Abstract
Methicillin-Resistant Staphylococcus aureus (MRSA) poses a serious threat to worldwide health. Historically, MRSA clones have strictly been associated with hospital settings and most hospital-associated MRSA (HA-MRSA) disease resulted from a limited number of virulent clones. Recently, MRSA has spread into the community causing disease in otherwise healthy people with no discernible contact with healthcare environments. These community-associated (CA-MRSA) are phylogenetically distinct from traditional HA-MRSA clones and CA-MRSA strains seem to exhibit hyper virulence and more efficient host:host transmission. Consequently, CA-MRSA clones belonging to the USA300 lineage have become dominant sources of MRSA infections in North America. The rise of this successful USA300 lineage represents an important step in the evolution of emerging pathogens and a great deal of effort has been exerted to understand how these clones evolved. Here we review much of the recent literature aimed at illuminating the source of USA300 success and broadly categorize these findings into three main categories: newly acquired virulence genes, altered expression of common virulence determinants and alterations in protein sequence that increase fitness. We argue that none of these evolutionary events alone account for the success of USA300, but rather their combination may be responsible for the rise and spread of CA-MRSA.
Multidrug-resistance in Staphylococcus aureus: the rise of MRSA
The Gram-positive pathogen Staphylococcus aureus remains one of the most problematic and costly sources of bacterial infection worldwide1. Disease typically presents as mild skin/soft tissue infections but can also be the source of more serious bacteremia, endocarditis, osteomyelitis and necrotizing pneumonia2. S. aureus asymptomatically colonizes the skin, and more commonly, the anterior nasal passages of healthy people3. Nasal colonization is the most significant predictor of invasive disease, however, in some studies, nearly half of patients carrying S. aureus are strictly colonized extranasally4. Thus, estimates of S. aureus carriage at ~25% of the human population may be an underestimate of true colonization levels. Given the near ubiquity of S. aureus among the human population combined with its virulence potential, it is no wonder this organism has been recognized as a significant healthcare burden for over a century. S. aureus was first described by Alexander Ogston in 1881 as the sole microorganism within the fluid drained from a severe knee abscess5. Then, he noted that “once established the micrococci are hard to kill…” underscoring the recalcitrant nature of S. aureus towards antiseptic treatment6. During this time, Joseph Lister’s influence on surgical procedures through the implementation of carbolic acid (phenol) to sterilize wounds and instruments, had greatly reduced the occurrence of post-operative infections7. However, it was subsequently shown that S. aureus was inherently resistant to phenol explaining its association with surgical infections despite good “sterile technique”8. Thus, S. aureus was recognized as an important hospital associated pathogen over 130 years ago in the pre-antibiotic era and little has changed to this day.
Perhaps because of its intimate association with hospitals and patients, S. aureus has always been among the first bacterial species reported to develop resistance to new antimicrobials, from sulfonamide-resistance in the early 1940s9 to the identification of penicillinase in 194410 just months after US penicillin production reached full scale. Interestingly, these progenitor β-lactamase positive S. aureus clones were isolated from patients that had not even been treated with penicillin. Nonetheless, Penicillin-Resistant S. aureus (PRSA) was here to stay, and became pandemic in hospitals during the late 1950s and early 1960s11. Subsequently, a penicillinase-resistant β-lactam derivative, methicillin (Celbenin, Beecham Pharmaceuticals), was approved for use in the US in 1959. Less than two years later, the first report of methicillin-resistant S. aureus (MRSA) was published documenting the isolation of MRSA clones from a patient and hospital staff in the UK, again none of which were treated with methicillin12. It was immediately recognized that methicillin-resistance was mechanistically different than penicillin-resistance in that the MRSA phenotype did not involve direct inactivation of the drug. Rather, resistance was mediated through the acquisition of an alternative penicillin-binding protein (PBP2a) with lowered affinity for β-lactam antibiotics. Within 20 years after the first discovery of MRSA, it became a leading cause of hospital-acquired infections13. Currently, it can still be responsible for nearly 60% of skin/soft-tissue infections presenting to US emergency rooms14.
The methicillin-resistance determining PBP2a is encoded by mecA harbored on a mobile genetic element, Staphylococcal Cassette Chromsome (SCCmec). A nearly identical homologue, now thought to be the ancestral mecA, was recently discovered in Staphylococcus fleuretti, an animal colonizing staphylococcal species15. Unlike a previously identified mecA homologue in Staphylococcus sciuri that does not confer methicillin-resistance16, S. fleuretti is fully resistant to β-lactam antibiotics. Interestingly, the S. fleuretti mecA homologue is not found on a mobile SCC, but rather in the core chromosome between the mevalonate biosynthetic and xylose utilization operons, explaining the presence of mva and xyl gene fragments in some S. aureus SCCmec elements15. These mobile islands have diversified considerably over the 50-year history of MRSA such that there are currently eight distinct SCCmec types circulating among S. aureus as well as some species of coagulase negative staphylococci17. SCCmec elements can vary greatly in size and composition with the largest (SCCmec type II) spanning 52 kb and additionally encoding erythromycin-, spectinomycin- and tobramycin-resistance determinants18. Depending on the particular SCCmec type, these mobile islands peppered with IS elements, transposons and integrated plasmids, can confer multidrug-resistance determinants that significantly diminish treatment options in a clinical setting. Thus, in addition to methicillin resistance, MRSA isolates have evolved multidrug-resistance leading to what the popular press refers to as an emerging superbug19.
Paradigm-shift: The rise of CA-MRSA
After 1961, MRSA spread worldwide causing significant morbidity and mortality almost entirely as hospital-acquired infections. Advances in molecular epidemiology allowed for in depth analyses of MRSA spread and expansion at the evolutionary level. For instance, spa-typing (polymorphisms in Protein A coding sequence) and SCCmec-typing discriminated unrelated clones and identified clusters of related MRSA lineages responsible for disease20,21. Multi-Locus Sequence Typing (MLST) involves the sequencing of fragments from seven “housekeeping” genes (arcC, aroE, glpF, gmk, pta, tpi and yqiL) yielding unique sequence types (STs)22. STs sharing identity at the majority of these loci are grouped into Clonal Complexes (CCs) encompassing related lineages of MRSA23. Another highly-discriminatory approach that can identify genomic rearrangements and insertions/deletions is Pulsed-Field Gel Electrophoresis (PFGE) whereby SmaI digested chromosomal DNA is separated and similarities in banding patterns reflect relatedness among lineages24,25. This allows for the classification of S. aureus strains into the now familiar PFGE-types USA100–1200. Employing these epidemiological approaches,researchers appreciated that most MRSA disease worldwide (nearly 70% of reported infections) was caused by five major CCs: CC5, CC8, CC22, CC30 and CC4524,26 (Figure 1). CC5 includes clones belonging to the USA100 PFGE-type (e.g. SCCmec-II New York/Japan clone) the most common source of US hospital acquired MRSA as well as USA800 (SCCmec-IV Pediatric clone). CC8 includes the archaic, or original MRSA clones as well as the related Iberian clone, the SCCmec-III Brazilian/Hungarian clone and the SCCmec-IV USA500 clones. CC22 includes the EMRSA-15 clones that dominated hospital infections in the UK during the 1990s along with strains from CC30 encompassing EMRSA-16 as well as the USA200 PFGE type. Finally, CC45 consists of clones belonging to USA600 PFGE type (e.g. Berlin clone) that caused widespread MRSA hospital infections in northern Europe. In essence, after 30 years of investigation, the scientific community began to understand the population structure of the MRSA clones responsible for the majority of hospital-acquired disease. The source of high-virulence potential inherent to these five CCs was never fully appreciated before everything we knew about MRSA epidemiology changed at the turn of the century.
Figure 1. Schematic representation of the evolution of MRSA.
Sequence Types (STs) belonging to established Clonal Complexes (CCs) are colored as follows: CC1, purple; CC5, green; CC8, red; CC22, orange; CC30, blue; CC45, black. ST59 has not been assigned to a CC. Roman numerals reflect acquired SCCmec type. Commonly used S. aureus strains are depicted around their relevant ST symbol.
Initially reported in 1993, patients without any contact with healthcare settings contracted invasive MRSA infections in Kimberly Australia, a region in the northern part of Western Australia27. It was later discovered that simultaneously, strains related to these “community-acquired” MRSA (CA-MRSA) clones were causing serious and fatal respiratory infections in Chicago, again in patients without direct contact with hospital environments28. Prior to these reports, MRSA infections were exclusively associated with healthcare settings. These new clones belong to CC1 (USA400 PFGE type), a clonal complex unrelated to the five traditional Hospital-Associated MRSA (HA-MRSA) complexes28. CC1 clones spread quickly through Australia, the mid- and northwestern United States as well as Canada and Alaska where they still cause significant CA-MRSA disease28–32. Recent studies show that USA400 can account for over 98% of MRSA infections in northern Canada33 and has been implicated in isolated MRSA disease in southern Europe34,35. However, about 10 years ago a new source of CA-MRSA arose from one of the “traditional” virulent clonal complexes, CC8. Descending from a USA500 clone through acquisition of various mobile genetic elements (MGEs)26,36, USA300 became the dominant CA-MRSA clone in US14,37,38, effectively replacing USA400 clones in most regions39,40, and has also been isolated from patients in Canada and Mexico41,42. The explosion of USA300 CA-MRSA across North America resulted from a very recent clonal expansion of a successful CA-MRSA clone as demonstrated by very low sequence divergence among geographically distinct USA300 isolates43.
Given the occurrence of multiple CA-MRSA clones in the population, a formal definition was put forth by the Centers for Disease Control and Prevention for CA-MRSA disease as that which is contracted within 48 hours of hospital admission by patients not having recently undergone surgery, hemodialysis, prolonged hospitalization, catheterization or MRSA colonization44. Currently in the US, MRSA disease fitting these criteria is almost always caused by USA300 clones, followed by USA400 and occasionally USA1000 and USA110014. To complicate matters further, USA300 clones have recently been implicated in causing significant HA-MRSA disease38,45–47, blurring the lines between the two disease onset environments38,45–47. In some studies, USA300 accounted for at least half of hospital acquired MRSA infections38,46. Thus, USA300 represents a highly successful S. aureus clone that emerged in the community and quickly spread throughout the North American continent to become the leading cause of MRSA infection even in healthcare settings. For now, USA300 seems to be primarily limited to North America, while in Europe, South America and Asia CA-MRSA disease is dominated by divergent clones unrelated to CC8 (e.g. ST30, ST80 and ST59)48. Given the rapid and efficient transmissibility of USA300 in North America49, it remains to be seen whether these clones will become the dominant source of MRSA disease worldwide.
