Abstract
Context
In many cancers, specific subpopulations of cells appear to be uniquely capable of initiating and maintaining tumors. The strongest support for this cancer stem cell model comes from transplantation assays in immune-deficient mice, which indicate that human acute myeloid leukemia (AML) is driven by self-renewing leukemia stem cells (LSC). This model has significant implications for the development of novel therapies, but its clinical relevance has yet to be determined.
Objective
To identify a leukemic stem cell gene expression signature and test its association with clinical outcomes in AML.
Design, Setting, and Patients
Global gene expression (microarray) profiles of LSC-enriched subpopulations from primary AML and normal patient samples were analyzed. Patient samples were obtained at Stanford University Medical Center between April 2005 and July 2007. Validation datasets of global transcriptional profiles of AML tumors from four independent cohorts totaling 1047 patients were analyzed retrospectively.
Main Outcome Measures
Identification of genes discriminating LSC-enriched from other subpopulations in AML tumors; association of the LSC-specific genes with overall, event-free, and relapse-free survival, and with therapeutic response.
Results
Expression levels of 52 genes distinguished LSC-enriched from other subpopulations in cell-sorted AML samples. An LSC score summarizing expression of these genes in bulk primary AML tumor samples was defined and found to be associated with clinical outcomes in four independent patient cohorts. High LSC scores were associated with worse overall (OS), event-free (EFS), and relapse-free (RFS) survival, among patients with either a normal karyotype (NKAML), or with chromosomal abnormalities. For the largest cohort of patients with NKAML (n=163), the LSC score was significantly associated with OS as a continuous variable (hazard ratios [HR] 1.15, 95% Confidence Interval [CI] 1.08-1.22, log-likelihood p<0.001). When patients were split into high and low LSC score groups, the absolute risk of death by 3 years was 57% (95% CI 43-67%) for the low LSC score group, versus 78% (95% CI 66-86%) in the high LSC score group (HR 1.9, 95% CI 1.3-2.7, log-rank p=0.002). In another cohort with available data on EFS for 70 patients with NKAML, the risk of an event by 3 years was 48% (95% CI 27-63%) in the low LSC score group vs. 81% (95% CI 60-91%) in the high LSC score group (HR 2.4, 95% CI 1.3-4.5, log-rank p=0.006). The LSC score was associated with poorer outcomes, independently of known prognostic factors including age, FLT3 or NPM1 mutations, and cytogenetic risk group, and added to their prognostic value. For OS in three cohorts that included patients with cytogenetic abnormalities, the HRs of the continuous LSC score in multivariate Cox regression with FLT3/NPM1 status, age, and cytogenetic risk group were respectively HR 1.07 (95%CI 1.01-1.13), p=0.02; HR 1.10 (95% CI 1.03-1.17), p=0.005; and HR 1.17 (95% CI 1.05-1.30), p=0.005.
Conclusions
High expression of a leukemic stem cell gene expression signature is independently associated with adverse outcomes in AML.
INTRODUCTION
Acute Myeloid Leukemia (AML) is an aggressive malignancy of the bone marrow characterized by accumulation of early myeloid blood cells that fail to mature and differentiate. The course of the disease is marked by poor prognosis, frequent relapse, and high disease-related mortality1-2. Recent clinical investigation has focused on the identification of prognostic subgroups in adult AML with the goal of guiding patients into risk-adapted therapies. Such investigation determined that cytogenetic abnormalities are prognostic, some favorable and others unfavorable3-4, yet up to 50% of patients have normal karyotype AML with a wide range of clinical outcomes. In these patients, the presence of specific molecular mutations can provide prognostic information, including internal tandem duplications within the FLT3 gene (FLT3-ITD), partial tandem duplication of the MLL gene (MLL-PTD), mis-localizing mutations of the NPM1 gene (NPM1c), mutations in the CEBPA and RAS genes, and increased expression of the BAALC and ERG genes5-6. However, these parameters, and others such as patient age, are only partially successful at capturing risk of relapse and patient outcomes following treatment.
A growing body of evidence suggests that specific cancer cell subpopulations possess the ability to initiate and maintain tumors 7-8. AML is the paradigm for which this cancer stem cell hypothesis has been advanced, and this model has major implications for the development of novel therapeutic agents 9. There is significant experimental evidence indicating that AML is organized as a hierarchy of malignant cells initiated and maintained by self-renewing leukemia stem cells (LSC) that comprise a subset of the total leukemic burden (eFigure 1)7, 10. These LSC are enriched in the CD34+CD38− fraction (hereon referred to as the LSC-enriched subpopulation), and in turn give rise to CD34+CD38+ leukemia progenitor cells (LPC), which further differentiate into the CD34− leukemic blast population10-11. A major implication of this cancer stem cell model is that in order to eradicate the cancer and cure the patient, the LSC must be eliminated7-8. While AML was the first human malignancy for which this model gained experimental support, its clinical significance has yet to be fully established.
We hypothesized that if the cancer stem cell model accurately reflects the biology of human AML, then patients with LSC enrichment should have worse clinical outcomes, even when accounting for known prognostic parameters, and that this association could be quantified by global gene expression profiling of bulk AML samples.
METHODS
Purification and Genomic Expression Profiling of Normal and Leukemic Cell Subpopulations
Seven human AML tumor samples were obtained at the Stanford University Medical Center between April 2005 and July 2007, according to an approved protocol of the Institutional Review Board after informed consent. Normal human bone marrow mononuclear cells were purchased from AllCells Inc. (Emeryville, CA), and human cord blood was obtained from Stanford University Medical Center. Normal and leukemic subpopulations were purified from peripheral blood and/or bone marrow by fluorescence-activated cell sorting using the antibodies shown in eFigure 1, as follows: AML LSC (n=7), AML leukemic progenitor cells (n=7), AML Blasts (n=7), normal hematopoietic stem cells (HSC; bone marrow and cord blood, n=7), normal multipotent progenitors (bone marrow and cord blood, n=7), normal common myeloid progenitors (bone marrow, n=4), normal granulocyte-monocyte progenitors (bone marrow, n=4), and megakaryocyte-erthythrocyte progenitors (bone marrow, n=4). Global transcriptional profiles were generated for each sample using Affymetrix U133 Plus 2.0 gene expression microarrays. Raw data were deposited at the NCBI Gene Expression Omnibus (GEO, accession GSE24006). Detailed experimental procedures for purification of cell subpopulations have been reported previously12.
Definition of LSC Signature Based on Gene Expression Microarray Analysis
Global gene expression profiles of 14 paired LSC-enriched and LPC-enriched subpopulations purified from the 7 AML patient samples described above were combined with 16 paired profiles (8 LSC-enriched and 8 LPC-enriched subpopulations) from an independent study 13 to produce one dataset of 30 samples for this analysis. All genes profiled on microarrays were ranked by the mean ratio of their expression between paired LSC-enriched and LPC-enriched subpopulations, and evaluated using Gene Set Enrichment Analysis14. This approach assessed whether any pre-defined groups of biologically-related genes were concordantly more highly expressed in LSC-enriched or LPC-enriched subpopulations. Individual genes expressed more highly in LSC-enriched compared to LPC-enriched subpopulations (or vice versa) were identified by using Significance Analysis of Microarrays15 (false discovery rate<10%). Ingenuity Pathways Analysis was used to identify interaction networks involving these genes.
The genes that were more highly expressed in LSC-enriched relative to LPC-enriched subpopulations were summarized by an LSC signature. The LSC signature was defined as a single number representing the relative expression of the LSC-enriched genes in a given sample compared to the other samples in the same dataset. The signature was computed as the first principal component of the gene expression data matrix whose rows were the LSC-enriched genes. Each column of the matrix represented one sample, and matrix entries were the gene expression values corresponding to each gene in each sample. By definition, the first principal component of such a data matrix is the weighted sum of the genes’ expression levels that explains the maximum possible amount of their total variation across all samples. Thus, for a group of n genes, each sample S is associated with one number: (LSC signature)S=a1g1S+a2g2S+…+angnS, where giS is the expression level of gene i in sample S, and is weighted by ai in the summation. The LSC signature was evaluated in the purified normal and leukemic subpopulations described above to investigate the expression of the LSC-enriched genes beyond the LSC-enriched and LPC-enriched subpopulations used to identify them.
