Abstract
In higher plants, the phloem plays a central role in the delivery of nutrients and signals from source to sink tissues. These signals likely coordinate different aspects of plant development, as well as its response to environmental cues. Although some phloem-transported proteins and RNAs may function as signaling molecules in plants, their mode of action remains poorly understood. Previous analysis of transcripts from CMV-infected pumpkin (Cucurbita maxima cv Big Max) identified a Translationally-Controlled Tumor Protein (TCTP) mRNA homolog, designated CmTCTP. In the present work this transcript was analyzed in terms of its expression pattern. This RNA accumulates, both in healthy and CMV-infected plants, in developing and mature phloem in petiole and roots, as well as in apices at high levels. The protein was present at lower levels in most cell types, and almost no signal was detected in apices, suggesting translational regulation of this RNA. Additionally, CmTCTP harbored by Agrobacterium rhizogenes is capable of inducing whole plant regeneration. These data suggest a role for CmTCTP in growth regulation, possibly through long-distance signaling.
Keywords: Translationally Controlled Tumor Protein, Phloem, Agrobacterium rhizogenes, plant regeneration, long-distance signaling
Introduction
TCTP constitutes a large family of exclusively eukaryotic proteins involved in the regulation of cell proliferation and differentiation in animals.1-3 It appears to display a guanine nucleotide exchange factor (GEF) activity, and therefore, is thought to regulate the activity of certain G proteins, although this has been disputed.2-6 Human TCTP is also an antagonist of the p53 tumor suppressor, which reinforces the notion that it is a positive modulator of cell proliferation.7 TCTP functions in a non-cell autonomous manner in several organisms. Indeed, human TCTP is identical to the histamine release factor (HRF) found circulating in serum, and Plasmodium falciparum also secretes a serum TCTP isoform.8,9 It may also have a role in DNA repair.10
There is evidence that TCTP in plants is a central regulator of growth and development. Arabidopsis AtTCTP1 (gene ID At3g16640) controls cell proliferation and cell size, and functions downstream of auxin.11 This gene is expressed constitutively, its mutant shows defects in the male gametophyte, and plants are much smaller in size. It appears that TCTP has a conserved function at least in plants and animals, since AtTCTP1 can rescue a Drosophila mutant and vice versa.12 More recently, a role in stomatal closure and thus response to water deficit has been shown for AtTCTP1.13
Pumpkin TCTP has been isolated from phloem and found to interact with CmPP16 and suggested to be necessary for its phloem root-ward transport.14 The isolation of a TCTP mRNA homolog from Lupinus phloem sap exudates indicates that this gene may also function in a non-cell autonomous fashion in plants as well.15
Phloem sap exudates contain a large array of mRNAs and proteins, which have been analyzed with more detail in pumpkin, cucumber, Brassica napus, and Lupinus; some of these macromolecules have an important function in development, such as leaf morphogenesis as well as cell-to-cell and long-distance transport of signaling molecules.15-19 The notion that a large proportion of the enucleate Sieve Element mRNAs acts in a non-cell autonomous manner is supported by the fact that several code for proteins involved in regulation of transcription and cell cycle progression such as transcription factors, MAP kinases, and cyclin-dependent protein kinases. Indeed, recently, a number of Aux/IAA RNAs have been found to be phloem-mobile and to influence root architecture.20,21 However, the function of most phloem-mobile mRNAs, as well as of their long-distance transport, is largely unknown.
In previous works we have found CmTCTP mRNA in pumpkin phloem sap exudates. Its overexpression in tobacco leads to increase in callus formation, as well as enhanced differentiation of transformed calli.22,23 To gain insight into the function of this gene in plants, we studied with more detail its localization in planta by in situ hybridization and immunolocalization. The effect of its overexpression in Agrobacterium rhizogenes-transformed tobacco tissue was also determined. Our results indicate that this gene is expressed mostly in vascular tissue in pumpkin. However, its mRNA accumulates mostly in the apex and the vascular regions of petioles, stems, and roots, while the protein is not particularly enriched in vascular tissue, in contrast to its mRNA. This is especially evident in the shoot apical meristem, where RNA accumulates to high levels, while the protein is barely detectable.
While no evident phenotype was observed in tobacco explants cocultivated with A. tumefaciens harboring CmTCTP under a strong constitutive promoter, tobacco explants cocultured with A. rhizogenes harboring the same construct led to whole plant regeneration. Furthermore, leaf explants from regenerated plants were able to produce embryos and shoots in the absence of external growth regulators. These results suggest that CmTCTP regulates differentiation, likely at a non-cell autonomous level.
Results
CmTCTP mRNA and protein are present in the phloem long-distance translocation stream of pumpkin
We had previously reported that only a minor number of mRNAs are induced in phloem sap exudates from CMV-infected plants relative to healthy controls, although their effect could be profound in plant development.22 A cDNA was identified that codes for a protein with high similarity to the family of Translationally Controlled Tumor Proteins (TCTP), and was thus termed CmTCTP (for Cucurbita maxima TCTP; accession number DQ304537). The sequence of this mRNA consists of 802 nucleotides, with an open reading frame of 507 nucleotides coding for a 168 aa protein. The predicted 3D structure of this protein is quite similar to that of the canonical member of this family, from Schizosaccharomyces pombe, and to that predicted for most members of this family.4,24 In general, the predicted structure of CmTCTP displays domains for tubulin and calcium binding, as well as a TCTP2, but not TCTP1, signature, among others. Thus, CmTCTP biochemical activity may be similar to mammalian and insect (Drosophila) TCTP.
