Abstract
NODULE INCEPTION (NIN) is a key regulator of the symbiotic nitrogen fixation pathway in legumes including Lotus japonicus. NIN-like proteins (NLPs), which are presumably present in all land plants, were recently identified as key transcription factors in nitrate signaling and responses in Arabidopsis thaliana, a non-leguminous plant. Here we show that both NIN and NLP1 of L. japonicus (LjNLP1) can bind to the nitrate-responsive cis-element (NRE) and promote transcription from an NRE-containing promoter as did the NLPs of A. thaliana (AtNLPs). However, differing from LjNLP1 and the AtNLPs that are activated by nitrate signaling through their N-terminal regions, the N-terminal region of NIN did not respond to nitrate. Thus, in the course of the evolution of NIN into a transcription factor that functions in nodulation in legumes, some mutations might arise that converted it to a nitrate-insensitive transcription factor. Because nodule formation is induced under nitrogen-deficient conditions, we speculate that the loss of the nitrate-responsiveness of NIN may be one of the evolutionary events necessary for the emergence of symbiotic nitrogen fixation in legumes.
Keywords: NIN-like protein, NODULE INCEPTION, nitrate response, nodulation, symbiosis, transcription factor, transcriptional control
Unlike other higher plants, legumes can use atmospheric nitrogen (N2) as a nitrogen source because they harbor symbiotic bacteria within nodules of their roots that have a nitrogen fixation ability.1 NODULE INCEPTION (NIN), which was identified in 1999 in Lotus japonicus, was the first plant gene essential for nodulation to be described.2 The symbiotic processes in the nin mutant are arrested at the bacterial recognition stage, which is accompanied by the incomplete formation of infection threads and aborted initiation of primordia.2,3 Although the nin mutant does not develop nodules, no morphological changes have been observed in its roots, shoots, leaves, flowers, or seeds, suggesting that the NIN gene is not required for general plant development but is specifically involved in the developmental program for nodulation.2 The product of NIN was proposed to be a putative transcription factor due to a putative DNA-binding domain identified in its secondary structure, and this domain was referred to as the RWP-RK domain.2
NIN-like proteins (NLPs), which are encoded by several gene copies in probably all plant genomes, are proteins having a homology to NIN not only in their C-terminal RWP-RK domain but also in their N-terminal regions.4 Recently, we showed that NLPs interact with the nitrate-responsive cis-elements (NREs) of nitrate-inducible genes5-7 and function as master regulators of the nitrate-inducible gene expression in Arabidopsis thaliana.8 We further reported that the RWP-RK domain of NLP is a DNA-binding domain whose activity is unrelated to nitrate signaling, whereas the N-terminal regions of NLPs function as a transcriptional activation domain that mediates this signaling.8
Symbiotic nitrogen fixation is unique to legumes, whereas the regulation of gene expression by nitrate, a key nutrient signal in plants, is essential for plant growth and is therefore common to all land plants.9,10 Thus, the NIN/NLP gene family appears to be associated with the control of nitrate-regulated genes in the plant kingdom, but a member of this gene family appears to have specifically evolved as the NIN gene with functional responsibility for symbiotic nitrogen fixation in legumes.8 Despite a high degree of homology between NIN and NLPs, these proteins apparently play non-redundant roles in planta, as the closest rice homolog of NIN, OsNLP1, does not rescue the nin mutant phenotype.11 To gain an insight into the molecular basis for the different roles of NIN and the NLPs, we compared the DNA-binding and transcriptional regulation activities of NIN and NLPs from L. japonicus in the current study.
