evidence shows that long non-coding RNA (LncRNA) play important regulatory roles in many biology process, including cell development, activation and oncogenesis. However, the roles of these LncRNAs in the development and activation of CD4+ T cells, which is an important component of immune response, remain unknown.
Results
To predict the function of LncRNA in the development and activation of CD4+ T cells, first, we examined the expression profiles of LncRNAs and mRNAs in CD4−CD8− (DN), CD4+CD8+ (DP), CD4+CD8−, and activated CD4+CD8− T cells in a microarray analysis and verified these results by real time PCRs (qPCR). We found that the expression of hundreds of LncRNAs significantly changed in each process of developmental transition, including DN into DP, DP into CD4+CD8−, and CD4+CD8− into activated CD4+ T cells. A Kendall distance analysis suggested that the expression of LncRNAs in DN, DP, CD4+CD8− T cells and activated CD4+ T cells were correlated with the expression of mRNAs in these T cells. The Blat algorithm and GO analysis suggested that LncRNAs may exert important roles in the development and activation of CD4+ T cells. These roles included proliferation, homeostasis, maturation, activation, migration, apoptosis and calcium ion transportation.
Conclusion
The present study found that the expression profiles of LncRNAs in different stages of CD4+ T cells are distinguishable. LncRNAs are involved in the key biological process in CD4+ T cell development and activation.
Introduction
It is well-known that CD4+ T cells play critical roles in the adaptive immune system. These mature CD4+ T cells develop from common lymphoid precursors (CLP) in bone marrow. Subsequently, these cells mature into CD4−CD8− double negative thymocytes (DN) and CD4+CD8+ double positive thymocytes (DP). These double positive cells develop into CD4+ and CD8+ single positive T cells. Once these mature CD4+ T cells encounter the antigen-loaded dendritic cells in periphery lymph organs or local sites, they can differentiate into Th1, Th2 or Th17 cells. Several of the cells can become regulatory T cells. Most of these polarized Th cells died. However, a minority of these activated Th cells become renewable memory CD4+ T cells that are able to rapidly mount a protective response upon encountering the same antigen because they are stimulated in the initiation stage [1]. A flurry of studies have been reported to explain how the CD4+ T cells develop from CLP to CD4+ single positive cells and how naïve CD4+ T cells can be polarized into different subsets of CD4+ T cells [1]–[5]. However, our understanding of the underlying molecular mechanism of CD4+ T cell differentiation and activation remains largely incomplete.
In addition to protein molecules, many studies already suggest that non-coding RNAs play important regulatory roles in many biological processes, including cell development, activation and oncogenesis [6]–[8]. Recently, long non-coding RNAs (LncRNAs) have been increasingly studied [9]–[12]. Genome-wide studies show that more than four thousand LncRNAs exist in mammalian species, such as mice and humans [13]–[15]. These LncRNAs function not only in normal development and homeostasis but also in some diseases [16], [17]. Because CD4+ T cells play pivotal roles in the immune system, we are therefore interested in studying the expression of LncRNA in CD4+ T cells during development and activation to provide new insights into the regulation of CD4+ T cells [18].
In the present study, we found that 7037, 9456, 8206 and 7847 LncRNAs were detected in DN, DP, CD4+CD8− and activated CD4+ T cells, respectively. The expression of hundreds of LncRNAs changed significantly in each process of developmental transition, including DN into DP, DP into CD4+CD8−, and CD4+CD8− into activated CD4+ T cells. A Kendall distance analysis suggested that the expression of LncRNAs in DN, DP, naive CD4+ T cells and activated CD4+ T cells were correlated with the expression of mRNAs in these T cells. The Blat algorithm and GO analysis suggested that LncRNAs may exert important roles in CD4+ T cells during development and activation such as proliferation, homeostasis, maturation, activation, migration, apoptosis and calcium ion transportation.
Results
Overview of the expression profiles of LncRNAs and mRNA during CD4+ T cell development and activation
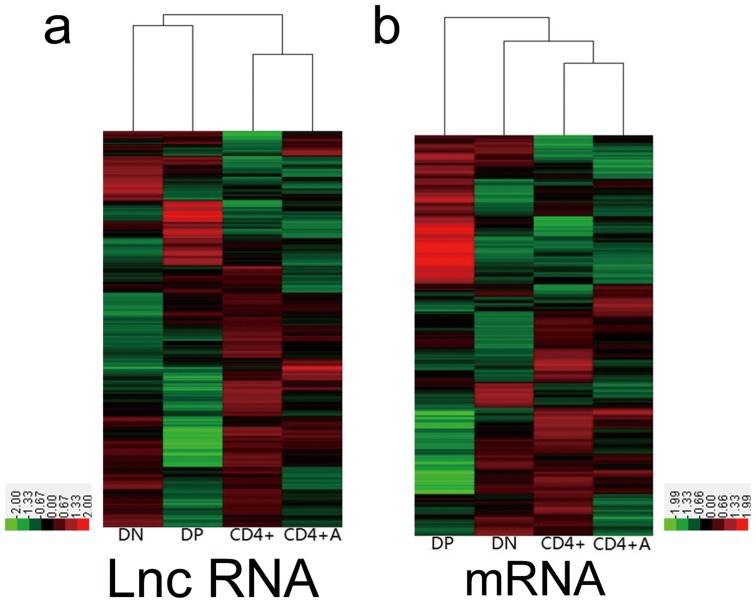
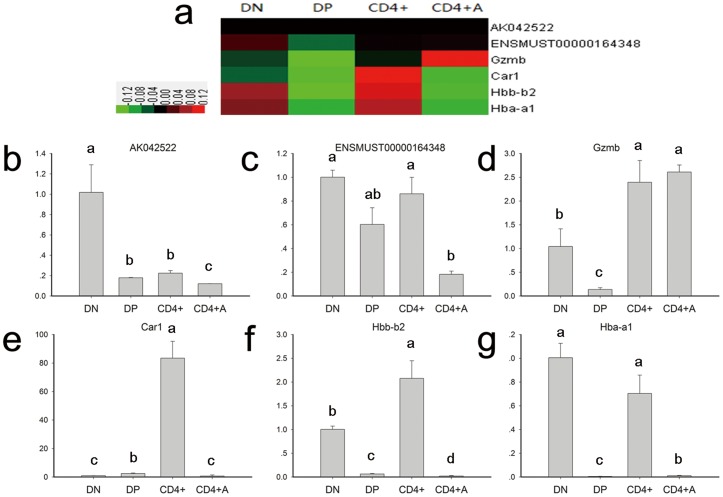
To explore the functions of LncRNAs in CD4+ T cell development and activation, we detected the expression of LncRNAs and mRNAs among the DN, DP, CD4+ and anti-CD3/anti-CD28 activated CD4+ T cells on a microarray. DN and DP cells were sorted from thymocytes, and CD4+ T cells were purified from the spleen. In total, 31,423 LncRNAs and 25,376 coding transcripts were detected on a second-generation LncRNA microarray (Table S1–S2). The pools of LncRNAs and mRNAs were carefully collected from databases such as RefSeq, UCSC Knowngenes, and Ensembl and from datasets published in previous studies. The organization of the expression profiles into heatmaps showed that the expression profiles of LncRNAs and mRNAs in different stages of CD4+ T cells were distinct (Figure 1). To examine the reliability of the array data, we randomly selected four mRNAs (Gzmb, Car1, Hbb-b2 and Hba-a1) and two LncRNAs (ENSMUST00000164348 and AK042522) to confirm their expression in different T cell subsets by using qPCR. The results of the qPCR study were consistent with the microarray data (Figure 2).