RT-PCR was performed from different tissues in order to assess the sites of accumulation of the CmTCTP mRNA. As shown in Figure 1A, this RNA accumulates mostly in vascular strips, stems, petioles, roots and shoot apices, with very low levels in leaves and flowers; the highest levels for this transcript were found in roots and apices. CmTCTP RNA was detected in phloem sap at considerably high levels, although lower than another phloem sap mRNA, CmPP16. Actin mRNA was detected after 30 cycles of PCR at much lower levels (Fig. 1C, lower panel). CmTCTP protein levels were highest in leaves, roots, shoot apex, and phloem sap. Interestingly, levels were low in vascular strips, compared with its mRNA (Fig. 1B). Importantly, CmTCTP protein is also present in phloem sap exudates. The lack of correspondence between protein and mRNA levels in certain tissues prompted us to analyze the distribution of both CmTCTP transcript and protein levels in more detail.
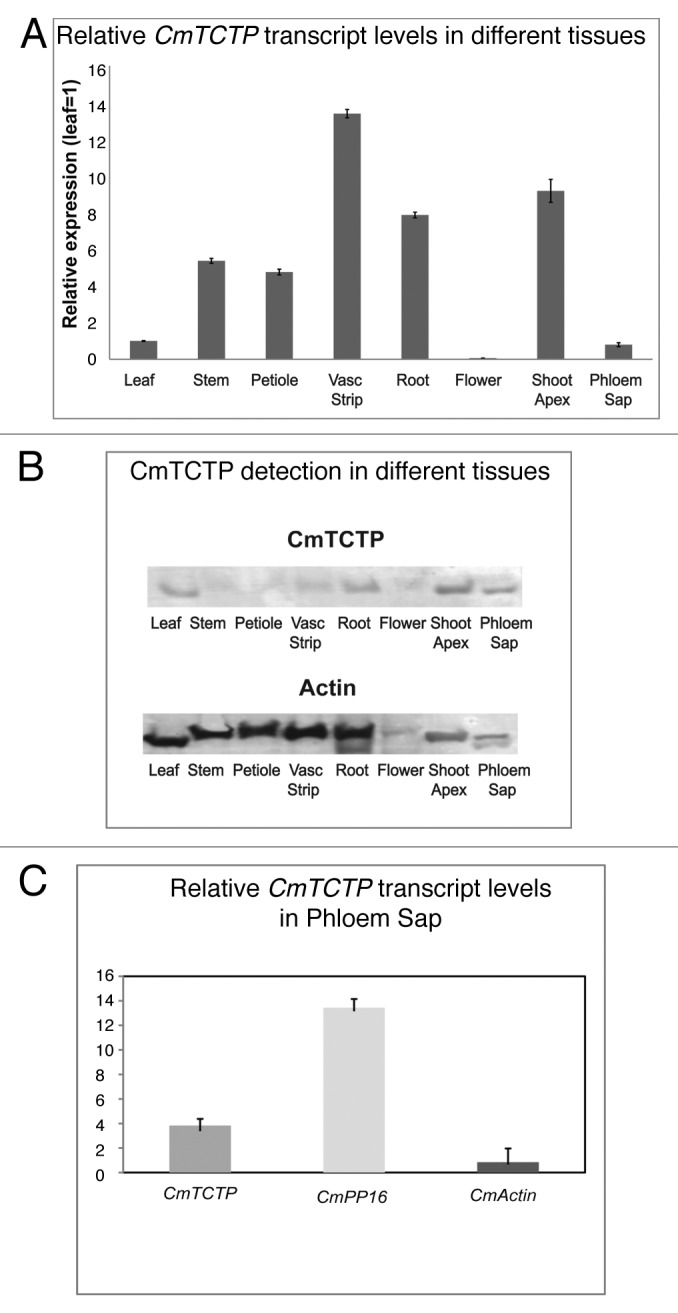
Figure 1. Highest levels of CmTCTP mRNA are found in vascular strips. (A) Real Time RT-PCR of CmTCTP in different tissues from pumpkin; levels were normalized to leaf tissue. (B) immunodetection of CmTCTP in different tissues from pumpkin. CmTCTP accumulates to higher levels in shoot apex. (C) Real Time RT-PCR of CmTCTP, CmPP16 and CmActin RNAs in phloem sap.
CmTCTP mRNA localizes to the phloem
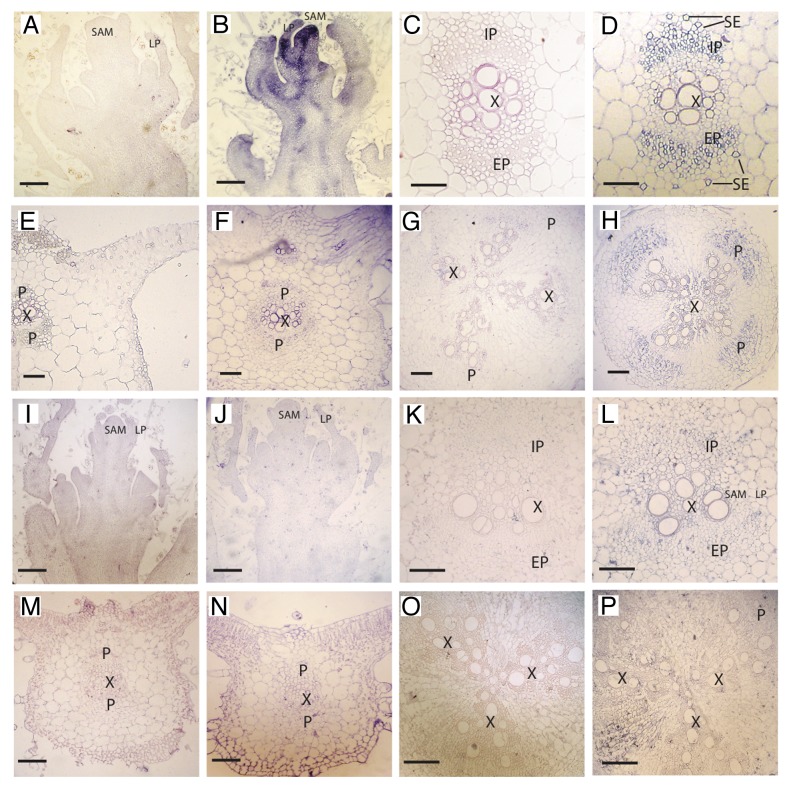
In situ localization of CmTCTP mRNA is shown in Figure 2. In agreement with the RT-PCR experiments, this transcript is present at high levels in all cell types in the apex in healthy plants, although in subapical sections the signal is more concentrated in what will become the vascular tissue (Fig. 2B). In other tissues, the accumulation of CmTCTP RNA was observed in mature and developing phloem in petioles and roots (Fig. 2D and H). In particular, this RNA was detected in what appear as mature sieve elements in internal and external phloem. A faint signal was observed around the vascular bundles in transversal sections of leaf blade (Fig. 2F). Transcript levels were roughly the same in mock-and CMV-infected plants, although signal in ectopic vascular bundles in leaf main vein of infected plants was observed (Fig. S1F). In general, RNA accumulation was observed in cell types other than the vascular tissue in CMV-infected plants, for example in roots (Fig. S1H). Interestingly, a strong, reproducible signal was observed when the CmTCTP sense probe was hybridized against apices of CMV-infected plants (Fig. S1A and B). No signal was detected in mock-inoculated apices with sense probe.
