Abstract
The patterns of DNA methylation in human cancer cells are highly abnormal and often involve the acquisition of DNA hypermethylation at hundreds or thousands of CpG islands that are usually unmethylated in normal tissues. The recent discovery of 5-hydroxymethylcytosine (5hmC) as an enzymatic oxidation product of 5-methylcytosine (5mC) has led to models and experimental data in which the hypermethylation and 5mC oxidation pathways may become connected. Key discoveries in this setting include the findings that several genes coding for proteins involved in the 5mC oxidation reaction are mutated in human tumors and that there is a broad loss of 5hmC across many types of cancer. In this review, we will summarize current knowledge and discuss models of the potential roles of 5hmC in human cancer biology.
Keywords: 5-hydroxymethylcytosine, 5-methylcytosine, DNA methylation, cancer epigenetics, TET protein
Introduction
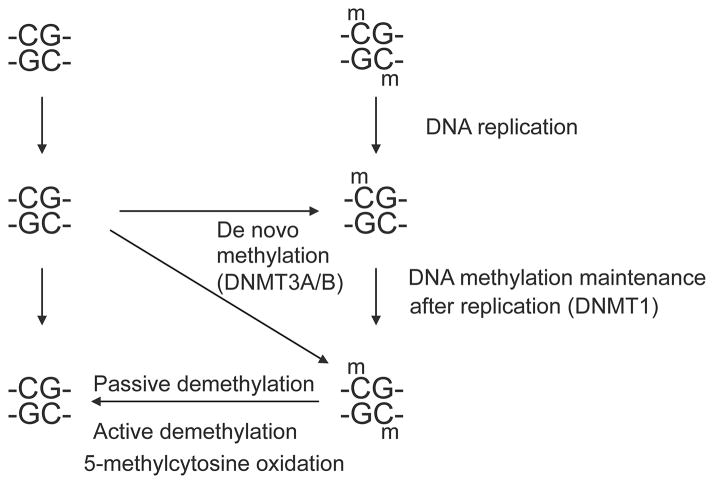
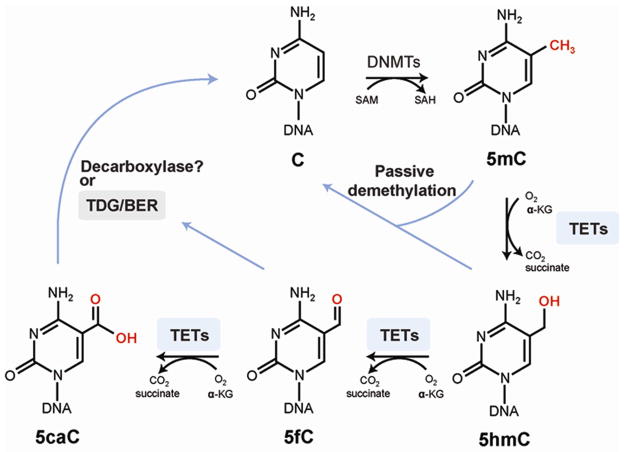
For many decades, 5-methylcytosine (5mC) has been recognized as the only confirmed mammalian modified DNA base. This base is created in a post-replicative enzymatic process by which DNA methyltransferases incorporate a methyl group into DNA at the 5-position of the cytosine ring. The reaction occurs predominantly at CpG dinucleotide sequences allowing for semi-conservative copying of this short palindromic, methylated sequence (Riggs, 1975). Methylation patterns are generally quite stable at their CpG target sequences but under certain conditions, an unmethylated CpG site can become de novo methylated by a DNA methyltransferase, mostly involving DNMT3A or DNMT3B, or a methylated sequence can become demethylated, loosing a methyl group on either one or on both DNA strands (Figure 1). The demethylation process may occur in a passive manner by which methylated CpG sites are replicated in absence of the maintenance DNA methyltransferase DNMT1. Complete removal of methylation on both DNA strands in the passive demethylation mode requires at least two cell divisions, and if methylation is to be completely lost in most cells of a population, many replication cycles are required. However, a more rapid, so-called active DNA methylation pathway also exists and occurs genome-wide at certain stages of embryo or germ cell development (Reik, et al., 2001). During active DNA demethylation, the 5mC bases on one or both DNA strands are converted to cytosine in the absence of DNA replication. Mechanisms for active DNA demethylation have remained mysterious and controversial for a long time (Ooi and Bestor, 2008, Wu and Zhang, 2010). However, today the most convincing pathway for active demethylation is thought to be the one involving 5mC oxidation (Kohli and Zhang, 2013, Pfeifer, et al., 2013). In 2009, the existence of a second modified cytosine base, 5-hydroxymethylcytosine (5hmC), was demonstrated in cells of the nervous system and in embryonic stem cells (Kriaucionis and Heintz, 2009, Tahiliani, et al., 2009). The levels of 5hmC were orders of magnitude higher than those that could have arisen from oxidative damage to 5mC alone. An enzyme converting 5mC to 5hmC was identified as a protein encoded by the Ten-eleven translocation 1 (TET1) gene (Tahiliani, et al., 2009), a gene previously implicated in a chromosomal translocation in leukemia patients (Ono, et al., 2002, Lorsbach, et al., 2003). The Tet1 protein was identified based on its homology to a trypanosomal protein, JBP1, known to oxidize thymine to 5-hydroxymethyluracil (Yu, et al., 2007). Soon after the first report on TET1, two additional 5mC oxidases, TET2 and TET3, were also described and characterized (Ito, et al., 2010). These enzymes are 2-oxoglutarate- and Fe(II)-dependent dioxygenases that have catalytic domains similar to other oxidases of that same category including, for example, many histone lysine demethylases (Hou and Yu, 2010). The TET proteins are capable of not only producing 5hmC in DNA but can drive the 5mC oxidation process further to produce 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) as reaction products (Figure 2) (He, et al., 2011, Ito, et al., 2011). This stepwise oxidation suggested that the oxidation of 5mC to 5caC produces a DNA base that could be decarboxylated to form cytosine, thus allowing for complete conversion of 5mC back to C in an oxidation-decarboxylation cycle (Figure 2). However, such an enzymatic decarboxylase activity has so far not been identified and 5caC is chemically quite stable (Munzel, et al., 2011). As a biochemical pathway, decarboxylation of 5caC would seem most logical inasmuch as such a mechanism avoids breakage of the sugar-phosphate backbone. Instead, however, cells use a multi-step base excision repair mechanism to remove 5caC from DNA (He, et al., 2011). The 5caC base is an excellent substrate for thymine DNA glycosylase (TDG) (He, et al., 2011, Maiti and Drohat, 2011, Zhang, et al., 2012), an enzyme previously implicated in removal of thymine (as a deaminated 5mC base) from G/T mispairs at CpG sequences (Neddermann, et al., 1996).
Figure 1. Stability and propagation of DNA methylation patterns.
A balance between DNA methylation and demethylation determines steady state levels of methylation at CpG sites.
Figure 2. Oxidation of 5-methylcytosine in a DNA demethylation pathway.
Enzymes and reaction intermediates are shown schematically.
It should be pointed out that several other pathways for DNA demethylation had been proposed earlier (Ooi and Bestor, 2008). These include, for example, a cytidine-deaminase-initiated process in which the deaminated base, i.e. thymine in case of 5mC deamination or 5-hydroxymethyluracil in case of 5hmC deamination, is removed by a DNA repair mechanism that might involve DNA glycosylases such as MBD4, TDG, or SMUG1 (Rai, et al., 2008, Guo, et al., 2011). Interestingly, plants do not contain TET homologues (Liu, et al., 2013b) and depend on a 5mC DNA glycosylase pathway mediated by ROS1, DME, or DML2/3 for active DNA demethylation (Zhu, 2009). These enzymes have no orthologues in mammalian cells.