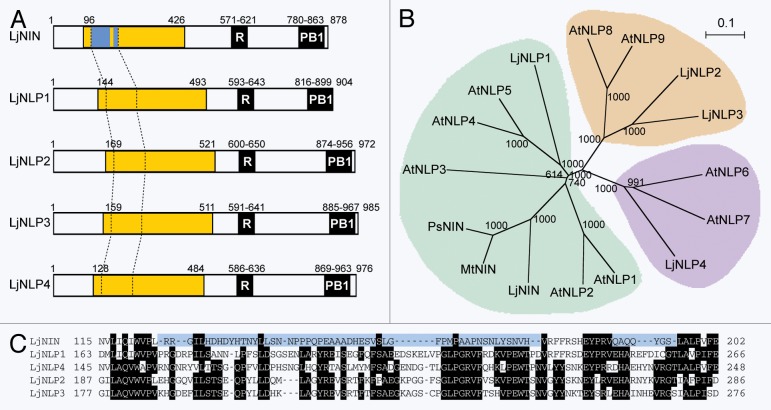
To identify all NLP genes in the genome of L. japonicus, we performed a blast search of a genomic database for L. japonicus (Lotus japonicus genome assembly build 2.5; http://www.kazusa.or.jp/lotus/). In addition to 2 NLP genes, LjNLP1 and LjNLP2,4 2 transcripts encoding an NLP (chr3.CM0091.230.r2.m and chr5.CM0148.170.r2.a) and an NLP-related transcript from a pseudogene (chr2.CM0904.210.r2.d) had been previously identified in L. japonicus.12 Because we did not find any additional sequence encoding an NLP and the L. japonicus genome database is now almost complete,13 it is likely that these 4 NLP gene copies represent the full complement in L. japonicus. We have designated the 2 NLP genes corresponding to the chr3.CM0091.230.r2.m and chr5.CM0148.170.r2.a transcripts as LjNLP3 and LjNLP4, respectively (Fig. 1A). Like all known NLPs,4,12 all LjNLP proteins have their N-terminal region homologous to NIN, an RWP-RK domain and a PB1 domain that mediates protein-protein interactions among PB1 domain-containing proteins4 (Fig. S1, Fig. 1A).
Figure 1.Phylogenic relationship between NIN and the NLPs of A. thaliana and L. japonicus. (A) NIN and the NLPs from L. japonicus. N-terminal conserved regions are indicated by yellow boxes. A region within the conserved region of NIN is denoted by a blue box because it is not homologous to the corresponding regions of NLPs. The C-terminal regions contain the RWP-WK DNA binding domains (R) and additionally conserved domains (PB1).4 Numbers indicate the positions of the amino acid residues at the ends of each region and at the C-terminus. (B) An un-rooted phylogenic tree of the NIN and NLPs generated using the Neighbor-Joining method by ClustalW (http://clustalw.ddbj.nig.ac.jp/)22 and the NJplot program.23 The numbers are bootstrap values from 1000 replicates. (C) A unique amino acid sequence of a region within the N-terminal conserved region of NIN. The unique amino acid sequence that is not found in the NLPs is indicated by blue shading. The amino acid sequences of the corresponding regions of NLPs are also indicated.
Phylogenic analysis using the entire amino acid sequences of NIN and NIN homologs of pea (PsNIN)14 and M. truncatula (MtNIN),15 and NLPs from L. japonicus and A. thaliana, provided an un-rooted phylogenic tree (Fig. 1B), which is similar to the tree generated in a previous report.12 Bootstrap values at nodes of this phylogenic tree suggested that NIN, LjNLP1, AtNLP1–5, PsNIN, and MtNIN form a clade, which includes a subgroup consisting of NIN and its homologs. Schauser et al. previously proposed 6 blocks of conservation in amino acid sequences of NIN and NLPs;4 consecutively arranged 4 blocks (I to V) form the N-terminal conserved region as a whole, whereas blocks V and VI correspond to the RWP-RK domain and the PB1 domain, respectively (Fig. S1). Furthermore, as the amino acid sequence in a region spanning block I to block II of NIN is different from those of NLPs, it has been also proposed that the unique amino acid sequence within the N-terminal conserved region may characterize NIN and its homologs.4 This unique amino acid was not found even in NjNLP1, but the corresponding sequences are conserved among 4 LjNLPs (Fig. 1C). Thus, the result of our phylogenic analysis suggests that that NIN and its homologs have higher homologies with LjNLP1 and AtNLP1–5 rather than other NLPs overall, but a subgroup of NIN, PsNIN, and MtNIN was formed probably due to the fact that NIN and its homologs contain a unique amino acid sequence in their N-terminal regions (Fig. 1C, Fig. S1). The results of our phylogenic analysis also suggest the NIN gene evolved to function in the commitment of the developmental program of nodulation in legumes after the formation of subgroups of the NIN/NLP gene family in the plant kingdom.