Figure 2.CmTCTP mRNA localizes to developing and mature phloem while CmTCTP protein is detected at low levels in several cell types. (A-H) in situ localization of CmTCTP mRNA in various tissues. (A, C, E, and G) sense probe; (B, D, F, and H) antisense probe. (A and B) shoot apex longitudinal section; (C and D) petiole cross section; (E and F) leaf blade transversal section; (G and H) main root cross section. (I-P) immunolocalization using CmTCTP-specific polyclonal antibodies. (I, K, M, and O) preimmune serum; (J, L, N, and P) immune serum. (I and J) apex transversal section (compare with apex in in situ hybridization); (K and L) petiole cross section; (M and N) leaf blade transversal section; (O and P) main root transversal section. All bars, 500μM. IP, internal phloem; SAM, shoot apical meristem; LP, leaf primordia; EP, external phloem; X, xylem; P phloem region; SE, sieve elements.
Serial sections were also utilized for CmTCTP protein localization. In contrast to the abundant mRNA signal in the apex, almost no protein was detected in this tissue (Fig. 2J; Fig S1J). It must be mentioned that CmTCTP was detected in subapical tissue of CMV-infected plants, but only in isolated cells (Fig. 2K and L); however, much higher levels were detected further from the apex, which could account for the abundance of the protein in the shoot apex, as determined by western blot. The strongest signal was observed in the vascular region in petioles, although still at low levels compared with its mRNA (Fig. 2L). Signal was also detectable in mesophyll in leaf midrib, and at much higher levels in CMV-infected plants in this same tissue (Fig. 2N; Fig. S1P). The levels of protein were very low in roots (Fig. 2P; Fig. S1R). In all, CmTCTP protein levels appear to be lower than those of the corresponding mRNA, although is present in more cell types than the RNA.
Agrobacterium rhizogenes harboring CmTCTP induces whole plant regeneration of tobacco leaf explants with
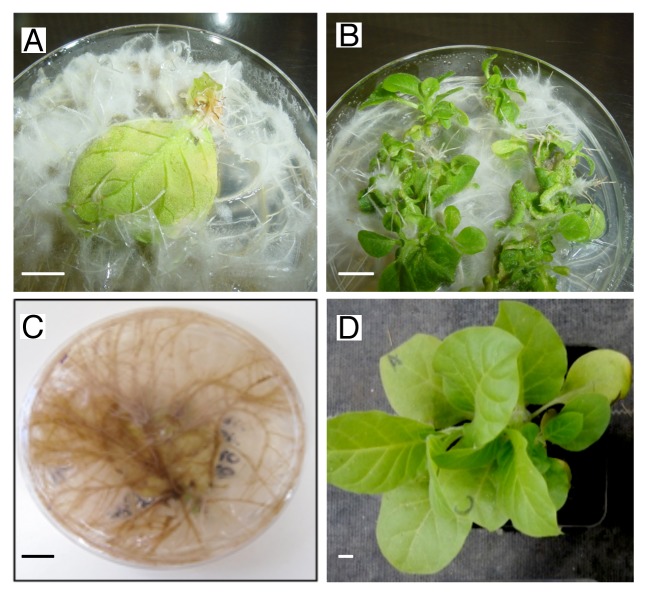
Previous work in our group, as well as from other groups, indicate that TCTP has an important role in promoting proliferation and differentiation in plants. However, given that several organisms harbor more than one TCTP gene, it is possible that there is a “division of labor” in this respect. CmTCTP was introduced into A. rhizogenes K599, a cucumopin strain, to determine whether an increase in root biomass could be detected, considering the higher expression levels of this gene in this tissue, and the role of the Arabidopsis homolog as a mitotic regulator. A construct containing an untranslatable version of CmTCTP (CmTCTP mut) was used as control. Sterilized leaf explants from tobacco were cocultured with either untransformed A. rhizogenes or harboring the different versions of this gene. As expected, leaf explants cocultured with A. rhizogenes induced extensive formation of roots (Fig. 3A and C); similar results were obtained with CmTCTP mut (not shown). Interestingly, CmTCTP, while also inducing tumor phenotype, it also led to the formation of shoot structures from which eventually whole plants arose (Fig. 3B and D). Resulting plants were viable and produced progeny. Table 1 shows the results of such analysis. Plant regeneration was quite efficient, reaching ca. 60% of total explants cocultured with A. rhizogenes. However, no such regeneration was observed when leaf explants were cocultured with A. tumefaciens harboring CmTCTP (not shown). This construct was also delivered biolistically onto tobacco leaf explants, and also failed to induce plant regeneration in 5 independent assays (not shown). Thus, this phenomenon presumably requires the concerted action of the aux and/or rol genes from A. rhizogenes Ri plasmid and CmTCTP.