Genome-wide DNA ‘demethylation’ in fertilized oocytes was long thought to result in unmethylated cytosines within hours after fertilization (Mayer, et al., 2000). However, in 2011 it was shown that this ‘demethylation’ in fact reflects genome-wide oxidation of 5mC catalyzed by Tet3 (Gu, et al., 2011, Iqbal, et al., 2011, Wossidlo, et al., 2011). The further oxidation products, 5fC and 5caC, can also be found in zygotes and in early cleavage-stage embryos (Inoue, et al., 2011). However, 5hmC itself seems fairly stable persisting throughout the first mitosis and first few cell divisions in early embryos (Iqbal, et al., 2011). Therefore, it is unlikely that a Tet3/Tdg pathway is operating at the whole genome level in zygotes or at other developmental stages. It seems intuitive that DNA breakage at millions of CpG sites during the Tdg-initiated base excision repair process would pose an enormous risk to genome integrity. For these reasons, it is more likely that Tet/Tdg-induced DNA demethylation operates gene-specifically at certain developmental or tissue differentiation stages rather than genome-wide. The other major wave of genome-wide DNA demethylation in mammals occurs in primordial germ cells. Whereas current evidence suggests that this demethylation is mostly passive and occurs during DNA replication (Seisenberger, et al., 2012, Kagiwada, et al., 2013), a specific role of Tet-mediated 5mC oxidation is also likely (Hackett, et al., 2013, Vincent, et al., 2013).
Clearly, there still is much to be learned about whether 5hmC represents a stable DNA base or rather is merely an intermediate in DNA demethylation, as outlined in Figure 2. If 5hmC were a stable epigenetic mark, it might be recognized by specific proteins, the so-called readers of the mark. Initial work showed that several proteins of the MBD family of methyl-CpG binding proteins are unable to interact with 5hmC suggesting that 5hmC may have a repelling effect on proteins that normally recognize 5mC (Jin, et al., 2010). However, MeCP2 and MBD3 are noticeable exceptions; these two MBD family proteins are able to bind to 5hmC (Yildirim, et al., 2011, Mellen, et al., 2012) although the data on MBD3 is controversial (Hashimoto, et al., 2012, Baubec, et al., 2013).
Soon after the discovery of 5hmC, its levels in various tissues and cell types have been characterized extensively (Kriaucionis and Heintz, 2009, Globisch, et al., 2010, Münzel, et al., 2010, Szwagierczak, et al., 2010) and its location in the genome has been mapped by various enrichment-based approaches or by single base resolution mapping techniques (Jin, et al., 2011b, Pastor, et al., 2011, Song, et al., 2011, Williams, et al., 2011, Nestor, et al., 2012, Yu, et al., 2012, Hahn, et al., 2013, Lister, et al., 2013, Thomson, et al., 2013b). The 5hmC base is most abundant in neuronal cells of the central nervous system but occurs in almost every tissue type at significant levels. These mapping techniques have found that 5hmC is present at promoters, enhancers and along gene bodies at various levels, also depending on the cell type (Laird, et al., 2013). When present in gene bodies, 5hmC levels are correlated positively with the level of expression of the 5hmC-marked genes (Jin, et al., 2011b, Song, et al., 2011). The mechanistic basis of this 5hmC-associated gene activation is still unknown.
It was also noted soon that 5hmC is strongly depleted in human cancers of many different histological origins (Jin, et al., 2011a, Yang, et al., 2013a). A few specific types of human cancer are characterized by mutations in one of the TET genes, TET2, but some tumor types can also carry mutations in the pathway producing the 2-oxo-glutarate cofactor, most commonly represented by mutations in isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2). However, the majority of human cancers do not contain TET1, TET2, TET3, IDH1 or IDH2 mutations and yet, they also show a dramatic loss of 5hmC when compared to corresponding normal tissue.
In this review, we will summarize current knowledge of the role of 5hmC in human cancer and speculate about possible mechanisms of its depletion in tumors as well as the interplay between aberrations in 5hmC pathways and alteration of 5mC patterns in human cancers.
Aberrant DNA methylation patterns in human cancer
It has been known for several decades that DNA methylation patterns in tumors differ drastically from those found in their normal tissue counterparts. Whereas DNA hypomethylation at a global, genome-wide level was recognized and described early on (Romanov and Vanyushin, 1981, Feinberg and Vogelstein, 1983b, Feinberg and Vogelstein, 1983a, Gama-Sosa, et al., 1983), the aberrant hypermethylation of CpG-rich DNA regions, the so-called CpG islands, was observed subsequently (Baylin, et al., 1986) and is now a major area of research in cancer epigenetics (Baylin and Jones, 2011). Hypermethylation of CpG islands is found in a variety of malignancies and is a pervasive change in tumors often affecting hundreds or even a few thousand independent CpG islands across the genome (Costello, et al., 2000, Rauch, et al., 2008). Methylation of specific CpG islands is of interest for development of disease biomarkers and for predicting treatment responses or survival of cancer patients (Laird, 2003, Ushijima, 2005). However, we are still very much in the dark when it comes to understanding the mechanistic pathways that leads to these methylation changes. A common observation is that a large fraction of the genes that become methylated in tumors are targets of Polycomb repression complexes in normal tissues or in embryonic stem cells. These genes most often include homeobox genes and other developmental transcription factors (Rauch, et al., 2006, Ohm, et al., 2007, Rauch, et al., 2007, Schlesinger, et al., 2007, Widschwendter, et al., 2007, Gal-Yam, et al., 2008, Hahn, et al., 2008). Such genes are not expressed or are expressed only at very low levels in normal somatic tissues and often are characterized by bivalent chromatin architecture that includes both active (H3K4me3) and repressive (H3K27me3) histone marks. Therefore, methylation of these Polycomb target genes at CpG dinucleotides along their promoters does not lead to a fundamental ‘downregulation’ of gene expression (Sproul and Meehan, 2013). Rather, DNA methylation is considered as a silencing event that is more permanent than that imposed by repressive histone modifications and is almost irreversible once it has occurred (although this may not hold true in light of Tet-induced DNA demethylation suggesting that DNA methylation is possibly more dynamic than previously thought). Current ideas about the role of CpG island hypermethylation in cancer include models in which the methylation events serve to silence differentiation-associated genes thus persistently locking the tumor cell population into an undifferentiated state (Wu, et al., 2010, Sproul, et al., 2012, Kalari, et al., 2013, Timp and Feinberg, 2013, Nejman, et al., 2014). In that sense, DNA hypermethylation can be considered as a pathway that reduces cellular plasticity of gene expression.
However, despite of decades of research, the mechanistic basis for CpG island methylation in cancer has remained unclear. The methylation state of CpG dinucleotides can be seen as a steady state level situation in which methylation and loss of methylation are balanced (Figure 1). In this scenario, hypermethylation can be viewed as a shift in the balance and can be promoted by increased methylation or by a failure of demethylation. Overexpression of DNA methyltransferases can be observed in tumors, but is thought to be mostly a consequence of enhanced cell division in the tumor cell population. Such overexpression also does not explain why certain CpG islands become hypermethylated and others never undergo this change. Interest in DNA demethylation processes, which have remained controversial for a long time (Ooi and Bestor, 2008, Wu and Zhang, 2010), has been revitalized by the discovery of an active, oxidation-dependent pathway in which 5mC is converted to 5hmC (Tahiliani, et al., 2009). Since this mechanism may have a potentially important role in shifting the methylation balance in tumors towards demethylation (when 5mC oxidation is over-active) or towards hypermethylation (when 5mC oxidation is impaired), there has been considerable interest in analyzing 5mC oxidation pathways in human cancer.