LjNLP1 is closer to NIN than LjNLP2–4 in the phylogenic tree. However, LjNLP1 as well as other LjNLPs probably plays a role in nitrate-regulated gene expression, because AtNLP4 and 5, rather than NIN, are closer to LjNLP1 (Fig. 1B). Different expression patterns of NIN and LjNLP1 genes also imply their different roles in L. japonicus. The expression of NIN is specific to cells involved in nodulation, consistent with a specific role of NIN in this process,2 while the expression of LjNLP1 in both leaves and roots is reported in a database of L. japonicus genes (http://cgi-www.cs.au.dk/cgi-compbio/Niels/index.cgi).
NIN and LjNLP1 were found to form a clade with AtNLP1–5 that bind to NRE8 (Fig. 1B). Thus, we investigated whether NIN and LjNLP1 can similarly bind to NRE using the RWP-RK domains of NIN (553–666 a.a.) and LjNLP1 (575–690 a.a.) that had been expressed as thioredoxin fusion proteins and purified from Escherichia coli. The recombinant proteins formed a DNA-protein complex with the non-labeled DNA probe containing an NRE, while a thioredoxin negative control did not form this association (Fig. 2A). Furthermore, a non-labeled DNA probe could inhibit the complex formation between NIN and the labeled DNA probe. Although the complex formation of NIN appeared not to be very effective compared with AtNLP6 and LjNLP1, these results indicated that the RWP-RK domain of NIN is sufficient for DNA binding and also that NIN and the NLPs can bind to NRE sites in a sequence-specific manner, at least in vitro.
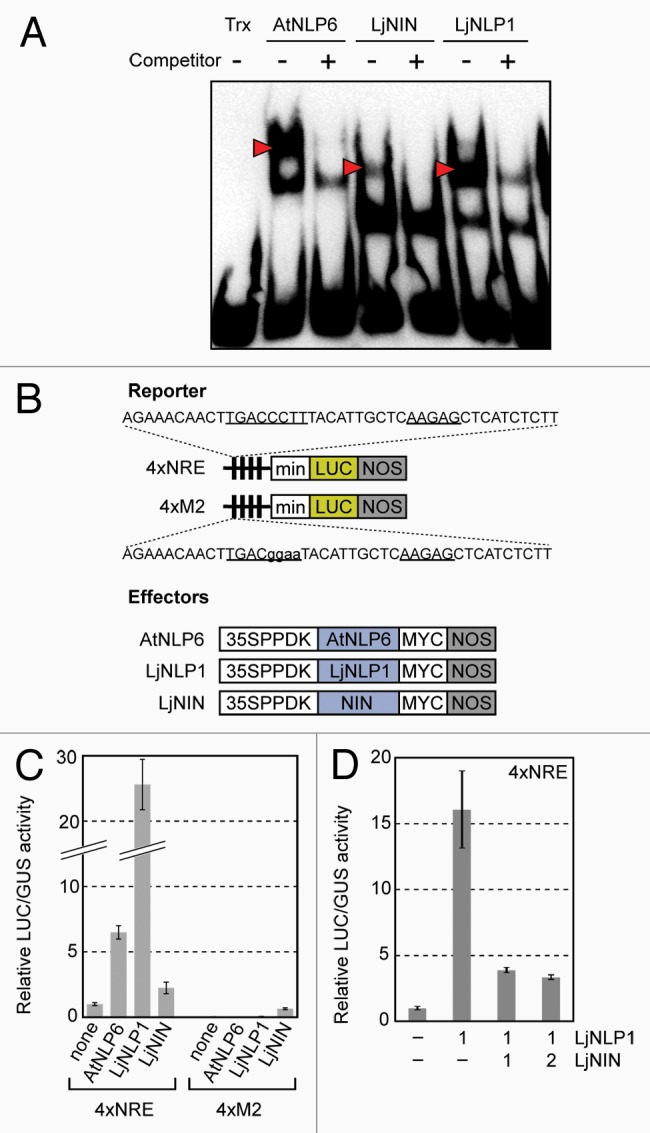
Figure 2.Interaction between NIN and the NRE in vivo and in vitro. (A) Sequence-specific DNA binding of NIN in vitro. Electrophoretic mobility shift assay was performed as described previously.8,24 Recombinant proteins were incubated with a biotin-labeled NRE probe in the presence or absence of a non-labeled DNA containing NRE fragment used as a competitor DNA at a 25-fold molar excess. Thioredoxin and thioredoxin-fused AtNLP6 were used as negative and positive controls, respectively. The positions of the DNA-protein complexes are indicated by red arrowheads. (B) Reporter and effector constructs used in protoplast transient assays. In the reporter constructs, 4 copies of the NRE (4xNRE) or a mutant version (4xM2)8 were placed upstream of the 35S minimal promoter truncated at position -72 (min) that was fused to the LUC reporter gene (LUC) and the NOS terminator (NOS). In the sequences of the wild-type and mutated NRE, the pseudopalindromic half-sites are underlined and mutated nucleotides are indicated in lowercase. Effector constructs were designed to express myc-tagged NIN or NLP under the control of a strong and constitutive promoter (the 35SPPDK promoter). (C) Activation of NRE-dependent transcription by NIN in vivo. Activation of an NRE-containing