Figure 1. The expression profiles of LncRNAs and mRNAs in DN, DP, CD4+ and CD4+A T cells.
The DN, DP, CD4+ and CD4+A T cells and their total RNA were prepared as described, the expression of (a) LncRNAs and (b) mRNAs were detected on an Agilent microarray. The raw data are displayed in a heatmap.
Figure 2. The expression profiles of two LncRNAs and four mRNAs in the T cell development and activation process.
(a) A heatmap of two LncRNAs and four mRNAs in the T cell development and activation process. (b–g) The expression patterns of two LncRNAs and four mRNAs in 4 cell lines as monitored by RT-qPCR. Unlike letters associated with bars on the histogram indicate a significant difference, (P<0.05).
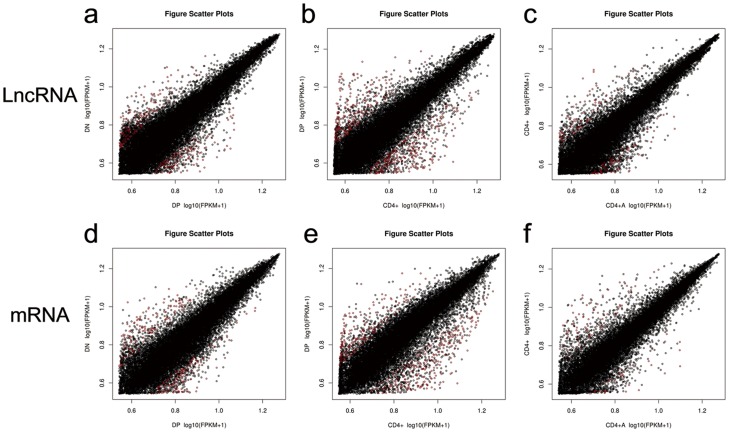
To analyze the expression changes of LncRNAs and mRNAs among DN, DP, CD4+ and activated CD4+ T cells, we grouped these cells as DN/DP, DP/CD4+, CD4+/activated CD4+. Additionally, we screened the LncRNA candidates and mRNAs (Table S3–S8) with the following criteria: t-test p-value ≤0.05 and fold-change≥1.5 or ≤2/3. The top 10 up- and down-regulated LncRNAs in each compare groups are shown in Table 1–3. The number of up-regulated and down-regulated LncRNAs and mRNAs in the different comparison groups are shown in Table 4. From Table 4, the number of up-regulated LncRNAs or down-regulated LncRNAs was similar to the number of mRNAs in the DN/DP and DP/CD4+. For example, 112 LncRNAs and 119 mRNAs were higher in expression in the DP group than those in the DN group; 131 LncRNAs and 128 mRNAs were lower in expression in the DP group than those in the DN group. We can observe this phenotype in a log-log scatter plot (Figure 3). These results may suggest that the expressions of LncRNA maintain a relationship with the expression of mRNAs in the development and activation process.
Table 1. The top 10 up- and down-regulated LncRNAs in the DN VS. DP group.
| DN VS. DP | |||||
| UP | DOWN | ||||
| Gene name | log2(DP/DN) | p-value | Gene name | log2(DP/DN) | p-value |
| uc009qpv.1 | 1.471739597 | 0.03004419 | uc008xuk.1 | −1.417225538 | 0.019526053 |
| ENSMUST00000129991 | 1.467481417 | 0.008212429 | AK020242 | −1.397376466 | 0.016699622 |
| ENSMUST00000165285 | 1.457457191 | 0.022426522 | AK007978 | −1.342522112 | 0.006499939 |
| uc007rul.1 | 1.430331255 | 0.002263296 | AK042522 | −1.291541257 | 0.047566101 |
| ENSMUST00000130391 | 1.385793398 | 0.041690832 | AK132374 | −1.250973337 | 0.029195609 |
| uc007rum.1 | 1.348379165 | 0.036369644 | uc007agf.1 | −1.248073538 | 0.033428332 |
| ENSMUST00000156387 | 1.252114567 | 0.021579499 | AK081565 | −1.21586253 | 0.006826746 |
| AK037315 | 1.234271805 | 0.028538955 | ENSMUST00000117178 | −1.196960609 | 0.032789358 |
| uc009axs.1 | 1.227981603 | 0.019878953 | uc009pzn.1 | −1.162297221 | 0.030087904 |
| ENSMUST00000130122 | 1.226793109 | 0.008816597 | uc007enj.1 | −1.15015982 | 0.028863781 |
Table 3. The top 10 up- and down-regulated LncRNAs in the CD4+ VS. CD4+A group.
