Abstract
A current need in the neuroscience field is a simple method to monitor autophagic activity in vivo in neurons. Until very recently, most reports have been based on correlative and static determinations of the expression levels of autophagy markers in the brain, generating conflicting interpretations. Autophagy is a fundamental process mediating the degradation of diverse cellular components, including organelles and protein aggregates at basal levels, whereas alterations in the process (i.e., autophagy impairment) operate as a pathological mechanism driving neurodegeneration in most prevalent diseases. We have recently described a new simple method to deliver and express an autophagy flux reporter through the peripheral and central nervous system of mice by the intracerebroventricular delivery of adeno-associated viruses (AAV) into newborn mice. We obtained a wide expression of a monomeric tandem mCherry-GFP-LC3 construct in neurons through the nervous system and demonstrated efficient and accurate measurements of LC3 flux after pharmacological stimulation of the pathway or in disease settings of axonal damage. Here we discuss the possible applications of this new method to assess autophagy activity in neurons in vivo.
Keywords: autophagy flux, LC3, neurons, and nervous system
Autophagy is a catabolic mechanism that sequesters cytoplasmatic components for lysosomal-mediated degradation, including protein inclusions and damaged organelles.1 Autophagy has a key role in diverse physiological processes such as adaptation to nutrient starvation, embryonic development, cell death, energy metabolism, and antigen presentation, among other events (reviewed in ref. 2). Moreover, autophagy has been highlighted as a key component of pathological processes such as cancer, pathogen infection, hypoxia, and neurodegeneration.1 Therefore, the estimation of the rate of autophagy in a quantitative manner, especially in in vivo models,3,4 has become an essential need in the field; however, to date this issue has not been fully solved.
Autophagy, like most biological processes, is highly dynamic and regulated, with the peculiarity that the mediating organelle, the autophagosome, is newly formed when the process is activated and then it is cleared after fusion with lysosomes. The occurrence of a dynamic ratio between formation and degradation of these vesicles makes the measurement of either the activation or the speed of the process very complex in a steady-state condition. The autophagy process includes different regulatory steps (induction, initiation, elongation, lysosomal fusion, and degradation);5 thus, when the pathway is induced it becomes important to differentiate if this activation continues to reach the final degradative step or if autophagy is inhibited or attenuated at specific points, leading to an incomplete autophagic process.6
Detection of MAP1LC3 levels (hereafter LC3) is one of the main parameters monitored to measure autophagy induction, where LC3 conversion into its lipidated form (LC3-II) by western blot or the presence of LC3-positive vesicles are mostly used to determine autophagy activation.7 However, considering the degradation of the marker itself or the possible generation of an incomplete autophagy process, other approaches are necessary to monitor the flux of autophagy vesicles or cargoes.4,6 The use of autophagy or lysosomal inhibitors has been widely used to evaluate the net flux through the autophagy pathway in mammals.3,8 In particular, the use of lysosomal inhibitors such as bafilomycin A1, chloroquine, or pepstatin A (the latter typically in combination with leupeptin or E-64), that promote the accumulation of autophagy substrates, can be highly toxic and not recommended for an in vivo approach. All these analyses are not sufficient by themselves to assess the process and most of the time it is necessary to complement the measurements with laborious electron microscopy determinations, or an analysis of long-lived protein degradation that requires the use of radioisotopes.4 Another tool used to monitor autophagy is the tandem monomeric mCherry-GFP-LC3 tagged protein, which has been used in in vitro systems to monitor autophagy flux through direct fluorescence microscopy.9,10 This approach is based in the sensitivity of the GFP fluorescent signal to acidic conditions of the lysosome lumen, leading to its inactivation compared with the mCherry fluorescence, which is more