Loss of 5hmC in human cancer
Oxidation of 5mC by TET family proteins produces substantial levels of 5hmC, which can easily be detected and quantitated in mammalian tissues (Globisch, et al., 2010). However, the further oxidation products, 5fC and 5caC, occur at rather low levels only and are close to undetectable altogether in many tissues (Globisch, et al., 2010). This could mean that either the TET-mediated oxidation process in most situations does not proceed further beyond 5hmC or, alternatively, the 5fC and 5caC bases are effectively removed from the genome. There is evidence that the latter situation may apply, at least in certain cell types or developmental stages (He, et al., 2011). We also know that 5hmC is a fairly stable DNA base and no excision repair activity has been described that would effectively remove 5hmC from DNA.
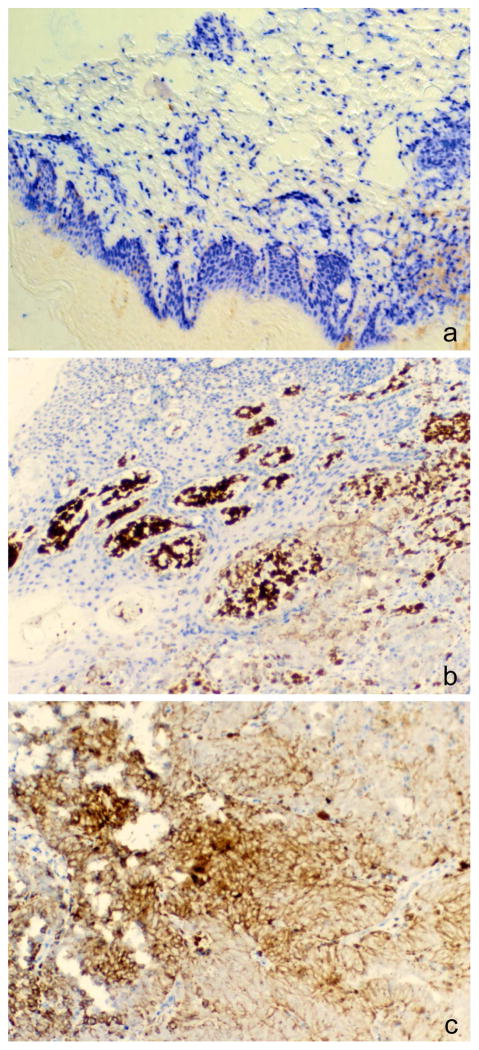
Several recent studies have reported that the levels of 5hmC are substantially reduced in human cancers (Haffner, et al., 2011, Jin, et al., 2011a, Kudo, et al., 2012, Lian, et al., 2012, Orr, et al., 2012, Gambichler, et al., 2013, Liu, et al., 2013a, Yang, et al., 2013a). In these studies, different methodologies have been used to quantitate 5hmC levels in tumor versus normal tissues. These methods include antibody-based immunofluorescence and immunohistochemistry approaches, immuno-dot blots, and liquid chromatography coupled with mass spectrometry. A clear tumor-specific loss of 5hmC was seen using all these different methodologies. Figure 3 shows an example where loss of 5hmC is seen in a melanoma tumor in comparison to adjacent normal skin sections using immunohistochemistry staining. Studies focusing on solid tumors have observed a loss of 5hmC ranging from ~50% to over 90% in tumors of almost all major histological types, including lung cancer, breast cancer, colorectal cancer, prostate cancer, liver cancer, glioma, ovarian cancer, melanoma, and several other tumor types (Haffner, et al., 2011, Jin, et al., 2011a, Kudo, et al., 2012, Lian, et al., 2012, Orr, et al., 2012, Gambichler, et al., 2013, Liu, et al., 2013a, Yang, et al., 2013a). A reduction of 5hmC also has been seen in certain cell types that are highly proliferative and carry stem cell character such as the crypt cells of the small intestine (Haffner, et al., 2011, Jin, et al., 2011a), basal cells of stratified epithelia (Haffner, et al., 2011) and proliferative neural progenitor cells in mouse embryo brain (Hahn, et al., 2013).
Figure 3. Reduced staining of 5-hydroxymethylcytosine in a melanoma biopsy.
Different sections of the same biopsy showing an area of normal skin (A), normal skin adjacent to a melanoma (B; tumor in lower right quadrant) and a melanoma tumor section (C) were stained with anti-5hmC antibody and AP-blue dye (Blue Alkaline Phosphatase Substrate Kit, Vector Laboratories).
The mechanistic basis for loss of 5hmC in cancer is currently unclear. One simple, and probably incomplete explanation is that tumor cells proliferate more rapidly than corresponding normal cells of the same tissue and cannot maintain 5hmC. In this situation, a 5hmC base when present on one DNA strand, would not undergo copying of the methylation and hydroxymethylation state by the maintenance DNA methyltransferase, DNMT1 in combination with a TET protein. In fact, in vitro experiments have shown that DNMT1 is incapable of methylating an unmethylated CpG site on the strand opposite to a hydroxymethylated CpG site (Valinluck and Sowers, 2007, Hashimoto, et al., 2012, Otani, et al., 2013). Also, the maintenance methylation cofactor UHFR1, which aids DNMT1 in the methylation reactions following DNA replication, does not have a good affinity to 5hmC-containing CpG sites (Otani, et al., 2013). However, the lack of maintenance of 5mC in proliferating cells at CpG sites containing 5hmC cannot be the only factor leading to loss of 5hmC in tumors. Also, we do not see a similar loss of total 5mC as we see for 5hmC in tumors, the loss of 5hmC always being much greater (Jin, et al., 2011a). Interestingly, changes in 5hmC occur early as a consequence of exposure of mammalian liver to a non-genotoxic carcinogen (Thomson, et al., 2012, Thomson, et al., 2013a) but it is unclear if other known carcinogens can impair 5hmC formation or maintenance.
Another possible explanation for the loss of 5hmC in cancer is an impairment of TET activity or function. The level of TET expression at the RNA level seems to be reduced in some but not all cancers examined (Jin, et al., 2011a, Lian, et al., 2012, Yang, et al., 2013a). Recent studies indicated that expression of TET family members is controlled by microRNAs which can be dysregulated leading to changes in TET expression in cancer (Cheng, et al., 2013, Fu, et al., 2013, Song, et al., 2013a, Song, et al., 2013b). Another possibility, discussed in more detail below, is that TET genes or components of co-factor pathways, i.e. IDH1/IDH2, are mutated in cancer leading to loss of function for 5mC oxidation. However, these mutations are found in only a few select tumor types including myeloid malignancies, gliomas and a few others, whereas the global reduction of 5hmC is seen universally in almost every tumor type analyzed.
One interesting observation is the recent recognition that optimal function of TET enzymes depends on the presence of ascorbic acid (vitamin C), which seems to be important for maintaining the TET oxidase cofactor Fe(II) in its reduced state (Blaschke, et al., 2013, Chen, et al., 2013, Minor, et al., 2013). Ascorbic acid is often lacking in cell culture media leading to very low levels of 5hmC in cultured proliferating cell lines. However, when ascorbic acid is added to these cell cultures, levels of 5hmC are strongly increased and return to baseline level after withdrawal of ascorbic acid from cell culture media (Blaschke, et al., 2013). This raises the question whether lack of ascorbic acid in cancer cells in vivo might impede the function of TET proteins leading to strongly decreased levels of 5hmC in tumors. Much further research is required to determine the exact mechanisms of 5hmC depletion in cancer.