Definition of LSC Score for Testing Association with AML Survival Outcomes
To test associations between the LSC-enriched genes and clinical outcomes, a retrospective training-validation scheme was adopted. Raw microarray data were obtained for four publicly available bulk AML gene expression studies with available clinical annotations16-20 from NCBI GEO (GSE12417, n=163 normal-karyotype AML only, with OS outcomes; GSE10358, n=184, OS and EFS; GSE14468, n=527, OS, EFS and RFS) and the National Cancer Institute caArray database (accession willm-00119, n=170, OS only). The largest NKAML dataset16 (n=163) was used as the training set, and the other three datasets were used for validation. The LSC signature was calculated in the training cohort and the same weights were then applied to the test cohorts. Because the genes weights were not recomputed in each test cohort to ensure unbiased validation, the resulting number for each sample was referred to as the LSC score. In the training set, the LSC signature and LSC score were identical by definition. The median LSC score in the training set was used to partition patients in all cohorts into high versus low LSC score groups.
Statistical Analysis of LSC Score on AML Survival Outcomes
The LSC score was tested for associations with survival outcomes as a continuous variable using Cox proportional hazards regression (log-likelihood test), and as a dichotomous stratification (high vs. low LSC score group) by Kaplan-Meier analysis (log-rank test), using the survival package (version 2.35) in R version 2.11. Patients with missing data were excluded from analyses. Absolute risk (AR) of events occurring by 3 years was determined from Kaplan-Meier analysis. Given that LSC have been experimentally demonstrated to be chemotherapy resistant13, 21, the LSC score was tested for associations with primary refractoriness to therapy and disease relapse (event-free and relapse-free survival). For relapse-free survival, only patients who had first achieved clinical remission from disease were included22.
The robustness of the association between the LSC score and outcomes was evaluated as follows. The training dataset was split in half. Gene weightings defining the LSC score were derived in one half of the data, and then applied to the other half to test associations with survival. Results from 1000 random splits of the training set were compared. Furthermore, the uniqueness of the prognostic value of the LSC-enriched genes was tested by comparing it to results obtained by repeating the analysis on 10000 sets of the same number of randomly selected genes.
Independent Prognostic Value of the LSC Score
Prior clinical investigation in adult AML has defined several important prognostic factors including age, karyotype (chromosomal rearrangements) and molecular mutations, particularly internal tandem duplications in the gene FLT3 (FLT3-ITD) and mis-localizing mutations in NPM1 (NPM1c)5, 23. In the current analysis, multivariate Cox regression was used to test whether the LSC score conferred prognostic value independent from these established clinical predictors. For further investigation of how the LSC score added to known prognostic factors, Area Under the Received Operating Characteristic curve (AUC-ROC) was conducted using the survivalROC package (version 1.0) in R 24. Because assignments to cytogenetic risk groups were inconsistent between different clinical groups, risk was compared in uniform fashion across datasets, by applying the refined Medical Research Council risk scheme (favorable, intermediate, adverse) based on metaphase karyotypes 23.
Since acute promyelocytic leukemia (APL) is a distinct disease entity, it was excluded from all survival analyses. Association of the LSC score with AML clinical subtypes was also assessed by ANOVA followed by Games-Howell post-hoc test (SPSS12, IBM Inc) for groups with unequal sizes and variances. All statistical tests were two-sided. P-values less than 0.05 were considered significant.
RESULTS
An LSC-Enriched Gene Expression Signature is Shared with Normal HSC
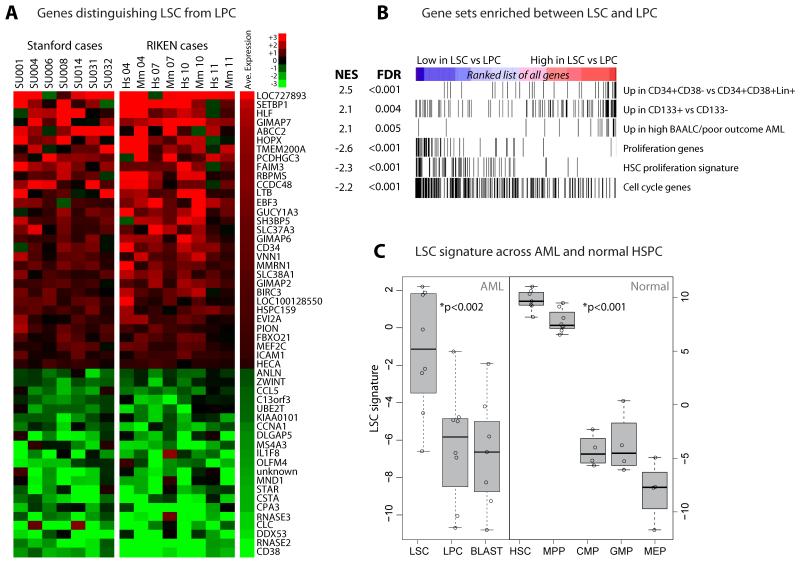
Global gene expression profiles of 15 AML LSC-enriched subpopulations were compared to 15 paired LPC-enriched subpopulations, collected from the same 15 AML samples. The samples were derived from patients representing a diversity of AML subtypes and clinical outcomes (Table 1). Significance Analysis of Microarrays 15 identified 31 genes as more highly expressed in the LSC-enriched than LPC-enriched subpopulations, and 21 genes as more highly expressed in LPC-enriched than LSC-enriched subpopulations (false discovery rate <10%; Figure 1A and eTable 1). Many of these genes were significantly associated with each other in a network-based analysis (eFigure 2). In addition to the CD34 and CD38 cell surface markers used to purify the samples, the group of genes included factors known to be differentially expressed in early hematopoiesis such as VNN1, RBPMS, SETBP1, GUCY1A3, and MEF2C (all gene names reported are the gene symbol assigned by the Human Genome Organization Gene Nomenclature Committee). Interestingly one other gene more highly expressed in the LSC-enriched subpopulation, the homeobox protein HOPX, has known interactions with the induced pluripotency factors SOX2, POU5F1, and NANOG, as well as the histone deacetylase HDAC2 (eFigure 3).
Table 1. Characteristics of Patient Samples Used to Identify LSC Signature Genes.
For Stanford patients 12, age (at initial diagnosis), gender, cytogenetic abnormalities, FAB subtype, FLT3-ITD status, time from diagnosis to last follow-up, and status at last follow-up are reported. For the RIKEN dataset13, only FAB subtype and gender was available. Unknown entries are dashed.
| Institution | Sample ID |
Age | Gender | De Novo/ Relapsed |
Cytogenetics | FAB | FLT3- ITD |
Time to last follow-up (days) |
Status at last follow- up |
|---|---|---|---|---|---|---|---|---|---|
| Stanford12 | SU001 | 59 | Female | Relapsed | Normal | M2 | Negative | 32 | DEAD |
| SU004 | 47 | Female | Relapsed | Normal | M5 | Positive | 74 | DEAD | |
| SU006 | 51 | Female | De Novo | - | M1 | Negative | 1196 | ALIVE | |
| SU008 | 64 | Male | De Novo | Normal | M1 | Positive | 1102 | ALIVE | |
| SU014 | 59 | Male | De Novo | Normal | n/a | Positive | 23 | ALIVE | |
| SU031 | 31 | Female | De Novo | Complex | M4 | Negative | 708 | ALIVE | |
| SU032 | 47 | Male | De Novo | Normal | M5 | Negative | 226 | ALIVE | |
| RIKEN13 | Hs04 | - | Male | De Novo | - | M2 | - | - | - |
| Hs07 | - | Female | De Novo | - | M4 | - | - | - | |
| Hs10 | - | Male | De Novo | - | M2 | - | - | - | |
| Hs11 | - | Male | De Novo | - | M1 | - |
Figure 1. LSC-Enriched Subpopulations Have a Distinct Gene Expression Signature That is Shared with Normal HSC.