Figure 3.A. rhizogenes harboring CmTCTP induces plant regeneration in tobacco explants. (A and C) tobacco leaf explants cocultured with A. rhizogenes without vector 4 and 8 weeks after transformation; (B and D) tobacco leaf explants cocultured with A. rhizogenes harboring CmTCTP 4 and 8 weeks after transformation. Bar, 1cm.
Table 1.CmTCTP induces whole plant regeneration in tobacco with high efficiency when harbored by Agrobacterium rhizogenes.
| Strain | % Root tumor formation | % Plant regeneration | Plants per leaf explant |
|---|---|---|---|
| K599 | 100% | 0% (0/350) | 0 |
| K599::CmTCTP | 90% | 58% (203/350) | 7.5 +/− 0.5 |
| K599::CmTCTP mut | 100% | 0% (0/350) | 0 |
Discussion
The phloem is involved in the delivery of nutrients from source to sink tissues, but an additional function is now well established, that of signaling between distant tissues.25 A large array of potentially signaling molecules has been identified in the phloem translocation stream, including proteins and different species of RNAs.26-32 Since it is likely that the enucleate sieve element does not engage in protein synthesis, such RNAs probably constitute signaling molecules. We have previously found several RNAs in phloem sap of pumpkin infected with CMV.22 One of these RNAs corresponds to the translationally controlled tumor protein (TCTP). Interestingly, this protein has been reported in the phloem proteome of Brassica napus and cucumber,18,19 as well as in the phloem transcriptome of lupin.15
The levels of this mRNA were higher in apices and in developing and mature phloem. Of note, signal was particularly high in mature sieve elements, supporting the notion that this mRNA functions in a non-cell autonomous manner. On the other hand the levels of this RNA, as determined by RT-PCR and in situ hybridization of paraffin-embedded sections, are similar between mock-inoculated and CMV-infected plants, except in leaves and apices. In general, the levels of the protein did not correspond to those of the mRNA; for example, the highest levels of protein were found in root, while the mRNA levels here were lower than in other tissues. On the other hand, the levels of mRNA in apical tissues were also high in both mock and infected plants, but the protein levels were much lower at least in the shoot apical meristem; translational regulation of this RNA could explain this discrepancy. The translational regulation of this gene in animals is well documented.1 Arabidopsis AtTCTP1 appears to be regulated in such way; in this case the 3′UTR has been suggested to be involved.11 Another explanation for this discrepancy would be that most of the RNA in the apex is transported from distant tissues. Run-on assays should help clarify this question.
The low levels of this protein in the apical meristem are also unexpected given the role of TCTP in cell proliferation. Also, the presence of potential antisense TCTP sequences in apices of CMV-infected plants only (as inferred from in situ hybridization) raises the possibility of regulation of this RNA through interaction with a potential microRNA. While there are no predicted microRNAs that target the Arabidopsis TCTP mRNAs (either AtTCTP1 or AtTCTP2), this possibility cannot be excluded in pumpkin and/or other plant species.
Our in situ hybridization results for CmTCTP mRNA are consistent with its localization in internal and external phloem, in both the CC-SE complex, as well as in cambium at lower levels. Since TCTP plays an important role in cell proliferation and regulation of cell cycle duration in both plants and animals,2,11,12 it is rather interesting that both the protein and the mRNA are found also in terminally differentiated cells (such as the CC and SE). Indeed, the localization of both mRNA and protein could indicate a role in long-distance signaling in response to either a genetic program or to an external stimulus, such as viral infection, as in the present case.
It has been shown that Arabidopsis AtTCTP1 mutants show a strong delay in development and decreased cell size in epidermis.11,12 The overexpression of CmTCTP in tobacco leaf explants led to an unexpected, additional phenotype in which whole plants were regenerated, instead of forming a tumor phenotype. Interestingly this was not observed when CmTCTP was harbored by A. tumefaciens, or when it was delivered biolistically. Therefore, plant regeneration induced by this gene probably also requires A. rhizogenes Ri plasmid aux and/or rol genes, either by creating a favorable milieu for cell reprogramming mediated by CmTCTP in roots, or by directly interacting with CmTCTP protein. It appears that root formation by the action of the aux or rol genes per se is not required for plant regeneration, since shoots can be observed before root formation. While the reprogramming observed in the present work is induced, it evidently underlies the biological activity of CmTCTP (and by extension, other plant TCTP genes); thus, it may not only have a role as mitotic integrator, but as a factor involved in differentiation. It should be noted that the Arabidopsis AtTCTP1 gene acts downstream of auxin.11 A. tumefaciens Ti encodes cytokinin-synthesis genes, while the Ri plasmid encodes genes that regulate auxin synthesis;33 thus, this could explain the absence of a contrasting phenotype in explants transformed with A. tumefaciens.
Given that the genome of pumpkin has not been elucidated yet, it is not known how many TCTP genes it harbors. However, other cucurbits, such as Citrullus lanatus (watermelon) and Cucumis sativus (cucumber), contain 2 genes each; it may be speculated that there is a division of labor between these genes, one involved in differentiation and another more related to cell proliferation. It is also possible that TCTP in other plant species may be involved in vegetative reproduction through specialized root and stem structures, such as rhizomes, tubers, and stolons. This could be of interest for the clonal propagation of commercially relevant crops.
Materials and Methods
Plant material and phloem sap collection
Pumpkin plants (Cucurbita maxima cv Big Max) were grown in greenhouse conditions. Arabidopsis was grown at 24 °C and 12h light/12h dark photoperiod in a controlled environment chamber (Conviron, Winnipeg, Canada).