Loss of 5hmC as a biomarker for cancer
The loss of 5hmC in cancer is observed almost universally across a spectrum of human malignant tumors. This raises the possibility that this loss could be exploited for developing a cancer biomarker. Some interesting data are already available suggesting that this type of approach may be feasible. For example, in several studies of human benign, pre-malignant and malignant melanocytic lesions, it was shown that benign nevi, dysplastic nevi and Spitz nevi show normal levels of 5hmC, but 5hmC is drastically reduced in malignant melanoma specimens (Lian, et al., 2012, Gambichler, et al., 2013, Larson, et al., 2014, Uchiyama, et al., 2014). A standard biomarker used in diagnostic pathology examinations to score for presence or absence of malignant cells in tissue biopsies is the proliferation marker Ki67, a nuclear antigen that is present in actively dividing cells. However, staining with Ki67 antibody can lead to false negative results with a rather high degree of discordance for staining results in different laboratories (Polley, et al., 2013). Therefore, an additional biomarker for cancer would be useful. In previous studies, we have observed that signals for Ki67 staining and for 5hmC staining are mutually exclusive (Jin, et al., 2011a) with Ki67-positive cells almost never being positive for 5hmC. This finding again emphasizes the fact that cell proliferation is incompatible with maintaining normal levels of 5hmC. However, another interesting observation we made was that not only the Ki67-positive cells in a tumor are lacking 5hmC, but in fact most cells of a tumor, including the Ki67-negative ones, have lost 5hmC (Jin, et al., 2011a). This finding indicates that cells with a past history of proliferation that make up the bulk of the tumor mass, but are currently not cycling, have undergone 5hmC loss. This fact may be of interest to pathologists because a tumor may be present in a biopsy, but is currently dormant having few proliferating, Ki67-positive cells. Such tumors should easily be detectable by anti-5hmC staining.
Relationship between 5hmC loss and DNA hypermethylation in cancer
The general reduction of 5hmC in human cancer tissues raises the question as to whether this reduction bears any relationship to the DNA methylation (5mC) changes so commonly observed in tumors. As mentioned earlier, the extent of loss of 5hmC does not parallel the magnitude of loss of 5mC, so DNA hypomethylation in tumors does not seem to have much to do with the 5mC oxidation pathway. However, loss of 5hmC may directly impinge upon DNA hypermethylation in cancer. There are examples for specific cancer types, in which TET or IDH genes are mutated; these will be discussed below. However, for most solid tumors, which contain wildtype TET or IDH genes, this possible connection has not yet been investigated. For example, one could imagine that specific CpG islands that undergo gain of 5mC in tumors are never occupied or modified by TET proteins. In that sense, TET-mediated oxidation of 5mC can be seen as a protective mechanism that safeguards CpG islands from occasional misdirected CpG methylation. Once such a spurious methylation event has occurred, the aberrant 5mC is oxidized by a TET protein and the resulting oxidized base is either diluted by passive methylation loss during DNA replication, or the 5hmC is further oxidized and is then removed by TDG-dependent base excision repair. Thus, TET oxidation of 5mC at CpG islands may serve as a methylation repair pathway as previously proposed (Jin, et al., 2011b, Pfeifer, et al., 2011, Williams, et al., 2011, Williams, et al., 2012). This mechanism may be defective at CpG islands that undergo cancer-associated hypermethylation, such as those also occupied by Polycomb complexes. On the other hand, those CpG islands that never become methylated in cancer, including those associated with many housekeeping genes, may have particularly active TET-mediated oxidation reactions taking place on aberrantly introduced methylated CpG sites. Further research is required to elucidate these points.
Mutations in the 5mC oxidation pathway in human tumors
The TET1 gene was initially identified as a translocation partner in a leukemia giving it its name, ten-eleven-translocation 1 (TET1) (Lorsbach, et al., 2003). In 2009, it was first reported that the TET2 gene is mutated in patients with myeloproliferative diseases including myelodysplastic syndrome (MDS), acute myeloid leukemia (AML) and chronic myelomonocytic leukemia (CMML) (Abdel-Wahab, et al., 2009, Delhommeau, et al., 2009, Jankowska, et al., 2009, Kosmider, et al., 2009, Langemeijer, et al., 2009). These TET2 mutations have been most commonly found along exons 3a and 10, the largest exons of the gene (Delhommeau, et al., 2009). It is likely that TET2 mutations represent a loss of function phenotype. For example, Kosmider et al reported that about half of the patients with TET2 mutations had two defective TET2 gene copies (Kosmider, et al., 2009). Thus, TET2 may play a tumor suppressive role. Several laboratories have confirmed such a functional role of Tet2 by creating Tet2 knockout mice in which important Tet2 exons have been deleted (Ko, et al., 2011, Li, et al., 2011, Moran-Crusio, et al., 2011, Quivoron, et al., 2011). Tet2-deficient mice show phenotypes similar to those seen in human CMML patients that have TET2 mutations (Moran-Crusio, et al., 2011). Hematopoietic stem cells derived from Tet2 knockout mice have a proliferative advantage. As expected, the levels of 5hmC are reduced in bone marrow and spleen cells of Tet2-deficient mice (Ko, et al., 2011, Quivoron, et al., 2011). Since TET2 functions as a 5mC oxidase, it was of interest to examine if TET2 mutations in patients have any effect on the levels and gene-specific patterns of 5mC in cancer cells of these same patients. In one study, TET2 mutation in AML were linked to genome-wide DNA hypermethylation (Figueroa, et al., 2010). However, other studies reported that bone marrow samples from patients with mutated TET2 show DNA hypomethylation (Ko, et al., 2010, Perez, et al., 2012). It is important to note that such correlative studies may not be straightforward at all because myeloid malignancies not only are characterized by mutations in TET2 but also frequently carry mutations in other epigenetic modifier genes including EZH2, IDH1, IDH2, MLL, DNMT3A, ASXL1, and others (Abdel-Wahab and Levine, 2013). Since mutations in several of these factors may directly or indirectly impinge upon DNA methylation patterns, the situation is very complex.
Mutations in TET cofactor pathways
2-oxoglutarate is a cofactor for iron-dependent dioxygenase enzymes including certain lysine demethylases and 5mC oxidases. 2-oxoglutarate is formed enzymatically in the tricarboxylic acid (TCA) cycle by the enzyme isocitrate dehydrogenase (IDH). IDH1 and IDH2 are metabolic enzymes that are mutated in some cancer genomes and thus represent examples where metabolism and cancer are linked directly through mutational events. IDH1 mutations are commonly observed in human brain tumors, specifically in grade II and III gliomas, in which they are present in up to 70 % of the patient population (Yan, et al., 2009). IDH1 or IDH2 are also mutated in myeloid leukemias and a few other malignancies (Figueroa, et al., 2010, Yen, et al., 2010, Pansuriya, et al., 2011). Tumor-specific IDH1 mutations are almost exclusively seen at arginine 132 suggesting a gain of function rather than loss of function phenotype in tumors. Interestingly, the common IDH1 mutant protein, in which arginine is substituted by histidine at position 132, is capable of producing 2-hydroxyglutarate (2HG) rather than the complete oxidation product 2-oxoglutarate (Dang, et al., 2009). Providing a possible mechanistic explanation for the selection of such specific IDH1 mutations in cancer, it was found that 2HG acts as a competitive inhibitor of 2-oxoglutarate-dependent enzymatic activities including TET proteins and lysine demethylases (Xu, et al., 2011).