Genes distinguishing leukemic stem cells (LSC) from leukemic progenitor cells (LPC). (A) Gene expression heatmap, with each column representing the difference in expression between LSC/LPC-enriched subpopulations isolated from the same AML patient12-13; ‘Hs’ denotes LSC/LPC profile purified from primary human patient specimen, and ‘Mm’ represents corresponding samples from mouse xenografts. 52 unique genes were identified as differentially expressed between LSC and LPC at 10% false discovery rate (eTable 1), with red indicating higher expression in LSC. (B) Enrichment analysis of relative expression between LSC and LPC of 17119 genes for the samples depicted in panel A (see eTable 2 for gene set definitions). Vertical bars in each of the six rows represent genes from each of the indicated gene sets. All nominal p-values were <0.001. NES: normalized enrichment score14; FDR: false discovery rate. (C) Expression of the LSC signature across AML subpopulations (left) and normal hematopoietic stem and progenitor cell (HSPC) populations involved in myeloid differentiation (right), including AML leukemic stem cell (LSC), leukemic progenitor cell (LPC), and leukemic blast (BLAST) populations, as well as normal hematopoietic stem cell (HSC), multipotent progenitor (MPP), common myeloid progenitor (CMP), granulocyte-monocyte progenitor (GMP), and megakaryocyte-erythrocyte progenitor (MEP). LSC and LPC samples are the same as those whose paired differences are depicted in panel A (Stanford cases). Boxes span the interquartile range, with median depicted by the thick horizontal bar. Each circle marks one sample. P-values were derived from Wilcoxon test comparing LSC to LPC/Blast, and for HSC/MPP compared to CMP/GMP/MEP.
Gene Set Enrichment Analysis14 showed that genes more highly expressed in LSC were enriched for those expressed in normal CD34+CD38− cells, which include normal hematopoietic stem cells (HSC), compared to normal CD34+CD38+ progenitors (Figure 1B and eTable 2). Notably, genes more highly expressed in LSC were also enriched for genes whose expression in AML has been correlated with high expression of the gene BAALC, an adverse prognostic factor25. Conversely, proliferation, cell cycle, and differentiation genes were systematically repressed in the LSC-enriched subpopulations when compared to more differentiated LPC-enriched subpopulations, consistent with a tendency for replicative quiescence10.
The 31 genes more highly expressed in LSC were combined to generate an LSC signature as described in the Methods. The LSC signature was computed in the purified subpopulations from primary AML patient samples and in subpopulations from the normal hierarchy of differentiating myeloid blood cells. By definition, the LSC signature was high in LSC compared to LPC, but also relative to downstream, more differentiated CD34− blasts (Figure 1C). Among normal hematopoietic cells from healthy individuals, the LSC signature was high in HSC and multipotent progenitors, compared to more mature myeloid cell populations (Figure 1C). These observations suggest that the LSC signature is shared with normal HSC, implying that it may reflect self-renewal ability and relative proliferative quiescence.
High LSC Score is Associated with Inferior Overall Survival
We next evaluated whether expression of LSC-enriched genes was associated with clinical outcomes using four public datasets of bulk AML expression profiles with available clinical annotations. Details of patient characteristics, primary therapies, clinical responses, remission rates, and outcomes have been reported previously16-19, 26, and are summarized in Table 2.
Table 2. Summary of Four Independent AML Cohorts with Public Gene Expression Data on Bulk Samples.
Summary indicates size of study, type of AML samples, age of patients (median and range) and follow-up periods. Microarray platform and database accession (GEO or caArray) are indicated. Dates of study reflect earliest patient enrollment to most recent follow-up as captured by clinical trial registration, enrollment, and publication dates. Primary therapyprotocol and survival data available for each study (response to therapy, OS, EFS, RFS) are summarized. *7 days of infusional cytarabine and 3 days of anthracycline.
| AML Cohort | ||||
|---|---|---|---|---|
| Metzeler16, 55 | Wouters19,26 | Tomasson17, 20 | Wilson18 | |
| Adult AML, Normal karyotype |
Adult AML, Mixed karyotypes |
Adult AML, Mixed karyotypes |
Adult AML, Mixed karyotypes |
|
|
Median age, yrs
(range) |
60 (17-85) | 46 (15-77) | 47 (16-81) | 65 (20-84) |
| Patients (n) | 163 | 526 | 188 | 170 |
|
Median follow-up,
months (inter quartile range) |
9 (3-27) | 17 (7-73) | 25 (10-55) | 10 (2-22) |
|
Cooperative
Group |
German AML Cooperative Group (AMLCG) |
Dutch-Belgian Hematology-Oncology Cooperative (HOVON) |
Washington University (WU) and CALGB |
Southwest Oncology Group (SWOG) |
|
Primary Therapy
Protocol(s) |
AMLCG 1999 | Multiple HOVON trials:PMIDs 9396403, 12930926, 15070662 |
WashU: Primarily 7+3*; CALGB 9621/9222/9191/9710 |
S9031/ S9333/ S9034/ S9500/ S9126 |
| Dates of study | 1999-2007 | 1992-2008 | 1993-2007 | 1993-2004 |
|
Patient Outcome
Data |
OS | Response to Primary Therapy; OS; EFS; RFS |
OS; EFS | Response to Primary Therapy; OS |
|
Microarray
Platform(s) |
Affymetrix HG- U133A&B |
Affymetrix HG-U133 Plus 2.0 |
Affymetrix HG-U133 Plus 2.0 |
Affymetrix HG- U95Av2 |
|
Dataset
Accession |
GSE12417 | GSE14468 | GSE10358 | NCI-caArray-willm- 00119 |
The LSC signature was calculated for a training set of 163 AML patients with no chromosomal abnormalities (NKAML)16, defining the weights for combining expression levels of the 31 LSC genes into a single measure for each sample (eTable 3). Because the same gene weights were applied to the independent test cohorts, we refer to this as the LSC score. In the training set, the LSC score ranged from 17.4 to 33.1 (median 24.9), and was associated with overall survival as a continuous variable (p<0.0001 log-likelihood, hazard ratio [HR] 1.15, 95% Confidence Interval [CI] 1.08-1.22), with higher LSC score associating with inferior outcome (Table 3). Stratification of patients into high versus low LSC score groups robustly separated survival curves (p=0.002 log-rank, HR 1.85, 95% CI 1.25-2.74; Table 3 and Figure 2A). The absolute risk of an event by 3 years (AR1) was 57% (95% CI 43-67%) in the low LSC score group versus 78% (95% CI 66-86%) in the high LSC score group. Association of the LSC-enriched genes with OS was supported by internal cross-validation in the training cohort (eFigure 4).
Table 3. The LSC Score As a Univariate Predictor of Survival in Four Independent AML Cohorts, Stratified by AML Subtype and Outcome Variable.
Prognostic value of the LSC score, FLT3-ITD mutation status, NPM1 mutation status, age, and cytogenetic risk are shown for OS, EFS, and RFS for the datasets described. Shown are the hazard ratios (HR) with 95% confidence intervals, and p-value (p), computed using log-likelihood test for continuous predictors, and log-rank test for discrete predictors. Units for variables are as follows: Age in years; continuous LSC score as log2 of gene expression intensity; dichotomous LSC score as high (1) vs low (0); FLT3 and NPM1 as mutated (1) vs wild-type (0); cytogenetic risk as favorable (1), intermediate (2), adverse (3) per the modified Medical Research Council scheme23. Patients with APL were excluded.