Cotyledons were mock-inoculated mechanically with phosphate buffer or infective sap from plants infected with a Brazilian isolate of CMV, a generous gift from Dr. Robert Gilbertson, UC Davis. Two weeks after inoculation, different tissues (apices, leaves, petioles, stems, and roots) were harvested from mock-and CMV-inoculated plants. Phloem exudates for RNA extraction were collected from these same plants through cut petioles as described in order to minimize contamination of xylem sap due to root pressure. Exudates were then transferred into tubes containing Trizol reagent (Invitrogen), 500 μl for 200 μl of phloem sap. Briefly, the aqueous phase was extracted twice and the RNA precipitated with 2 volumes of absolute ethanol and 1μg of linear acrylamide (Ambion, Midlands TX), washed with 70% ethanol, resuspended in ddH2O, and stored at −80 °C until further use. Phloem sap exudates for western blot analysis were collected essentially as described.34
Total RNA extraction
Total RNA was extracted by the guanidine-hydrochloride method35 from 1g of tissue. For phloem sap RNA isolation, phloem exudates in Trizol were treated essentially according to the manufacturer’s recommendations (Invitrogen), starting with 200–400 μl of collected phloem sap. RNA was centrifuged at full speed at 4 °C and the resulting pellet washed with 70% ethanol and resuspended in ddH2O and stored at −80 °C for further analysis.
RT-PCR and Real Time RT-PCR
For cDNA synthesis, RNA from phloem sap, leaves, apices, stems, and roots was used as template. Superscript Reverse Transcriptase II (Invitrogen) was used according to the manufacturer’s recommendations. Reactions were performed with 10 ng total RNA, which was mixed with 1μl 10 μM SMART and dTGAGA primers. One μl of cDNA was then employed for PCR reactions that included 2.5U Taq DNA polymerase (Takara), and the corresponding primers. For the pumpkin actin mRNA, degenerate primers, designed from alignment of several plant actin sequences, were used (CmACT5deg forward: 5′-AAYTTGGAYG AYATGGARAA R-3′ and CmACT3deg reverse: RAANGTYTCR AACATDATYT GNGTCAT). For CmPP16 and CmTCTP, specific primers were used (CmPP16 forward: 5′-ATGGGGATGG GAATGATGGA GGTCCAT-3′ and CmPP16 reverse: 5′-TAGTTTTCCC ATGGGTAACA TCCTCC-3′; (CmTCTP Forward: 5′-ATGTTGGTTT ATCAAGACCT TCTCACT-3′ and CmTCTP reverse 5′-GCACTTGACT TCCTTCAAAG CCTGGCGC-3′). The PCR conditions used were as follows: 35 cycles of denaturation (94 °C for 30s), annealing (58 °C for 30s) and extension (72 °C for 1 min) and a final extension step of 7 min at 72 °C. The amplified product was analyzed by agarose gel electrophoresis.
For real time RT-PCR, a commercial system was used following the manufacturer’s recommendations (SYBR Green qRT-PCR system, Invitrogen). The same primers at 10 pM were used with 50 ng total RNA in a 10 μL reaction mix. The Real Time RT-PCR reactions were incubated in a Rotor Gene 3000 apparatus (Corbett Research, Australia). Conditions for amplification were 40 min at 42 °C for first strand synthesis, followed by 40 cycles at the annealing temperatures corresponding to each primer pair. To verify that no additional products were amplified in the reaction, a dissociation curve was generated through progressive sample heating (60–95 °C). The Ct value for each product was determined by triplicate in each treatment. Actin mRNA was used to normalize gene expression. Relative quantification for transcript accumulation was performed according to the method described by Livak and Schmittgen.36
In situ hybridization
Tissues from healthy and infected pumpkin, 3 weeks after mechanical inoculation, were cut into small sections and fixed with FAA solution (3.7% (v/v) formaldehyde, 5% (v/v) acetic acid, and 50% (v/v) ethanol) for 48h at room temperature. Fixed tissue was embedded in paraffin, sectioned with a microtome (Microm, Germany) and mounted on slides coated with Vectabond reagent (VECTOR laboratories) for hybridization with in vitro digoxigenin-labeled transcripts. In vitro transcriptions were performed using the T7/SP6 MEGAscript kit (Ambion, Midlands, TX) according to the manufacturer’s recommendations. Hybridizations were carried overnight at 42 °C in standard buffer essentially as described.37,38 Slides were mounted with Permount reagent (Fisher Scientific) and viewed and photographed with a Nikon Optiphot-2 microscope (model HFX-DX).
Protein expression and purification
The CmTCTP open reading frame was PCR amplified from the cloned CmTCTP cDNA (GenBank: DQ304537) with Pfu polymerase (Stratagene), and cloned in frame with a 6X His tag-encoding sequence (in the N end of the resulting protein) in the pProExB-C30 bacterial expression vector (Novagen) and propagated in E. coli DH5α. Bacteria were grown in 2TY medium and protein induction was performed at 37 °C by addition of 1mM IPTG. When the bacterial culture reached 0.6 (A590), the bacteria were harvested, lysed, and the protein was purified in a nickel affinity column. Total protein was loaded onto a nickel-Sepharose High Performance resin (Amersham Biosciences) following the manufacturer’s recommendations. The column was preequilibrated with EQ buffer (20mM sodium phosphate, 0.5M NaCl, and 20mM imidazole, supplemented with 8M urea, pH 7.4), and bound proteins washed with 15 ml buffer EQ supplemented with 8M urea and 50mM imidazole, pH 7.4; different fractions were eluted with EQ buffer containing 8mM urea and 250mM imidazole, pH 7.4. The protein fractions were denatured by heating for 5 min at 95 °C, and analyzed by SDS-PAGE in a 15% gel and visualized by Coomassie staining. Fractions containing rCmTCTP protein free from contaminating proteins were dialyzed against PBS 1X buffer (10X PBS: 1.3M NaCl, 2.7mM KCl, 4.3mM Na2HPO4. 7H2O and 1.4mM KH2PO4, pH 7) and samples were stored at –20 °C.