Several studies have examined whether human cancers that carry IDH1 mutations have aberrant DNA methylation patterns. Gliomas with IDH1 mutations are clearly associated with widespread hypermethylation of CpG islands (Noushmehr, et al., 2010), the so-called CpG island methylator (CIMP) phenotype (Toyota, et al., 1999). Furthermore, introduction of DNA encoding a codon 132 mutant IDH1 protein into human astrocytes or generation of knock-in mice in which a wildtype copy of the Idh1 gene is replaced with a mutant copy, produces a DNA hypermethylation phenotype (Sasaki, et al., 2012, Turcan, et al., 2012). The mechanism of IDH1-mutant-induced DNA hypermethylation is not yet clear. In studies of human grade II/III gliomas, we did not observe a substantial difference in levels of 5hmC between IDH1/2 mutant and wildtype brain tumors (Jin, et al., 2011a). The codon 132 IDH1 mutant, through its enzymatic reaction product 2HG, will not only affect TET enzymes that produce 5hmC, but 2HG also is known to inhibit many lysine demethylases (Xu, et al., 2011). Because of the strong interplay between histone modifications and DNA methylation, inhibition of lysine demethylases may impact upon DNA methylation patterns at CpG islands indirectly leading to a CpG island hypermethylation phenotype.
Two other genes encode proteins of the TCA cycle that are mutated in a subset of human cancers: fumarate hydratase (FH) and succinate dehydrogenase (SDH). Their substrates, fumarate and succinate respectively, accumulate under conditions were FH or SDH are mutated (Yang, et al., 2013b). It has been shown that both fumarate and succinate are competitive inhibitors of 2-oxoglutarate-dependent dioxygenases including TET enzymes (Xiao, et al., 2012). In gastrointestinal stromal tumors, SDH-mutated tumors showed an order of magnitude higher DNA hypermethylation relative to the same category of tumors but with KIT gene mutations (Killian, et al., 2013). Similarly, SDH mutations were linked to a DNA hypermethylation phenotype in paraganglioma (Letouze, et al., 2013). Mutations in FH and SDH were linked to lower levels of 5hmC (Mason and Hornick, 2013, Castro-Vega, et al., 2014). These data indicate that altered epigenomic patterns are common consequences of metabolic pathway aberrations. The distinct role of 5mC oxidation versus other dioxygenase (eg. lysine oxidation) pathways in producing the altered DNA methylation patterns in these types of cancer warrants further investigation.
Perspective
During the past five years, we have witnessed many exciting new findings regarding modified cytosines in mammalian DNA spurred by the discovery of 5hmC and TET proteins as 5mC oxidases in 2009 (Kriaucionis and Heintz, 2009, Tahiliani, et al., 2009). There is evidence suggesting that the 5mC oxidation pathway is important for human cancer biology. Mouse knockout models for Tet2, a gene mutated in certain human hematological malignancies, have proven a cancer-causing role for dysfunctionality of the 5mC oxidation pathway. However, depletion of 5hmC is a universal occurrence in human cancer, not just those with TET2 or IDH1 gene mutations. More research efforts are required to understand the mechanisms of 5hmC loss in tumors and the biological consequences of this loss for tumor progression.
Acknowledgments
Work of the authors was supported by NIH grants ES006070 and CA160965 to G.P.P.
References
- Abdel-Wahab O, Levine RL. Mutations in epigenetic modifiers in the pathogenesis and therapy of acute myeloid leukemia. Blood. 2013;121:3563–3572. doi: 10.1182/blood-2013-01-451781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Wahab O, Mullally A, Hedvat C, Garcia-Manero G, Patel J, Wadleigh M, Malinge S, Yao J, Kilpivaara O, Bhat R, Huberman K, Thomas S, Dolgalev I, Heguy A, Paietta E, Le Beau MM, Beran M, Tallman MS, Ebert BL, Kantarjian HM, Stone RM, Gilliland DG, Crispino JD, Levine RL. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114:144–147. doi: 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baubec T, Ivanek R, Lienert F, Schubeler D. Methylation-dependent and - independent genomic targeting principles of the MBD protein family. Cell. 2013;153:480–492. doi: 10.1016/j.cell.2013.03.011. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Hoppener JW, de Bustros A, Steenbergh PH, Lips CJ, Nelkin BD. DNA methylation patterns of the calcitonin gene in human lung cancers and lymphomas. Cancer Res. 1986;46:2917–2922. [PubMed] [Google Scholar]
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke K, Ebata KT, Karimi MM, Zepeda-Martinez JA, Goyal P, Mahapatra S, Tam A, Laird DJ, Hirst M, Rao A, Lorincz MC, Ramalho-Santos M. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013;500:222–226. doi: 10.1038/nature12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Vega LJ, Buffet A, De Cubas AA, Cascon A, Menara M, Khalifa E, Amar L, Azriel S, Bourdeau I, Chabre O, Curras-Freixes M, Franco-Vidal V, Guillaud-Bataille M, Simian C, Morin A, Leton R, Gomez-Grana A, Pollard PJ, Rustin P, Robledo M, Favier J, Gimenez-Roqueplo AP. Germline mutations in FH confer predisposition to malignant pheochromocytomas and paragangliomas. Human molecular genetics. 2014 doi: 10.1093/hmg/ddt639. [DOI] [PubMed] [Google Scholar]
- Chen J, Guo L, Zhang L, Wu H, Yang J, Liu H, Wang X, Hu X, Gu T, Zhou Z, Liu J, Mao SQ, Mo K, Li Y, Lai K, Qi J, Yao H, Pan G, Xu GL, Pei D. Vitamin C modulates TET1 function during somatic cell reprogramming. Nat Genet. 2013;45:1504–1509. doi: 10.1038/ng.2807. [DOI] [PubMed] [Google Scholar]
- Cheng J, Guo S, Chen S, Mastriano SJ, Liu C, D’Alessio AC, Hysolli E, Guo Y, Yao H, Megyola CM, Li D, Liu J, Pan W, Roden CA, Zhou XL, Heydari K, Chen J, Park IH, Ding Y, Zhang Y, Lu J. An extensive network of TET2-targeting MicroRNAs regulates malignant hematopoiesis. Cell Rep. 2013;5:471–481. doi: 10.1016/j.celrep.2013.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello JF, Fruhwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomaki P, Lang JC, Schuller DE, Yu L, Bloomfield CD, Caligiuri MA, Yates A, Nishikawa R, Su Huang H, Petrelli NJ, Zhang X, O’Dorisio MS, Held WA, Cavenee WK, Plass C. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, Kosmider O, Le Couedic JP, Robert F, Alberdi A, Lecluse Y, Plo I, Dreyfus FJ, Marzac C, Casadevall N, Lacombe C, Romana SP, Dessen P, Soulier J, Viguie F, Fontenay M, Vainchenker W, Bernard OA. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983a;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. Hypomethylation of ras oncogenes in primary human cancers. Biochem Biophys Res Commun. 1983b;111:47–54. doi: 10.1016/s0006-291x(83)80115-6. [DOI] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, Tallman MS, Sun Z, Wolniak K, Peeters JK, Liu W, Choe SE, Fantin VR, Paietta E, Lowenberg B, Licht JD, Godley LA, Delwel R, Valk PJ, Thompson CB, Levine RL, Melnick A. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Jin L, Wang X, Luo A, Hu J, Zheng X, Tsark WM, Riggs AD, Ku HT, Huang W. MicroRNA-26a targets ten eleven translocation enzymes and is regulated during pancreatic cell differentiation. Proc Natl Acad Sci U S A. 2013;110:17892–17897. doi: 10.1073/pnas.1317397110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Yam EN, Egger G, Iniguez L, Holster H, Einarsson S, Zhang X, Lin JC, Liang G, Jones PA, Tanay A. Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc Natl Acad Sci U S A. 2008;105:12979–12984. doi: 10.1073/pnas.0806437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Sosa MA, Slagel VA, Trewyn RW, Oxenhandler R, Kuo KC, Gehrke CW, Ehrlich M. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 1983;11:6883–6894. doi: 10.1093/nar/11.19.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambichler T, Sand M, Skrygan M. Loss of 5-hydroxymethylcytosine and ten-eleven translocation 2 protein expression in malignant melanoma. Melanoma Res. 2013;23:218–220. doi: 10.1097/CMR.0b013e32835f9bd4. [DOI] [PubMed] [Google Scholar]
