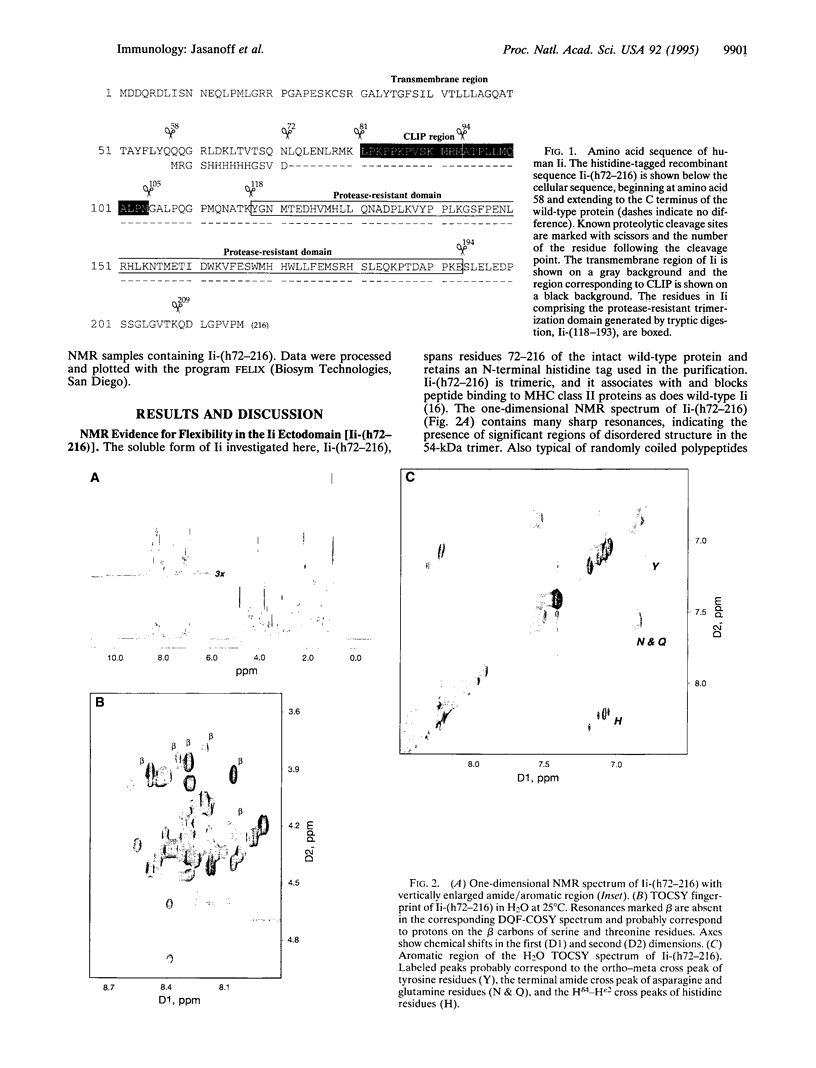
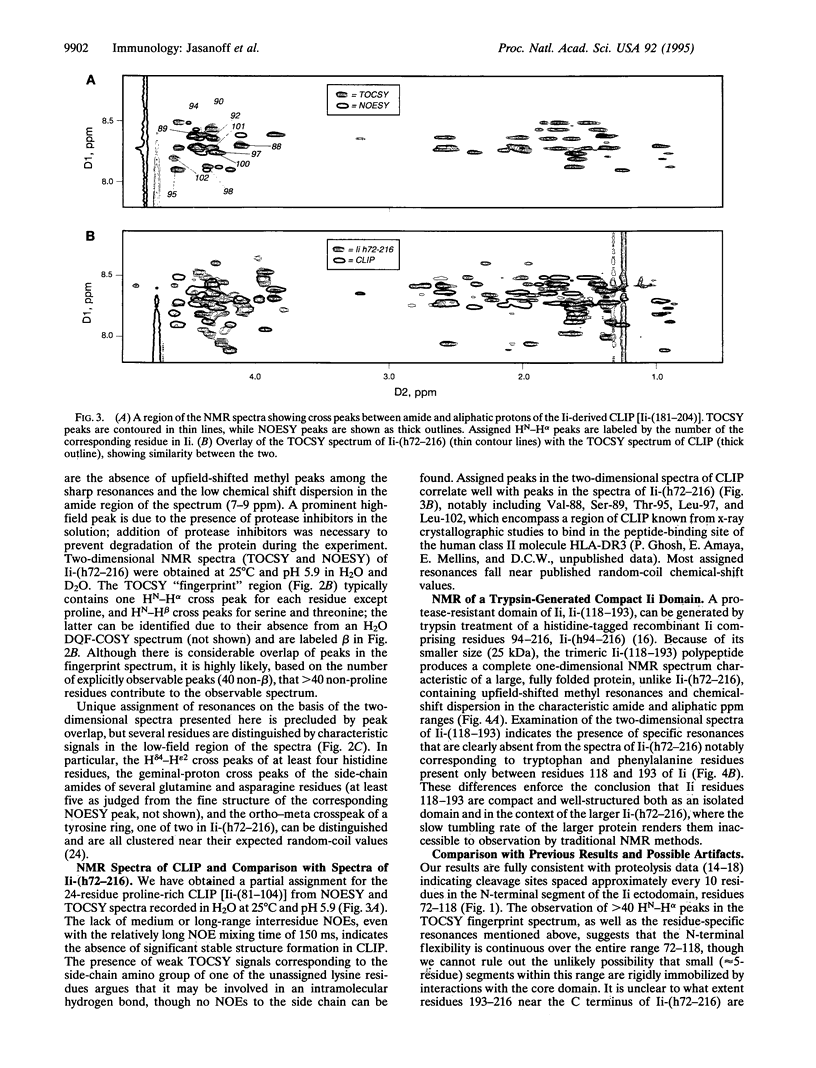
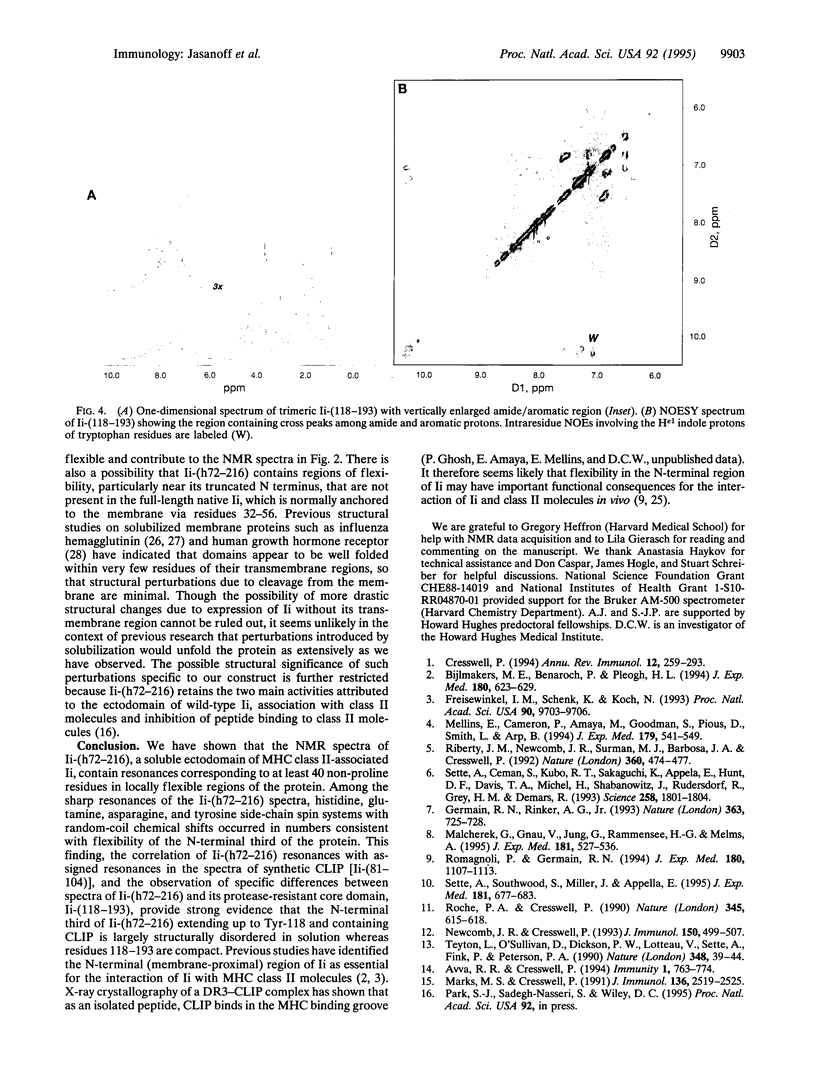
Abstract
Invariant chain (Ii) is a trimeric membrane protein which binds and stabilizes major histocompatibility complex class II heterodimers in the endoplasmic reticulum and lysosomal compartments of antigen-presenting cells. In concert with an intracellular class II-like molecule, HLA-DM, Ii seems to facilitate loading of conventional class II molecules with peptides before transport of the class II-peptide complex to the cell surface for recognition by T cells. The interaction of Ii with class II molecules is thought to be mediated in large part through a region of 24 amino acids (the class II-associated Ii peptide, CLIP) which binds as a cleaved moiety in the antigenic peptide-binding groove of class II molecules in HLA-DM-deficient cell lines. Here we use nuclear magnetic resonance techniques to demonstrate that a soluble recombinant Ii ectodomain contains significant disordered regions which probably include CLIP.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avva R. R., Cresswell P. In vivo and in vitro formation and dissociation of HLA-DR complexes with invariant chain-derived peptides. Immunity. 1994 Dec;1(9):763–774. doi: 10.1016/s1074-7613(94)80018-9. [DOI] [PubMed] [Google Scholar]
- Bijlmakers M. J., Benaroch P., Ploegh H. L. Mapping functional regions in the lumenal domain of the class II-associated invariant chain. J Exp Med. 1994 Aug 1;180(2):623–629. doi: 10.1084/jem.180.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresswell P. Assembly, transport, and function of MHC class II molecules. Annu Rev Immunol. 1994;12:259–293. doi: 10.1146/annurev.iy.12.040194.001355. [DOI] [PubMed] [Google Scholar]