| CD4+ VS. CD4+A | |||||
| UP | DOWN | ||||
| Gene name | log2(CD4+A/CD4+) | p-value | Gene name | log2(CD4+A/CD4+) | p-value |
| AK144448 | 1.249066165 | 0.047781106 | ENSMUST00000100689 | −1.535201274 | 0.040108269 |
| AK052414 | 1.239167117 | 0.01849099 | AK050225 | −1.490038968 | 0.005443587 |
| uc007axg.1 | 1.124892531 | 0.018913152 | ENSMUST00000171480 | −1.452712194 | 0.031353875 |
| uc007hdw.1 | 1.093175797 | 0.002535733 | MM9LINCRNAEXON12067- | −1.178812357 | 0.002233869 |
| NR_033499 | 1.035562581 | 0.005349267 | uc009mjt.1 | −1.166556654 | 0.006021145 |
| ENSMUST00000111862 | 1.027297815 | 0.012138723 | AK033429 | −1.163091814 | 0.012025539 |
| ENSMUST00000117414 | 1.020519677 | 0.04108955 | AK046352 | −1.145642967 | 0.043572889 |
| NR_001586 | 0.992395853 | 0.010219202 | ENSMUST00000121631 | −1.137694171 | 0.031992465 |
| ENSMUST00000148383 | 0.988554886 | 0.010784593 | uc.256+ | −1.093056264 | 0.041327858 |
| uc007vms.1 | 0.974917911 | 0.046199718 | AK008541 | −1.068485138 | 0.00621642 |
Table 4. The number of up- and down-regulated LncRNAs and mRNAs in the different comparison groups.
| DN VS. DP | DP VS. CD4+ | CD4+ VS. CD4+A | ||||
| LncRNA | mRNA | LncRNA | mRNA | LncRNA | mRNA | |
| UP | 112 | 119 | 224 | 228 | 68 | 36 |
| DOWN | 131 | 128 | 185 | 169 | 68 | 66 |
| TOTAL | 243 | 247 | 409 | 397 | 136 | 102 |
Figure 3. A log-log scatterplot comparing the global mRNA and LncRNA expression profiles between four subsets of CD4+ T cells.
(a,d) A log-log scatter plot between DN and DP, (b,e) a log-log scatter plot between DP and CD4+, and (c,f) a log-log scatter plot between CD4+ and CD4+A. Red dots indicate significant differences and the black dots indicate no significant differences.
Table 2. The top 10 up- and down-regulated LncRNAs in the DP VS. CD4+ group.
| DP VS. CD4+ | |||||
| UP | DOWN | ||||
| Gene name | log2(CD4+/DP) | p-value | Gene name | log2(CD4+/DP) | p-value |
| AK050225 | 1.899682846 | 0.020961628 | ENSMUST00000144984 | −1.921669086 | 0.00194036 |
| AK013439 | 1.697413174 | 0.038629194 | ENSMUST00000129991 | −1.90189424 | 0.008526756 |
| AK088994 | 1.623751317 | 0.019923999 | uc008xof.1 | −1.883039124 | 0.003175766 |
| AK135581 | 1.611038693 | 0.001436439 | uc007rum.1 | −1.845952524 | 0.037784114 |
| uc009sfx.1 | 1.580810976 | 0.012595544 | ENSMUST00000130391 | −1.815547 | 0.017109652 |
| uc007dot.1 | 1.560820411 | 0.03895783 | uc007kwe.1 | −1.812476625 | 0.001288994 |
| MM9LINCRNAEXON12112-_P1 | 1.546387496 | 0.021190725 | NR_015488 | −1.740748633 | 0.012579072 |
| AK040987 | 1.534823411 | 0.04997947 | uc009qpw.1 | −1.679873533 | 0.019139803 |
| AK079251 | 1.526350541 | 0.00054264 | ENSMUST00000149186 | −1.679190796 | 0.011900309 |
| AK020242 | 1.505769055 | 0.01577034 | uc007rul.1 | −1.67397823 | 0.002808748 |
Correlation analysis of LncRNAs and mRNAs
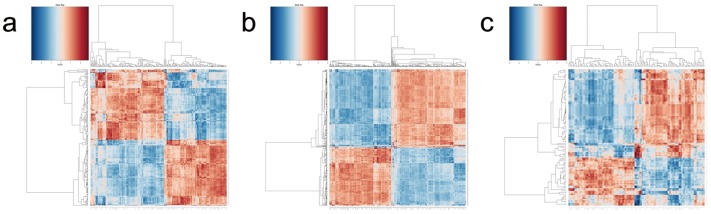
To understand the relationships among selected LncRNAs and mRNAs, a Kendall distance analysis was employed to evaluate the correlation between LncRNAs and mRNAs in different comparison groups (Figure 4). In this analysis, tau is the correlation coefficient, the p-value is the significant index and a p-value≤0.05 was considered significant. By this method, we analyzed the correlation between selected LncRNAs and mRNAs in each group. We found that there were 6390, 27890, 1274 positively correlated LncRNA-mRNAs pairs in the DN/DP, DP/CD4+ and CD4+/CD4+A groups, respectively (p-value≤0.05), and 5699, 24824, 817 negatively correlated LncRNA-mRNAs pairs in the DN/DP, DP/CD4+ and CD4+/CD4+A groups, respectively (p-value≤0.05). These results suggested that the expression of LncRNAs display high correlations with mRNAs in the DP developing into single positive CD4+ T cell process and CD4+ T cells during development and activation.
Figure 4. A correlation heatmap between the differentially expressed LncRNAs and genes.
Rows represent differentially expressed genes, and the columns represent differentially expressed LncRNAs. A pink color indicates a positive correlation, while blue-green indicates a negative correlation. (a) DP VS. DN; (b) CD4+ VS. DP; and (c) CD4+A VS. CD4+.
