Abstract
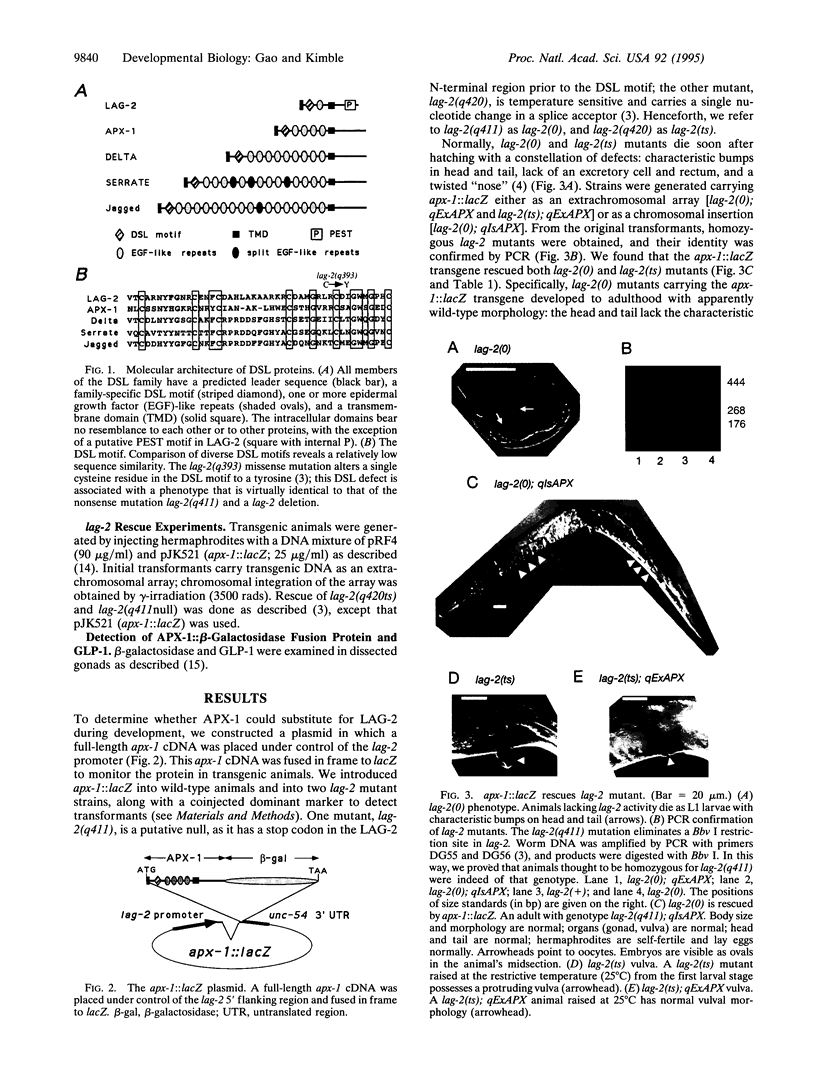
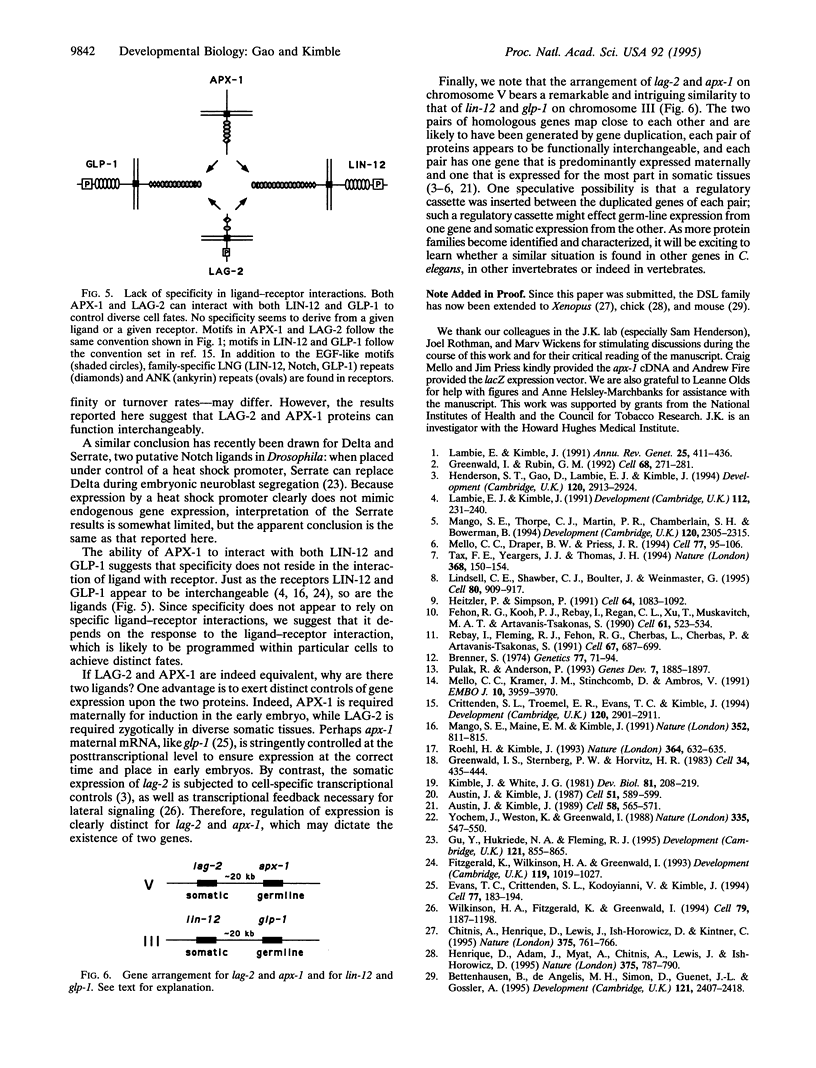
The homologous LAG-2 and APX-1 membrane proteins are putative signaling ligands in the GLP-1/LIN-12 signal-transduction pathway in Caenorhabditis elegans. Normally, LAG-2 and APX-1 mediate distinct cell interactions. Here, we demonstrate that APX-1, which normally interacts with GLP-1 in the early embryo, can substitute for LAG-2 throughout development. When expressed under control of the lag-2 promoter, an apx-1 cDNA can completely rescue a lag-2 null mutant. To substitute for LAG-2, APX-1 must be able to interact with both GLP-1 and LIN-12 receptors and to mediate a variety of cell interactions during development. Therefore, APX-1 and LAG-2 are essentially equivalent in their ability to influence receptor activity. On the basis of this result, we suggest that the existence of multiple-signaling ligands in the LIN-12/GLP-1 signal transduction pathway does not reflect the evolution of functionally distinct proteins but rather the imposition of distinct controls of gene expression upon functionally similar proteins. Finally, we propose that the specification of distinct cell fates by the LIN-12/GLP-1 signal-transduction pathway relies on activities functioning downstream of the ligand and receptor, rather than on specific ligand-receptor interactions.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Austin J., Kimble J. Transcript analysis of glp-1 and lin-12, homologous genes required for cell interactions during development of C. elegans. Cell. 1989 Aug 11;58(3):565–571. doi: 10.1016/0092-8674(89)90437-6. [DOI] [PubMed] [Google Scholar]
- Austin J., Kimble J. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell. 1987 Nov 20;51(4):589–599. doi: 10.1016/0092-8674(87)90128-0. [DOI] [PubMed] [Google Scholar]
- Bettenhausen B., Hrabe de Angelis M., Simon D., Guénet J. L., Gossler A. Transient and restricted expression during mouse embryogenesis of Dll1, a murine gene closely related to Drosophila Delta. Development. 1995 Aug;121(8):2407–2418. doi: 10.1242/dev.121.8.2407. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974 May;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis A., Henrique D., Lewis J., Ish-Horowicz D., Kintner C. Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature. 1995 Jun 29;375(6534):761–766. doi: 10.1038/375761a0. [DOI] [PubMed] [Google Scholar]
- Crittenden S. L., Troemel E. R., Evans T. C., Kimble J. GLP-1 is localized to the mitotic region of the C. elegans germ line. Development. 1994 Oct;120(10):2901–2911. doi: 10.1242/dev.120.10.2901. [DOI] [PubMed] [Google Scholar]