- Freisewinkel I. M., Schenck K., Koch N. The segment of invariant chain that is critical for association with major histocompatibility complex class II molecules contains the sequence of a peptide eluted from class II polypeptides. Proc Natl Acad Sci U S A. 1993 Oct 15;90(20):9703–9706. doi: 10.1073/pnas.90.20.9703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain R. N., Rinker A. G., Jr Peptide binding inhibits protein aggregation of invariant-chain free class II dimers and promotes surface expression of occupied molecules. Nature. 1993 Jun 24;363(6431):725–728. doi: 10.1038/363725a0. [DOI] [PubMed] [Google Scholar]
- Landry S. J., Zeilstra-Ryalls J., Fayet O., Georgopoulos C., Gierasch L. M. Characterization of a functionally important mobile domain of GroES. Nature. 1993 Jul 15;364(6434):255–258. doi: 10.1038/364255a0. [DOI] [PubMed] [Google Scholar]
- Malcherek G., Gnau V., Jung G., Rammensee H. G., Melms A. Supermotifs enable natural invariant chain-derived peptides to interact with many major histocompatibility complex-class II molecules. J Exp Med. 1995 Feb 1;181(2):527–536. doi: 10.1084/jem.181.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks M. S., Blum J. S., Cresswell P. Invariant chain trimers are sequestered in the rough endoplasmic reticulum in the absence of association with HLA class II antigens. J Cell Biol. 1990 Sep;111(3):839–855. doi: 10.1083/jcb.111.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks M. S., Cresswell P. Invariant chain associates with HLA class II antigens via its extracytoplasmic region. J Immunol. 1986 Apr 1;136(7):2519–2525. [PubMed] [Google Scholar]
- Mellins E., Cameron P., Amaya M., Goodman S., Pious D., Smith L., Arp B. A mutant human histocompatibility leukocyte antigen DR molecule associated with invariant chain peptides. J Exp Med. 1994 Feb 1;179(2):541–549. doi: 10.1084/jem.179.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb J. R., Cresswell P. Characterization of endogenous peptides bound to purified HLA-DR molecules and their absence from invariant chain-associated alpha beta dimers. J Immunol. 1993 Jan 15;150(2):499–507. [PubMed] [Google Scholar]