stable. Thus, colocalization of GFP and mCherry fluorescence (yellow puncta) indicates that the tandem protein is not localized in compartments fused with a lysosome (i.e., on the phagophore or within the autophagosome). In contrast, detection of the mCherry signal without GFP (red puncta), indicates that the protein is located in the autolysosome. The use of this fusion protein serves as a method to monitor basal states of autophagy and the activation of the route without any potentially toxic inhibitor. To date, this method is one of the most-used strategies to monitor autophagy flux, however, this strategy has not been successfully developed in a mammalian system in vivo.1,8
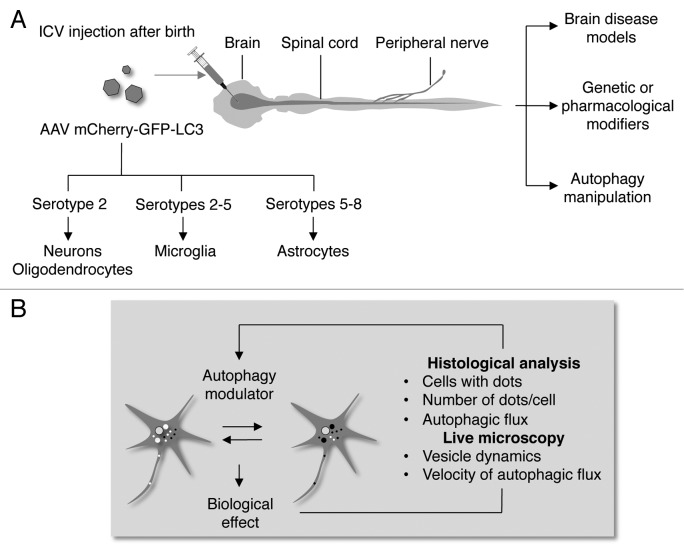
Although GFP-LC3 transgenic mice have been used to monitor autophagosome content in neurons,11,12 assays to quantify autophagy flux in neurons in vivo were missing until very recently mostly due to the low permeability of lysosomal inhibitors to the blood-brain barrier. This issue is particularly relevant because autophagy stimulation with pharmacological strategies is becoming an attractive therapeutic strategy for the treatment of neurodegenerative diseases, whereas alterations of the pathway are also reported in pathophysiologies such as Huntington, Parkinson, and Alzheimer diseases and amyotrophic lateral sclerosis (ALS).13-15 We recently described a simple method to deliver the autophagy reporter mCherry-GFP-LC3 through the central and peripheral nervous system to quantify autophagy fluxes in neurons in vivo in different settings. The methodology presented in Castillo et al. uses a combination of strategies to measure autophagy flux in the brain, spinal cord, and sciatic nerves: i) the generation of adeno-associated virus that allows an extended transduction and stable expression of transgenes, ii) the use of the dynamic fluorescent reporter mCherry-GFP-LC3 and iii) the delivery of the viral particles using intracerebroventricular (ICV) injection in newborn animals (Fig. 1).16 This approach led to a widespread transduction along the central nervous system, including pyramidal cortical and hippocampal neurons, Purkinje cells, and motor neurons in the spinal cord. With this method of AAV delivery, the cerebellum was the most efficiently transduced brain region. We also detected positive fibers in the peripheral nervous system, specifically at the sciatic nerves. This method is based on the manual injection of concentrated AAVs into the ventricle of newborn mice, which led to the diffusion of the viral particles through the cerebrospinal fluid, generating a massive and global transduction of neurons of the nervous system.17,18 The specificity of the expression was obtained through the use of a particular serotype of AAVs, serotype 2, which has a high tropism for neurons,19,20 but it also can transduce oligodendrocytes to a lower extent.21 Importantly, there are many different AAV serotypes available that could be used to transduce other cell types of the nervous system (Fig. 1A) including astrocytes (preferably transduced by serotypes 5 and 822,23), microglia (preferably transduced by serotype 524) and oligodendrocytes (preferably transduced by serotype 224,25), and even selective subpopulations of neurons of the brain.22 These viruses are not pathogenic, they do not induce an immune or inflammatory response, they are maintained as episomal DNA and the expression of the transgene can last for months and even years.
Figure 1. Schematic representation of the fluorescent reporter viral delivery strategy and the autophagic flux analysis. (A) AAV-mediated delivery of the autophagy flux reporter mCherry-GFP-LC3 into neonatal mice by intracerebroventricular (ICV) injection. This methodology allows wide transduction and stable expression of the transgene along the central nervous system, including pyramidal cortical and hippocampal neurons, Purkinje cells, and motoneurons. This methodology could be used to evaluate the role of autophagy in several pharmacological or genetic models of neurodegenerative diseases or injury conditions. This experimental strategy would also allow monitoring the effects of drug treatments designed to modulate autophagy in the central nervous system. Cell type-specific delivery of AAV could also be achieved by using different virus serotypes. (B) LC3 flux quantifications can be done using different approaches, through either manual or automatic counting of responding cells.