- Evans T. C., Crittenden S. L., Kodoyianni V., Kimble J. Translational control of maternal glp-1 mRNA establishes an asymmetry in the C. elegans embryo. Cell. 1994 Apr 22;77(2):183–194. doi: 10.1016/0092-8674(94)90311-5. [DOI] [PubMed] [Google Scholar]
- Fehon R. G., Kooh P. J., Rebay I., Regan C. L., Xu T., Muskavitch M. A., Artavanis-Tsakonas S. Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. Cell. 1990 May 4;61(3):523–534. doi: 10.1016/0092-8674(90)90534-l. [DOI] [PubMed] [Google Scholar]
- Fitzgerald K., Wilkinson H. A., Greenwald I. glp-1 can substitute for lin-12 in specifying cell fate decisions in Caenorhabditis elegans. Development. 1993 Dec;119(4):1019–1027. doi: 10.1242/dev.119.4.1019. [DOI] [PubMed] [Google Scholar]
- Greenwald I. S., Sternberg P. W., Horvitz H. R. The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell. 1983 Sep;34(2):435–444. doi: 10.1016/0092-8674(83)90377-x. [DOI] [PubMed] [Google Scholar]
- Greenwald I., Rubin G. M. Making a difference: the role of cell-cell interactions in establishing separate identities for equivalent cells. Cell. 1992 Jan 24;68(2):271–281. doi: 10.1016/0092-8674(92)90470-w. [DOI] [PubMed] [Google Scholar]
- Gu Y., Hukriede N. A., Fleming R. J. Serrate expression can functionally replace Delta activity during neuroblast segregation in the Drosophila embryo. Development. 1995 Mar;121(3):855–865. doi: 10.1242/dev.121.3.855. [DOI] [PubMed] [Google Scholar]
- Heitzler P., Simpson P. The choice of cell fate in the epidermis of Drosophila. Cell. 1991 Mar 22;64(6):1083–1092. doi: 10.1016/0092-8674(91)90263-x. [DOI] [PubMed] [Google Scholar]
- Henderson S. T., Gao D., Lambie E. J., Kimble J. lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development. 1994 Oct;120(10):2913–2924. doi: 10.1242/dev.120.10.2913. [DOI] [PubMed] [Google Scholar]
- Henrique D., Adam J., Myat A., Chitnis A., Lewis J., Ish-Horowicz D. Expression of a Delta homologue in prospective neurons in the chick. Nature. 1995 Jun 29;375(6534):787–790. doi: 10.1038/375787a0. [DOI] [PubMed] [Google Scholar]
- Kimble J. E., White J. G. On the control of germ cell development in Caenorhabditis elegans. Dev Biol. 1981 Jan 30;81(2):208–219. doi: 10.1016/0012-1606(81)90284-0. [DOI] [PubMed] [Google Scholar]
- Lambie E. J., Kimble J. Genetic control of cell interactions in nematode development. Annu Rev Genet. 1991;25:411–436. doi: 10.1146/annurev.ge.25.120191.002211. [DOI] [PubMed] [Google Scholar]