USA300 Virulence
Animal models of S. aureus infection have repeatedly demonstrated the hypervirulence associated with USA300 compared to other MRSA strains36,50–52. USA300 strains exhibited enhanced production of dermonecrotic lesions in skin abscess models when compared to HA-MRSA clones36,50,51 and USA300 was more lethal in a rat model of pneumonia compared with a USA400 isolate52. Furthermore, USA300 strains were more lethal in septic infections compared to archaic and Iberian clones as well as ST239 clones (Brazilian clones)36. When compared to other CA-MRSA clones, USA300 isolates generally exhibit increased virulence with the exception of ST80 and USA1000, which also possess enhanced virulence51. In contrast, nearly every clone of HA-MRSA tested was significantly less virulent than USA300 with the only exception being USA500 HA-MRSA36,51. This is of particular interest in that USA300 clones descended from USA500 via the acquisition of a prophage containing Panton-Valentine Leukotoxin (PVL), a mobile Arginine Catabolic Mobile Element (ACME) and enterotoxins K and Q (see below)36. Thus, the source of USA300 hypervirulence may have originally evolved in the HA-MRSA isolates belonging to USA500. However, for unknown reasons, despite exhibiting hypervirulence in animal infection models, USA500 clones remain relegated to healthcare settings and do not cause significant CA-MRSA disease. Whether CA-MRSA USA300 clones exhibit hypervirulence in human disease has been difficult to directly discern, however, recent population based clinical data are beginning to corroborate conclusions drawn from laboratory animal model experiments.
In humans, USA300 S. aureus primarily causes skin infections of which, it can account for up to 98% of all MRSA presenting as skin/soft tissue infections to US emergency rooms14. In addition, USA300 can also cause more invasive disease such as bacteremia53, endocarditis54 and necrotizing fasciitis55, a condition almost never associated with S. aureus. In particular, pulmonary infections caused by USA300 S. aureus can lead to aggressive and often fatal necrotizing pneumonia56–58. The populations most at risk for contracting USA300 CA-MRSA are military personnel59, athletes60–62, prisoners63–65, African Americans58,66, daycare attendees67,68 and men who have sex with men69. Patients contracting CA-MRSA are, on average, younger than those with HA-MRSA and otherwise generally healthy70,71. Furthermore, CA-MRSA is often associated with worse clinical outcomes. For instance, USA300 infections were associated with increased in-hospital mortality and a higher occurrence of severe sepsis than HA-MRSA infections66,72. USA300-related strains were also more prone to spread from the initial infection site and caused more severe infections than HA-MRSA in patients suffering from pneumonia with pulmonary emboli73,74. However, other reports describe better clinical outcomes associated with USA300 infections45,75. Although some studies that reported more positive clinical outcomes with CA-MRSA also described hypervirulent CA-MRSA trends, such as increased risk of being admitted into intensive care, that merely lack full statistical significance (OR = 1.8, p = 0.09)46. Additionally, effective treatment, which is easier to achieve when treating CA-MRSA infections given their inherent susceptibility to clindamycin, tetracyclines, rifampicin and trimethoprim/sulfonamide, can reduce the severity of CA-MRSA disease outcomes in population-based studies76. Unfortunately, this trend of increased antibiotic susceptibility may be diminishing as new reports show increased antibiotic resistance among USA300 isolates, possibly through direct acquisition of resistance determinants from multidrug-resistant HA-MRSA strains77. Thus, the future clinical outlook appears grim with respect to USA300 infections given their increased prevalence in both hospital- and community-acquired infections, their propensity to acquire new antibiotic resistance determinants and the steady decline in positive clinical outcomes associated with USA300 infections.
Genetic Determinants Contributing to USA300 Success
Given the recent impact of USA300 on human health, significant research effort has been exerted to elucidate the source of USA300 success. Here we review these findings and broadly categorize them into three main classes: 1. Newly acquired genes that promote virulence and/or fitness, 2. Altered regulation of core genes resulting in elevated virulence and/or fitness and 3. Non-synonomous mutations in core genes that enhance virulence and/or fitness.
Newly Acquired Genes
Many different lineages of CA-MRSA (USA400, USA1000, and USA1100) cause outbreaks and invasive infections, but in North America, none are as prevalent as epidemic USA300. These clones have acquired many genes in the form of Mobile Genetic Elements (MGEs) that may confer a selective advantage over other CA-MRSA strains. Several groups have investigated many of these MGEs with the goal of elucidating factors (if any) that have contributed to the overwhelming success of USA300.
Enterotoxins K and Q
USA300 CA-MRSA isolates contain genes encoding enterotoxins K and Q (sek2 and seq2) in a unique pathogenicity island SaPI5 78. Sek2 and Seq2 are thought to contribute to pathogenesis by stimulating T-cells through binding of the Vβ chain of αβ T-cell receptors. Sek2 and Seq2 share 98% amino acid homology with enterotoxins (Sek and Seq) found on SaPI3 in S. aurueus COL an archaic HA-MRSA clone belonging to ST250 that is less virulent than CA-MRSA isolates79. USA400 isolates (e.g. MW2) harbor νSA3, a pathogenicity island that shares similarity to SaPI3 of COL and SaPI5 of USA300, however, νSA3 does not contain the genes for Sek or Seq78. Thus, the acquisition of these toxins by USA300 and not US400 may potentially explain the differences in pathogenicity although direct demonstration of this has not been reported.
SCCmecIVa
The mecA gene encodes a penicillin-binding protein and is located on a mobile genetic element known as the Staphylococcal Cassette Chromosome mec (SCCmec). There are currently eight recognized SCCmec types (I VIII). SCCmec types I, II and III contain additional drug resistance determinants, whereas types IV, V, VI, and VII cause resistance only to β-lactams80. Initial sequence comparisons show that both USA400 and USA300 strains contain a nearly identical SSCmecIVa78,81. As it turns out, SCCmecIV is the most common form of SCCmec found across divergent S. aureus lineages in addition to ST8 (USA300) including ST1 (USA400), ST80, ST72 (USA700) and ST8 (USA500)82,83. It has been shown that SSCmecIV does not impose a fitness cost in vitro or in vivo, whereas acquisition of the SSCmec types I, II and III resulted in decreased in vitro growth rates84–86. Thus, it is thought that harboring SSCmecIV as opposed to other SCCmec types imparts CA-MRSA with an advantage in its ability to cause infection in healthy individuals. However, though SSCmecIV may provide a selective advantage to CA-MRSA over other SCCmec types, the fact that nearly all CA-MRSA isolates contain SSCmecIVa suggests that it is not a major contributing factor to the dominance of USA300 among CA-MRSA isolates.
Panton-Valentine Leukocidin (PVL)
The Panton-Valentine leukocidin (PVL) is a bicomponent pore-forming toxin that induces necrosis and apoptosis in leukocytes87. PVL is encoded by the genes lukS-PV and lukF-PV located on the prophage ϕSA2 pvl78. This phage is highly associated with CA-MRSA clones in that nearly all USA300, USA400 and USA1100 clinical isolates are positive for PVL as are many USA1000 strains88,89. Furthermore, epidemiological and clinical reports indicate a strong correlation between PVL production and severe skin/soft tissue infections, as well as necrotizing pneumonia and fasciitis, suggesting PVL may be a major contributor to the virulence of CA-MRSA90–92. Moreover, PVL can be directly detected in human skin abscesses at levels known to result in rapid neutrophil lysis93,94. Thus, PVL is significantly correlated with invasive CA-MRSA disease, however, recent clinical studies demonstrate that CA-MRSA strains lacking PVL can still cause disease outbreaks95–97.
Until recently, demonstrating a direct role for PVL in model disease has proven difficult. This likely stems from the host specificity of PVL in that it is rapidly leukocidal for rabbit and human neutrophils, but much less active against murine, rat or simian PMNs98. Consequently, a virulence effect of PVL in murine or rat pneumonia, sepsis and skin infection models has never been reproducibly defined99–104. Moreover, there was no demonstrable role for PVL in a pneumonia model involving nonhuman primates105. In contrast, using PVL susceptible rabbit models, isogenic USA300 strains lacking PVL were less virulent in pneumonia, osteomyelitis and skin abscess models106–109. However, the attenuation of mutants lacking PVL in rabbit skin lesions was not nearly as striking as a mutant lacking α-hemolysin or phenol-soluble modulin production underscoring the contributory nature of PVL towards S. aureus pathogenesis108,110. Furthermore, the nearly ubiquitous presence of PVL among CA-MRSA isolates clearly suggests that this toxin cannot explain the particular success of the USA300 lineage.
Arginine Catabolic Mobile Element (ACME)
Of all the genetic elements acquired by CA-MRSA isolates, only the arginine catabolic mobile element (ACME) is completely unique to USA30078. The type 1.02 ACME carried by USA300 is juxtaposed to the SCCmecIV island and was acquired from S. epidermidis through horizontal gene transfer via a mechanism likely involving the SCCmec-related CcrAB recombinases78,84,111. The physical linkage of ACME with SCCmecIVa is mirrored by an epidemiological linkage in that nearly all USA300 strains harboring SCCmecIVa also carry ACME, while USA300 clones with other SCCmec islands, with rare exceptions, do not83,112. The ACME of USA300 contains a complete arginine deaminase (arc) system that converts L-arginine to L-ornithine for both ATP and ammonia production. The island also encodes a putative oligopeptide permease, a zinc-containing alcohol dehydrogenase, and a spermine/spermidine acetlytransferase (SpeG) as well as several hypothetical proteins78. While a role for ACME in USA300 virulence was demonstrated in a rabbit sepsis model84, no effect of ACME was observed in murine pneumonia or skin abscess models113. Thus, it has been proposed that ACME aids primarily in USA300 colonization, in part, through the Arc mediated ammonification of the acidic skin environment, though this has never been experimentally verified84,114.
We have additionally observed a peculiar phenotype in S. aureus suggestive of a selective advantage afforded by the ACME cassette. Polyamines, including spermine, spermidine and putrescine are a group of polycationic compounds reportedly synthesized from L-arginine by all living organisms. Not only does S. aureus lack the ability to synthesize polyamines de novo, but spermine and spermidine are bactericidal to this organism at levels found within mammalian tissue115,116. Polyamine-sensitivity was apparent in all tested strains except those belonging to USA300, and in these isolates polyamine-resistance was dependent on speG encoding a spermine/spermidine aceytltrasferase harbored on ACME. Could speG provide USA300 with a selective advantage by nullifying the staphylocidal effects of host polyamines? While no direct measure of host polyamine levels during S. aureus infections have been reported, several indirect lines of evidence may suggest that polyamines do affect the outcome of staphylococcal disease and/or colonization.