| Cohort | Wouters19, 26 (test) | Tomasson17, 20 (test) | Wilson18 (test) | Metzeler16, 55 (training) | |||||
|---|---|---|---|---|---|---|---|---|---|
| Predictor | HR (95% CI) | p | HR (95% CI) | p | HR (95% CI) | p | HR (95% CI) | p | |
| OS, NKAML | Number of patients | 99 | 70 | 65 | 163 | ||||
| LSC score (continuous) | 1.17 (1.07-1.28) | 0.0007 | 1.13 (1.04-1.22) | 0.003 | 1.18 (1.04-1.34) | 0.011 | 1.15 (1.08-1.22) | <0.001 | |
| LSC score (dichotomous) | 1.86 (1.15-3.02) | 0.01 | 2.70 (1.43-5.10) | 0.002 | 2.55 (1.44-4.51) | <0.001 | 1.85 (1.25-2.74) | 0.002 | |
| FLT3-ITD | 1.84 (1.14-2.98) | 0.012 | 2.68 (1.42-5.07) | 0.002 | 1.28 (0.73-2.23) | 0.39 | 2.22 (1.49-3.31) | <0.001 | |
| NPM1c | 0.76 (0.47-1.23) | 0.26 | 1.55 (0.86-2.79) | 0.14 | 0.67 (0.38-1.17) | 0.16 | 0.79 (0.54-1.17) | 0.24 | |
| Age | 1.01 (0.98-1.03) | 0.6 | 1.02 (1.00-1.04) | 0.06 | 1.03 (1.00-1.05) | 0.029 | 1.03 (1.01-1.04) | <0.001 | |
| OS, all non-APL | Number of patients | 219 | 137 | 170 | |||||
| LSC score (continuous) | 1.07 (1.02-1.13) | 0.009 | 1.10 (1.04-1.17) | 0.001 | 1.15 (1.07-1.25) | <0.001 | |||
| LSC score (dichotomous) | 1.36 (0.98-1.88) | 0.07 | 2.01 (1.27-3.18) | 0.003 | 1.99 (1.43-2.79) | <0.001 | |||
| FLT3-ITD | 1.82 (1.28-2.59) | <0.001 | 1.82 (1.09-3.02) | 0.019 | 1.12 (0.78-1.62) | 0.53 | |||
| NPM1c | 0.86 (0.60-1.22) | 0.39 | 1.49 (0.95-2.32) | 0.079 | 0.79 (0.55-1.14) | 0.2 | |||
| Age | 1.01 (1.00-1.03) | 0.082 | 1.03 (1.01-1.04) | <0.001 | 1.03 (1.02-1.05) | <0.001 | |||
| Cytogenetic Risk Group | 2.04 (1.54-2.69) | <0.001 | 1.97 (1.34-2.90) | <0.001 | 2.16 (1.52-3.06) | <0.001 | |||
| EFS, NKAML | Number of patients | 99 | 70 | ||||||
| LSC score (continuous) | 1.15 (1.06-1.26) | 0.001 | 1.11 (1.03-1.21) | 0.007 | |||||
| LSC score (dichotomous) | 1.69 (1.07-2.69) | 0.02 | 2.39 (1.26-4.52) | 0.006 | |||||
| FLT3-ITD | 2.12 (1.33-3.38) | 0.001 | 2.44 (1.23-4.82) | 0.008 | |||||
| NPM1c | 0.92 (0.58-1.47) | 0.74 | 1.29 (0.70-2.37) | 0.41 | |||||
| Age | 1.00 (0.98-1.02) | 0.96 | 1.01 (0.99-1.03) | 0.25 | |||||
| RFS, NKAML | Number of patients | 85 | |||||||
| LSC score (continuous) | 1.13 (1.01-1.27) | 0.03 | |||||||
| LSC score (dichotomous) | 1.78 (0.99-3.25) | 0.055 | |||||||
| FLT3-ITD | 2.80 (1.53-5.14) | 0.0005 | |||||||
| NPM1c | 1.09 (0.59-2.02) | 0.79 | |||||||
| Age | 0.99 (0.97-1.02) | 0.63 | |||||||
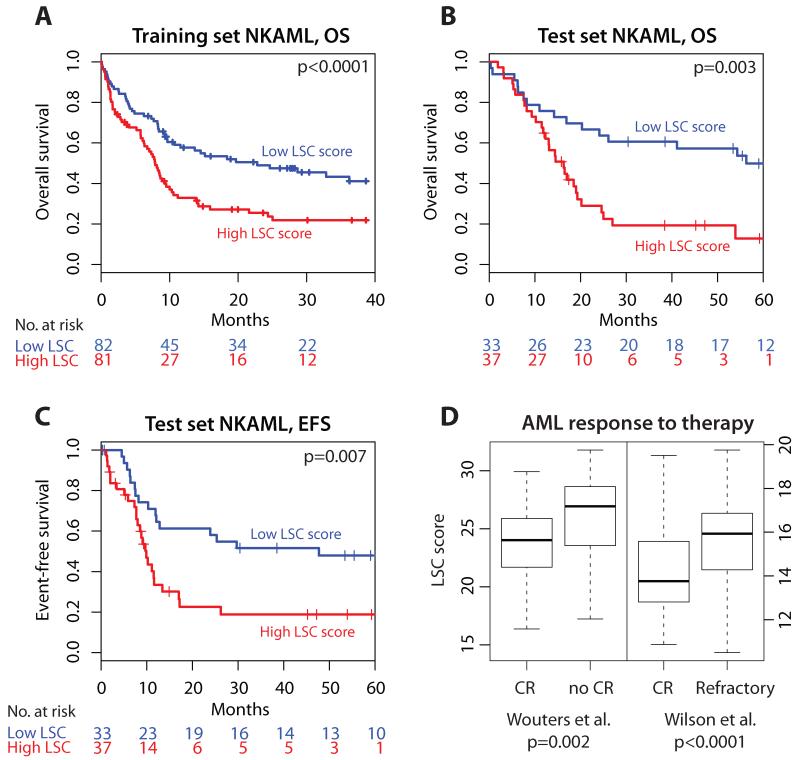
Figure 2. Higher LSC Score is Associated with Worse Outcomes.
Kaplan-Meier analysis of the association between the LSC score and survival outcomes in normal karyotype AML (NKAML). Excluding those with acute promyelocytic leukemia (APL), patients were split into high versus low LSC score groups according to the median value of the LSC score in the training cohort. Stratification of outcomes using this approach is depicted for OS of NKAML patients in the training set16 (A), in NKAML from one of the validation sets17 for OS (B), and for EFS (C). Vertical ticks on curves indicate censored events, and p-values shown are for the LSC score as a continuous predictor of survival (log-likelihood test; log-rank estimates provided in Table 3). Similar results were obtained in additional independent datasets (eFigure 5 and Table 3). (D) The LSC score was significantly associated with initial therapeutic response as determined by the ability to achieve clinical remission in two datasets for which this information was available18-19 (p-values derived from t-test). Boxes indicate the interquartile range, with median shown as the thick horizontal bar. Numbers within boxes indicate the sample sizes. OS=overall survival; EFS=event-free survival; CR=clinical remission.
The LSC score was calculated for each sample in the three independent test cohorts. For the NKAML cases in these cohorts, high LSC score was associated with inferior OS as a continuous variable (p<0.012 in all cases; with HR 1.17, 95% CI 1.07-1.28; HR 1.13, 95% CI 1.04-1.22; and HR 1.18, 95% CI 1.04-1.34 in the three cohorts; Table 3). Using the median LSC score from the training set as a pre-specified threshold, stratification of patients into high versus low LSC score groups significantly separated survival curves in each dataset (Table 3, Figure 2B and eFigure 5). For example, in NKAML patients from one well-characterized cohort of adult AML patients with diverse karyotypes primarily treated with induction regimens including cytarabine and an anthracycline (Tomasson et al.)17, 20, the LSC score ranged from 16.6 to 31.0 and was significantly associated with OS (Table 3 and Figure 2B; see Table 4 for medians and interquartile ranges of the LSC score in all cohorts). This association was significant whether the LSC score was evaluated as a continuous predictor (p=0.003; HR 1.13, 95% CI 1.04-1.22), or a high versus low LSC score split (p=0.002; HR 2.7, 95% CI 1.4-5.1), with those in the low LSC group having a median OS of 56.3 months (AR 0.39, 95% CI 0.20-0.54) compared to 16.3 months (AR 0.81, 95% CI 0.61-0.90) for those in the high LSC group (Table 4). The set of genes comprising the LSC score was significant in its prognostic utility when compared to 10000 randomly selected gene sets of the same size (eFigure 6), supporting the conclusion that the association with clinical outcomes was not a false positive result.
Table 4. Range of LSC Scores Across AML Cohorts Subjected to Survival Analyses.