- Lambie E. J., Kimble J. Two homologous regulatory genes, lin-12 and glp-1, have overlapping functions. Development. 1991 May;112(1):231–240. doi: 10.1242/dev.112.1.231. [DOI] [PubMed] [Google Scholar]
- Lindsell C. E., Shawber C. J., Boulter J., Weinmaster G. Jagged: a mammalian ligand that activates Notch1. Cell. 1995 Mar 24;80(6):909–917. doi: 10.1016/0092-8674(95)90294-5. [DOI] [PubMed] [Google Scholar]
- Mango S. E., Maine E. M., Kimble J. Carboxy-terminal truncation activates glp-1 protein to specify vulval fates in Caenorhabditis elegans. Nature. 1991 Aug 29;352(6338):811–815. doi: 10.1038/352811a0. [DOI] [PubMed] [Google Scholar]
- Mango S. E., Thorpe C. J., Martin P. R., Chamberlain S. H., Bowerman B. Two maternal genes, apx-1 and pie-1, are required to distinguish the fates of equivalent blastomeres in the early Caenorhabditis elegans embryo. Development. 1994 Aug;120(8):2305–2315. doi: 10.1242/dev.120.8.2305. [DOI] [PubMed] [Google Scholar]
- Mello C. C., Draper B. W., Priess J. R. The maternal genes apx-1 and glp-1 and establishment of dorsal-ventral polarity in the early C. elegans embryo. Cell. 1994 Apr 8;77(1):95–106. doi: 10.1016/0092-8674(94)90238-0. [DOI] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991 Dec;10(12):3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulak R., Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 1993 Oct;7(10):1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- Rebay I., Fleming R. J., Fehon R. G., Cherbas L., Cherbas P., Artavanis-Tsakonas S. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell. 1991 Nov 15;67(4):687–699. doi: 10.1016/0092-8674(91)90064-6. [DOI] [PubMed] [Google Scholar]
- Roehl H., Kimble J. Control of cell fate in C. elegans by a GLP-1 peptide consisting primarily of ankyrin repeats. Nature. 1993 Aug 12;364(6438):632–635. doi: 10.1038/364632a0. [DOI] [PubMed] [Google Scholar]
- Tax F. E., Yeargers J. J., Thomas J. H. Sequence of C. elegans lag-2 reveals a cell-signalling domain shared with Delta and Serrate of Drosophila. Nature. 1994 Mar 10;368(6467):150–154. doi: 10.1038/368150a0. [DOI] [PubMed] [Google Scholar]
- Wilkinson H. A., Fitzgerald K., Greenwald I. Reciprocal changes in expression of the receptor lin-12 and its ligand lag-2 prior to commitment in a C. elegans cell fate decision. Cell. 1994 Dec 30;79(7):1187–1198. doi: 10.1016/0092-8674(94)90010-8. [DOI] [PubMed] [Google Scholar]
- Yochem J., Weston K., Greenwald I. The Caenorhabditis elegans lin-12 gene encodes a transmembrane protein with overall similarity to Drosophila Notch. Nature. 1988 Oct 6;335(6190):547–550. doi: 10.1038/335547a0. [DOI] [PubMed] [Google Scholar]