Antibody production
Before starting immunization, the rabbits were bled and the serum tested for non-reactivity against recombinant CmTCTP (rCmTCTP) and total plant protein. Subcutaneous doses (approximately 100 μg/rabbit) were injected each 15 d over a 2-mo period and blood collected. Rabbit serum was purified by ammonium sulfate precipitation in 2 steps, fraction 1: 0–25% and fraction 2: 25–50% salt-saturated. The proteins were then purified by affinity chromatography with protein A-agarose (Sigma). Unbound proteins were washed off with 10 column volumes of buffer A (0.02M NaH2PO4, 0.15M NaCl, pH 8); bound immunoglobulin was eluted with 0.2M Na2HPO4 and 0.1M citric acid pH 3, and immediately neutralized and dialyzed with 1X PBS. Immunoglobulin concentration was determined by absorbance at 280nm, and protein profile antibody analyzed by SDS-PAGE. The specificity of the polyclonal antisera was determined by western blot assays as described below.
Total protein extraction and western blot
Total protein was extracted essentially as described.39 Briefly, plant tissue was ground in liquid nitrogen, homogenized, and centrifuged at 13000g for 5min. Supernatants were recovered and dissolved in SDS-PAGE sample buffer. Protein concentration was determined with the BCA Protein Assay Kit (Pierce) or the Bradford method followed by 15% SDS-PAGE. Total proteins were transferred for 1h at 100 V to polyvinylidene difluoride (PVDF) membranes (Whatman) which were blocked for 2h in blocking solution (PBS 1X, 5% non-fat milk, and 0.1% Tween 20) followed by rinsing with 1X PBS, and incubated overnight at 4 °C with the polyclonal antiserum (diluted 1:2000 in 1X PBS, 5% non-fat milk, and 0.1% Tween 20). Membranes were washed (1X PBS, 0.1% Tween 20), incubated with peroxidase-conjugated goat anti-rabbit IgG (Zymax CA) at 1:4000 in PBS 1X containing 2.5% skim milk. After washes the signal was detected with HRP color development reagent (Biorad) or with the ECL detection system (Amersham Pharmacia).
Immunolocalization
This was performed essentially as previously described, healthy and infected pumpkin tissues were fixed, embedded, sectioned, and rehydrated.37,38 After blocking with 5% skim milk, sections were incubated at 4 °C overnight with anti-CmTCTP antibody, and then with alkaline phosphatase-coupled anti-rabbit antibody (Zymax, CA). Finally, alkaline phosphatase activity was visualized by incubation with 5-bromo-4-chloro-3-indoxyl phosphate/nitroblue tetrazolium chloride (NBT; Sigma).
Explant transformation
Cultures containing A. rhizogenes strain K599 harboring the recombinant plasmids were grown on selective liquid media (Luria Bertani with Spectinomycin 100 mg/L) during 36hrs at 120 rpm; the cultures were concentrated (3,000 rpm/1 min) and resulting pellets were resuspended in the same amount of fresh media. Leaves from N. tabacum 15–20 dag (days after germination) plants were used as explants, and sterilized by treatment with ethanol 70% (1 min) followed by soaking in 10% sodium hypochlorite for 30 min and several washes with sterile water. Explants were placed in an A. rhizogenes suspension for 2–5 min at 25 °C. After co-cultivation, the explants were placed on MS medium and incubated for 3 d in a growth chamber in the dark at 25 °C, then transferred into fresh MS solid medium containing vancomycin (250µg/ml) and cefatoxime (500µg/ml) to avoid contamination, and placed in a controlled environment growth chamber under long-day conditions (16 h light/8 h dark).
For tobacco transformation with A. rhizogenes harboring CmTCTP, the PCR product of the reading frame was cloned in the pCR8/GW/TOPO vector (Invitrogen) and then transferred through recombination to the p*7FWG2 binary vector (Plant Genetic Systems, Ghent, Belgium). The resulting construct was introduced into Agrobacterium rhizogenes K599 by electroporation. Tobacco leaves were surface-sterilized and incubated with A. rhizogenes. As control, an untranslatable version of CmTCTP (CmTCTP mut) was constructed by PCR in which the 5′ primer used contained a stop codon replacing the start codon (5′- CACCGAGTAA ATTGGGCAAT CTGATTGGTT TATCAAGACC-3′, underlined). This was also cloned in the p7WG2D binary vector (Plant Genetic Systems), in which the CaMV 35S promoter drives the expression of the transgene, and coexpresses GFP independently, and introduced to tobacco as described above. A version of the CmTCTP ORF lacking stop codon was obtained by PCR from the original CmTCTP cDNA, cloned into pCR8/GW/TOPO, and then introduced into the p*7FWG2 binary vector with GFP fused to the C-end. These constructs were introduced into A. rhizogenes K599 via electroporation.
Cultures containing A. rhizogenes strain K599 harboring the recombinant plasmids were grown on selective liquid media (Luria Bertani with Spectinomycin 100 mg/L) during 36h at 120 rpm; the cultures were concentrated (3,000 rpm/1 min) and resulted pellets were re-suspended in the same amount of fresh media. Leaves from N. tabaccum 15–20 dag (days after germination) plants were used as explants. For tissue sterilization, leaves were pre-washed with sterile water and then treated with ethanol 70% (1 min) followed by soaked in 10% sodium hypochlorite for 30 min and several washes with sterile water. A sterile scalpel was used to tear gently each explant, thereafter explants were placed in a solution of A. rhizogenes containing the recombinant plasmid for 2–5 min at 25 °C. After co-cultivation the explants were placed on MS medium (1.0 MS salts, 1.5% sucrose, and 0.4% agar [Gelrite]) and incubated for 3 d in a growth chamber under dark conditions at 25 °C, to finally been transferred into fresh solid MS medium containing vancomycin (250µg/ml) and cefatoxime (500µg/ml) and been placed in a controlled environment growth chamber under long-day conditions (16 h light/8 h dark).