The median and interquartile range (IQR) of the LSC score is reported for each of the four AML cohorts, separately for NKAML and non-APL subtypes. The dataset of Wilson et al. does not have probes for some of the LSC genes, hence the range is different from the other three cohorts. Also shown is median survival for specified endpoints in low and high LSC score groups, together with the comparative risk (percentage of patients having an event) by 36 months in the low and high LSC score groups. Corresponding hazard ratios and p-values are presented in Table 3.
| AML Cohort |
n | LSC score, median (IQR) |
End- point |
Median survival, months |
Comparative absolute risk of event by 3 years, % |
|||
|---|---|---|---|---|---|---|---|---|
| Low LSC score group |
High LSC score group |
Low LSC score group (95% CI) |
High LSC score group (95% CI) |
|||||
| NKAML | Metzeler 16, 55 | 163 | 24.9 (22.6-27.0) |
OS | 22.8 | 7.9 | 57 (43-67) | 78 (66-86) |
| Tomasson 17, 20 | 74 | 25.2 (22.3-27.6) |
OS | 56.3 | 16.3 | 39 (20-54) | 81 (61-90) | |
| EFS | 47.7 | 9.9 | 48 (27-63) | 81 (60-91) | ||||
| Wouters 19, 26 | 181 | 25.0 (22.6-27.0) |
OS | 31.3 | 8.4 | 52 (36-63) | 73 (57-84) | |
| EFS | 14.0 | 7.4 | 61 (46-72) | 80 (64-89) | ||||
| RFS | 65.6 | 10.4 | 43 (26-56) | 68 (46-81) | ||||
| Wilson 18 | 65 | 14.0 (12.8-15.6) |
OS | 23.7 | 7.3 | 58 (39-72) | 93 (72-98) | |
| non-APL AML | Tomasson 17, 20 | 143 | 25.7 (23.1-28.4) |
OS | 56.3 | 16.5 | 45 (30-57) | 75 (64-83) |
| Wouters 19, 26 | 392 | 25.6 (23.4-28.2) |
OS | 25.0 | 14.5 | 55 (44-64) | 69 (59-76) | |
| Wilson18 | 170 | 14.7 (13.4-16.3) |
OS | 15.9 | 6.6 | 67 (56-76) | 93 (84-97) | |
The LSC score ranged from 16.6 to 35.5 among all non-APL AML patients, including those with cytogenetic abnormalities, in the Tomasson et al. cohort (Table 4), and was again associated with OS (p=0.001, HR 1.10, 95% CI 1.04-1.17) as a continuous variable. Patients in the low LSC score group had a median OS of 56.3 months (AR 0.45, 95% CI 0.30-0.57) compared to 16.5 months (AR 0.75, 95% CI 0.64-0.83) in the high LSC score group (p=0.003, HR 2.0, 95% CI 1.3-3.2). Investigation of the LSC score in patients from two additional cohorts including patients with chromosomal abnormalities confirmed its association with adverse OS in both (eFigure 5 and Table 3).
Higher LSC Score is Associated with Inferior EFS, Refractoriness to Treatment, and Disease Relapse
Higher LSC scores were consistently associated with inferior EFS in patients with NKAML from two cohorts with available data (p=0.001, HR 1.15, 95% CI 1.06-1.26; and p=0.007, HR 1.11, 95% CI 1.03-1.21 as a continuous variable). As with OS, the LSC score-high group had inferior EFS, with a median of 10 months (AR 0.81, 95% CI 0.60-0.91) compared to 48 months (AR 0.48, 95% CI 0.27-0.63) in the low LSC score group of the Tomasson et al. cohort17 (p=0.006, HR 2.4, 95% CI 1.3-4.5; Figure 2C, and Tables 3 and 4). In the second cohort with available EFS data19, high versus low LSC score groupings separated survival curves (eFigure 5; p=0.02; HR 1.7, 95% CI 1.1-2.7; AR 61%, 95% CI 46-72% in the low LSC score group vs. AR 80%, 95% CI 64-89% in the high group). For the latter dataset19, LSC score was also associated with RFS in NKAML patients who had achieved an initial clinical remission (p=0.03, HR 1.13, 95% CI 1.01-1.27 as a continuous variable), with median RFS of 66 months (AR 43%, 95% CI 26-56%) in the low LSC score cases, compared to only 10 months (AR 68%, 95% CI 46-81%) in the high group (p<0.06, HR 1.8, 95% CI 1.0-3.3; eFigure 5). This finding is consistent with the demonstrated chemo-resistance of LSC13.
Concordantly, the rate of clinical remission (CR) was superior among AML patients with low LSC score compared to those with high LSC score, both in an older cohort (median age 65y, 56% CR for low LSC score vs. 29% for high LSC score, p<0.001 by Fisher exact test)18, and a younger one (median age 43y, 88% vs. 76%, p=0.02)19, 26. Furthermore, LSC scores were significantly higher in patients failing to achieve clinical remission compared to those who reached CR (p<0.001, Figure 2D), a distinction most evident for those patients in whom such remissions were durable.
The LSC Score is Independently Associated with Clinical Outcomes
The LSC score carried prognostic value independent of other known clinical factors including age, FLT3/NPM1 mutations, and cytogenetic risk group. It was significant independent of these factors in multivariate Cox regression, with high LSC score again being associated with adverse OS, EFS, and RFS, in all but one instance (Table 5). Comparisons of Area Under the Receiver Operator Characteristic curves (AUC-ROC, a measure of the accuracy of a prognostic model) showed that the LSC score added to the prognostic value of age, FLT3-ITD, NPM1c, and cytogenetic risk in predicting OS at 2 years in all cohorts for both NKAML and non-APL AML (eTable 4). Models that incorporated the LSC score in addition to other known prognostic factors had consistently higher AUC-ROC, when compared to models that did not include the LSC score (see eSupplement for further details).
Table 5. Multivariate Survival Analysis Including the LSC Score in Four Independent AML Cohorts, Stratified by AML Subtype and Outcome Variable.
The LSC score was tested as a multivariate predictor in combination with age, FLT3-ITD status, NPM1 status, age, and cytogenetic risk group using Cox regression. Hazard ratios (HR) and p-values (p), using log-likelihood test, are reported for each variable within the multivariate model. The overall log-likelihood p-value for the model is also indicated. The number of patients (n) differs from those in Table 3, depending on whether information on all covariates was available in each case. Units are as in Table 3. Patients with APL were excluded
| Wouters19, 26 (test) | Tomasson17, 20 (test) | Wilson18 (test) | Metzeler16, 55 (training) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Predictor | HR (95% CI) | p | HR (95% CI) | p | HR (95% CI) | p | HR (95% CI) | p | |
| OS, NKAML | Patients (n) | 99 | 70 | 63 | 162 | ||||
| LSC score | 1.16 (1.05-1.27) | 0.003 | 1.15 (1.06-1.26) | 0.002 | 1.14 (0.97-1.34) | 0.1 | 1.10 (1.03-1.17) | 0.006 | |
| FLT3-ITD | 1.94 (1.15-3.27) | 0.013 | 3.00 (1.50-6.00) | 0.002 | 2.05 (1.10-3.85) | 0.025 | 2.19 (1.42-3.37) | <0.001 | |
| NPM1c | 0.73 (0.42-1.27) | 0.27 | 1.58 (0.83-3.01) | 0.17 | 0.82 (0.42-1.61) | 0.57 | 0.87 (0.58-1.30) | 0.49 | |
| Age | 1.02 (1.00-1.04) | 0.087 | 1.02 (1.00-1.04) | 0.14 | 1.03 (1.00-1.06) | 0.026 | 1.03 (1.01-1.04) | <0.001 | |
| Overall | <0.001 | <0.001 | 0.01 | <0.001 | |||||
| OS, all non-APL | Patients (n) | 219 | 137 | 136 | |||||
| LSC score | 1.07 (1.01-1.13) | 0.02 | 1.10 (1.03-1.17) | 0.005 | 1.17 (1.05-1.30) | 0.005 | |||
| FLT3-ITD | 1.98 (1.35-2.91) | <0.001 | 2.00 (1.18-3.37) | 0.01 | 1.45 (0.91-2.30) | 0.12 | |||
| NPM1c | 0.70 (0.46-1.06) | 0.094 | 1.64 (1.01-2.65) | 0.045 | 0.93 (0.55-1.60) | 0.8 | |||
| Age | 1.02 (1.00-1.03) | 0.023 | 1.02 (1.01-1.04) | 0.007 | 1.03 (1.01-1.04) | 0.002 | |||
| Cytogenetic Risk Group |
2.02 (1.53-2.67) | <0.001 | 1.86 (1.26-2.76) | 0.002 | 1.99 (1.37-2.89) | <0.001 | |||
| Overall | <0.001 | <0.001 | <0.001 | ||||||
| EFS, NKAML | Patients (n) | 99 | 70 | ||||||
| LSC score | 1.14 (1.04-1.24) | 0.004 | 1.13 (1.04-1.23) | 0.005 | |||||
| FLT3-ITD | 2.15 (1.30-3.55) | 0.003 | 2.86 (1.37-5.94) | 0.005 | |||||
| NPM1c | 0.88 (0.52-1.48) | 0.62 | 1.27 (0.67-2.42) | 0.46 | |||||
| Age | 1.01 (0.99-1.03) | 0.35 | 1.01 (0.99-1.03) | 0.38 | |||||
| Overall | <0.001 | 0.003 | |||||||
| RFS, NKAML | Patients (n) | 85 | |||||||
| LSC score | 1.12 (1.00-1.25) | 0.05 | |||||||
| FLT3-ITD | 2.83 (1.47-5.45) | 0.002 | |||||||
| NPM1c | 0.95 (0.47-1.91) | 0.89 | |||||||
| Age | 1.00 (0.98-1.03) | 0.78 | |||||||
| Overall | 0.004 | ||||||||
In NKAML, higher LSC score associated with inferior OS in NPM1-wild-type and NPM1-mutant cases, despite the fact that the latter are frequently CD34-negative, and in patients with both wild-type FLT3 and wild-type NPM1 (eTable 5). Furthermore, when all analyses (including derivation of LSC score gene weightings) were performed excluding CD34-negative cases (defined either as NPM1 mutant, or as the 40% of samples with lowest CD34 expression), similar results were obtained. Exclusion of CD34 from the model-building and validation resulted in an LSC score with similar gene weightings and prognostic value (eTable 3). Therefore, the LSC score was not simply a proxy for CD34 status.