Using the approach described in Figure 1A, the detection of mCherry-GFP-LC3-positive dots and the measurement of autophagy flux was reproducible and sensitive, and LC3 was easily detected with different microscopy methods, including direct epifluorescence, confocal microscopy, a super zoom microscope, and spinning-disk microscopy. Using this combinatorial strategy, a robust LC3 flux was detected in neurons in vivo in animals treated with the drug rapamycin or an MTOR-independent inducer of the pathway, trehalose. This complementary approach also allows for following autophagic flux in models of mechanical injury in mice in the spinal cord and sciatic nerve. The quantifications of LC3 flux can be done by using manual or automatic counting of cells responding (percentage of cells that are positive for LC3 dots), the amplitude of the response (total dots per cell in each channel) or autophagy flux (the ratio between yellow and red puncta) (Fig. 1B). In addition, just by measuring total fluorescence of tissue sections or total fluorescence per cell, we were able to corroborate the induction of LC3 flux by the drug treatment regimens used in the study. This experimental setting represents the first approach to monitor the autophagy activity in vivo in the central nervous system. However, the analysis was still static because it was based on the quantification of LC3 puncta in tissue sections. Real-time measurements of autophagy vesicle dynamics were determined (speed of movement) in explants of dissociated sciatic nerve fibers (ex vivo) after treatment with rapamycin or trehalose,16 using regimens we recently published.26 Thus, the method described herein can be applied both in vivo and ex vivo, which makes it a reliable strategy to be applied in a wide range of experimental conditions.16 Another projection in terms of applications of this method is the monitoring of autophagy activity in living animals (i.e., in vivo live microscopy) using 2-photon microscopy in the brain,27 which could conclusively reveal autophagic flux adding new information about temporal and spatial aspects of the autophagy process. Pharmacology could be also used locally to manipulate autophagy levels in the brain, making it possible to avoid an undesirable nonspecific effect of global and systemic treatments.
This methodology could be used in any pharmacological or genetic model of neurodegenerative diseases (Fig. 1), and also will be useful to assess the effects of systemic or local treatments with drugs that modulate autophagy in different cell types. This method is simple and can be used by any laboratory since it does not require any sophisticated method for brain injection like stereotaxis,17,18 and the AAV particles can be obtained at a reasonable cost and in large quantities. In addition, the in vivo model does not require housing costs of transgenic lines since any rodent model can be injected manually with AAVs through an ICV route. We propose that our combinatorial strategy represents a significant technical advantage compared with other in vivo autophagy flux assays used for peripheral tissues, such as the measure of the amino acid exchange at arterious/venous levels at fed and fasted conditions or treatment of mice with protease/lysosomal inhibitors.4 Those alternative methods may alter the activity of the ubiquitin-proteasome system or lead to massive disturbances of lysosomal function.28
To apply our methodological principles, it is also important to consider the post-tissue removal process in order to maintain stable pH conditions and prevent re-emission of the quenched GFP fluorescence. However, this problem could only underestimate the experimental results. It is also important to account for the possible toxicity caused by the aggregation of overexpressed proteins, especially considering red fluorophores.29 However, we did not detect any signs of neuronal loss in the original characterization of the method. Besides, in vitro studies have shown that the mCherry-GFP-LC3 construct can aggregate after high overexpression, generating false positive signals.4 One alternative is the replacement of mCherry for a Dendra family member fluorophore that conserves the monomeric state and does not aggregate.30 After irradiation with a short-wavelength of visible light, Dendra fluorescent protein undergoes an irreversible conformational change and emits red fluorescence that can be tracked until the activated molecules are cleared. Dendra2-LC3 has been used to monitor autophagy flux in cellular models of Huntington disease.31 Beside, as a negative control, a lipidation-deficient mutant form of LC3 (G120A) could be used to determine the extent of mCherry-GFP-LC3 aggregation.