promoter by NIN in protoplast transient assays. (D) Competition assay in vivo. Effector constructs for the expression of LjNLP1 and LjNIN were co-transfected into protoplasts at the indicated ratio. As an internal control plasmid containing the GUS gene under the control of the A. thaliana ubiquitin promoter (UBQ10-GUS)25 was also co-transfected in each case in (C) and (D), the relative LUC activities were calculated using GUS activity levels. Relative LUC activities are shown with SD (n = 3) in (C) and (D).
To investigate whether NIN can bind to NRE in vivo and thereby regulate transcription via this interaction, we performed co-transfection experiments using effector plasmids containing NIN, LjNLP1, or AtNLP6 cDNA under the control of a constitutive strong promoter, a reporter plasmid containing the luciferase reporter gene (LUC) downstream of the NRE-containing promoter, and an internal control plasmid containing the β-glucuronidase reporter gene (GUS) fused to A. thaliana ubiquitin promoter (Fig. 2B). As shown in Figure 2C, NIN and LjNLP1 promoted the expression of LUC in A. thaliana mesophyll protoplasts, as did AtNLP6. Such activation was not evident using promoter with mutations in its NRE, suggesting the sequence-specific interaction of NIN and LjNLP1 in vivo. However, the fold activation by NIN was much lower than that by LjNLP1. Such weak transcriptional activation by NIN could be due to its lower expression, lower affinity to NRE in vivo, or lower transcriptional activation ability in mesophyll protoplasts, compared with LjNLP1. To clarify the reason for the lower activation ability of NIN, we performed an in vivo competition assay. When the expression vector for LjNLP1 was co-transfected into mesophyll protoplasts together with the expression vector for NIN, transcriptional activation by LjNLP1 was found to be reduced (Fig. 2D). Hence, NIN appears to be expressed at appreciable levels and to successfully bind to NRE, but to function as a weaker transcriptional activator than LjNLP1 in mesophyll protoplasts. The reason that NIN than LjNLP1 more dominantly exerted its effects on NRE-dependent transcription is currently uncertain. However, this phenomenon might be caused by effective expression of NIN protein than LjNLP1 protein in mesophyll protoplasts. Alternatively, as NIN and NLPs have a PB1 domain that may enable dimerization between PB1 domain-containing proteins,4 NIN and LjNLP1 might form heterodimers of which transcriptional activation ability is comparable to that of NIN homodimers but much weaker than that of LjNLP1 homodimers. Taken together, our results suggest that the DNA-binding specificities of NIN and LjNLP1 are similar to that of the AtNLPs in vivo and in vitro and also that NIN and the NLPs can enhance transcription through their interaction with similar DNA sequences.