- Oswald R. E., Bogusky M. J., Bamberger M., Smith R. A., Dobson C. M. Dynamics of the multidomain fibrinolytic protein urokinase from two-dimensional NMR. Nature. 1989 Feb 9;337(6207):579–582. doi: 10.1038/337579a0. [DOI] [PubMed] [Google Scholar]
- Perham R. N., Duckworth H. W., Roberts G. C. Mobility of polypeptide chain in the pyruvate dehydrogenase complex revealed by proton NMR. Nature. 1981 Jul 30;292(5822):474–477. doi: 10.1038/292474a0. [DOI] [PubMed] [Google Scholar]
- Riberdy J. M., Newcomb J. R., Surman M. J., Barbosa J. A., Cresswell P. HLA-DR molecules from an antigen-processing mutant cell line are associated with invariant chain peptides. Nature. 1992 Dec 3;360(6403):474–477. doi: 10.1038/360474a0. [DOI] [PubMed] [Google Scholar]
- Roche P. A., Cresswell P. Invariant chain association with HLA-DR molecules inhibits immunogenic peptide binding. Nature. 1990 Jun 14;345(6276):615–618. doi: 10.1038/345615a0. [DOI] [PubMed] [Google Scholar]
- Roche P. A., Cresswell P. Proteolysis of the class II-associated invariant chain generates a peptide binding site in intracellular HLA-DR molecules. Proc Natl Acad Sci U S A. 1991 Apr 15;88(8):3150–3154. doi: 10.1073/pnas.88.8.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche P. A., Marks M. S., Cresswell P. Formation of a nine-subunit complex by HLA class II glycoproteins and the invariant chain. Nature. 1991 Dec 5;354(6352):392–394. doi: 10.1038/354392a0. [DOI] [PubMed] [Google Scholar]