Functional prediction of LncRNAs in CD4+ T cells during development and activation
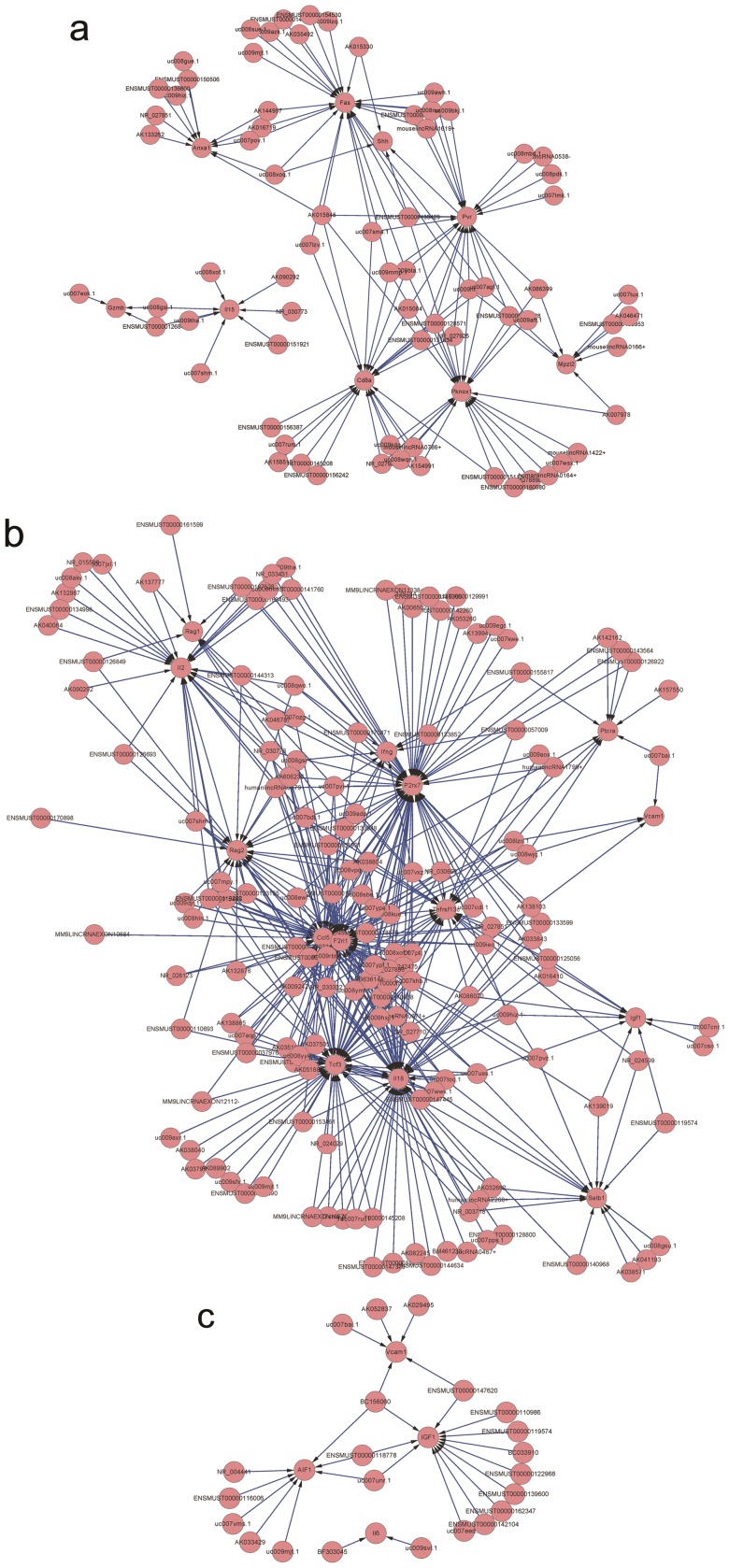
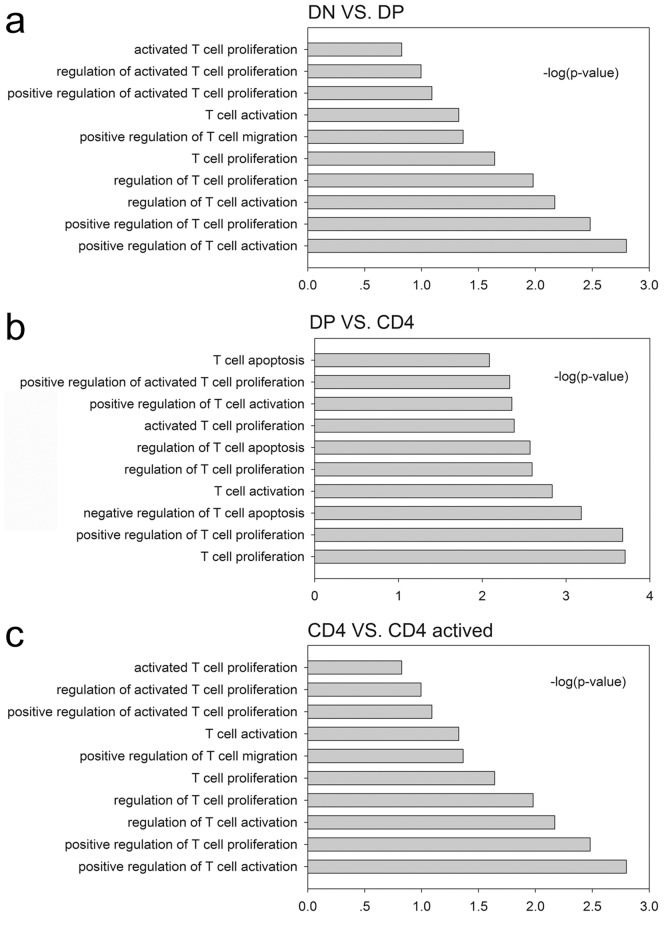
The functional prediction of LncRNA is obscure. Numerous efforts have been pursued to predict the potential function of LncRNAs. Matching the sequence is one of the often used methods to predict the function of LncRNAs: if the sequence of a LncRNA matches a mRNA, then this LncRNA may function on this mRNA [19]. Using this hypothesis, we compared the sequences of differently expressed LncRNA 1 Kb away (upstream or downstream) from the mRNAs and the entire mRNA gene in a positively and negatively correlated group, respectively (Table S9–S26). The Blat algorithm was used for the comparison. If the LncRNA sequence matched one of the sequences 1Kb upstream or downstream of the mRNA or the mRNA gene itself (and the expression of LncRNAs and mRNAs were positively or negatively correlated), then this LncRNA potentially regulated the related mRNA. The topological graph in figure 5 demonstrates this regulation network between LncRNAs and mRNAs. For example, in the CD4+/CD4+A group, the expression of Vcam1 may be regulated by numerous LncRNAs such as uc007bai.1, AK052837, AK029495, BC156060 and ENSMUST00000147620. Meanwhile, BC156060 can regulate the expression of numerous mRNAs such as Vcam1, IGF1 and AIF1. To further predict the function of a LncRNA in CD4+ T cell development and activation, we performed a GO analysis with the different mRNAs that were related with LncRNAs in DN/DP, DP/CD4+ and CD4+/CD4+A group. In all groups, we found that LncRNA primarily regulate T cell proliferation and activation. In the DN/DP and CD4+/CD4+A groups, LncRNAs were primarily involved in the regulation of T cell migration. In the DP/CD4+ group, LncRNAs were primarily involved in the regulation of T cell apoptosis (Figure 6).