Upon wounding, the host response in the skin is proinflammatory and dominated by cytokines such as IL-1, INF-γ and TNF- α117. The resulting inflammation is mediated, among other effectors, by the production of reactive oxygen and nitrogen species, the latter of which, nitric oxide (NO·) is synthesized from L-arginine by the inducible NO·-synthase (iNOS, Figure 2). This enzyme competes for available L-arginine with host enzymes such as Arginase-1 (Figure 2) as well as with arginine-auxotrophic S. aureus118. Once tissue damage signals resulting from the primary inflammation outweigh pathogen-associated signals, the host response shifts away from proinflammatory mediators and initiates the profibrotic response117. This phase is dependent on the production of TH2-like anti-inflammatory cytokines such as IL-4, IL-10, IL-13 and TGFβ and results in induction of host fibrotic response involving Arginase-1 expression. At this stage the L-ornithine produced by Arginase-1 can be converted to staphylocidal polyamines that will additionally promote fibroblast proliferation, collagen deposition and inhibition of inflammation (e.g. blocking iNOS translation)119. It therefore may be during this TH2-dominant fibrotic phase that host polyamines exert their effects on invading S. aureus thereby selecting for ACME encoded SpeG. Indeed, inhibiting IL-4 signaling in mice increased organism burdens during S. aureus sepsis while INF-γ -/- mice (lacking robust inflammatory wound response) survived better than WT mice120. Thus, TH2-dependent signaling, as opposed to an inflammatory TH1 response, proved critical to the host’s ability to control S. aureus infections. Recently, protection against chronic implant infections was also highly dependent on an effective TH2/Treg response121. Furthermore, polymorphisms in the human IL-4 gene associated with reduced IL-4 production are significantly linked with increased S. aureus colonization122. These data are consistent with the TH2 anti-inflammatory fibrotic response as being critical for controlling S. aureus infection. Whether this is directly due to the induction of polyamine synthesis has yet to be reported, but the acquisition of speG-encoding ACME would counter increased spermine levels in fibrotic tissue perhaps explaining the association of USA300 CA-MRSA with severe skin/soft tissue infections.
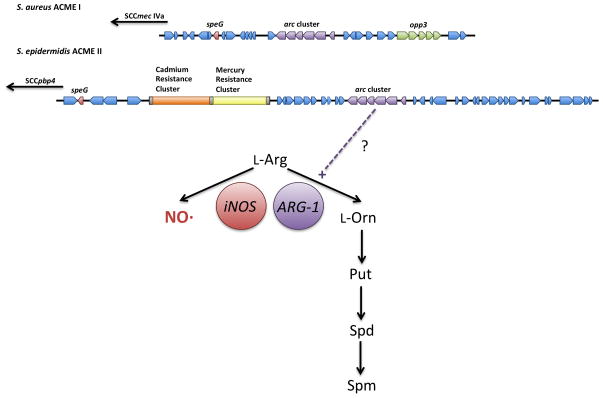
Figure 2. Association between arc gene cluster and speG in ACME. TOP.
ACME type I, found in USA300 S. aureus and also found in many S. epidermidis isolates, and ACME type II, found primarily in S. epidermidis, both harbor arc gene clusters as well as speG. ACME type III (not shown) lacks an identifiable arc gene cluster but does contain an opp-3 locus. BOTTOM: Fate of host arginine depends on competition between iNOS and Arginase-1 enzyme activities. The net production of ornithine by Arc-expressing S. aureus may skew the fate of host arginine down the polyamine synthesis pathway thereby necessitating speG.
How do we reconcile a significant role for SpeG in S. aureus pathogenesis with the lack of a strong ACME phenotype in most model infections84,113? One explanation could be that the observed increase in α-hemolysin and Protein A expression upon ACME inactivation in USA300 could overcompensate for the resulting polyamine-sensitivity84. Another possibility is that the Arc operon on ACME actually drives excess polyamine production necessitating SpeG-mediated spermine detoxification. The Arc operon consists of genes that convert L-arginine to L-ornithine and CO2 while producing ATP and ammonia. The resulting L-ornithine is exchanged for extracellular L-arginine by the L-arginine/L-ornithine antiporter ArcD effectively converting extracellular L-arginine to L-ornithine. Thus, the Arc operon could skew the flux of host L-arginine away from iNOS towards polyamine synthesis rendering speG essential (Figure 2). Deleting all of ACME might allow the host to partition available L-arginine towards NO·-production, an immune effector that S. aureus is known to effectively resist123–125. This is consistent with the presence of speG on ACME islands that harbor the auxiliary arc gene cluster (Figure 2). While this hypothesis could explain the modularity of ACME that results in ΔspeG attenuation, it has several aspects that require experimental attention. First, all strains of S. aureus already encode an Arc operon on the core chromosome that could also result in excess host polyamine synthesis, yet SpeG is only associated with ACME-positive USA300 S. aureus. This could be explained by the fact that the chromosomal Arc operon is only expressed under conditions of low oxygen and low glucose and little is known about ACME Arc expression in S. aureus126. Second, a dominant MRSA clone of ST22 lineage in Irish hospitals harbors an ACME island with an arc gene cluster but appears to lack a speG homologue112. Another issue is that significant CA-MRSA disease in Latin America is caused by USA300 clones that lack ACME127. Thus, ACME may contribute to colonization and virulence, but it cannot fully explain the predominance of USA300 in CA-MRSA disease in North America.
Enhanced Virulence Gene Expression
S. aureus elaborates a wide variety of toxins and proteases that have proven critical for efficient dissemination, inflammation and disease progression128–130. For instance, α-toxin or α-hemolysin (Hla) is a potent heptameric pore-forming toxin known to be critical for virulence in nearly every tested disease model from skin lesions and endocarditis to murine mastitis131–133. Upon interacting with susceptible cells, which include leukocytes, keratinocytes, platelets and endothelial cells, it forms a 100 Å deep pore in the plasma membrane resulting in rapid cell lysis134,135. Recently, a number of reports have shown that Hla expression is highly elevated in USA300 clones compared with other S. aureus isolates36,50–52. Moreover, deletion of hla abrogates USA300 virulence in murine and rabbit skin lesion models as well as pneumonia43,100,136. However, it should be noted that hla mutants in almost any S. aureus background are attenuated133,137–140, thus the loss of virulence in USA300 hla mutants is consistent with α-toxin in general being a critical pathogenicity factor to S. aureus. δ-toxin (encoded by hld) and related α-type phenol-soluble modulins (αPSMs) are amphipathic α-helical peptides with potent leukocidal and chemotactic properties141. They have been shown to be overproduced by CA-MRSA clones with respect to most HA-MRSA isolates36,51,141. Their abundant production is essential for full virulence in murine and rabbit skin models of infection as well as murine sepsis108,141. Interestingly, they have recently been shown to exert potent antimicrobial activity against multiple Gram-positive bacterial species142. This property may prove critical for efficient colonization of non-sterile sites such as skin and nasal passages, thereby providing CA-MRSA with a selective advantage during transmission. Finally, S. aureus expresses a number of secreted proteases that, while antagonistic to in vitro biofilm formation, likely mediate the breakdown of host fibrotic tissue synthesized to confine S. aureus-containing lesions thereby promoting bacterial dissemination and disease progression. As with α-toxin and αPSMs, USA300 clones are also known to excrete proteases in excess, potentially limiting the host’s ability to control minor skin and soft-tissue infections143. Thus, several groups have consistently reported the robust expression of numerous virulence determinants in USA300 compared with other clinical isolates. It has therefore been hypothesized that this over-production of toxins/proteases confers the selective advantage that explains the overwhelming success of USA300 clones. If true, the regulatory mechanisms explaining these virulence trait expression phenomena are poorly defined.
Agr Quorum Sensing System
S. aureus expresses a peptide-based quorum sensing system known as Agr for Accessory Gene Regulator129,144. Signaling is mediated through a peptide form of AgrD (processed by the combined activity of the AgrB endopeptidase and a type I signal peptidase, SpsB145) that stimulates the two-component system sensor kinase, AgrC. The resulting activation of the response regulator AgrA leads to induction of the agrBDCA operon as well as the divergently transcribed RNAIII. While RNAIII encodes δ-toxin, the RNA molecule itself mediates a significant proportion of Agr regulation by affecting the expression of α-toxin146, protein A147, repressor of toxins (Rot)148 and others149. Active AgrA is also known to directly control the expression of other virulence determinants including the PSMs150. Thus, the reported overproduction of Hla, Hld and PSMs in USA300 clones may be explained by a hyperactive Agr system in these clones. Indeed, the RNAIII molecule was shown to be expressed to a higher level in USA300 clones than in other S. aureus isolates explaining the overabundance of δ-hemolysin production51,52. Additionally, the overactive USA300 Agr system was the source of excess PSM and protease production associated with these clones and was partially responsible for excessive Hla expression50. Consistent with these data, Δagr mutants in USA300 are highly attenuated in murine sepsis, pneumonia and skin abscess models50,108,151. Though, given the importance of Agr in virulence gene regulation, it is not surprising that mutants exhibit such attenuation. Moreover, overproduction of PSMs was reported for USA400 CA-MRSA clones implying that the greater success of USA300 cannot be fully attributed to overactive Agr51,141. In fact, USA500 clones, thought to be ancestral to USA300 also exhibit phenotypes with hyperactive Agr as well as being highly virulent in murine model infections36,51. Thus, the high virulence potential of USA300, including high Agr-activity, likely evolved in the HA-MRSA clones belonging to USA500. Still, Δagr mutants of USA300 are highly attenuated and exhibit no increased virulence relative to non-USA300 agr mutants underscoring its importance in the evolution of USA30050.
SaeR Two-Component System
The S. aureus exoprotein expression (Sae) locus contains four genes, saePQRS the latter of which comprise a two-component regulatory system152–154. The response regulator/sensor kinase genes (saeRS) are preceded by genes encoding a membrane protein (SaeQ) and a lipoprotein (SaeP) of unknown function. All four genes are cotranscribed from a promoter that is strongly induced by active SaeR155. A second promoter drives the expression of saeRS alone and is modestly repressed by these regulatory gene products155. Activation of the Sae system seems to involve sensing changes in the overall integrity of the cell envelope and is highly stimulated by hydrogen peroxide and cationic peptides including α-defensins155,156. Active SaeR promotes the induction of a number of virulence genes in S. aureus through binding of a consensus sequence found upstream of promoters for hla, sbi, efb, lukS-PVL, splA and saeP157. Additionally, expression of β-hemolysin, fibrinogen-binding proteins, lactose catabolizing enzymes and the chromosomal arginine deiminase operon are all highly affected by Sae158. It has been shown that SaeRS expression is higher in USA300 than in USA400 clones52,155_ENREF_52, which may be a result of overactive Agr system (see above) since RNAIII is known to positively regulate Sae expression156. Deletion of saeRS resulted in almost complete loss of Hla expression and a significant drop in PVL levels as well151,157. Moreover, Δsae USA300 was attenuated in murine sepsis, peritonitis, dermonecrosis and pneumonia models151,157–159. This was surprising given that in USA400, Sae was only essential for sepsis and peritonitis and not for survival within skin abscesses158,159. However, USA400 clones do not induce the same level of dermonecrosis and do not express high levels of Hla as in USA300 infections51,52. Thus, it appears as though some of the hypervirulence attributed to USA300 clones in skin/soft tissue infections is likely due to Sae-mediated Hla overproduction. However, HA-MRSA USA500 clones also exhibit severe dermonecrosis during skin infections and overproduce Hla and PSMs yet have not disseminated as widely as USA300.