Conclusion
In this work we have characterized the pumpkin translationally controlled tumor protein CmTCTP mRNA and protein. The mRNA accumulates in the phloem as well as in the apex, while the protein was detected in more cell types but at lower levels. Also, our results indicate that CmTCTP induces, when harbored by A. rhizogenes, whole plant regeneration, suggesting a role in differentiation. The accumulation of its mRNA in the CC-SE complex also suggests that this may occur through a non-cell autonomous mechanism.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by CONACYT grant 50769 to RR-M and 105985 to BX-C. JJH-M, RT-M, and FAR-O were supported by CONACyT doctoral fellowships.
Glossary
Abbreviations:
- CC
companion cell
- SE
sieve element
Supplementary Material
Supplementary material may be found here: https://www.landesbioscience.com/journals/psb/article/26477/
References
- 1.Bommer UA, Thiele BJ. The translationally controlled tumour protein (TCTP) Int J Biochem Cell Biol. 2004;36:379–85. doi: 10.1016/S1357-2725(03)00213-9. [DOI] [PubMed] [Google Scholar]
- 2.Hsu YC, Chern JJ, Cai Y, Liu M, Choi KW. Drosophila TCTP is essential for growth and proliferation through regulation of dRheb GTPase. Nature. 2007;445:785–8. doi: 10.1038/nature05528. [DOI] [PubMed] [Google Scholar]
- 3.Chen SH, Wu PS, Chou CH, Yan YT, Liu H, Weng SY, Yang-Yen HF. A knockout mouse approach reveals that TCTP functions as an essential factor for cell proliferation and survival in a tissue- or cell type-specific manner. Mol Biol Cell. 2007;18:2525–32. doi: 10.1091/mbc.E07-02-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hinojosa-Moya J, Xoconostle-Cázares B, Piedra-Ibarra E, Méndez-Tenorio A, Lucas WJ, Ruiz-Medrano R. Phylogenetic and structural analysis of translationally controlled tumor proteins. J Mol Evol. 2008;66:472–83. doi: 10.1007/s00239-008-9099-z. [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Fonseca BD, Tang H, Liu R, Elia A, Clemens MJ, Bommer UA, Proud CG. Re-evaluating the roles of proposed modulators of mammalian target of rapamycin complex 1 (mTORC1) signaling. J Biol Chem. 2008;283:30482–92. doi: 10.1074/jbc.M803348200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durán RV, Hall MN. Regulation of TOR by small GTPases. EMBO Rep. 2012;13:121–8. doi: 10.1038/embor.2011.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amson R, Pece S, Lespagnol A, Vyas R, Mazzarol G, Tosoni D, Colaluca I, Viale G, Rodrigues-Ferreira S, Wynendaele J, et al. Reciprocal repression between P53 and TCTP. Nat Med. 2011;18:91–9. doi: 10.1038/nm.2546. [DOI] [PubMed] [Google Scholar]
- 8.MacDonald SM, Bhisutthibhan J, Shapiro TA, Rogerson SJ, Taylor TE, Tembo M, Langdon JM, Meshnick SR. Immune mimicry in malaria: Plasmodium falciparum secretes a functional histamine-releasing factor homolog in vitro and in vivo. Proc Natl Acad Sci U S A. 2001;98:10829–32. doi: 10.1073/pnas.201191498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacDonald SM, Rafnar T, Langdon J, Lichtenstein LM. Molecular identification of an IgE-dependent histamine-releasing factor. Science. 1995;269:688–90. doi: 10.1126/science.7542803. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J, de Toledo SM, Pandey BN, Guo G, Pain D, Li H, Azzam EI. Role of the translationally controlled tumor protein in DNA damage sensing and repair. Proc Natl Acad Sci U S A. 2012;109:E926–33. doi: 10.1073/pnas.1106300109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berkowitz O, Jost R, Pollmann S, Masle J. Characterization of TCTP, the translationally controlled tumor protein, from Arabidopsis thaliana. Plant Cell. 2008;20:3430–47. doi: 10.1105/tpc.108.061010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brioudes F, Thierry AM, Chambrier P, Mollereau B, Bendahmane M. Translationally controlled tumor protein is a conserved mitotic growth integrator in animals and plants. Proc Natl Acad Sci U S A. 2010;107:16384–9. doi: 10.1073/pnas.1007926107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim YM, Han YJ, Hwang OJ, Lee SS, Shin AY, Kim SY, Kim JI. Overexpression of Arabidopsis translationally controlled tumor protein gene AtTCTP enhances drought tolerance with rapid ABA-induced stomatal closure. Mol Cells. 2012;33:617–26. doi: 10.1007/s10059-012-0080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aoki K, Suzui N, Fujimaki S, Dohmae N, Yonekura-Sakakibara K, Fujiwara T, Hayashi H, Yamaya T, Sakakibara H. Destination-selective long-distance movement of phloem proteins. Plant Cell. 2005;17:1801–14. doi: 10.1105/tpc.105.031419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodríguez-Medina C, Atkins CA, Mann AJ, Jordan ME, Smith PM. Macromolecular composition of phloem exudate from white lupin (Lupinus albus L.) BMC Plant Biol. 2011;11:36. doi: 10.1186/1471-2229-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haywood V, Yu TS, Huang NC, Lucas WJ. Phloem long-distance trafficking of GIBBERELLIC ACID-INSENSITIVE RNA regulates leaf development. Plant J. 2005;42:49–68. doi: 10.1111/j.1365-313X.2005.02351.x. [DOI] [PubMed] [Google Scholar]