A large number of papers suggest that impairment of autophagy is critical in the progression of neurodegenerative diseases. For this reason, this methodology offers the possibility to study the net activity of autophagy in these proteopathies in most transgenic mouse models of disease and also allows the testing of therapeutic strategies to manipulate the pathway. For example, treatment of mice with rapamycin provides a protective effect for Huntington,32,33 Parkinson,34 and Alzheimer diseases.35 However, rapamycin treatment in some ALS models exacerbates the progression of symptoms and the disease onset.36 This surprising result may be due to impairment of autophagy flux as a pathological mechanism of disease and to the pleiotropic effects of MTOR signaling inhibition on many different processes including regulation of mRNA translation, transcription, cell growth and metabolism, and inflammation, among other functions.37 Moreover, another study did not detect any effect of rapamycin administration on an ALS mouse model.38 Therefore, our combinatorial strategy to monitor LC3 flux in neurons could be applied to solve these controversies and track autophagy activity in animal models of diseases affecting the nervous system (Fig. 1). In addition, the method offers a simple opportunity to test the effectiveness of drugs to modulate autophagy in the brain, a challenging requirement in the field of neurodegeneration. In summary, we have developed a new simple strategy to deliver and express an autophagy flux reporter through the peripheral and central nervous system of mice, allowing accurate measurements of LC3 flux after pharmacological or pathological stimulation of the pathway in vivo.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgements
This work was funded by FONDECYT 11121524 (SM); The Muscular Dystrophy Association and ALS Therapy Alliance, Millennium Institute no. P09-015-F, FONDECYT no. 1140549, ACT1109; CONICYT-EEUU grant USA2013-0003, Alzheimer Disease Association, ECOS CONICYT C13S02, and FONDEF D11I1007 (CH). VV received a CONICYT PhD fellowship no. 21120411.
Glossary
Abbreviations:
- AAV
adeno-associated virus
- LC3
microtubule-associated protein 1 light chain 3
- LC3-II
microtubule-associated protein 1 light chain 3 lipidated form
- ICV
intracerebroventricular
References
- 1.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–73. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 3.Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007;6:304–12. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- 4.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–52. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 6.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–26. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 2005;1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- 8.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–60. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 10.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 11.Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA, Small S, Spencer B, Rockenstein E, Levine B, et al. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 2008;118:2190–9. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mizushima N, Kuma A. Autophagosomes in GFP-LC3 Transgenic Mice. Methods Mol Biol. 2008;445:119–24. doi: 10.1007/978-1-59745-157-4_7. [DOI] [PubMed] [Google Scholar]
- 13.Mariño G, Madeo F, Kroemer G. Autophagy for tissue homeostasis and neuroprotection. Curr Opin Cell Biol. 2011;23:198–206. doi: 10.1016/j.ceb.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 14.Menzies FM, Moreau K, Rubinsztein DC. Protein misfolding disorders and macroautophagy. Curr Opin Cell Biol. 2011;23:190–7. doi: 10.1016/j.ceb.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nassif M, Valenzuela V, Rojas-Rivera D, Vidal R, Matus S, Castillo K, et al. Pathogenic role of Beclin 1/BECN1 in the development of amyotrophic lateral sclerosis. Autophagy. 2014 doi: 10.4161/auto.28784. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castillo K, Valenzuela V, Matus S, Nassif M, Oñate M, Fuentealba Y, Encina G, Irrazabal T, Parsons G, Court FA, et al. Measurement of autophagy flux in the nervous system in vivo. Cell Death Dis. 2013;4:e917. doi: 10.1038/cddis.2013.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Passini MA, Wolfe JH. Widespread gene delivery and structure-specific patterns of expression in the brain after intraventricular injections of neonatal mice with an adeno-associated virus vector. J Virol. 2001;75:12382–92. doi: 10.1128/JVI.75.24.12382-12392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glascock JJ, Osman EY, Coady TH, Rose FF, Shababi M, Lorson CL. Delivery of therapeutic agents through intracerebroventricular (ICV) and intravenous (IV) injection in mice. J Vis Exp. 2011 doi: 10.3791/2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartlett JS, Samulski RJ, McCown TJ. Selective and rapid uptake of adeno-associated virus type 2 in brain. Hum Gene Ther. 1998;9:1181–6. doi: 10.1089/hum.1998.9.8-1181. [DOI] [PubMed] [Google Scholar]