We previously showed that AtNLP6, and probably other NLPs from A. thaliana, is post-translationally activated in response to nitrate signals through the N-terminal region flanking the RWP-RK DNA-binding domain.8 Since NIN and the NLPs share homology even at the N-terminal region (Fig. 1A, Fig. S1), we tested the nitrate responsiveness of NIN and LjNLP1. In our previous analysis of the nitrate responsiveness of AtNLP6, effector constructs were introduced into an A. thaliana transgenic line harboring a reporter construct, and the resultant stable transformants were treated with or without nitrate, and reporter activity was measured.8 However, as this assay system is time-consuming, we newly developed an alternative assay system for our present study using a biolistic particle delivery system, in which particles coated by effector plasmids are bombarded onto the roots of the A. thaliana transgenic line harboring the GUS reporter gene. The activity of the effectors is then evaluated by the numbers of cells showing GUS activity. In our current experiments, we used a GUS reporter gene under the control of the 35S minimal promoter fused to 8 copies of the LexA-binding site rather than the NRE (Fig. 3A). This was because NRE itself responds to nitrate due to the activity of endogenous NLPs, making it difficult to then evaluate the nitrate-responsiveness of particular effectors. We at first evaluated feasibility of this newly developed system by the use of LexA-VP16 fusion protein that promoted transcription independently of nitrate treatment and AtNLP6-LexA fusion protein that promoted transcription only after nitrate treatment.8 As the results indicated that we could evaluate nitrate-responsiveness of transcription factors using this system (Fig. 3B), we examined nitrate-responsiveness of the N-terminal regions of NIN and LjNLP1. Similar to the N-terminal region of AtNLP6, the N-terminal region of LjNLP1 (1–585 a.a.) fused to LexA was found to enhance the expression of the GUS reporter gene in a nitrate-dependent manner. However, the N-terminal region of NIN (1–563 a.a.) fused to LexA was found to transactivate transcription independently of nitrate (Fig. 3B). Although NIN was observed to be a weaker transcription factor than LjNLP1 in leaf mesophyll protoplasts (Fig. 2C and D), NIN appeared to function as a transcriptional activator at a comparable or even stronger degree of potency than LjNLP1 in root cells.
Figure 3. Different nitrate responses of the N-terminal regions of NIN and LjNLP1. (A) The reporter construct used in which 8 copies of the LexA-binding site were placed upstream of the 35S minimal promoter (min) that was fused to the GUS reporter gene (GUS) and the NOS terminator (NOS).8 (B) Analysis using a biolistic particle delivery system. Effector constructs for expression of the N-terminal region of AtNLP6, LiNLP1, or NIN fused to LexA were bombarded into the roots of 5-d old A. thaliana seedlings harboring the reporter construct. The seedlings were then incubated in the presence of 10 mM nitrate or a control compound (KCl). The roots were fixed in 90% acetone and subsequently stained with X-Gluc buffer at 37 °C overnight as described previously.26 Nitrate-induced expression was evaluated by counting the numbers of cells showing GUS staining, and GUS-positive stained cells per bombardment are shown with SD (n = 3). Typical images obtained for each construct shown side by side with the bar graphs. Bar, 200 µm.
In our current study, we show that LjNLP1 and NIN are transcription factors with comparable DNA binding and transcriptional regulatory activities but different nitrate responsiveness, although they are encoded by genes belonging to the same clade of the NIN/NLP gene family. Because NIN is an essential regulator during nodule formation,2,3 its nitrate-insensitivity is an important characteristic which is tightly associated with its physiological role. Nitrate inhibits nodulation, nodule development, and also symbiotic nitrogen assimilation in accordance with the fact that symbiotic nitrogen fixation is important in a nitrogen-deficient environment.16-18 If the activity of NIN required nitrate, as is the case with the NLPs, it could not regulate nodule formation in the absence of nitrate. Hence, the loss of nitrate-responsiveness of NIN appears to have been an essential molecular event in its evolution as a specific regulator of nodulation. We speculated that the unique amino acid sequence within the N-terminal region of NIN (Fig. 1C) may be associated with the loss of nitrate-responsiveness of NIN, because NIN and LjNLP1 showed similar ability for transcriptional activation but different nitrate-responsiveness.