- Globisch D, Munzel M, Muller M, Michalakis S, Wagner M, Koch S, Bruckl T, Biel M, Carell T. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, Iqbal K, Shi YG, Deng Z, Szabo PE, Pfeifer GP, Li J, Xu GL. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, Surani MA. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339:448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffner MC, Chaux A, Meeker AK, Esopi DM, Gerber J, Pellakuru LG, Toubaji A, Argani P, Iacobuzio-Donahue C, Nelson WG, Netto GJ, De Marzo AM, Yegnasubramanian S. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget. 2011;2:627–637. doi: 10.18632/oncotarget.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MA, Hahn T, Lee DH, Esworthy RS, Kim BW, Riggs AD, Chu FF, Pfeifer GP. Methylation of polycomb target genes in intestinal cancer is mediated by inflammation. Cancer Res. 2008;68:10280–10289. doi: 10.1158/0008-5472.CAN-08-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MA, Qiu R, Wu X, Li AX, Zhang H, Wang J, Jui J, Jin SG, Jiang Y, Pfeifer GP, Lu Q. Dynamics of 5-hydroxymethylcytosine and chromatin marks in Mammalian neurogenesis. Cell Rep. 2013;3:291–300. doi: 10.1016/j.celrep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X, Cheng X. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 2012;40:4841–4849. doi: 10.1093/nar/gks155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C, Xu GL. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H, Yu H. Structural insights into histone lysine demethylation. Curr Opin Struct Biol. 2010;20:739–748. doi: 10.1016/j.sbi.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Shen L, Dai Q, He C, Zhang Y. Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res. 2011;21:1670–1676. doi: 10.1038/cr.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci U S A. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowska AM, Szpurka H, Tiu RV, Makishima H, Afable M, Huh J, O’Keefe CL, Ganetzky R, McDevitt MA, Maciejewski JP. Loss of heterozygosity 4q24 and TET2 mutations associated with myelodysplastic/myeloproliferative neoplasms. Blood. 2009;113:6403–6410. doi: 10.1182/blood-2009-02-205690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SG, Jiang Y, Qiu R, Rauch TA, Wang Y, Schackert G, Krex D, Lu Q, Pfeifer GP. 5-Hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res. 2011a;71:7360–7365. doi: 10.1158/0008-5472.CAN-11-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010;38:e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SG, Wu X, Li AX, Pfeifer GP. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res. 2011b;39:5015–5024. doi: 10.1093/nar/gkr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagiwada S, Kurimoto K, Hirota T, Yamaji M, Saitou M. Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. The EMBO journal. 2013;32:340–353. doi: 10.1038/emboj.2012.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalari S, Jung M, Kernstine KH, Takahashi T, Pfeifer GP. The DNA methylation landscape of small cell lung cancer suggests a differentiation defect of neuroendocrine cells. Oncogene. 2013;32:3559–3568. doi: 10.1038/onc.2012.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killian JK, Kim SY, Miettinen M, Smith C, Merino M, Tsokos M, Quezado M, Smith WI, Jr, Jahromi MS, Xekouki P, Szarek E, Walker RL, Lasota J, Raffeld M, Klotzle B, Wang Z, Jones L, Zhu Y, Wang Y, Waterfall JJ, O’Sullivan MJ, Bibikova M, Pacak K, Stratakis C, Janeway KA, Schiffman JD, Fan JB, Helman L, Meltzer PS. Succinate dehydrogenase mutation underlies global epigenomic divergence in gastrointestinal stromal tumor. Cancer Discov. 2013;3:648–657. doi: 10.1158/2159-8290.CD-13-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Bandukwala HS, An J, Lamperti ED, Thompson EC, Hastie R, Tsangaratou A, Rajewsky K, Koralov SB, Rao A. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci U S A. 2011;108:14566–14571. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, Liu XS, Aravind L, Agarwal S, Maciejewski JP, Rao A. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmider O, Gelsi-Boyer V, Cheok M, Grabar S, Della-Valle V, Picard F, Viguie F, Quesnel B, Beyne-Rauzy O, Solary E, Vey N, Hunault-Berger M, Fenaux P, Mansat-De Mas V, Delabesse E, Guardiola P, Lacombe C, Vainchenker W, Preudhomme C, Dreyfus F, Bernard OA, Birnbaum D, Fontenay M. TET2 mutation is an independent favorable prognostic factor in myelodysplastic syndromes (MDSs) Blood. 2009;114:3285–3291. doi: 10.1182/blood-2009-04-215814. [DOI] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo Y, Tateishi K, Yamamoto K, Yamamoto S, Asaoka Y, Ijichi H, Nagae G, Yoshida H, Aburatani H, Koike K. Loss of 5-hydroxymethylcytosine is accompanied with malignant cellular transformation. Cancer Sci. 2012;103:670–676. doi: 10.1111/j.1349-7006.2012.02213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird A, Thomson JP, Harrison DJ, Meehan RR. 5-hydroxymethylcytosine profiling as an indicator of cellular state. Epigenomics. 2013;5:655–669. doi: 10.2217/epi.13.69. [DOI] [PubMed] [Google Scholar]
- Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, Stevens-Linders E, van Hoogen P, van Kessel AG, Raymakers RA, Kamping EJ, Verhoef GE, Verburgh E, Hagemeijer A, Vandenberghe P, de Witte T, van der Reijden BA, Jansen JH. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009;41:838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- Larson AR, Dresser KA, Zhan Q, Lezcano C, Woda BA, Yosufi B, Thompson JF, Scolyer RA, Mihm MC, Jr, Shi YG, Murphy GF, Lian CG. Loss of 5-hydroxymethylcytosine correlates with increasing morphologic dysplasia in melanocytic tumors. Mod Pathol. 2014 doi: 10.1038/modpathol.2013.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letouze E, Martinelli C, Loriot C, Burnichon N, Abermil N, Ottolenghi C, Janin M, Menara M, Nguyen AT, Benit P, Buffet A, Marcaillou C, Bertherat J, Amar L, Rustin P, De Reynies A, Gimenez-Roqueplo AP, Favier J. SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer Cell. 2013;23:739–752. doi: 10.1016/j.ccr.2013.04.018. [DOI] [PubMed] [Google Scholar]