Figure 5. A topological graph displaying the regulation network between the LncRNAs and mRNAs.
(a) DP VS. DN; (b) CD4+ VS. DP; and (c) CD4+A VS. CD4+.
Figure 6. Results of the GO analysis of the genes with sequences matching LncRNAs. –log10(P) indicates the GO score related to the genes with a biological process P value.
(a) T cell function related biological processes of the LncRNA sequence matched genes in the DP VS. DN group; (b) T cell function related biological processes of the LncRNA sequence matched genes in the CD4+ VS. DP group; (c) T cell function related biological processes of the LncRNA sequence matched genes in the CD4+A VS. CD4+ group.
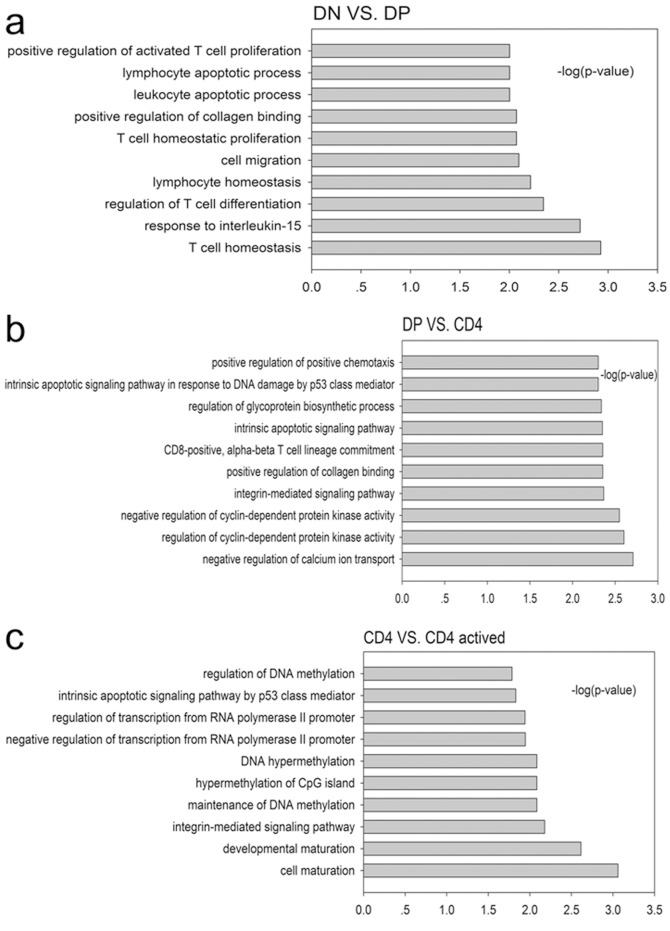
Accumulating evidence has shown that the expression of LncRNAs can regulate the expression of neighboring mRNAs, and their expressions were correlated [20], [21]. Based on this, we selected the mRNAs that neighbor LncRNAs and correlate with their expression, and we performed a GO analysis of these mRNAs in different groups. Figure 7 shows that the highest enriched biological processes were T cell homeostasis in the DN/DP group, the negative regulation of calcium ion transport in the DP/CD4+ group and cell maturation in the CD4/CD4+A group. This analysis suggested that LncRNA may be involved in these biological functions of T cells.
Figure 7. Results of the GO analysis of the genes neighboring LncRNA. –log10(P) indicates the GO score related to genes with a biological process P value.
(a) The main biological processes of the LncRNA neighboring genes in the DP VS. DN group; (b) the main biological processes of the LncRNA neighboring genes in the CD4+ VS. DP group; and (c) the main biological processes of the LncRNA neighboring genes in the CD4+A VS. CD4+ group.
Discussion
Accumulating evidence suggests that non-coding RNAs play critical regulatory roles in biological processes. Most of these studies focused on short RNAs, including small interfering RNAs (siRNAs), microRNAs (miRNAs), and PIWI-interacting RNAs. These RNAs regulate gene expression at transcriptional and/or posttranscriptional levels [22]–[24]. Recently, long non-coding RNAs have also been recognized as an important regulator in biological functions. However, our knowledge of long non-coding RNAs in immune systems is limited. Ten years ago, Deanna Haasch first showed that transcriptional activation of the BIC proto-oncogene is an early and sustained T cell activation event that suggests an important role of non-coding mRNAs in T cell function [5]. Afterward, Murad JM and colleagues demonstrated that a 200 nt ncRNA bound to PKR RNA binding site regulated gene expression in activated lymphocytes [25]. John S. Mattick's group was the first to discover long ncRNAs expressed in CD8+ T cells and suggested that many of these transcripts likely play roles in adaptive immunity [26]. All of these studies suggest that non-coding RNAs exert important roles in T cell activation.