As found for the A. thaliana NLPs, NIN and LjNLP1 can bind to an NRE and thereby promote transcription. Very recently, it was shown that NIN binds to the promoters of pectate lyase gene19 and NF-Y subunit gene.20 Although the binding site on the pectate lyase gene promoter has not yet been identified, the binding sites on the NF-Y subunit gene promoters have been proposed with sequences that are similar to the NRE. Thus, NIN and LjNLP1 likely bind to similar sequences through their highly conserved RWP-RK domain in planta. It is currently uncertain whether the targets of NIN and NLPs are completely different or partially overlapping. Nitrate signaling oppositely regulates NIN and NLP activities. Nitrate represses the Nod factor-induced expression of the NIN gene21 and induces the post-translational activation of the NLPs.8 Therefore, it is unlikely that NIN and the NLPs regulate the same genes in the same cells at the same time. However, we cannot yet rule out the possibility that NIN and the NLPs regulate the expression of common genes in different cells. Further analysis of the target genes of NIN and the NLPs may help to elucidate how NIN, a member of a transcription factor family responsible for the regulation of nitrate-regulated gene expression, came to play a key role in symbiotic nodulation.
Supplementary Material
Acknowledgments
We thank the National Bioresource Project (Legume Base, The University of Miyazaki, Japan) for providing the NIN cDNA clone. This work was supported in part by grants (KAKENHI No. 22380043 and No. 25252014) from the Japan Society for the Promotion of Science (JSPS) and a Grant-in-Aid for Scientific Research on Innovative Areas (No. 21114004) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary Materials
Supplementary materials may be found here: www.landesbioscience.com/journals/psb/article/25975
References
- 1.Mylona P, Pawlowski K, Bisseling T. Symbiotic nitrogen fixation. Plant Cell. 1995;7:869–85. doi: 10.1105/tpc.7.7.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schauser L, Roussis A, Stiller J, Stougaard J. A plant regulator controlling development of symbiotic root nodules. Nature. 1999;402:191–5. doi: 10.1038/46058. [DOI] [PubMed] [Google Scholar]
- 3.Madsen LH, Tirichine L, Jurkiewicz A, Sullivan JT, Heckmann AB, Bek AS, Ronson CW, James EK, Stougaard J. The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat Commun. 2010;1:10. doi: 10.1038/ncomms1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schauser L, Wieloch W, Stougaard J. Evolution of NIN-like proteins in Arabidopsis, rice, and Lotus japonicus. J Mol Evol. 2005;60:229–37. doi: 10.1007/s00239-004-0144-2. [DOI] [PubMed] [Google Scholar]
- 5.Konishi M, Yanagisawa S. Identification of a nitrate-responsive cis-element in the Arabidopsis NIR1 promoter defines the presence of multiple cis-regulatory elements for nitrogen response. Plant J. 2010;63:269–82. doi: 10.1111/j.1365-313X.2010.04239.x. [DOI] [PubMed] [Google Scholar]
- 6.Konishi M, Yanagisawa S. The regulatory region controlling the nitrate-responsive expression of a nitrate reductase gene, NIA1, in Arabidopsis. Plant Cell Physiol. 2011;52:824–36. doi: 10.1093/pcp/pcr033. [DOI] [PubMed] [Google Scholar]
- 7.Konishi M, Yanagisawa S. Roles of the transcriptional regulation mediated by the nitrate-responsive cis-element in higher plants. Biochem Biophys Res Commun. 2011;411:708–13. doi: 10.1016/j.bbrc.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 8.Konishi M, Yanagisawa S. Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat Commun. 2013;4:1617. doi: 10.1038/ncomms2621. [DOI] [PubMed] [Google Scholar]
- 9.Stitt M. Nitrate regulation of metabolism and growth. Curr Opin Plant Biol. 1999;2:178–86. doi: 10.1016/S1369-5266(99)80033-8. [DOI] [PubMed] [Google Scholar]