Taken together, these data indicate that higher LSC score is associated with inferior survival outcomes independent of age, FLT3-ITD, NPM1 mutations, CD34 expression, and cytogenetic risk group, and adds to their prognostic utility.
Lower LSC Score is Associated with Prognostically Favorable AML Subtypes
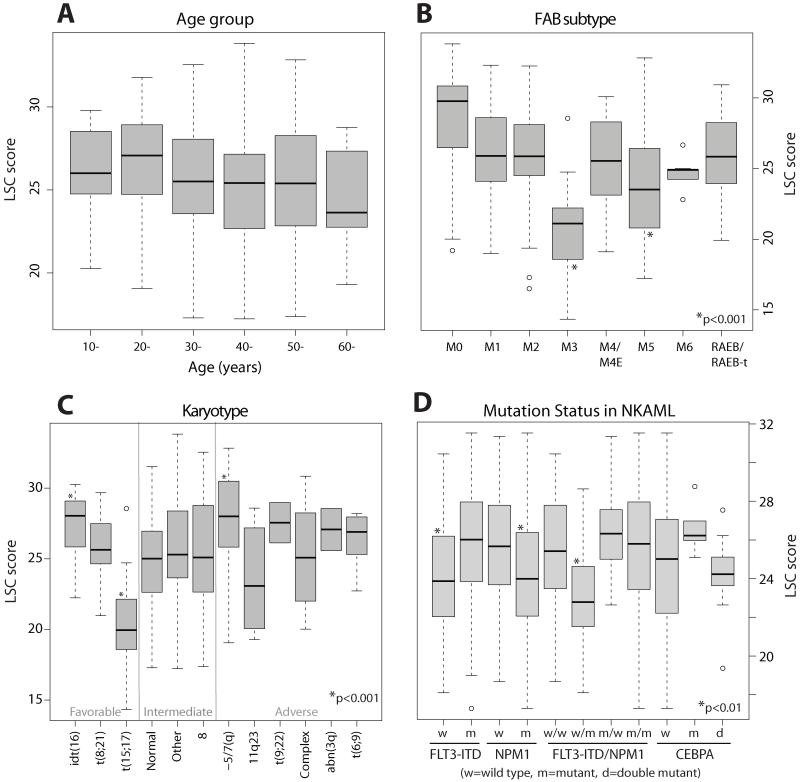
Though similar across most age groups and morphological subtypes, LSC scores were higher in cases with minimally differentiated myeloblasts (French-American British M0), which typically have poor prognosis27, consistent with previous reports of high LSC prevalence in this subtype (eFigure 7)28. In general, the LSC score was similar in favorable, intermediate, and adverse cytogenetic risk groups, and was not a direct proxy for this factor (eFigure 7C). This is consistent with our findings from survival analysis that the LSC score confers independent prognostic value. When considering specific cytogenetic subgroups, the LSC score had higher than average values in patients with unfavorable −5 or 7(q) abnormalities, and lower than average values among AML harboring anomalies involving 11q23/MLL. Recent studies have reported that self-renewing cells from AML mouse models carrying MLL anomalies reside in more mature cells29.
We also investigated the relationship of the LSC score to molecular mutations in the largest single cytogenetic subgroup of AML, NKAML. LSC scores were significantly lower in those harboring NPM1c mutations (eFigures 7D,8), in agreement with recent observations that leukemia initiating cells in NPM1 mutant AML are frequently CD34 negative30. Furthermore, LSC scores were significantly lower within the subgroup of patients with wild type FLT3 but mutant NPM1c, a combination conferring a distinctly favorable prognosis in NKAML (eFigure 7D)6. LSC scores were also lower in NKAML with double CEBPA mutations, again associated with favorable outcomes 19, relative to cases with single mutants, but not relative to wild-type CEBPA. Similar findings were observed in all four independent datasets totaling 1047 patients (eFigure 8 and 9). Of note, no significant differences in LSC scores were observed when patients with AML were stratified according to less common recurrent somatic mutations, including those in the tyrosine kinase domain of FLT3 (FLT3-TKD), or activating mutations in NRAS, KRAS, or IDH1.
COMMENT
Clinical evidence supporting the significance of the cancer stem cell model for human AML has been lacking despite ample experimental evidence from transplantation assays in immune-compromised mice. In this study, we show that a gene expression score associated with the LSC-enriched subpopulation is an independent prognostic factor in AML, with high score associated with adverse outcomes in multiple independent cohorts. Specifically, high LSC score is associated with poor OS, EFS, and RFS in NKAML, and inferior OS in patients with chromosomal abnormalities. Additionally, the LSC score was associated with primary response to induction chemotherapy, as high scores strongly correlated with lower remission rates. Multivariate analysis demonstrated that the score associated with poor outcomes independently of age, FLT3 or NPM1 mutations, and cytogenetic risk group. These findings support the clinical relevance of the cancer stem cell model for AML.
AML stem cells were originally identified by prospectively separating primary leukemic specimens into subpopulations based on expression of CD34 and CD38, surface markers that are differentially expressed in normal hematopoiesis (eFigure 1)10. When the function of these tumor subpopulations was assessed by transplantation into immune-deficient mice, leukemia-initiating activity was demonstrated exclusively in the CD34+CD38− fraction11. The majority of recent studies indicate that AML LSC activity is enriched in the CD34+CD38− subpopulation, although recent reports have challenged whether this is exclusive 30-31. The clinical significance of the leukemia stem cell model is suggested by two prior studies, the first of which identified an inverse correlation between the frequency of CD34+CD38− cells at diagnosis and the duration of relapse-free survival32. The second study reported that the relative ability of AML cells to successfully engraft in immune-deficient mice (a property associated with LSC) correlated with adverse clinical features33. While suggestive, neither of these studies investigated large cohorts of patients with long-term follow-up and diverse clinical features.
Notably, the LSC signature was highly expressed in purified HSC, and much lower in more differentiated myeloid progenitor cells, suggesting that it may be reflective of self-renewal ability. Despite the observed similarities between the LSC signature and HSC gene expression programs, therapeutic targeting of leukemic stem cells is still possible without toxicity toward normal HSC. Indeed, markers distinguishing LSC from HSC exist and are amenable to targeted therapies, including antibodies to CD47, CLL-1, and CD12334-36. Future work is needed to prospectively validate the prognostic ability of the LSC score by evaluating its component genes using RT-PCR in an independent patient cohort. It will also be pertinent to examine the relationship of the LSC score to other gene expression signatures that have been proposed for predicting survival in AML16, 26, 37.