Source of overactive Agr
While it has not been directly tested, it is tempting to hypothesize that the overactive Agr system inherent to USA300 results in excessive PSMs and Sae expression, the latter of which leads to high Hla expression. However, the mechanism driving high Agr activity in USA300 is not defined. Agr activity can be modulated through the actions of a number of trans-acting regulators including SarA160, Stk1161, MgrA162, SigB143, CodY163, CcpA164, Sar-family proteins other than SarA165–168, ArlRS169, Rsr161 and SrrAB170. Many of these regulators are presumed to affect Agr expression indirectly, however some (CodY171, SrrA172 and SarA173) have been shown to directly bind to the Agr locus. It is intriguing that many of these regulators are involved in modulating metabolic adaptation to various environments (CodY, CcpA, Rsr and SrrAB) given the apparent increase in fitness associated with USA300174 (see below). Though any one of these or other unknown regulatory systems may be responsible for enhanced Agr activity in USA300, therefore investigations into strain-specific differences in activity among these regulators may prove enlightening. For instance, SarA positively affects Agr expression160,175, and deletion of sarA in USA300 lead to drastic reductions in Hla and PSM levels176,177. However, recently it was demonstrated that the loss of cytolytic expoprotein expression in the ΔsarA mutant was attributed to the resulting overproduction of extracellular proteases and not due to altered exoprotein gene transcription177.
While trans-acting regulators may prove to be major influences on USA300 Agr activity, cis-acting polymorphisms may also be involved. RNAIII transcripts among sequenced ST8 isolates are 100% conserved, but there is a single nucleotide polymorphism (SNP) 3 bp upstream of a known AgrA binding site within the RNAIII promoter that is only found among USA300 isolates. While this is the only SNP among ST8 and ST1 clones specific to USA300, other sites of variation exist when compared to USA100 and USA200 promoter sequences. SNPs in the Hla promoter were recently shown to drive its overexpression in bovine isolates by modulating SarZ binding178. It remains to be determined whether SNPs in the RNAIII promoter region of USA300 isolates affect expression leading to high Agr activity. Regardless of the mechanism behind hyperactive toxin production in USA300, it is important to remember that similar high-level expression is observed in the HA-MRSA progenitor clone, USA500. Thus, while the high virulence potentials of USA300 and USA500 may result from overproduction of exoproteins, this phenomenon alone cannot fully explain the enormous success of USA300 in human disease.
Non-synonymous Mutations in Core Genes
The evolutionary forces that drive diversification in S. aureus have been recently examined, in part, due to the availability of more than 15 published S. aureus genome sequences. While a significant level of divergence is achieved through acquisition of mobile genetic elements (MGE), variability within the S. aureus core genome (~2000 orthologous genes shared among most S. aureus strains) is primarily generated through mutation179,180. The most common forms of mutation are single nucleotide polymorphisms (SNPs) or short insertion/deletions (indels) that have been estimated to be ~15-fold more attributable to de novo mutation than to recombination179. However, recent reports contend that the contribution of homologous recombination to core diversity in S. aureus may be underestimated181. Nevertheless, mutation is a significant driving force in S. aureus diversification allowing for evolutionary classification of strains into ST types (see above)22. Most SNPs are within coding regions reflecting the fact that ~80% of the core genome encodes protein182. Synonymous SNPs, those that do not result in amino acid changes, by far outweigh amino acid substituting non-synonymous SNPs in S. aureus183–186. This is likely because nonsynonymous mutations are more often detrimental and are therefore subject to evolutionary loss via purifying selection. Consequently, the relative ratio of nonsynonymous to synonymous substitution rate (dN/dS) among staphylococci is generally less than 1. In contrast, a recent report comparing the complete genome sequences of 10 newly isolated USA300 clones with the published FPRF3757 USA300 sequence revealed an unusually high ratio of nonsynonymous:synonymous SNPs (as high as 2.6:1, much higher than reported in comparisons of non-USA300 S. aureus lineages)43. This discrepancy can be rationalized by assuming a recent clonal expansion of the USA300 lineage such that new isolates still harbor nonsynonymous SNPs that have not yet undergone purifying selection187. To be sure, the unusually high dN/dS ratio of USA300 clones is inconsistent with evolutionary convergence among distantly related clones, an event that would only be consistent with normal to low dN/dS ratios if the converging progenitors were of sufficiently diverse origins43.
It is important to note that overall low dN/dS ratios are not necessarily constant across all functional gene families. For instance, while housekeeping and metabolic genes generally exhibit low dN/dS ratios, genes encoding surface associated or secreted proteins can often have elevated dN/dS ratios188,189. This is indicative of forward selective pressures driving variability in these genes either to promote functional differences (e.g. an adhesin adapting to a host receptor molecule) or immune avoidance through changes in antigenicity. Indeed, comparisons among divergent S. aureus clones reveal higher dN/dS ratios for genes encoding components of the cell envelope and secreted proteins than genes encoding housekeeping or metabolic enzymes182,184,185. USA300 clones however seem to be an exception to this rule. A recent comparison of genome sequences from USA200, USA300 and a distantly related S. aureus strain revealed high dN/dS ratios indicative of forward selection in a large number of USA300 metabolic genes190. The largest subset of USA300 genes predicted to be under positive selection (45%) were involved with metabolism whereas only 7% encoded components of the cell envelope. This phenomenon cannot be explained by the fact that metabolic genes make up a large proportion of the core genome because this same study showed that in USA200, the most prominent class of genes undergoing positive selection were those encoding cell envelope components (a third of all genes with elevated dN/dS)186,190. An independent study verified that all of the metabolic genes in USA300 exhibiting forward selection were completely conserved among 10 sequenced USA300 genomes43. Moreover, data from this same study showed that, while relatively few SNPs were found among ten different USA300 genomes, genes encoding cell envelope proteins more commonly exhibited high dN/dS ratios (57% of all genes with multiple nonsynonymous substitutions)43. Thus, the peculiar overrepresentation of S. aureus metabolic genes among those undergoing positive selection is only evident when comparing USA300 with non-USA300 genomes implying that USA300 clones in general seem to be adapting to disproportionately high selective pressures at the metabolic level.
It is possible that the resulting adaptive mutations in the overall metabolism of USA300 directly contribute to the evolutionary success of this clone. For instance, it has been observed that USA300 clones simply grow faster than any other tested S. aureus isolate174. Taken together, it would appear that USA300 is more metabolically fit and/or adaptable than other S. aureus lineages. This may provide an advantage when competing for limiting nutrients with endogenous microflora as well as contribute to severe disease given a rapid growth rate within sterile sites of the body. Further inspection in our laboratory revealed that USA300 clones have growth advantages when metabolizing many different carbon sources (Table 1). In general, USA300 clones exhibited higher growth rates than other clones when cultivated on nutrients that are abundant in human sweat and skin191, consistent with the high prevalence of skin/soft tissue infections associated with USA300 clones. But, can a relatively small set of amino acid changes in metabolic genes really account for such drastic growth differences? Laboratory adaptation of E. coli to growth on lactate resulted in strains that exhibited nearly twice the growth rate on lactate alone192. These adapted strains exhibited major alterations in metabolic flux capacity through gluconeogenic and pyruvate catabolic pathways, yet none of these changes were due to altered gene expression. This would be consistent with subtle changes in protein sequence (nonsynonymous SNPs) that alter enzyme activity or response to allosteric regulation. Furthermore, a laboratory adapted clone of Caulobacter crescentus exhibited a ~20% greater growth rate than its progenitor strain and this entire phenotype was explained by a single SNP altering the expression of glucose-6-phosphate dehydrogenase (zwf)193. This enzyme controls the primary flux between energy generating glycolysis and the precursor generating pentose-phosphate pathway (PPP). It was shown that lower flux through PPP with concomitant increased glycolytic activity lead to higher growth rates in lab-adapted C. crescentus193. Interestingly, one of the very genes exhibiting signs of positive selection in USA300 was zwf along with two glycolytic genes (pgm and pfkA) potentially linked to the USA300 growth advantage on numerous carbon sources190. Whether or not SNPs within these metabolic genes account for enhanced USA300 growth rates and whether that contributes to the success of this clone remain to be proven, however the unusual SNP distribution among metabolic genes in USA300 combined with its enhanced growth rate suggest there may be more to USA300 virulence than newly acquired or overexpression of virulence genes.
Table 1. Maximal Growth rates of S. aureus strains on various carbon sources.
Rates (μ, h−1) were calculated from at least 4 independent curves and mean ± S.D. are reported. Chemically defined medium194 was used varying only the primary carbon source. Red font indicates rates significantly lower than those of USA300 strain SF8300 (p ≤ 0.05, 2-tailed Students T-test).
| 0.5% Glucose | 1.0% Lactate | 1.0% Pyruvate | 1.0% Glycerol | 1.0% Cas A.A. | 1.0% Tryptone | |
|---|---|---|---|---|---|---|
| SF8300 (USA300) | 0.92 ± 0.02 | 0.60 ± 0.01 | 0.60 ± 0.01 | 0.90 ± 0.03 | 0.74 ± 0.02 | 0.65 ± 0.02 |
| LAC (USA300) | 0.89 ± 0.03 | 0.57 ± 0.02 | 0.57 ± 0.01 | 0.89 ± 0.03 | 0.72 ± 0.01 | 0.63 ± 0.02 |
| Newman (ST8 MSSA) | 0.71 ± 0.01 | 0.43 ± 0.02 | 0.46 ± 0.02 | 0.73 ± 0.02 | 0.43 ± 0.02 | 0.36 ± 0.01 |
| MW2 (USA400) | 0.85 ± 0.04 | 0.41 ± 0.02 | 0.52 ± 0.02 | 0.76 ± 0.04 | 0.75 ± 0.02 | 0.51 ± 0.05 |
| UAMS-1 (USA200) | 0.73 ± 0.02 | 0.48 ± 0.01 | 0.52 ± 0.01 | 0.79 ± 0.01 | 0.57 ± 0.01 | 0.68 ± 0.02 |
Conclusions
The overwhelming success of USA300 in North America as the dominant source of CA-MRSA infections represents a fascinating example of a pathogenic variant emerging as a new threat to human health. The adaptations acquired by USA300 clones in the form of novel genetic components, altered gene regulation and sequence polymorphisms likely act in concert to provide these strains with a selective advantage. It appears as though USA300 hypervirulence, as assayed in animal models of infection, correlates with increases in virulence gene expression and is apparent in HA-MRSA progenitors as well as other unrelated CA-MRSA lineages. Whether this is due to hyperactive Agr resulting in elevated PSM production and Sae expression (which in turn could lead to excess Hla and other exoprotein excretion) remains to be proven. In contrast to overt virulence, traits that affect transmission and colonization efficiency are inherently difficult to model in the laboratory. It may prove, however that this aspect of USA300 biology is as critical to its success as is high virulence potential. It remains to be determined whether newly acquired genetic components (e.g. ACME) and/or sequence polymorphisms contribute to the rapid transmission and success of USA300 in the community. In the end, we may appreciate that none of the three evolutionary events (gene acquisitions, altered gene regulation, protein sequence divergence) outlined here can alone explain the success of USA 300. Rather, the amalgamation of all these events created the highly successful pathogen that we must contend with today.
Acknowledgments
This work was supported by funding from the NIH (AI088158 to A.R.R.)