- 10.Wang R, Tischner R, Gutiérrez RA, Hoffman M, Xing X, Chen M, Coruzzi G, Crawford NM. Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol. 2004;136:2512–22. doi: 10.1104/pp.104.044610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yokota K, Soyano T, Kouchi H, Hayashi M. Function of GRAS proteins in root nodule symbiosis is retained in homologs of a non-legume, rice. Plant Cell Physiol. 2010;51:1436–42. doi: 10.1093/pcp/pcq124. [DOI] [PubMed] [Google Scholar]
- 12.Yokota K, Hayashi M. Function and evolution of nodulation genes in legumes. Cell Mol Life Sci. 2011;68:1341–51. doi: 10.1007/s00018-011-0651-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sato S, Nakamura Y, Kaneko T, Asamizu E, Kato T, Nakao M, Sasamoto S, Watanabe A, Ono A, Kawashima K, et al. Genome structure of the legume, Lotus japonicus. DNA Res. 2008;15:227–39. doi: 10.1093/dnares/dsn008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borisov AY, Madsen LH, Tsyganov VE, Umehara Y, Voroshilova VA, Batagov AO, Sandal N, Mortensen A, Schauser L, Ellis N, et al. The Sym35 gene required for root nodule development in pea is an ortholog of Nin from Lotus japonicus. Plant Physiol. 2003;131:1009–17. doi: 10.1104/pp.102.016071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marsh JF, Rakocevic A, Mitra RM, Brocard L, Sun J, Eschstruth A, Long SR, Schultze M, Ratet P, Oldroyd GE. Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 2007;144:324–35. doi: 10.1104/pp.106.093021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Streeter J. Inhibition of legume nodule formation and N2 fixation by nitrate. Crit Rev Plant Sci. 1988;7:1–23. doi: 10.1080/07352688809382257. [DOI] [Google Scholar]
- 17.Zahran HH. Rhizobium-legume symbiosis and nitrogen fixation under severe conditions and in an arid climate. Microbiol Mol Biol Rev. 1999;63:968–89. doi: 10.1128/mmbr.63.4.968-989.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stougaard J. Regulators and regulation of legume root nodule development. Plant Physiol. 2000;124:531–40. doi: 10.1104/pp.124.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie F, Murray JD, Kim J, Heckmann AB, Edwards A, Oldroyd GE, Downie JA. Legume pectate lyase required for root infection by rhizobia. Proc Natl Acad Sci U S A. 2012;109:633–8. doi: 10.1073/pnas.1113992109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soyano T, Kouchi H, Hirota A, Hayashi M. Nodule inception directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus. PLoS Genet. 2013;9:e1003352. doi: 10.1371/journal.pgen.1003352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barbulova A, Rogato A, D’Apuzzo E, Omrane S, Chiurazzi M. Differential effects of combined N sources on early steps of the Nod factor-dependent transduction pathway in Lotus japonicus. Mol Plant Microbe Interact. 2007;20:994–1003. doi: 10.1094/MPMI-20-8-0994. [DOI] [PubMed] [Google Scholar]
- 22.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 23.Perrière G, Gouy M. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–9. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- 24.Konishi M, Yanagisawa S. Ethylene signaling in Arabidopsis involves feedback regulation via the elaborate control of EBF2 expression by EIN3. Plant J. 2008;55:821–31. doi: 10.1111/j.1365-313X.2008.03551.x. [DOI] [PubMed] [Google Scholar]
- 25.Yanagisawa S, Yoo SD, Sheen J. Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature. 2003;425:521–5. doi: 10.1038/nature01984. [DOI] [PubMed] [Google Scholar]
- 26.Konishi M, Yanagisawa S. Sequential activation of two Dof transcription factor gene promoters during vascular development in Arabidopsis thaliana. Plant Physiol Biochem. 2007;45:623–9. doi: 10.1016/j.plaphy.2007.05.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.