- 20.Zincarelli C, Soltys S, Rengo G, Rabinowitz JE. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol Ther. 2008;16:1073–80. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 21.Valenzuela V, Collyer E, Armentano D, Parsons GB, Court FA, Hetz C. Activation of the unfolded protein response enhances motor recovery after spinal cord injury. Cell Death Dis. 2012;3:e272. doi: 10.1038/cddis.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davidson BL, Stein CS, Heth JA, Martins I, Kotin RM, Derksen TA, Zabner J, Ghodsi A, Chiorini JA. Recombinant adeno-associated virus type 2, 4, and 5 vectors: transduction of variant cell types and regions in the mammalian central nervous system. Proc Natl Acad Sci U S A. 2000;97:3428–32. doi: 10.1073/pnas.97.7.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harding TC, Dickinson PJ, Roberts BN, Yendluri S, Gonzalez-Edick M, Lecouteur RA, Jooss KU. Enhanced gene transfer efficiency in the murine striatum and an orthotopic glioblastoma tumor model, using AAV-7- and AAV-8-pseudotyped vectors. Hum Gene Ther. 2006;17:807–20. doi: 10.1089/hum.2006.17.807. [DOI] [PubMed] [Google Scholar]
- 24.Cucchiarini M, Ren XL, Perides G, Terwilliger EF. Selective gene expression in brain microglia mediated via adeno-associated virus type 2 and type 5 vectors. Gene Ther. 2003;10:657–67. doi: 10.1038/sj.gt.3301925. [DOI] [PubMed] [Google Scholar]
- 25.Chen H, McCarty DM, Bruce AT, Suzuki K. Oligodendrocyte-specific gene expression in mouse brain: use of a myelin-forming cell type-specific promoter in an adeno-associated virus. J Neurosci Res. 1999;55:504–13. doi: 10.1002/(SICI)1097-4547(19990215)55:4<504::AID-JNR10>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 26.Castillo K, Nassif M, Valenzuela V, Rojas F, Matus S, Mercado G, Court FA, van Zundert B, Hetz C. Trehalose delays the progression of amyotrophic lateral sclerosis by enhancing autophagy in motoneurons. Autophagy. 2013;9:1308–20. doi: 10.4161/auto.25188. [DOI] [PubMed] [Google Scholar]
- 27.Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–8. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- 28.Bedford L, Lowe J, Dick LR, Mayer RJ, Brownell JE. Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat Rev Drug Discov. 2011;10:29–46. doi: 10.1038/nrd3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strack RL, Keenan RJ, Glick BS. Noncytotoxic DsRed derivatives for whole-cell labeling. Methods Mol Biol. 2011;699:355–70. doi: 10.1007/978-1-61737-950-5_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gurskaya NG, Verkhusha VV, Shcheglov AS, Staroverov DB, Chepurnykh TV, Fradkov AF, Lukyanov S, Lukyanov KA. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol. 2006;24:461–5. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- 31.Tsvetkov AS, Arrasate M, Barmada S, Ando DM, Sharma P, Shaby BA, Finkbeiner S. Proteostasis of polyglutamine varies among neurons and predicts neurodegeneration. Nat Chem Biol. 2013;9:586–92. doi: 10.1038/nchembio.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O’Kane CJ, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–95. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 33.Berger Z, Ravikumar B, Menzies FM, Oroz LG, Underwood BR, Pangalos MN, Schmitt I, Wullner U, Evert BO, O’Kane CJ, et al. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum Mol Genet. 2006;15:433–42. doi: 10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- 34.Malagelada C, Jin ZH, Jackson-Lewis V, Przedborski S, Greene LA. Rapamycin protects against neuron death in in vitro and in vivo models of Parkinson’s disease. J Neurosci. 2010;30:1166–75. doi: 10.1523/JNEUROSCI.3944-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spilman P, Podlutskaya N, Hart MJ, Debnath J, Gorostiza O, Bredesen D, Richardson A, Strong R, Galvan V. Inhibition of mTOR by rapamycin abolishes cognitive deficits and reduces amyloid-beta levels in a mouse model of Alzheimer’s disease. PLoS One. 2010;5:e9979. doi: 10.1371/journal.pone.0009979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, Li L, Chen S, Yang D, Wang Y, Zhang X, Wang Z, Le W. Rapamycin treatment augments motor neuron degeneration in SOD1(G93A) mouse model of amyotrophic lateral sclerosis. Autophagy. 2011;7:412–25. doi: 10.4161/auto.7.4.14541. [DOI] [PubMed] [Google Scholar]
- 37.Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell. 2010;40:310–22. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhattacharya A, Bokov A, Muller FL, Jernigan AL, Maslin K, Diaz V, Richardson A, Van Remmen H. Dietary restriction but not rapamycin extends disease onset and survival of the H46R/H48Q mouse model of ALS. Neurobiol Aging. 2012;33:1829–32. doi: 10.1016/j.neurobiolaging.2011.06.002. [DOI] [PubMed] [Google Scholar]