Literature Cited
- 1.Diekema DJ, et al. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997–1999. Clin Infect Dis. 2001;32(Suppl 2):S114–132. doi: 10.1086/320184. [DOI] [PubMed] [Google Scholar]
- 2.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 3.Foster TJ. Colonization and infection of the human host by staphylococci: adhesion, survival and immune evasion. Vet Dermatol. 2009;20:456–470. doi: 10.1111/j.1365-3164.2009.00825.x. [DOI] [PubMed] [Google Scholar]
- 4.Schechter-Perkins EM, et al. Prevalence and predictors of nasal and extranasal staphylococcal colonization in patients presenting to the emergency department. Ann Emerg Med. 2011;57:492–499. doi: 10.1016/j.annemergmed.2010.11.024. [DOI] [PubMed] [Google Scholar]
- 5.Ogston A. Report upon Micro-Organisms in Surgical Diseases. Br Med J. 1881;1:369, b362–375. doi: 10.1136/bmj.1.1054.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newsom SW. Ogston’s coccus. J Hosp Infect. 2008;70:369–372. doi: 10.1016/j.jhin.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Lister J. On the Antiseptic Principle in the Practice of Surgery. Br Med J. 1867;2:246–248. doi: 10.1136/bmj.2.351.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reddish GF. The Resistance to Phenol of Staphylococcus Aureus. Am J Public Health (N Y) 1925;15:534–538. doi: 10.2105/ajph.15.6.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landy M, Larkum NW, Oswald EJ, Streightoff F. INCREASED SYNTHESIS OF p-AMINOBENZOIC ACID ASSOCIATED WITH THE DEVELOPMENT OF SULFONAMIDE RESISTANCE IN STAPHYLOCOCCUS AUREUS. Science. 1943;97:265–267. doi: 10.1126/science.97.2516.265. [DOI] [PubMed] [Google Scholar]
- 10.Kirby WM. Extraction of a Highly Potent Penicillin Inactivator from Penicillin Resistant Staphylococci. Science. 1944;99:452–453. doi: 10.1126/science.99.2579.452. [DOI] [PubMed] [Google Scholar]
- 11.Rountree PM, Freeman BM. Infections caused by a particular phage type of Staphylococcus aureus. Med J Aust. 1955;42:157–161. [PubMed] [Google Scholar]
- 12.Jevons MP. “Celbenin” - resistant Staphylococci. Br Med J. 1961;1:124–125. [Google Scholar]
- 13.Archer GL, Mayhall CG. Comparison of epidemiological markers used in the investigation of an outbreak of methicillin-resistant Staphylococcus aureus infections. J Clin Microbiol. 1983;18:395–399. doi: 10.1128/jcm.18.2.395-399.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Talan DA, et al. Comparison of Staphylococcus aureus from skin and soft-tissue infections in US emergency department patients, 2004 and 2008. Clin Infect Dis. 2011;53:144–149. doi: 10.1093/cid/cir308. [DOI] [PubMed] [Google Scholar]
- 15.Tsubakishita S, Kuwahara-Arai K, Sasaki T, Hiramatsu K. Origin and molecular evolution of the determinant of methicillin resistance in staphylococci. Antimicrob Agents Chemother. 2010;54:4352–4359. doi: 10.1128/AAC.00356-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Couto I, et al. Ubiquitous presence of a mecA homologue in natural isolates of Staphylococcus sciuri. Microb Drug Resist. 1996;2:377–391. doi: 10.1089/mdr.1996.2.377. [DOI] [PubMed] [Google Scholar]
- 17.Center for Disease Control and Prevention, C. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother. 2009;53:4961–4967. doi: 10.1128/AAC.00579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katayama Y, Ito T, Hiramatsu K. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2000;44:1549–1555. doi: 10.1128/aac.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKenna M. Superbug : the fatal menace of MRSA. Free Press; 2010. 1st Free Press hardcover edn. [Google Scholar]
- 20.Okuma K, et al. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J Clin Microbiol. 2002;40:4289–4294. doi: 10.1128/JCM.40.11.4289-4294.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shopsin B, et al. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol. 1999;37:3556–3563. doi: 10.1128/jcm.37.11.3556-3563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enright MC, et al. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA) Proc Natl Acad Sci U S A. 2002;99:7687–7692. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDougal LK, et al. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol. 2003;41:5113–5120. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bannerman TL, Hancock GA, Tenover FC, Miller JM. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J Clin Microbiol. 1995;33:551–555. doi: 10.1128/jcm.33.3.551-555.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson DA, Enright MC. Evolutionary models of the emergence of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2003;47:3926–3934. doi: 10.1128/AAC.47.12.3926-3934.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Udo EE, Pearman JW, Grubb WB. Genetic analysis of community isolates of methicillin-resistant Staphylococcus aureus in Western Australia. J Hosp Infect. 1993;25:97–108. doi: 10.1016/0195-6701(93)90100-e. [DOI] [PubMed] [Google Scholar]
- 28.From the Centers for Disease Control and Prevention. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus--Minnesota and North Dakota 1997–1999. Jama. 1999;282:1123–1125. [PubMed] [Google Scholar]
- 29.Van De Griend P, et al. Community-associated methicillin-resistant Staphylococcus aureus, Iowa, USA. Emerg Infect Dis. 2009;15:1582–1589. doi: 10.3201/eid1510.080877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coombs GW, et al. Genetic diversity among community methicillin-resistant Staphylococcus aureus strains causing outpatient infections in Australia. J Clin Microbiol. 2004;42:4735–4743. doi: 10.1128/JCM.42.10.4735-4743.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.David MZ, Rudolph KM, Hennessy TW, Boyle-Vavra S, Daum RS. Molecular epidemiology of methicillin-resistant Staphylococcus aureus, rural southwestern Alaska. Emerg Infect Dis. 2008;14:1693–1699. doi: 10.3201/eid1411.080381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mulvey MR, et al. Community-associated methicillin-resistant Staphylococcus aureus, Canada. Emerg Infect Dis. 2005;11:844–850. doi: 10.3201/eid1106.041146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Golding GR, et al. High rates of Staphylococcus aureus USA400 infection, Northern Canada. Emerg Infect Dis. 2011;17:722–725. doi: 10.3201/eid1704.100482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neocleous C, Damani A, Gerogianni I, Gourgoulianis K, Petinaki E. Necrotizing pneumonia in Greece caused by a USA400 (ST1) Staphylococcus aureus harboring SSCmec type V. Infection. 2010;38:76–77. doi: 10.1007/s15010-009-9199-8. [DOI] [PubMed] [Google Scholar]
- 35.Vignaroli C. Methicillin-resistant Staphylococcus aureus USA400 Clone, Italy. Emerg Infect Dis. 2009;15:995–996. doi: 10.3201/eid1506.081632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li M, et al. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci U S A. 2009;106:5883–5888. doi: 10.1073/pnas.0900743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moran GJ, et al. Methicillin-resistant S. aureus infections among patients in the emergency department. N Engl J Med. 2006;355:666–674. doi: 10.1056/NEJMoa055356. [DOI] [PubMed] [Google Scholar]
- 38.Hulten KG, et al. Hospital-acquired Staphylococcus aureus infections at Texas Children’s Hospital, 2001–2007. Infect Control Hosp Epidemiol. 2010;31:183–190. doi: 10.1086/649793. [DOI] [PubMed] [Google Scholar]
- 39.Como-Sabetti K, et al. Community-associated methicillin-resistant Staphylococcus aureus: trends in case and isolate characteristics from six years of prospective surveillance. Public Health Rep. 2009;124:427–435. doi: 10.1177/003335490912400312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simor AE, et al. Methicillin-resistant Staphylococcus aureus colonization or infection in Canada: National Surveillance and Changing Epidemiology, 1995–2007. Infect Control Hosp Epidemiol. 2010;31:348–356. doi: 10.1086/651313. [DOI] [PubMed] [Google Scholar]
- 41.Nichol KA, et al. Comparison of community-associated and health care-associated methicillin-resistant Staphylococcus aureus in Canada: results of the CANWARD 2007–2009 study. Diagn Microbiol Infect Dis. 2011;69:320–325. doi: 10.1016/j.diagmicrobio.2010.10.028. [DOI] [PubMed] [Google Scholar]
- 42.Velazquez-Meza ME, et al. First report of community-associated methicillin-resistant Staphylococcus aureus (USA300) in Mexico. J Clin Microbiol. 2011;49:3099–3100. doi: 10.1128/JCM.00533-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kennedy AD, et al. Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification. Proc Natl Acad Sci U S A. 2008;105:1327–1332. doi: 10.1073/pnas.0710217105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morrison MA, Hageman JC, Klevens RM. Case definition for community-associated methicillin-resistant Staphylococcus aureus. J Hosp Infect. 2006;62:241. doi: 10.1016/j.jhin.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 45.Moore CL, et al. Comparative evaluation of epidemiology and outcomes of methicillin-resistant Staphylococcus aureus (MRSA) USA300 infections causing community- and healthcare-associated infections. Int J Antimicrob Agents. 2009;34:148–155. doi: 10.1016/j.ijantimicag.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 46.Popovich KJ, Weinstein RA, Hota B. Are community-associated methicillin-resistant Staphylococcus aureus (MRSA) strains replacing traditional nosocomial MRSA strains? Clin Infect Dis. 2008;46:787–794. doi: 10.1086/528716. [DOI] [PubMed] [Google Scholar]
- 47.Jenkins TC, et al. Epidemiology of healthcare-associated bloodstream infection caused by USA300 strains of methicillin-resistant Staphylococcus aureus in 3 affiliated hospitals. Infect Control Hosp Epidemiol. 2009;30:233–241. doi: 10.1086/595963. [DOI] [PubMed] [Google Scholar]
- 48.Deleo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated meticillin-resistant Staphylococcus aureus. Lancet. 2010;375:1557–1568. doi: 10.1016/S0140-6736(09)61999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan ES, et al. Population dynamics of nasal strains of methicillin-resistant Staphylococcus aureus--and their relation to community-associated disease activity. J Infect Dis. 2005;192:811–818. doi: 10.1086/432072. [DOI] [PubMed] [Google Scholar]
- 50.Cheung GY, Wang R, Khan BA, Sturdevant DE, Otto M. Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infect Immun. 2011;79:1927–1935. doi: 10.1128/IAI.00046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li M, et al. Comparative analysis of virulence and toxin expression of global community-associated methicillin-resistant Staphylococcus aureus strains. J Infect Dis. 2010;202:1866–1876. doi: 10.1086/657419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Montgomery CP, et al. Comparison of virulence in community-associated methicillin-resistant Staphylococcus aureus pulsotypes USA300 and USA400 in a rat model of pneumonia. J Infect Dis. 2008;198:561–570. doi: 10.1086/590157. [DOI] [PubMed] [Google Scholar]
- 53.Seybold U, et al. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of health care-associated blood stream infections. Clin Infect Dis. 2006;42:647–656. doi: 10.1086/499815. [DOI] [PubMed] [Google Scholar]