In addition to the markers CD34 and CD38 which were employed for their purification, LSC were distinguished from LPC by the expression of several other genes known to be differentially expressed during early myelopoiesis. These included three members (GIMAP2, GIMAP6, and GIMAP7) of a small family of immune-associated nucleotide-binding proteins implicated in survival of hematopoietic cells and leukemia38; however, no prior associations with AML have been described. Two genes (HOPX, GUCY1A3) in this signature, which have previously been incorporated into AML prognostic models16, 37, are notable for their distinctive pattern of expression and histone modification in self-renewing cells39. HOPX is an unusual homeodomain protein known to directly recruit histone deacetylase activity without directly binding DNA40, and to be directly repressed in vivo in malignant cells in response to administration of the histone deacetylase inhibitor panobinostat41. The latter is currently being studied in clinical trials for patients with AML. GUCY1A3, which encodes a component of the soluble guanylate cyclase enzyme catalyzing the conversion of GTP to cGMP, is repressed during replicative senescence42, and cGMP has been reported to stimulate HSC proliferation43.
The cancer stem cell model has been studied in solid tumors in addition to leukemia7. Investigation of gene expression in human breast cancer stem cells identified a signature prognostic of metastasis-free and overall survival in multiple carcinomas, suggesting the clinical significance of the cancer stem cell model in these solid tumors44. Among other human malignancies, we and others have described prognostic significance of distinctive signatures of self-renewing populations including embryonic stem cells45-46 and HSC and progenitor cells47-48. However, the current work represents the first to directly define a signature of enriched AML-initiating cells, and to relate this signature to expression profiles of diagnostic specimens, allowing a link to corresponding clinical and pathological features of patients. Ultimately, this model has major implications for cancer therapy, most notably that in order to achieve cure, the cancer stem cells must be eliminated7. To accomplish this in AML, novel therapies targeting LSC must be developed. Several such therapies are being investigated including small molecules21, 49-51 and monoclonal antibodies34-35, 52, which hold promise for improving therapeutic efficacy beyond current conventional treatments.
Our LSC prognostic model requires validation in a prospective study of AML patients treated with a standardized protocol incorporating strict eligibility criteria, a uniform treatment plan, uniform sample collection and handling, and well-defined primary endpoints. Moreover, microarrays are not currently broadly employed in clinical decision making 53. While surrogate methods such as real-time PCR have demonstrated clinical utility54, their application requires performance assessment in independent laboratories. Flow cytometric analysis of the predictive power of the proteins comprising the LSC score, and comparison to RNA-based models, may help to determine the best platform for future clinical application. However, monoclonal antibodies useful for flow cytometry are not presently available for the full set of encoded proteins.
CONCLUSION
High expression of a leukemic stem cell gene expression signature is independently associated with adverse outcomes in AML. If prospectively validated, the described LSC score may be incorporated into routine clinical practice for predicting prognosis in patients with AML, and used in clinical trials incorporating risk-based stratification or randomization strategies.
Supplementary Material
Figure 3.
Acknowledgements
The authors would like to thank Dr. Irving Weissman, MD (Stanford University), and Dr. Ronald Levy, MD (Stanford) for support and discussion; and Dr. Rob Tibshirani, PhD (Stanford) for advice on statistical analyses. We also thank the following Stanford clinicians for critical reading of the manuscript: Dr Bruno Medeiros, MD, Dr Jason Gotlib, MD, Dr Linda Boxer, MD, PhD, and Dr Beverly Mitchell, MD. We are indebted to the patients and their physicians including the Stanford Hematology Tissue Bank, the German AML Cooperative Group, the Haemato-Oncology Foundation for Adults in the Netherlands, the Southwest Oncology Group, the German AML Study Group/Ulm, Washington University School of Medicine, and the Cancer and Leukemia Group B, for sharing raw data and clinical information from their studies. We also thank Dr. Fumihiko Ishikawa, MD, PhD, and Mr. Atsushi Hijikata, MSc (RIKEN) for providing access to their microarray data from purified AML subsets. No persons or groups acknowledged here received compensation.
Funding/Support: Dr. Majeti holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund, and Dr. Alizadeh holds a Career Development Award from the Leukemia & Lymphoma Society. This research is supported by the Integrative Cancer Biology Program through National Institutes of Health awards 1U54CA149145 and U56-CA112973 (Dr. Plevritis).
Role of the Sponsors: The funding organizations had no role in the design and conduct of the study, in the collection, analysis, and interpretation of the data, or in the preparation, review, or approval of the manuscript.
Footnotes
Throughout the text, absolute risk is reported at 3 years.
Author Contributions: All authors designed the experiments, conducted the analyses, wrote the manuscript, and endorse the full content of this work.
Data Access Statement: AJG and AAA had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis
Financial Disclosures: AJG, RM, and AAA hereby disclose that a patent application for development of the LSC score as a diagnostic assay has been submitted.
REFERENCES
- 1.Estey E, Dohner H. Acute myeloid leukaemia. Lancet. 2006 Nov 25;368(9550):1894–1907. doi: 10.1016/S0140-6736(06)69780-8. [DOI] [PubMed] [Google Scholar]
- 2.Lowenberg B, Downing JR, Burnett A. Acute myeloid leukemia. N Engl J Med. 1999 Sep 30;341(14):1051–1062. doi: 10.1056/NEJM199909303411407. [DOI] [PubMed] [Google Scholar]
- 3.Byrd JC, Mrozek K, Dodge RK, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002 Dec 15;100(13):4325–4336. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 4.Grimwade D, Walker H, Oliver F, et al. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children’s Leukaemia Working Parties. Blood. 1998 Oct 1;92(7):2322–2333. [PubMed] [Google Scholar]
- 5.Mrozek K, Marcucci G, Paschka P, Whitman SP, Bloomfield CD. Clinical relevance of mutations and gene-expression changes in adult acute myeloid leukemia with normal cytogenetics: are we ready for a prognostically prioritized molecular classification? Blood. 2007 Jan 15;109(2):431–448. doi: 10.1182/blood-2006-06-001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlenk RF, Dohner K, Krauter J, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008 May 1;358(18):1909–1918. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 7.Jordan CT, Guzman ML, Noble M. Cancer Stem Cells. N Engl J Med. 2006 Sep 21;355(12):1253–1261. doi: 10.1056/NEJMra061808. 2006. [DOI] [PubMed] [Google Scholar]
- 8.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001 Nov 1;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 9.Weissman I. Stem cell research: paths to cancer therapies and regenerative medicine. JAMA. 2005 Sep 21;294(11):1359–1366. doi: 10.1001/jama.294.11.1359. [DOI] [PubMed] [Google Scholar]
- 10.Dick JE. Stem cell concepts renew cancer research. Blood. 2008 Dec 15;112(13):4793–4807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 11.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997 Jul;3(7):730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 12.Majeti R, Becker MW, Tian Q, et al. Dysregulated gene expression networks in human acute myelogenous leukemia stem cells. Proc Natl Acad Sci U S A. 2009 Mar 3;106(9):3396–3401. doi: 10.1073/pnas.0900089106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishikawa F, Yoshida S, Saito Y, et al. Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nat Biotechnol. 2007 Nov;25(11):1315–1321. doi: 10.1038/nbt1350. [DOI] [PubMed] [Google Scholar]