In the present study, we explored the expression of long non-coding RNAs to systemically evaluate their function in CD4+ T cell development and activation. By using the Agilent long-coding RNA array platform, we found that the expression profiles of long non-coding RNA are significantly distinct among different stages of CD4+ T cells. Although expression is not necessarily indicative for function, several lines of evidence presented in this study support the likelihood that many of the LncRNAs expressed in CD4+ T cells are functional. First, many LncRNAs are dynamically regulated during either differentiation or activation. Second, by using a Kendall distance analysis to evaluate the correlation between the LncRNAs and mRNAs in different comparison groups (Figure 4), we found that the expression of LncRNA are highly correlated with the expression of mRNAs in CD4+ T cells development and activation. Notably, this high correlation was displayed in DP developing into single positive CD4+ T cells. Third, by using the Blat algorithm to compare the sequence match between LncRNA and mRNAs to predict the function of the LncRNA [19], we found that many LncRNAs may exert their function through certain mRNAs that play pivotal roles in T cell development and activation. For example, the sequences of several LncRNAs, including ENSMUST00000126849, ENSMUST00000161599, ENSMUST00000167528, ENSMUST00000144313, AK137777, AK046787, AK038804, and NR_030773, matched with RAG1, an important gene in T cells development. This result suggests that these LncRNAs may affect the expression of RAG1 and therefore be involved in the regulation of the arrangement of T cell receptors. The sequences of UC008xof.1, AK090292, NR_030773, ENSMUST00000151921, UC007shm.1, UC009tha.1, and UC008gsi.1 matched the sequence of IL-15 which is an important gene for T cell homeostasis, differentiation and proliferation [27]. Meanwhile, most of the evidence suggests that the expression of LncRNAs can regulate and were correlated with the expression of neighboring mRNAs [20], [21]. Based on this, we selected mRNAs, which neighbor LncRNAs and have high correlations with the expression of LncRNAs. We performed a GO analysis with these selected mRNAs for different groups. Therefore, LncRNAs may be involved in T cell maturation, homeostasis, differentiation, and apoptosis (among others) by regulating neighboring genes, including the following gene and LncRNA pairs: Runx3 and AK040461, IL-2rα and UC008iiP.1, Bax and Ak046787, Stat5b and UC0071mk.1, Myb and UC007eok.1, BCL6 and AK149396, and BCL2 and ENSMUST00000170471. However, all of these predictions only provide a starting point for the study of long non-coding RNAs in CD4+ T cells development and activation. The exact functions of many LncRNAs identified in this article are required experimental verification.
The present study found that the expression profiles of LncRNAs in different stages of CD4+ were significantly different. A bioinformatic analysis suggests that these LncRNAs may play an important role in CD4+ T cell development and activation. These roles may include proliferation, homeostasis, maturation, activation, migration, apoptosis and calcium ion transportation.
Experimental Procedures
Mice
Six- to eight-week-old C57/BL6 mice were purchased from Shanghai Laboratory Animal Company SLAC. All of these mice were maintained in the barrier facility at the Soochow University. All animal experiments were approved by the Institutional Animal Care and Use Committee of Soochow University.
Cell preparation
Six- to eight-week-old C57/BL6 mice were sacrificed, and the thymus and spleen were harvested. The red blood cells were removed by lysis with ACK buffer. Thymus single-cell suspensions were stained with fluorescent coupled antibodies (anti-CD4 and anti-CD8) on ice for 30 min. Then, these stained samples were subjected to FACS sorting to collect the CD4+CD8+ cells as DP cells and CD4−CD8− cells as DN cells. CD4+CD8− T cells were purified from splenocytes using anti-CD4 microbeads according to the manufacturer's protocol (Miltenyi Biotec Inc., Auburn, California, USA). Purified CD4+ T cells were activated with plate-coated anti-CD3 antibodies (5 µg/mL) and anti-CD28 antibodies (2.5 µg/mL). Cells were cultured with RPMI1640 medium with 10% FCS. After 48 hr, cells were collected for further experiments.
Antibodies
The anti-CD3 (145.11), anti-CD28 (37.51), anti-CD4-APC (GK-1.5) and anti-CD8-PE-cy7 (53–6.7) antibodies were purchased from Biolegend Inc. (San Diego, CA).
Microarray analysis
Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's description. Total RNA from each sample was quantified using a NanoDrop ND-1000, and the RNA integrity was assessed using a standard denaturing agarose gel electrophoresis assay. For the microarray analysis, the Agilent Array platform was employed. The sample preparation and microarray hybridization were performed based on the manufacturer's standard protocols with minor modifications. Briefly, mRNA was purified from the total RNA after removing rRNA (Eukaryotic mRNA Isolation Kit, Epicentre). Each sample was amplified and transcribed into fluorescent cRNA along the entire length of the transcripts. This method avoided a 3′ bias by utilizing a reaction with random primers. The labeled cRNAs were hybridized onto the Mouse LncRNA Array v2.0 (8×60K, Arraystar). After washing the slides, the arrays were scanned on an Agilent Scanner G2505C.
Agilent Feature Extraction software (version 11.0.1.1) was used to analyze the acquired array images. Quantile normalization and subsequent data processing were performed using the GeneSpring GX v11.5.1 software package (Agilent Technologies). After the quantile normalization of the raw data, LncRNAs and mRNAs that for at least 1 out of 8 samples have flags in Present or Marginal (“All Targets Value”) were selected to remove batch effects by the Combat Software. The microarray analysis was performed by KangChen Bio-tech (Shanghai, P.R. China).
Real time PCR
Total RNA was purified using TRIzol reagent (Invitrogen) and reverse transcribed using a reverse transcription system (Promega, Madison, WI, USA) according to the manufacturer's instructions. Real-time PCR was performed using the FastStart Universal SYBR Green Master (Roche Diagnostics, Rotkreuz, Switzerland) system and analyzed with an Eppendorf Real-Time Detection System (Eppendorf, Hauppauge, NY). The primer pairs used for Gzmb, Car1, Hbb-b2, Hba-a1, ENSMUST00000164348, AK042522 and GAPDH are shown in Table S27. The PCR parameters were as follows: 95°C for 5 min, followed by 40 cycles of 95°C for 15 sec, 60°C for 30 sec and 72°C for 30 sec. The relative expression level was calculated as 2− (CTgapdh—CTgene) [28]. The mRNA level was normalized with the house keeping gene GAPDH. We used unpaired t-test to perform statistic analyses for the expression levels in different stages and p ≤0.05 were considered significant.
Cluster analysis
Unsupervised hierarchical clustering was performed with average linkage and uncentered correlation as the similarity metrics in Cluster3.0 [29]. Heatmaps were generated in Java Treeview. The data from each raw probe on the microarrays of all four cell lines were averaged. Then, the respective data from the four cell lines were transformed as the provider divided by the average (mean). The relative expression of each gene was calculated as the ratio between the cell line microarray value and the average value of the four microarrays [30]. To draw a simple and perspicuity figure with the software, the relative expression of each gene was described as the log10(ratio) in the heatmap figures in Cluster3.0.
Selection of differentially expressed mRNAs and LncRNAs
DN, DP, naïve CD4+ T cells and CD4+ activated T cells were grouped and then compared to each other to screen for the significantly differentially expressed LncRNAs and mRNAs among these four type cells. We used a paired-sample t-test and fold-change to find differentially expressed genes and LncRNAs by comparing expression levels in each group. Those mRNAs and LncRNAs with a p≤0.05 and fold-change more than 1.5 were selected.