- Li Z, Cai X, Cai CL, Wang J, Zhang W, Petersen BE, Yang FC, Xu M. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118:4509–4518. doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, Xu W, Tan L, Hu Y, Zhan Q, Lee CW, Hu D, Lian BQ, Kleffel S, Yang Y, Neiswender J, Khorasani AJ, Fang R, Lezcano C, Duncan LM, Scolyer RA, Thompson JF, Kakavand H, Houvras Y, Zon LI, Mihm MC, Jr, Kaiser UB, Schatton T, Woda BA, Murphy GF, Shi YG. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150:1135–1146. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, Yu M, Tonti-Filippini J, Heyn H, Hu S, Wu JC, Rao A, Esteller M, He C, Haghighi FG, Sejnowski TJ, Behrens MM, Ecker JR. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Liu L, Chen X, Shen J, Shan J, Xu Y, Yang Z, Wu L, Xia F, Bie P, Cui Y, Bian XW, Qian C. Decrease of 5-hydroxymethylcytosine is associated with progression of hepatocellular carcinoma through downregulation of TET1. PLoS One. 2013a;8:e62828. doi: 10.1371/journal.pone.0062828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Dunwell TL, Pfeifer GP, Dunwell JM, Ullah I, Wang Y. Detection of oxidation products of 5-methyl-2′-deoxycytidine in Arabidopsis DNA. PLoS One. 2013b;8:e84620. doi: 10.1371/journal.pone.0084620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, Downing JR. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23) Leukemia. 2003;17:637–641. doi: 10.1038/sj.leu.2402834. [DOI] [PubMed] [Google Scholar]
- Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J Biol Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason EF, Hornick JL. Succinate dehydrogenase deficiency is associated with decreased 5-hydroxymethylcytosine production in gastrointestinal stromal tumors: implications for mechanisms of tumorigenesis. Mod Pathol. 2013;26:1492–1497. doi: 10.1038/modpathol.2013.86. [DOI] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor EA, Court BL, Young JI, Wang G. Ascorbate induces ten-eleven translocation (Tet) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine. J Biol Chem. 2013;288:13669–13674. doi: 10.1074/jbc.C113.464800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, Perna F, Pandey S, Madzo J, Song C, Dai Q, He C, Ibrahim S, Beran M, Zavadil J, Nimer SD, Melnick A, Godley LA, Aifantis I, Levine RL. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münzel M, Globisch D, Bruckl T, Wagner M, Welzmiller V, Michalakis S, Muller M, Biel M, Carell T. Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angew Chem Int Ed Engl. 2010;49:5375–5377. doi: 10.1002/anie.201002033. [DOI] [PubMed] [Google Scholar]
- Munzel M, Lischke U, Stathis D, Pfaffeneder T, Gnerlich FA, Deiml CA, Koch SC, Karaghiosoff K, Carell T. Improved synthesis and mutagenicity of oligonucleotides containing 5-hydroxymethylcytosine, 5-formylcytosine and 5-carboxylcytosine. Chemistry. 2011;17:13782–13788. doi: 10.1002/chem.201102782. [DOI] [PubMed] [Google Scholar]
- Neddermann P, Gallinari P, Lettieri T, Schmid D, Truong O, Hsuan JJ, Wiebauer K, Jiricny J. Cloning and expression of human G/T mismatch-specific thymine-DNA glycosylase. J Biol Chem. 1996;271:12767–12774. doi: 10.1074/jbc.271.22.12767. [DOI] [PubMed] [Google Scholar]
- Nejman D, Straussman R, Steinfeld I, Ruvolo M, Roberts D, Yakhini Z, Cedar H. Molecular rules governing de novo methylation in cancer. Cancer Res. 2014;74:1475–1483. doi: 10.1158/0008-5472.CAN-13-3042. [DOI] [PubMed] [Google Scholar]
- Nestor CE, Ottaviano R, Reddington J, Sproul D, Reinhardt D, Dunican D, Katz E, Dixon JM, Harrison DJ, Meehan RR. Tissue type is a major modifier of the 5-hydroxymethylcytosine content of human genes. Genome Res. 2012;22:467–477. doi: 10.1101/gr.126417.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, Verhaak RG, Hoadley KA, Hayes DN, Perou CM, Schmidt HK, Ding L, Wilson RK, Van Den Berg D, Shen H, Bengtsson H, Neuvial P, Cope LM, Buckley J, Herman JG, Baylin SB, Laird PW, Aldape K. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, Berman DM, Jenuwein T, Pruitt K, Sharkis SJ, Watkins DN, Herman JG, Baylin SB. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono R, Taki T, Taketani T, Taniwaki M, Kobayashi H, Hayashi Y. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23) Cancer Res. 2002;62:4075–4080. [PubMed] [Google Scholar]
- Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Orr BA, Haffner MC, Nelson WG, Yegnasubramanian S, Eberhart CG. Decreased 5-hydroxymethylcytosine is associated with neural progenitor phenotype in normal brain and shorter survival in malignant glioma. PLoS One. 2012;7:e41036. doi: 10.1371/journal.pone.0041036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani J, Kimura H, Sharif J, Endo TA, Mishima Y, Kawakami T, Koseki H, Shirakawa M, Suetake I, Tajima S. Cell cycle-dependent turnover of 5-hydroxymethyl cytosine in mouse embryonic stem cells. PLoS One. 2013;8:e82961. doi: 10.1371/journal.pone.0082961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pansuriya TC, van Eijk R, d’Adamo P, van Ruler MA, Kuijjer ML, Oosting J, Cleton-Jansen AM, van Oosterwijk JG, Verbeke SL, Meijer D, van Wezel T, Nord KH, Sangiorgi L, Toker B, Liegl-Atzwanger B, San-Julian M, Sciot R, Limaye N, Kindblom LG, Daugaard S, Godfraind C, Boon LM, Vikkula M, Kurek KC, Szuhai K, French PJ, Bovee JV. Somatic mosaic IDH1 and IDH2 mutations are associated with enchondroma and spindle cell hemangioma in Ollier disease and Maffucci syndrome. Nat Genet. 2011;43:1256–1261. doi: 10.1038/ng.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, Tahiliani M, Daley GQ, Liu XS, Ecker JR, Milos PM, Agarwal S, Rao A. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez C, Martinez-Calle N, Martin-Subero JI, Segura V, Delabesse E, Fernandez-Mercado M, Garate L, Alvarez S, Rifon J, Varea S, Boultwood J, Wainscoat JS, Cruz Cigudosa J, Calasanz MJ, Cross NC, Prosper F, Agirre X. TET2 mutations are associated with specific 5-methylcytosine and 5-hydroxymethylcytosine profiles in patients with chronic myelomonocytic leukemia. PLoS One. 2012;7:e31605. doi: 10.1371/journal.pone.0031605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer GP, Kadam S, Jin SG. 5-hydroxymethylcytosine and its potential roles in development and cancer. Epigenetics Chromatin. 2013;6:10. doi: 10.1186/1756-8935-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer GP, Rauch TA, Tommasi S, Besaratinia A, Hahn MA, Iqbal K, Szabó PE, Wu X, Jin S-G. Profiling of modified cytosines in normal and malignant cells. Princess Takamatsu Symp. 2011;41:69–72. [Google Scholar]
- Polley MY, Leung SC, McShane LM, Gao D, Hugh JC, Mastropasqua MG, Viale G, Zabaglo LA, Penault-Llorca F, Bartlett JM, Gown AM, Symmans WF, Piper T, Mehl E, Enos RA, Hayes DF, Dowsett M, Nielsen TO. An international Ki67 reproducibility study. J Natl Cancer Inst. 2013;105:1897–1906. doi: 10.1093/jnci/djt306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivoron C, Couronne L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, Do Cruzeiro M, Delhommeau F, Arnulf B, Stern MH, Godley L, Opolon P, Tilly H, Solary E, Duffourd Y, Dessen P, Merle-Beral H, Nguyen-Khac F, Fontenay M, Vainchenker W, Bastard C, Mercher T, Bernard OA. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch T, Li H, Wu X, Pfeifer GP. MIRA-assisted microarray analysis, a new technology for the determination of DNA methylation patterns, identifies frequent methylation of homeodomain-containing genes in lung cancer cells. Cancer Res. 2006;66:7939–7947. doi: 10.1158/0008-5472.CAN-06-1888. [DOI] [PubMed] [Google Scholar]