- Romagnoli P., Germain R. N. The CLIP region of invariant chain plays a critical role in regulating major histocompatibility complex class II folding, transport, and peptide occupancy. J Exp Med. 1994 Sep 1;180(3):1107–1113. doi: 10.1084/jem.180.3.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sant A. J., Miller J. MHC class II antigen processing: biology of invariant chain. Curr Opin Immunol. 1994 Feb;6(1):57–63. doi: 10.1016/0952-7915(94)90034-5. [DOI] [PubMed] [Google Scholar]
- Sette A., Ceman S., Kubo R. T., Sakaguchi K., Appella E., Hunt D. F., Davis T. A., Michel H., Shabanowitz J., Rudersdorf R. Invariant chain peptides in most HLA-DR molecules of an antigen-processing mutant. Science. 1992 Dec 11;258(5089):1801–1804. doi: 10.1126/science.1465617. [DOI] [PubMed] [Google Scholar]
- Sette A., Southwood S., Miller J., Appella E. Binding of major histocompatibility complex class II to the invariant chain-derived peptide, CLIP, is regulated by allelic polymorphism in class II. J Exp Med. 1995 Feb 1;181(2):677–683. doi: 10.1084/jem.181.2.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teyton L., O'Sullivan D., Dickson P. W., Lotteau V., Sette A., Fink P., Peterson P. A. Invariant chain distinguishes between the exogenous and endogenous antigen presentation pathways. Nature. 1990 Nov 1;348(6296):39–44. doi: 10.1038/348039a0. [DOI] [PubMed] [Google Scholar]
- Wiley D. C., Wilson I. A., Skehel J. J. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981 Jan 29;289(5796):373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- Xu M., Capraro G. A., Daibata M., Reyes V. E., Humphreys R. E. Cathepsin B cleavage and release of invariant chain from MHC class II molecules follow a staged pattern. Mol Immunol. 1994 Jul;31(10):723–731. doi: 10.1016/0161-5890(94)90146-5. [DOI] [PubMed] [Google Scholar]
- de Vos A. M., Ultsch M., Kossiakoff A. A. Human growth hormone and extracellular domain of its receptor: crystal structure of the complex. Science. 1992 Jan 17;255(5042):306–312. doi: 10.1126/science.1549776. [DOI] [PubMed] [Google Scholar]