- 54.Haque NZ, et al. Infective endocarditis caused by USA300 methicillin-resistant Staphylococcus aureus (MRSA) Int J Antimicrob Agents. 2007;30:72–77. doi: 10.1016/j.ijantimicag.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 55.Miller LG, et al. Necrotizing fasciitis caused by community-associated methicillin-resistant Staphylococcus aureus in Los Angeles. N Engl J Med. 2005;352:1445–1453. doi: 10.1056/NEJMoa042683. [DOI] [PubMed] [Google Scholar]
- 56.Hageman JC, et al. Severe community-acquired pneumonia due to Staphylococcus aureus, 2003–04 influenza season. Emerg Infect Dis. 2006;12:894–899. doi: 10.3201/eid1206.051141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Francis JS, et al. Severe community-onset pneumonia in healthy adults caused by methicillin-resistant Staphylococcus aureus carrying the Panton-Valentine leukocidin genes. Clin Infect Dis. 2005;40:100–107. doi: 10.1086/427148. [DOI] [PubMed] [Google Scholar]
- 58.Klevens RM, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. Jama. 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 59.Ellis MW, et al. Presence and molecular epidemiology of virulence factors in methicillin-resistant Staphylococcus aureus strains colonizing and infecting soldiers. J Clin Microbiol. 2009;47:940–945. doi: 10.1128/JCM.02352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Center for Disease Control and Prevention C. Outbreaks of community-associated methicillin-resistant Staphylococcus aureus skin infections--Los Angeles County, California, 2002–2003. MMWR Morb Mortal Wkly Rep. 2003;52:88. [PubMed] [Google Scholar]
- 61.Center for Disease Control and Prevention C. Methicillin-resistant staphylococcus aureus infections among competitive sports participants--Colorado, Indiana, Pennsylvania, and Los Angeles County, 2000–2003. MMWR Morb Mortal Wkly Rep. 2003;52:793–795. [PubMed] [Google Scholar]
- 62.Center for Disease Control and Prevention C. Methicillin-resistant Staphylococcus aureus among players on a high school football team--New York City, 2007. MMWR Morb Mortal Wkly Rep. 2009;58:52–55. [PubMed] [Google Scholar]
- 63.Maree CL, et al. Risk factors for infection and colonization with community-associated methicillin-resistant Staphylococcus aureus in the Los Angeles County jail: a case-control study. Clin Infect Dis. 2010;51:1248–1257. doi: 10.1086/657067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Center for Disease Control and Prevention C. Methicillin-resistant Staphylococcus aureus skin or soft tissue infections in a state prison--Mississippi, 2000. MMWR Morb Mortal Wkly Rep. 2001;50:919–922. [PubMed] [Google Scholar]
- 65.Center for Disease Control and Prevention C. Methicillin-resistant Staphylococcus aureus infections in correctional facilities---Georgia, California, and Texas, 2001–2003. MMWR Morb Mortal Wkly Rep. 2003;52:992–996. [PubMed] [Google Scholar]
- 66.Kempker RR, Farley MM, Ladson JL, Satola S, Ray SM. Association of methicillin-resistant Staphylococcus aureus (MRSA) USA300 genotype with mortality in MRSA bacteremia. J Infect. 2010;61:372–381. doi: 10.1016/j.jinf.2010.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Buckingham SC, et al. Emergence of community-associated methicillin-resistant Staphylococcus aureus at a Memphis, Tennessee Children’s Hospital. Pediatr Infect Dis J. 2004;23:619–624. doi: 10.1097/01.inf.0000131981.67342.c4. [DOI] [PubMed] [Google Scholar]
- 68.Kaplan SL, et al. Three-year surveillance of community-acquired Staphylococcus aureus infections in children. Clin Infect Dis. 2005;40:1785–1791. doi: 10.1086/430312. [DOI] [PubMed] [Google Scholar]
- 69.Sztramko R, et al. Community-associated methicillin-resistant Staphylococcus aureus infections in men who have sex with men: A case series. Can J Infect Dis Med Microbiol. 2007;18:257–261. doi: 10.1155/2007/592684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nair N, et al. Molecular Epidemiology of Methicillin-Resistant Staphylococcus aureus (MRSA) among Patients Admitted to Adult Intensive Care Units: The STAR*ICU Trial. Infect Control Hosp Epidemiol. 2011;32:1057–1063. doi: 10.1086/662178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Whitby CR, et al. Staphylococcus aureus sinus infections in children. Int J Pediatr Otorhinolaryngol. 2011;75:118–121. doi: 10.1016/j.ijporl.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 72.Kreisel KM, et al. USA300 methicillin-resistant Staphylococcus aureus bacteremia and the risk of severe sepsis: is USA300 methicillin-resistant Staphylococcus aureus associated with more severe infections? Diagn Microbiol Infect Dis. 2011;70:285–290. doi: 10.1016/j.diagmicrobio.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ganga R, et al. Role of SCCmec type in outcome of Staphylococcus aureus bacteremia in a single medical center. J Clin Microbiol. 2009;47:590–595. doi: 10.1128/JCM.00397-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hota B, et al. Predictors of clinical virulence in community-onset methicillin-resistant Staphylococcus aureus infections: the importance of USA300 and pneumonia. Clin Infect Dis. 2011;53:757–765. doi: 10.1093/cid/cir472. [DOI] [PubMed] [Google Scholar]
- 75.Lalani T, et al. Associations between the genotypes of Staphylococcus aureus bloodstream isolates and clinical characteristics and outcomes of bacteremic patients. J Clin Microbiol. 2008;46:2890–2896. doi: 10.1128/JCM.00905-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bassetti M, et al. Risk factors and mortality of healthcare-associated and community-acquired Staphylococcus aureus bacteraemia. Clin Microbiol Infect. 2011 doi: 10.1111/j.1469-0691.2011.03679.x. [DOI] [PubMed] [Google Scholar]
- 77.McDougal LK, et al. Emergence of resistance among USA300 methicillin-resistant Staphylococcus aureus isolates causing invasive disease in the United States. Antimicrob Agents Chemother. 2010;54:3804–3811. doi: 10.1128/AAC.00351-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Diep BA, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 79.Yarwood JM, et al. Characterization and expression analysis of Staphylococcus aureus pathogenicity island 3. Implications for the evolution of staphylococcal pathogenicity islands. J Biol Chem. 2002;277:13138–13147. doi: 10.1074/jbc.M111661200. [DOI] [PubMed] [Google Scholar]
- 80.Carvalho KS, Mamizuka EM, Gontijo Filho PP. Methicillin/Oxacillin-resistant Staphylococcus aureus as a hospital and public health threat in Brazil. Braz J Infect Dis. 2010;14:71–76. doi: 10.1590/s1413-86702010000100014. [DOI] [PubMed] [Google Scholar]
- 81.Baba T, et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–1827. doi: 10.1016/s0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 82.Daum RS, et al. A novel methicillin-resistance cassette in community-acquired methicillin-resistant Staphylococcus aureus isolates of diverse genetic backgrounds. J Infect Dis. 2002;186:1344–1347. doi: 10.1086/344326. [DOI] [PubMed] [Google Scholar]
- 83.Goering RV, et al. Epidemiologic distribution of the arginine catabolic mobile element among selected methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates. J Clin Microbiol. 2007;45:1981–1984. doi: 10.1128/JCM.00273-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Diep BA, et al. The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J Infect Dis. 2008;197:1523–1530. doi: 10.1086/587907. [DOI] [PubMed] [Google Scholar]
- 85.Ender M, McCallum N, Adhikari R, Berger-Bachi B. Fitness cost of SCCmec and methicillin resistance levels in Staphylococcus aureus. Antimicrob Agents Chemother. 2004;48:2295–2297. doi: 10.1128/AAC.48.6.2295-2297.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee SM, et al. Fitness cost of staphylococcal cassette chromosome mec in methicillin-resistant Staphylococcus aureus by way of continuous culture. Antimicrob Agents Chemother. 2007;51:1497–1499. doi: 10.1128/AAC.01239-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Coulter SN, et al. Staphylococcus aureus genetic loci impacting growth and survival in multiple infection environments. Mol Microbiol. 1998;30:393–404. doi: 10.1046/j.1365-2958.1998.01075.x. [DOI] [PubMed] [Google Scholar]
- 88.Coombs GW, et al. Differentiation of clonal complex 59 community-associated methicillin-resistant Staphylococcus aureus in Western Australia. Antimicrob Agents Chemother. 2010;54:1914–1921. doi: 10.1128/AAC.01287-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Diep BA, Carleton HA, Chang RF, Sensabaugh GF, Perdreau-Remington F. Roles of 34 virulence genes in the evolution of hospital- and community-associated strains of methicillin-resistant Staphylococcus aureus. J Infect Dis. 2006;193:1495–1503. doi: 10.1086/503777. [DOI] [PubMed] [Google Scholar]
- 90.Cribier B, et al. Staphylococcus aureus leukocidin: a new virulence factor in cutaneous infections? An epidemiological and experimental study. Dermatology. 1992;185:175–180. doi: 10.1159/000247443. [DOI] [PubMed] [Google Scholar]
- 91.Gillet Y, et al. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet. 2002;359:753–759. doi: 10.1016/S0140-6736(02)07877-7. [DOI] [PubMed] [Google Scholar]
- 92.Lina G, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29:1128–1132. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 93.Badiou C, et al. Panton-Valentine leukocidin is expressed at toxic levels in human skin abscesses. Clin Microbiol Infect. 2008;14:1180–1183. doi: 10.1111/j.1469-0691.2008.02105.x. [DOI] [PubMed] [Google Scholar]
- 94.Badiou C, et al. Rapid detection of Staphylococcus aureus Panton-Valentine leukocidin in clinical specimens by enzyme-linked immunosorbent assay and immunochromatographic tests. J Clin Microbiol. 2010;48:1384–1390. doi: 10.1128/JCM.02274-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Diep BA, et al. Contribution of Panton-Valentine leukocidin in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. PLoS One. 2008;3:e3198. doi: 10.1371/journal.pone.0003198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Otter JA, French GL. The emergence of community-associated methicillin-resistant Staphylococcus aureus at a London teaching hospital, 2000–2006. Clin Microbiol Infect. 2008;14:670–676. doi: 10.1111/j.1469-0691.2008.02017.x. [DOI] [PubMed] [Google Scholar]
- 97.Zhang K, McClure JA, Elsayed S, Tan J, Conly JM. Coexistence of Panton-Valentine leukocidin-positive and -negative community-associated methicillin-resistant Staphylococcus aureus USA400 sibling strains in a large Canadian health-care region. J Infect Dis. 2008;197:195–204. doi: 10.1086/523763. [DOI] [PubMed] [Google Scholar]
- 98.Loffler B, et al. Staphylococcus aureus panton-valentine leukocidin is a very potent cytotoxic factor for human neutrophils. PLoS Pathog. 2010;6:e1000715. doi: 10.1371/journal.ppat.1000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brown EL, et al. The Panton-Valentine leukocidin vaccine protects mice against lung and skin infections caused by Staphylococcus aureus USA300. Clin Microbiol Infect. 2009;15:156–164. doi: 10.1111/j.1469-0691.2008.02648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bubeck Wardenburg J, Bae T, Otto M, Deleo FR, Schneewind O. Poring over pores: alpha-hemolysin and Panton-Valentine leukocidin in Staphylococcus aureus pneumonia. Nat Med. 2007;13:1405–1406. doi: 10.1038/nm1207-1405. [DOI] [PubMed] [Google Scholar]