- Rauch T, Wang Z, Zhang X, Zhong X, Wu X, Lau SK, Kernstine KH, Riggs AD, Pfeifer GP. Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc Natl Acad Sci U S A. 2007;104:5527–5532. doi: 10.1073/pnas.0701059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch TA, Zhong X, Wu X, Wang M, Kernstine KH, Wang Z, Riggs AD, Pfeifer GP. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc Natl Acad Sci U S A. 2008;105:252–257. doi: 10.1073/pnas.0710735105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14:9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- Romanov GA, Vanyushin BF. Methylation of reiterated sequences in mammalian DNAs. Effects of the tissue type, age, malignancy and hormonal induction. Biochim Biophys Acta. 1981;653:204–218. doi: 10.1016/0005-2787(81)90156-8. [DOI] [PubMed] [Google Scholar]
- Sasaki M, Knobbe CB, Munger JC, Lind EF, Brenner D, Brustle A, Harris IS, Holmes R, Wakeham A, Haight J, You-Ten A, Li WY, Schalm S, Su SM, Virtanen C, Reifenberger G, Ohashi PS, Barber DL, Figueroa ME, Melnick A, Zuniga-Pflucker JC, Mak TW. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488:656–659. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, Bergman Y, Simon I, Cedar H. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W, Reik W. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Molecular cell. 2012;48:849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, Wang J, Zhang L, Looney TJ, Zhang B, Godley LA, Hicks LM, Lahn BT, Jin P, He C. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SJ, Ito K, Ala U, Kats L, Webster K, Sun SM, Jongen-Lavrencic M, Manova-Todorova K, Teruya-Feldstein J, Avigan DE, Delwel R, Pandolfi PP. The oncogenic microRNA miR-22 targets the TET2 tumor suppressor to promote hematopoietic stem cell self-renewal and transformation. Cell stem cell. 2013a;13:87–101. doi: 10.1016/j.stem.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SJ, Poliseno L, Song MS, Ala U, Webster K, Ng C, Beringer G, Brikbak NJ, Yuan X, Cantley LC, Richardson AL, Pandolfi PP. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell. 2013b;154:311–324. doi: 10.1016/j.cell.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproul D, Kitchen RR, Nestor CE, Dixon JM, Sims AH, Harrison DJ, Ramsahoye BH, Meehan RR. Tissue of origin determines cancer-associated CpG island promoter hypermethylation patterns. Genome biology. 2012;13:R84. doi: 10.1186/gb-2012-13-10-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproul D, Meehan RR. Genomic insights into cancer-associated aberrant CpG island hypermethylation. Briefings in functional genomics. 2013;12:174–190. doi: 10.1093/bfgp/els063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010;38:e181. doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JP, Hunter JM, Lempiainen H, Muller A, Terranova R, Moggs JG, Meehan RR. Dynamic changes in 5-hydroxymethylation signatures underpin early and late events in drug exposed liver. Nucleic Acids Res. 2013a;41:5639–5654. doi: 10.1093/nar/gkt232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JP, Hunter JM, Nestor CE, Dunican DS, Terranova R, Moggs JG, Meehan RR. Comparative analysis of affinity-based 5-hydroxymethylation enrichment techniques. Nucleic Acids Res. 2013b;41:e206. doi: 10.1093/nar/gkt1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JP, Lempiainen H, Hackett JA, Nestor CE, Muller A, Bolognani F, Oakeley EJ, Schubeler D, Terranova R, Reinhardt D, Moggs JG, Meehan RR. Non-genotoxic carcinogen exposure induces defined changes in the 5-hydroxymethylome. Genome biology. 2012;13:R93. doi: 10.1186/gb-2012-13-10-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timp W, Feinberg AP. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat Rev Cancer. 2013;13:497–510. doi: 10.1038/nrc3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, Thompson CB, Kaufman A, Guryanova O, Levine R, Heguy A, Viale A, Morris LG, Huse JT, Mellinghoff IK, Chan TA. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama R, Uhara H, Uchiyama A, Ogawa E, Takazawa Y, Ashida A, Koga H, Hayashi K, Kiniwa Y, Okuyama R. 5-Hydroxymethylcytosine as a useful marker to differentiate between malignant melanomas and benign melanocytic nevi. J Dermatol Sci. 2014;73:161–163. doi: 10.1016/j.jdermsci.2013.09.008. [DOI] [PubMed] [Google Scholar]
- Ushijima T. Detection and interpretation of altered methylation patterns in cancer cells. Nat Rev Cancer. 2005;5:223–231. doi: 10.1038/nrc1571. [DOI] [PubMed] [Google Scholar]
- Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 2007;67:946–950. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- Vincent JJ, Huang Y, Chen PY, Feng S, Calvopina JH, Nee K, Lee SA, Le T, Yoon AJ, Faull K, Fan G, Rao A, Jacobsen SE, Pellegrini M, Clark AT. Stage-specific roles for tet1 and tet2 in DNA demethylation in primordial germ cells. Cell stem cell. 2013;12:470–478. doi: 10.1016/j.stem.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, Laird PW. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- Williams K, Christensen J, Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO reports. 2012;13:28–35. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, Walter J. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Rauch TA, Zhong X, Bennett WP, Latif F, Krex D, Pfeifer GP. CpG island hypermethylation in human astrocytomas. Cancer Res. 2010;70:2718–2727. doi: 10.1158/0008-5472.CAN-09-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, Liu L, Liu Y, Yang C, Xu Y, Zhao S, Ye D, Xiong Y, Guan KL. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012;26:1326–1338. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Xiao MT, Liu LX, Jiang WQ, Liu J, Zhang JY, Wang B, Frye S, Zhang Y, Xu YH, Lei QY, Guan KL, Zhao SM, Xiong Y. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Liu Y, Bai F, Zhang JY, Ma SH, Liu J, Xu ZD, Zhu HG, Ling ZQ, Ye D, Guan KL, Xiong Y. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene. 2013a;32:663–669. doi: 10.1038/onc.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Soga T, Pollard PJ. Oncometabolites: linking altered metabolism with cancer. J Clin Invest. 2013b;123:3652–3658. doi: 10.1172/JCI67228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen KE, Bittinger MA, Su SM, Fantin VR. Cancer-associated IDH mutations: biomarker and therapeutic opportunities. Oncogene. 2010;29:6409–6417. doi: 10.1038/onc.2010.444. [DOI] [PubMed] [Google Scholar]
- Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, Weng Z, Rando OJ, Fazzio TG. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, Song CX, Jin P, Ren B, He C. Tet-assisted bisulfite sequencing of 5-hydroxymethylcytosine. Nat Protoc. 2012;7:2159–2170. doi: 10.1038/nprot.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Genest PA, ter Riet B, Sweeney K, DiPaolo C, Kieft R, Christodoulou E, Perrakis A, Simmons JM, Hausinger RP, van Luenen HG, Rigden DJ, Sabatini R, Borst P. The protein that binds to DNA base J in trypanosomatids has features of a thymidine hydroxylase. Nucleic Acids Res. 2007;35:2107–2115. doi: 10.1093/nar/gkm049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Lu X, Lu J, Liang H, Dai Q, Xu GL, Luo C, Jiang H, He C. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat Chem Biol. 2012;8:328–330. doi: 10.1038/nchembio.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annual review of genetics. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]