Analysis of the correlation between the expression of LncRNAs and mRNAs
Correlations between differently expressed LncRNAs and mRNAs in different groups were analyzed. A Kendall distance was used to calculate the correlation between LncRNA and mRNAs and the tau is the correlation coefficient. Correlations with p≤0.05 were considered significant and computed via R [31].
Functional prediction of LncRNAs
We compared the sequence of differentially expressed LncRNAs with the sequence of mRNAs that also contained the 1 kb upstream and downstream flanking sequences. In each significant LncRNA-mRNAs correlation group, the Blat algorithm was used in the comparison. The mRNAs that matched LncRNA were further explored by a Gene Ontology (GO) analysis. We selected the LncRNA-mRNA pairs with genes functions related to T cells to draw a regulation network using Cytoscape. The –log10(P-value) of the GO-results were shown in the histogram. The histogram was analyzed by SigmaPlot (Systat Software, Inc., San Jose, CA, USA). We performed a GO analysis on mRNAs that were both adjacent to LncRNAs and significantly positively or negatively correlated to the expression of LncRNAs.
Supporting Information
LncRNA expression profiling data.
(XLS)
mRNA expression profiling data.
(XLS)
DP VS. DN difference changed mRNA.
(XLS)
DP VS. DN difference changed LncRNA.
(XLS)
CD4+ VS. DP difference changed mRNA.
(XLS)
CD4+ VS. DP difference changed LncRNA.
(XLS)
CD4+A VS. CD4+ difference changed mRNA.
(XLS)
CD4+A VS. CD4+ difference changed LncRNA.
(XLS)
DP VS. DN LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were positively and LncRNA sequence matched upstream of the mRNA.
(CSV)
DP VS. DN LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were positively and LncRNA sequence matched genebody of the mRNA.
(CSV)
DP VS. DN LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were positively and LncRNA sequence matched downstream of the mRNA.
(CSV)
DP VS. DN LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were negatively and LncRNA sequence matched upstream of the mRNA.
(CSV)
DP VS. DN LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were negatively and LncRNA sequence matched genebody of the mRNA.
(CSV)
DP VS. DN LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were negatively and LncRNA sequence matched downstream of the mRNA.
(CSV)
CD4+ VS. DP LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were positively and LncRNA sequence matched upstream of the mRNA.
(CSV)
CD4+ VS. DP LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were positively and LncRNA sequence matched genebody of the mRNA.
(CSV)
CD4+ VS. DP LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were positively and LncRNA sequence matched downstream of the mRNA.
(CSV)
CD4+ VS. DP LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were negatively and LncRNA sequence matched upstream of the mRNA.
(CSV)
CD4+ VS. DP LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were negatively and LncRNA sequence matched genebody of the mRNA.
(CSV)
CD4+ VS. DP LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were negatively and LncRNA sequence matched downstream of the mRNA.
(CSV)
CD4+A VS. CD4 LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were positively and LncRNA sequence matched upstream of the mRNA.
(CSV)
CD4+A VS. CD4 LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were positively and LncRNA sequence matched genebody of the mRNA.
(CSV)
CD4+A VS. CD4 LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were positively and LncRNA sequence matched downstream of the mRNA.
(CSV)
CD4+A VS. CD4 LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were negatively and LncRNA sequence matched upstream of the mRNA.
(CSV)
CD4+A VS. CD4 LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were negatively and LncRNA sequence matched genebody of the mRNA.
(CSV)
CD4+A VS. CD4 LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were negatively and LncRNA sequence matched downstream of the mRNA.
(CSV)
Primers used in real time PCRs for detecting LncRNAs and mRNAs expressions.
(DOC)
Acknowledgments
The authors wish to thank Jingping Hu, Xuemei Zhu, Bin Bai (Genergy Bio tech. (Shanghai) Co.Ltd) and Guan Wang (Shanghai BioGenius Bio technology Co., Ltd; Genergy Bio tech. (Shanghai) Co.Ltd) for their technical assistance.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work is Supported by grants from the Priority Academic Development Program of Jiangsu Higher Education Institutions (PAPD), Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT, IRT1075), Jiangsu Key Laboratory of Infection and Immunity, Institutes of Biology and Medical Sciences of Soochow University, Jiangsu Provincial Innovative Research Team, and intramural research funding to Jinping Zhang (Q413401810, Q313406711) from Soochow University, National Natural Science Foundation of China to Jinping Zhang (No: 31270939), Fei Xia (No:31200660) and Fulu Dong (81300553), Natural Science Foundation of Jiangsu Province to Jinping Zhang (BK2012617) and Major Natural Science Foundation of Jiangsu High education to Jinping Zhang (13KJA310004). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Wang R, Xie H, Huang Z, Ma J, Fang X, et al. (2011) Transcription factor network regulating CD(+)CD8(+) thymocyte survival. Crit Rev Immunol 31: 447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ma CS, Deenick EK, Batten M, Tangye SG (2012) The origins, function, and regulation of T follicular helper cells. J Exp Med 209: 1241–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fink PJ (2013) The biology of recent thymic emigrants. Annu Rev Immunol 31: 31–50. [DOI] [PubMed] [Google Scholar]
- 4. Naito T, Tanaka H, Naoe Y, Taniuchi I (2011) Transcriptional control of T-cell development. Int Immunol 23: 661–668. [DOI] [PubMed] [Google Scholar]
- 5. Haasch D, Chen YW, Reilly RM, Chiou XG, Koterski S, et al. (2002) T cell activation induces a noncoding RNA transcript sensitive to inhibition by immunosuppressant drugs and encoded by the proto-oncogene, BIC. Cell Immunol 217: 78–86. [DOI] [PubMed] [Google Scholar]
- 6. Prasanth KV, Spector DL (2007) Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev 21: 11–42. [DOI] [PubMed] [Google Scholar]
- 7. Amaral PP, Dinger ME, Mercer TR, Mattick JS (2008) The eukaryotic genome as an RNA machine. Science 319: 1787–1789. [DOI] [PubMed] [Google Scholar]
- 8. Rinn JL, Chang HY (2012) Genome regulation by long noncoding RNAs. Annu Rev Biochem 81: 145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ponting CP, Oliver PL, Reik W (2009) Evolution and functions of long noncoding RNAs. Cell 136: 629–641. [DOI] [PubMed] [Google Scholar]