- 101.Bubeck Wardenburg J, Palazzolo-Ballance AM, Otto M, Schneewind O, DeLeo FR. Panton-Valentine leukocidin is not a virulence determinant in murine models of community-associated methicillin-resistant Staphylococcus aureus disease. J Infect Dis. 2008;198:1166–1170. doi: 10.1086/592053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Labandeira-Rey M, et al. Staphylococcus aureus Panton-Valentine leukocidin causes necrotizing pneumonia. Science. 2007;315:1130–1133. doi: 10.1126/science.1137165. [DOI] [PubMed] [Google Scholar]
- 103.Villaruz AE, et al. A point mutation in the agr locus rather than expression of the Panton-Valentine leukocidin caused previously reported phenotypes in Staphylococcus aureus pneumonia and gene regulation. J Infect Dis. 2009;200:724–734. doi: 10.1086/604728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Voyich JM, et al. Is Panton-Valentine leukocidin the major virulence determinant in community-associated methicillin-resistant Staphylococcus aureus disease? J Infect Dis. 2006;194:1761–1770. doi: 10.1086/509506. [DOI] [PubMed] [Google Scholar]
- 105.Olsen RJ, et al. Lack of a major role of Staphylococcus aureus Panton-Valentine leukocidin in lower respiratory tract infection in nonhuman primates. Am J Pathol. 2010;176:1346–1354. doi: 10.2353/ajpath.2010.090960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cremieux AC, et al. Panton-valentine leukocidin enhances the severity of community-associated methicillin-resistant Staphylococcus aureus rabbit osteomyelitis. PLoS One. 2009;4:e7204. doi: 10.1371/journal.pone.0007204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Diep BA, et al. Polymorphonuclear leukocytes mediate Staphylococcus aureus Panton-Valentine leukocidin-induced lung inflammation and injury. Proc Natl Acad Sci U S A. 2010;107:5587–5592. doi: 10.1073/pnas.0912403107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kobayashi SD, et al. Comparative analysis of USA300 virulence determinants in a rabbit model of skin and soft tissue infection. J Infect Dis. 2011;204:937–941. doi: 10.1093/infdis/jir441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lipinska U, et al. Panton-Valentine leukocidin does play a role in the early stage of Staphylococcus aureus skin infections: a rabbit model. PLoS One. 2011;6:e22864. doi: 10.1371/journal.pone.0022864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hongo I, et al. Phenol-soluble modulin alpha 3 enhances the human neutrophil lysis mediated by Panton-Valentine leukocidin. J Infect Dis. 2009;200:715–723. doi: 10.1086/605332. [DOI] [PubMed] [Google Scholar]
- 111.Miragaia M, et al. Genetic diversity of arginine catabolic mobile element in Staphylococcus epidermidis. PLoS One. 2009;4:e7722. doi: 10.1371/journal.pone.0007722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shore AC, et al. Characterization of a novel arginine catabolic mobile element (ACME) and staphylococcal chromosomal cassette mec composite island with significant homology to Staphylococcus epidermidis ACME type II in methicillin-resistant Staphylococcus aureus genotype ST22-MRSA-IV. Antimicrob Agents Chemother. 2011;55:1896–1905. doi: 10.1128/AAC.01756-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Montgomery CP, Boyle-Vavra S, Daum RS. The arginine catabolic mobile element is not associated with enhanced virulence in experimental invasive disease caused by the community-associated methicillin-resistant Staphylococcus aureus USA300 genetic background. Infect Immun. 2009;77:2650–2656. doi: 10.1128/IAI.00256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Otto M. Basis of virulence in community-associated methicillin-resistant Staphylococcus aureus. Annu Rev Microbiol. 2010;64:143–162. doi: 10.1146/annurev.micro.112408.134309. [DOI] [PubMed] [Google Scholar]
- 115.Baze PE, Milano G, Verrando P, Renee N, Ortonne JP. Distribution of polyamines in human epidermis. Br J Dermatol. 1985;112:393–396. doi: 10.1111/j.1365-2133.1985.tb02311.x. [DOI] [PubMed] [Google Scholar]
- 116.Joshi GS, Spontak JS, Klapper DG, Richardson AR. Arginine catabolic mobile element encoded speG abrogates the unique hypersensitivity of Staphylococcus aureus to exogenous polyamines. Mol Microbiol. 2011;82:9–20. doi: 10.1111/j.1365-2958.2011.07809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mahdavian Delavary B, van der Veer WM, van Egmond M, Niessen FB, Beelen RH. Macrophages in skin injury and repair. Immunobiology. 2011;216:753–762. doi: 10.1016/j.imbio.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 118.Emmett M, Kloos WE. The nature of arginine auxotrophy in cutaneous populations of staphylococci. J Gen Microbiol. 1979;110:305–314. doi: 10.1099/00221287-110-2-305. [DOI] [PubMed] [Google Scholar]
- 119.Maeno Y, Takabe F, Inoue H, Iwasa M. A study on the vital reaction in wounded skin: simultaneous determination of histamine and polyamines in injured rat skin by high-performance liquid chromatography. Forensic Sci Int. 1990;46:255–268. doi: 10.1016/0379-0738(90)90311-l. [DOI] [PubMed] [Google Scholar]
- 120.Sasaki S, et al. Interleukin-4 and interleukin-10 are involved in host resistance to Staphylococcus aureus infection through regulation of gamma interferon. Infect Immun. 2000;68:2424–2430. doi: 10.1128/iai.68.5.2424-2430.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Prabhakara R, et al. Suppression of the Inflammatory Immune Response Prevents the Development of Chronic Biofilm Infection Due to Methicillin-Resistant Staphylococcus aureus. Infect Immun. 2011;79:5010–5018. doi: 10.1128/IAI.05571-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Emonts M, et al. Host polymorphisms in interleukin 4, complement factor H, and C-reactive protein associated with nasal carriage of Staphylococcus aureus and occurrence of boils. J Infect Dis. 2008;197:1244–1253. doi: 10.1086/533501. [DOI] [PubMed] [Google Scholar]
- 123.Hochgrafe F, et al. Nitric oxide stress induces different responses but mediates comparable protein thiol protection in Bacillus subtilis and Staphylococcus aureus. J Bacteriol. 2008;190:4997–5008. doi: 10.1128/JB.01846-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Richardson AR, Dunman PM, Fang FC. The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol Microbiol. 2006;61:927–939. doi: 10.1111/j.1365-2958.2006.05290.x. [DOI] [PubMed] [Google Scholar]
- 125.Richardson AR, Libby SJ, Fang FC. A nitric oxide-inducible lactate dehydrogenase enables Staphylococcus aureus to resist innate immunity. Science. 2008;319:1672–1676. doi: 10.1126/science.1155207. [DOI] [PubMed] [Google Scholar]
- 126.Makhlin J, et al. Staphylococcus aureus ArcR controls expression of the arginine deiminase operon. J Bacteriol. 2007;189:5976–5986. doi: 10.1128/JB.00592-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Reyes J, et al. Dissemination of methicillin-resistant Staphylococcus aureus USA300 sequence type 8 lineage in Latin America. Clin Infect Dis. 2009;49:1861–1867. doi: 10.1086/648426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Arvidson S. Gram-positive pathogens. 2. ASM Press; 2006. pp. 478–485. [Google Scholar]
- 129.Bohach GA. Gram-positive pathogens. 2. ASM Press; 2006. pp. 464–477. [Google Scholar]
- 130.Dubin G. Extracellular proteases of Staphylococcus spp. Biol Chem. 2002;383:1075–1086. doi: 10.1515/BC.2002.116. [DOI] [PubMed] [Google Scholar]
- 131.Bayer AS, et al. Hyperproduction of alpha-toxin by Staphylococcus aureus results in paradoxically reduced virulence in experimental endocarditis: a host defense role for platelet microbicidal proteins. Infect Immun. 1997;65:4652–4660. doi: 10.1128/iai.65.11.4652-4660.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jonsson P, Lindberg M, Haraldsson I, Wadstrom T. Virulence of Staphylococcus aureus in a mouse mastitis model: studies of alpha hemolysin, coagulase, and protein A as possible virulence determinants with protoplast fusion and gene cloning. Infect Immun. 1985;49:765–769. doi: 10.1128/iai.49.3.765-769.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.O’Reilly M, de Azavedo JC, Kennedy S, Foster TJ. Inactivation of the alpha-haemolysin gene of Staphylococcus aureus 8325-4 by site-directed mutagenesis and studies on the expression of its haemolysins. Microb Pathog. 1986;1:125–138. doi: 10.1016/0882-4010(86)90015-x. [DOI] [PubMed] [Google Scholar]
- 134.Gouaux E. alpha-Hemolysin from Staphylococcus aureus: an archetype of beta-barrel, channel-forming toxins. J Struct Biol. 1998;121:110–122. doi: 10.1006/jsbi.1998.3959. [DOI] [PubMed] [Google Scholar]
- 135.Song L, et al. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 136.Kennedy AD, et al. Targeting of alpha-hemolysin by active or passive immunization decreases severity of USA300 skin infection in a mouse model. J Infect Dis. 2010;202:1050–1058. doi: 10.1086/656043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bubeck Wardenburg J, Patel RJ, Schneewind O. Surface proteins and exotoxins are required for the pathogenesis of Staphylococcus aureus pneumonia. Infect Immun. 2007;75:1040–1044. doi: 10.1128/IAI.01313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Patel AH, Nowlan P, Weavers ED, Foster T. Virulence of protein A-deficient and alpha-toxin-deficient mutants of Staphylococcus aureus isolated by allele replacement. Infect Immun. 1987;55:3103–3110. doi: 10.1128/iai.55.12.3103-3110.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bramley AJ, Patel AH, O’Reilly M, Foster R, Foster TJ. Roles of alpha-toxin and beta-toxin in virulence of Staphylococcus aureus for the mouse mammary gland. Infect Immun. 1989;57:2489–2494. doi: 10.1128/iai.57.8.2489-2494.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.McElroy MC, et al. Alpha-toxin damages the air-blood barrier of the lung in a rat model of Staphylococcus aureus-induced pneumonia. Infect Immun. 1999;67:5541–5544. doi: 10.1128/iai.67.10.5541-5544.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang R, et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13:1510–1514. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- 142.Joo HS, Cheung GY, Otto M. Antimicrobial activity of community-associated methicillin-resistant Staphylococcus aureus is caused by phenol-soluble modulin derivatives. J Biol Chem. 2011;286:8933–8940. doi: 10.1074/jbc.M111.221382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lauderdale KJ, Boles BR, Cheung AL, Horswill AR. Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect Immun. 2009;77:1623–1635. doi: 10.1128/IAI.01036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Thoendel M, Kavanaugh JS, Flack CE, Horswill AR. Peptide signaling in the staphylococci. Chem Rev. 2011;111:117–151. doi: 10.1021/cr100370n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kavanaugh JS, Thoendel M, Horswill AR. A role for type I signal peptidase in Staphylococcus aureus quorum sensing. Mol Microbiol. 2007;65:780–798. doi: 10.1111/j.1365-2958.2007.05830.x. [DOI] [PubMed] [Google Scholar]