- 14.Mootha V, Lindgren C, Eriksson K, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature Genetics. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 15.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001 Apr 24;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Metzeler KH, Hummel M, Bloomfield CD, et al. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood. 2008 Nov 15;112(10):4193–4201. doi: 10.1182/blood-2008-02-134411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomasson MH, Xiang Z, Walgren R, et al. Somatic mutations and germline sequence variants in the expressed tyrosine kinase genes of patients with de novo acute myeloid leukemia. Blood. 2008 May 1;111(9):4797–4808. doi: 10.1182/blood-2007-09-113027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson CS, Davidson GS, Martin SB, et al. Gene expression profiling of adult acute myeloid leukemia identifies novel biologic clusters for risk classification and outcome prediction. Blood. 2006 Jul 15;108(2):685–696. doi: 10.1182/blood-2004-12-4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wouters BJ, Lowenberg B, Erpelinck-Verschueren CA, van Putten WL, Valk PJ, Delwel R. Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood. 2009 Mar 26;113(13):3088–3091. doi: 10.1182/blood-2008-09-179895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009 Sep 10;361(11):1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guzman ML, Rossi RM, Karnischky L, et al. The sesquiterpene lactone parthenolide induces apoptosis of human acute myelogenous leukemia stem and progenitor cells. Blood. 2005 Jun 1;105(11):4163–4169. doi: 10.1182/blood-2004-10-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dohner H, Estey EH, Amadori S, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010 Jan 21;115(3):453–474. doi: 10.1182/blood-2009-07-235358. 2010. [DOI] [PubMed] [Google Scholar]
- 23.Grimwade D, Hills RK. Independent prognostic factors for AML outcome. Hematology Am Soc Hematol Educ Program. 2009:385–395. doi: 10.1182/asheducation-2009.1.385. [DOI] [PubMed] [Google Scholar]
- 24.Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000 Jun;56(2):337–344. doi: 10.1111/j.0006-341x.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 25.Langer C, Radmacher MD, Ruppert AS, et al. High BAALC expression associates with other molecular prognostic markers, poor outcome, and a distinct gene-expression signature in cytogenetically normal patients younger than 60 years with acute myeloid leukemia: a Cancer and Leukemia Group B (CALGB) study. Blood. 2008 Jun 1;111(11):5371–5379. doi: 10.1182/blood-2007-11-124958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valk PJ, Verhaak RG, Beijen MA, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004 Apr 15;350(16):1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 27.Lee EJ, Pollak A, Leavitt RD, Testa JR, Schiffer CA. Minimally differentiated acute nonlymphocytic leukemia: a distinct entity. Blood. 1987 Nov;70(5):1400–1406. [PubMed] [Google Scholar]
- 28.Costello R, Mallet F, Chambost H, et al. The immunophenotype of minimally differentiated acute myeloid leukemia (AML-M0): reduced immunogenicity and high frequency of CD34+/CD38− leukemic progenitors. Leukemia. 1999 Oct;13(10):1513–1518. doi: 10.1038/sj.leu.2401519. [DOI] [PubMed] [Google Scholar]
- 29.Cleary ML. Regulating the leukaemia stem cell. Best Practice & Research Clinical Haematology. 2009;22(4):483–487. doi: 10.1016/j.beha.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taussig DC, Vargaftig J, Miraki-Moud F, et al. Leukemia initiating cells from some acute myeloid leukemia patients with mutated nucleophosmin reside in the CD34-fraction. Blood. 2010 Jan 6;115(10):1976–1984. doi: 10.1182/blood-2009-02-206565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taussig DC, Miraki-Moud F, Anjos-Afonso F, et al. Anti-CD38 antibody-mediated clearance of human repopulating cells masks the heterogeneity of leukemia-initiating cells. Blood. 2008 Aug 1;112(3):568–575. doi: 10.1182/blood-2007-10-118331. [DOI] [PubMed] [Google Scholar]
- 32.van Rhenen A, Feller N, Kelder A, et al. High stem cell frequency in acute myeloid leukemia at diagnosis predicts high minimal residual disease and poor survival. Clin Cancer Res. 2005 Sep 15;11(18):6520–6527. doi: 10.1158/1078-0432.CCR-05-0468. [DOI] [PubMed] [Google Scholar]
- 33.Pearce DJ, Taussig D, Zibara K, et al. AML engraftment in the NOD/SCID assay reflects the outcome of AML: implications for our understanding of the heterogeneity of AML. Blood. 2006 Feb 1;107(3):1166–1173. doi: 10.1182/blood-2005-06-2325. 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin L, Lee EM, Ramshaw HS, et al. Monoclonal antibody-mediated targeting of CD123, IL-3 receptor alpha chain, eliminates human acute myeloid leukemic stem cells. Cell Stem Cell. 2009 Jul 2;5(1):31–42. doi: 10.1016/j.stem.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 35.Majeti R, Chao MP, Alizadeh AA, et al. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. 2009 Jul 23;138(2):286–299. doi: 10.1016/j.cell.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Rhenen A, van Dongen GAMS, Kelder A, et al. The novel AML stem cell associated antigen CLL-1 aids in discrimination between normal and leukemic stem cells. Blood. 2007 Oct 1;110(7):2659–2666. doi: 10.1182/blood-2007-03-083048. 2007. [DOI] [PubMed] [Google Scholar]
- 37.Bullinger L, Dohner K, Bair E, et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med. 2004 Apr 15;350(16):1605–1616. doi: 10.1056/NEJMoa031046. [DOI] [PubMed] [Google Scholar]
- 38.Nitta T, Takahama Y. The lymphocyte guard-IANs: regulation of lymphocyte survival by IAN/GIMAP family proteins. Trends in Immunology. 2007;28(2):58–65. doi: 10.1016/j.it.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 39.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A Chromatin Landmark and Transcription Initiation at Most Promoters in Human Cells. Cell. 2007;130(1):77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kook H, Lepore JJ, Gitler AD, et al. Cardiac hypertrophy and histone deacetylase-dependent transcriptional repression mediated by the atypical homeodomain protein Hop. The Journal of Clinical Investigation. 2003;112(6):863–871. doi: 10.1172/JCI19137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ellis L, Pan Y, Smyth GK, et al. Histone Deacetylase Inhibitor Panobinostat Induces Clinical Responses with Associated Alterations in Gene Expression Profiles in Cutaneous T-Cell Lymphoma. Clinical Cancer Research. 2008 Jul 15;14(14):4500–4510. doi: 10.1158/1078-0432.CCR-07-4262. 2008. [DOI] [PubMed] [Google Scholar]
- 42.Lodygin D, Menssen A, Hermeking H. Induction of the Cdk inhibitor p21 by LY83583 inhibits tumor cell proliferation in a p53-independent manner. The Journal of Clinical Investigation. 2002;110(11):1717–1727. doi: 10.1172/JCI16588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oshita A, Rothstein G, Lonngi G. cGMP stimulation of stem cell proliferation. Blood. 1977 Apr 1;49(4):585–591. 1977. [PubMed] [Google Scholar]
- 44.Liu R, Wang X, Chen G, et al. The prognostic role of a gene signature from tumorigenic breast-cancer cells. The New England journal of medicine. 2007;356(3):217. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- 45.Gentles A, Alizadeh A, Lee S, et al. A pluripotency signature predicts histologic transformation and influences survival in follicular lymphoma patients. Blood. 2009;114(15):3158. doi: 10.1182/blood-2009-02-202465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wong D, Liu H, Ridky T, Cassarino D, Segal E, Chang H. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2(4):333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krivtsov AV, Twomey D, Feng Z, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442(7104):818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 48.McWeeney SK, Pemberton LC, Loriaux MM, et al. A gene expression signature of CD34+ cells to predict major cytogenetic response in chronic-phase chronic myeloid leukemia patients treated with imatinib. Blood. 2010 Jan 14;115(2):315–325. doi: 10.1182/blood-2009-03-210732. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guzman ML, Li X, Corbett CA, et al. Rapid and selective death of leukemia stem and progenitor cells induced by the compound 4-benzyl, 2-methyl, 1,2,4-thiadiazolidine, 3,5 dione (TDZD-8) Blood. 2007 Dec 15;110(13):4436–4444. doi: 10.1182/blood-2007-05-088815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hahn CK, Berchuck JE, Ross KN, et al. Proteomic and genetic approaches identify Syk as an AML target. Cancer Cell. 2009 Oct 6;16(4):281–294. doi: 10.1016/j.ccr.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hassane DC, Guzman ML, Corbett C, et al. Discovery of agents that eradicate leukemia stem cells using an in silico screen of public gene expression data. Blood. 2008 Jun 15;111(12):5654–5662. doi: 10.1182/blood-2007-11-126003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med. 2006 Oct;12(10):1167–1174. doi: 10.1038/nm1483. [DOI] [PubMed] [Google Scholar]
- 53.Koscielny S. Why Most Gene Expression Signatures of Tumors Have Not Been Useful in the Clinic. Science Translational Medicine. 2010;2(14):14ps12. doi: 10.1126/scitranslmed.3000313. [DOI] [PubMed] [Google Scholar]
- 54.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. New England Journal of Medicine. 2004;351(27):2817. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 55.Dufour A, Schneider F, Metzeler K, et al. Acute myeloid leukemia with biallelic CEBPA gene mutations and normal karyotype represents a distinct genetic entity associated with a favorable clinical outcome. Journal of Clinical Oncology. 2010;28(4):570. doi: 10.1200/JCO.2008.21.6010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.