- 10. Zhang H, Chen Z, Wang X, Huang Z, He Z, et al. (2013) Long non-coding RNA: a new player in cancer. J Hematol Oncol 6: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hauptman N, Glavac D (2013) Long non-coding RNA in cancer. Int J Mol Sci 14: 4655–4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wierzbicki AT (2012) The role of long non-coding RNA in transcriptional gene silencing. Curr Opin Plant Biol 15: 517–522. [DOI] [PubMed] [Google Scholar]
- 13. Jia H, Osak M, Bogu GK, Stanton LW, Johnson R, et al. (2010) Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA 16: 1478–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Washietl S, Hofacker IL, Lukasser M, Huttenhofer A, Stadler PF (2005) Mapping of conserved RNA secondary structures predicts thousands of functional noncoding RNAs in the human genome. Nat Biotechnol 23: 1383–1390. [DOI] [PubMed] [Google Scholar]
- 15. Louro R, El-Jundi T, Nakaya HI, Reis EM, Verjovski-Almeida S (2008) Conserved tissue expression signatures of intronic noncoding RNAs transcribed from human and mouse loci. Genomics 92: 18–25. [DOI] [PubMed] [Google Scholar]
- 16. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, et al. (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, et al. (2011) A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472: 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Toes RE, Ossendorp F, Offringa R, Melief CJ (1999) CD4 T cells and their role in antitumor immune responses. J Exp Med 189: 753–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guil S, Esteller M (2012) Cis-acting noncoding RNAs: friends and foes. Nat Struct Mol Biol 19: 1068–1075. [DOI] [PubMed] [Google Scholar]
- 20. Guttman M, Amit I, Garber M, French C, Lin MF, et al. (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, et al. (2011) Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25: 1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aravin AA, Hannon GJ, Brennecke J (2007) The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318: 761–764. [DOI] [PubMed] [Google Scholar]
- 23. Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM, et al. (2011) Biological functions of microRNAs: a review. J Physiol Biochem 67: 129–139. [DOI] [PubMed] [Google Scholar]
- 24. Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R (2006) Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev 20: 515–524. [DOI] [PubMed] [Google Scholar]
- 25. Murad JM, Tone LG, de Souza LR, De Lucca FL (2005) A point mutation in the RNA-binding domain I results in decrease of PKR activation in acute lymphoblastic leukemia. Blood Cells Mol Dis 34: 1–5. [DOI] [PubMed] [Google Scholar]
- 26. Pang KC, Dinger ME, Mercer TR, Malquori L, Grimmond SM, et al. (2009) Genome-wide identification of long noncoding RNAs in CD8+ T cells. J Immunol 182: 7738–7748. [DOI] [PubMed] [Google Scholar]
- 27. Lodolce JP, Boone DL, Chai S, Swain RE, Dassopoulos T, et al. (1998) IL-15 receptor maintains lymphoid homeostasis by supporting lymphocyte homing and proliferation. Immunity 9: 669–676. [DOI] [PubMed] [Google Scholar]
- 28. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 29. de Hoon MJ, Imoto S, Nolan J, Miyano S (2004) Open source clustering software. Bioinformatics 20: 1453–1454. [DOI] [PubMed] [Google Scholar]
- 30. Sun L, Xie H, Mori MA, Alexander R, Yuan B, et al. (2011) Mir193b-365 is essential for brown fat differentiation. Nat Cell Biol 13: 958–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liao Q, Liu C, Yuan X, Kang S, Miao R, et al. (2011) Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co-expression network. Nucleic Acids Res 39: 3864–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
LncRNA expression profiling data.
(XLS)
mRNA expression profiling data.
(XLS)
DP VS. DN difference changed mRNA.
(XLS)
DP VS. DN difference changed LncRNA.
(XLS)
CD4+ VS. DP difference changed mRNA.
(XLS)
CD4+ VS. DP difference changed LncRNA.
(XLS)
CD4+A VS. CD4+ difference changed mRNA.
(XLS)
CD4+A VS. CD4+ difference changed LncRNA.
(XLS)
DP VS. DN LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were positively and LncRNA sequence matched upstream of the mRNA.
(CSV)
DP VS. DN LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were positively and LncRNA sequence matched genebody of the mRNA.
(CSV)
DP VS. DN LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were positively and LncRNA sequence matched downstream of the mRNA.
(CSV)
DP VS. DN LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were negatively and LncRNA sequence matched upstream of the mRNA.
(CSV)
DP VS. DN LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were negatively and LncRNA sequence matched genebody of the mRNA.
(CSV)
DP VS. DN LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were negatively and LncRNA sequence matched downstream of the mRNA.
(CSV)
CD4+ VS. DP LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were positively and LncRNA sequence matched upstream of the mRNA.
(CSV)
CD4+ VS. DP LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were positively and LncRNA sequence matched genebody of the mRNA.
(CSV)
CD4+ VS. DP LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were positively and LncRNA sequence matched downstream of the mRNA.
(CSV)
CD4+ VS. DP LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were negatively and LncRNA sequence matched upstream of the mRNA.
(CSV)
CD4+ VS. DP LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were negatively and LncRNA sequence matched genebody of the mRNA.
(CSV)
CD4+ VS. DP LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were negatively and LncRNA sequence matched downstream of the mRNA.
(CSV)
CD4+A VS. CD4 LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were positively and LncRNA sequence matched upstream of the mRNA.
(CSV)
CD4+A VS. CD4 LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were positively and LncRNA sequence matched genebody of the mRNA.
(CSV)
CD4+A VS. CD4 LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were positively and LncRNA sequence matched downstream of the mRNA.
(CSV)
CD4+A VS. CD4 LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were negatively and LncRNA sequence matched upstream of the mRNA.
(CSV)
CD4+A VS. CD4 LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were negatively and LncRNA sequence matched genebody of the mRNA.
(CSV)
CD4+A VS. CD4 LncRNA-mRNAs pairs with the expression of LncRNAs and mRNAs were negatively and LncRNA sequence matched downstream of the mRNA.
(CSV)
Primers used in real time PCRs for detecting LncRNAs and mRNAs expressions.
(DOC)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.