- 146.Novick RP, et al. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. Embo J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Vandenesch F, Kornblum J, Novick RP. A temporal signal, independent of agr, is required for hla but not spa transcription in Staphylococcus aureus. J Bacteriol. 1991;173:6313–6320. doi: 10.1128/jb.173.20.6313-6320.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Geisinger E, Adhikari RP, Jin R, Ross HF, Novick RP. Inhibition of rot translation by RNAIII, a key feature of agr function. Mol Microbiol. 2006;61:1038–1048. doi: 10.1111/j.1365-2958.2006.05292.x. [DOI] [PubMed] [Google Scholar]
- 149.Vanderpool CK, Balasubramanian D, Lloyd CR. Dual-function RNA regulators in bacteria. Biochimie. 2011;93:1943–1949. doi: 10.1016/j.biochi.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Queck SY, et al. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol Cell. 2008;32:150–158. doi: 10.1016/j.molcel.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Montgomery CP, Boyle-Vavra S, Daum RS. Importance of the global regulators Agr and SaeRS in the pathogenesis of CA-MRSA USA300 infection. PLoS One. 2010;5:e15177. doi: 10.1371/journal.pone.0015177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Adhikari RP, Novick RP. Regulatory organization of the staphylococcal sae locus. Microbiology. 2008;154:949–959. doi: 10.1099/mic.0.2007/012245-0. [DOI] [PubMed] [Google Scholar]
- 153.Giraudo AT, Calzolari A, Cataldi AA, Bogni C, Nagel R. The sae locus of Staphylococcus aureus encodes a two-component regulatory system. FEMS Microbiol Lett. 1999;177:15–22. doi: 10.1111/j.1574-6968.1999.tb13707.x. [DOI] [PubMed] [Google Scholar]
- 154.Giraudo AT, Martinez GL, Calzolari A, Nagel R. Characterization of a Tn925-induced mutant of Staphylococcus aureus altered in exoprotein production. J Basic Microbiol. 1994;34:317–322. doi: 10.1002/jobm.3620340507. [DOI] [PubMed] [Google Scholar]
- 155.Geiger T, Goerke C, Mainiero M, Kraus D, Wolz C. The virulence regulator Sae of Staphylococcus aureus: promoter activities and response to phagocytosis-related signals. J Bacteriol. 2008;190:3419–3428. doi: 10.1128/JB.01927-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Novick RP, Jiang D. The staphylococcal saeRS system coordinates environmental signals with agr quorum sensing. Microbiology. 2003;149:2709–2717. doi: 10.1099/mic.0.26575-0. [DOI] [PubMed] [Google Scholar]
- 157.Nygaard TK, et al. SaeR binds a consensus sequence within virulence gene promoters to advance USA300 pathogenesis. J Infect Dis. 2010;201:241–254. doi: 10.1086/649570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Voyich JM, et al. The SaeR/S gene regulatory system is essential for innate immune evasion by Staphylococcus aureus. J Infect Dis. 2009;199:1698–1706. doi: 10.1086/598967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Watkins RL, Pallister KB, Voyich JM. The SaeR/S gene regulatory system induces a pro-inflammatory cytokine response during Staphylococcus aureus infection. PLoS One. 2011;6:e19939. doi: 10.1371/journal.pone.0019939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Cheung AL, Projan SJ. Cloning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of agr. J Bacteriol. 1994;176:4168–4172. doi: 10.1128/jb.176.13.4168-4172.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tamber S, Schwartzman J, Cheung AL. Role of PknB kinase in antibiotic resistance and virulence in community-acquired methicillin-resistant Staphylococcus aureus strain USA300. Infect Immun. 2010;78:3637–3646. doi: 10.1128/IAI.00296-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ingavale S, van Wamel W, Luong TT, Lee CY, Cheung AL. Rat/MgrA, a regulator of autolysis, is a regulator of virulence genes in Staphylococcus aureus. Infect Immun. 2005;73:1423–1431. doi: 10.1128/IAI.73.3.1423-1431.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Majerczyk CD, et al. Staphylococcus aureus CodY negatively regulates virulence gene expression. J Bacteriol. 2008;190:2257–2265. doi: 10.1128/JB.01545-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Seidl K, et al. Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob Agents Chemother. 2006;50:1183–1194. doi: 10.1128/AAC.50.4.1183-1194.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Tamber S, Cheung AL. SarZ promotes the expression of virulence factors and represses biofilm formation by modulating SarA and agr in Staphylococcus aureus. Infect Immun. 2009;77:419–428. doi: 10.1128/IAI.00859-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Manna AC, Cheung AL. Transcriptional regulation of the agr locus and the identification of DNA binding residues of the global regulatory protein SarR in Staphylococcus aureus. Mol Microbiol. 2006;60:1289–1301. doi: 10.1111/j.1365-2958.2006.05171.x. [DOI] [PubMed] [Google Scholar]
- 167.Manna AC, Cheung AL. sarU, a sarA homolog, is repressed by SarT and regulates virulence genes in Staphylococcus aureus. Infect Immun. 2003;71:343–353. doi: 10.1128/IAI.71.1.343-353.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Schmidt KA, Manna AC, Gill S, Cheung AL. SarT, a repressor of alpha-hemolysin in Staphylococcus aureus. Infect Immun. 2001;69:4749–4758. doi: 10.1128/IAI.69.8.4749-4758.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Liang X, et al. Global regulation of gene expression by ArlRS, a two-component signal transduction regulatory system of Staphylococcus aureus. J Bacteriol. 2005;187:5486–5492. doi: 10.1128/JB.187.15.5486-5492.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yarwood JM, McCormick JK, Schlievert PM. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J Bacteriol. 2001;183:1113–1123. doi: 10.1128/JB.183.4.1113-1123.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Majerczyk CD, et al. Direct targets of CodY in Staphylococcus aureus. J Bacteriol. 2010;192:2861–2877. doi: 10.1128/JB.00220-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pragman AA, Yarwood JM, Tripp TJ, Schlievert PM. Characterization of virulence factor regulation by SrrAB, a two-component system in Staphylococcus aureus. J Bacteriol. 2004;186:2430–2438. doi: 10.1128/JB.186.8.2430-2438.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Heinrichs JH, Bayer MG, Cheung AL. Characterization of the sar locus and its interaction with agr in Staphylococcus aureus. J Bacteriol. 1996;178:418–423. doi: 10.1128/jb.178.2.418-423.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Herbert S, et al. Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infect Immun. 2010;78:2877–2889. doi: 10.1128/IAI.00088-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Reyes D, et al. Coordinated regulation by AgrA, SarA, and SarR to control agr expression in Staphylococcus aureus. J Bacteriol. 2011;193:6020–6031. doi: 10.1128/JB.05436-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Weiss EC, et al. Impact of sarA on daptomycin susceptibility of Staphylococcus aureus biofilms in vivo. Antimicrob Agents Chemother. 2009;53:4096–4102. doi: 10.1128/AAC.00484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zielinska AK, et al. Defining the strain-dependent impact of the Staphylococcal accessory regulator (sarA) on the alpha-toxin phenotype of Staphylococcus aureus. J Bacteriol. 2011;193:2948–2958. doi: 10.1128/JB.01517-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Liang X, et al. Identification of single nucleotide polymorphisms associated with hyperproduction of alpha-toxin in Staphylococcus aureus. PLoS One. 2011;6:e18428. doi: 10.1371/journal.pone.0018428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Feil EJ, et al. How clonal is Staphylococcus aureus? J Bacteriol. 2003;185:3307–3316. doi: 10.1128/JB.185.11.3307-3316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kuhn G, Francioli P, Blanc DS. Evidence for clonal evolution among highly polymorphic genes in methicillin-resistant Staphylococcus aureus. J Bacteriol. 2006;188:169–178. doi: 10.1128/JB.188.1.169-178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Chan CX, Beiko RG, Ragan MA. Lateral transfer of genes and gene fragments in Staphylococcus extends beyond mobile elements. J Bacteriol. 2011;193:3964–3977. doi: 10.1128/JB.01524-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Highlander SK, et al. Subtle genetic changes enhance virulence of methicillin resistant and sensitive Staphylococcus aureus. BMC Microbiol. 2007;7:99. doi: 10.1186/1471-2180-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Gill SR, et al. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol. 2005;187:2426–2438. doi: 10.1128/JB.187.7.2426-2438.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Herron LL, et al. Genome sequence survey identifies unique sequences and key virulence genes with unusual rates of amino Acid substitution in bovine Staphylococcus aureus. Infect Immun. 2002;70:3978–3981. doi: 10.1128/IAI.70.7.3978-3981.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Herron-Olson L, Fitzgerald JR, Musser JM, Kapur V. Molecular correlates of host specialization in Staphylococcus aureus. PLoS One. 2007;2:e1120. doi: 10.1371/journal.pone.0001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Sivaraman K, Cole AM. Pathogenesis gene families in the common minimal genome of Staphylococcus aureus are hypervariable. FEBS Lett. 2009;583:1304–1308. doi: 10.1016/j.febslet.2009.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Holden MT, et al. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci U S A. 2004;101:9786–9791. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Jordan IK, Rogozin IB, Wolf YI, Koonin EV. Microevolutionary genomics of bacteria. Theor Popul Biol. 2002;61:435–447. doi: 10.1006/tpbi.2002.1588. [DOI] [PubMed] [Google Scholar]
- 189.Rocha EP, Danchin A. An analysis of determinants of amino acids substitution rates in bacterial proteins. Mol Biol Evol. 2004;21:108–116. doi: 10.1093/molbev/msh004. [DOI] [PubMed] [Google Scholar]
- 190.Holt DC, et al. A very early-branching Staphylococcus aureus lineage lacking the carotenoid pigment staphyloxanthin. Genome Biol Evol. 2011;3:881–895. doi: 10.1093/gbe/evr078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Harvey CJ, LeBouf RF, Stefaniak AB. Formulation and stability of a novel artificial human sweat under conditions of storage and use. Toxicol In Vitro. 2010;24:1790–1796. doi: 10.1016/j.tiv.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 192.Hua Q, Joyce AR, Palsson BO, Fong SS. Metabolic characterization of Escherichia coli strains adapted to growth on lactate. Appl Environ Microbiol. 2007;73:4639–4647. doi: 10.1128/AEM.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Marks ME, et al. The genetic basis of laboratory adaptation in Caulobacter crescentus. J Bacteriol. 2010;192:3678–3688. doi: 10.1128/JB.00255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Pattee PA. Genetic linkage of chromosomal tetracycline resistance and pigmentation to a purine auxotrophic marker and the isoleucine-valine-leucine structural genes in Staphylococcus aureus. J Bacteriol. 1976;127:1167–1172. doi: 10.1128/jb.127.3.1167-1172.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]