- 17.Aoki K, Kragler F, Xoconostle-Cazares B, Lucas WJ. A subclass of plant heat shock cognate 70 chaperones carries a motif that facilitates trafficking through plasmodesmata. Proc Natl Acad Sci U S A. 2002;99:16342–7. doi: 10.1073/pnas.252427999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin MK, Lee YJ, Lough TJ, Phinney BS, Lucas WJ. Analysis of the pumpkin phloem proteome provides insights into angiosperm sieve tube function. Mol Cell Proteomics. 2009;8:343–56. doi: 10.1074/mcp.M800420-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Giavalisco P, Kapitza K, Kolasa A, Buhtz A, Kehr J. Towards the proteome of Brassica napus phloem sap. Proteomics. 2006;6:896–909. doi: 10.1002/pmic.200500155. [DOI] [PubMed] [Google Scholar]
- 20.Notaguchi M, Wolf S, Lucas WJ. Phloem-mobile Aux/IAA transcripts target to the root tip and modify root architecture. J Integr Plant Biol. 2012;54:760–72. doi: 10.1111/j.1744-7909.2012.01155.x. [DOI] [PubMed] [Google Scholar]
- 21.Chen H, Ahmad M, Rim Y, Lucas WJ, Kim JY. Evolutionary and molecular analysis of Dof transcription factors identified a conserved motif for intercellular protein trafficking. New Phytol. 2013;198:1250–60. doi: 10.1111/nph.12223. [DOI] [PubMed] [Google Scholar]
- 22.Hinojosa-Moya J, Xoconostle-Cázares B, Lucas WJ, Ruiz-Medrano R. Differential accumulation of a Translationally Controlled Tumor Protein mRNA from Cucurbita maxima (pumpkin) in response to CMV infection. In: Sánchez F, Quinto C, López-Lara y I, Geiger O, eds. Biology of Plant-Microbe Interactions. Vol 5. St. Paul, MN: International Society for Molecular Plant-Microbe Interactions, 2006:242-246 [Google Scholar]
- 23.Ruiz-Medrano R, Hinojosa Moya JJ, Xoconostle-Cázares B, Lucas WJ. Influence of cucumber mosaic virus infection on the mRNA population present in the phloem translocation stream of pumpkin plants. Funct Plant Biol. 2007;34:292–301. doi: 10.1071/FP06300. [DOI] [PubMed] [Google Scholar]
- 24.Thaw P, Baxter NJ, Hounslow AM, Price C, Waltho JP, Craven CJ. Structure of TCTP reveals unexpected relationship with guanine nucleotide-free chaperones. Nat Struct Biol. 2001;8:701–4. doi: 10.1038/90415. [DOI] [PubMed] [Google Scholar]
- 25.Lucas WJ, Groover A, Lichtenberger R, Furuta K, Yadav SR, Helariutta Y, He XQ, Fukuda H, Kang J, Brady SM, et al. The plant vascular system: evolution, development and functions. J Integr Plant Biol. 2013;55:294–388. doi: 10.1111/jipb.12041. [DOI] [PubMed] [Google Scholar]
- 26.Lin MK, Belanger H, Lee YJ, Varkonyi-Gasic E, Taoka K, Miura E, Xoconostle-Cázares B, Gendler K, Jorgensen RA, Phinney B, et al. FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. Plant Cell. 2007;19:1488–506. doi: 10.1105/tpc.107.051920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoo BC, Kragler F, Varkonyi-Gasic E, Haywood V, Archer-Evans S, Lee YM, Lough TJ, Lucas WJ. A systemic small RNA signaling system in plants. Plant Cell. 2004;16:1979–2000. doi: 10.1105/tpc.104.023614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pant BD, Buhtz A, Kehr J, Scheible WR. MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 2008;53:731–8. doi: 10.1111/j.1365-313X.2007.03363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banerjee AK, Chatterjee M, Yu Y, Suh SG, Miller WA, Hannapel DJ. Dynamics of a mobile RNA of potato involved in a long-distance signaling pathway. Plant Cell. 2006;18:3443–57. doi: 10.1105/tpc.106.042473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin A, Adam H, Díaz-Mendoza M, Zurczak M, González-Schain ND, Suárez-López P. Graft-transmissible induction of potato tuberization by the microRNA miR172. Development. 2009;136:2873–81. doi: 10.1242/dev.031658. [DOI] [PubMed] [Google Scholar]
- 31.Navarro C, Abelenda JA, Cruz-Oró E, Cuéllar CA, Tamaki S, Silva J, Shimamoto K, Prat S. Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature. 2011;478:119–22. doi: 10.1038/nature10431. [DOI] [PubMed] [Google Scholar]
- 32.Kovalchuk I, Kovalchuk O, Kalck V, Boyko V, Filkowski J, Heinlein M, Hohn B. Pathogen-induced systemic plant signal triggers DNA rearrangements. Nature. 2003;423:760–2. doi: 10.1038/nature01683. [DOI] [PubMed] [Google Scholar]
- 33.Georgiev MI, Agostini E, Ludwig-Müller J, Xu J. Genetically transformed roots: from plant disease to biotechnological resource. Trends Biotechnol. 2012;30:528–37. doi: 10.1016/j.tibtech.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 34.Xoconostle-Cázares B, Xiang Y, Ruiz-Medrano R, Wang H-L, Monzer J, Yoo B-C, McFarland KC, Franceschi VR, Lucas WJ. Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science. 1999;283:94–8. doi: 10.1126/science.283.5398.94. [DOI] [PubMed] [Google Scholar]
- 35.Logemann J, Schell J, Willmitzer L. Improved method for the isolation of RNA from plant tissues. Anal Biochem. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Ruzin SE. Plant Microtechnique and Microscopy. USA: Oxford University Press Inc, 1999 [Google Scholar]
- 38.Ruiz-Medrano R, Xoconostle-Cázares B, Lucas WJ. Phloem long-distance transport of CmNACP mRNA: implications for supracellular regulation in plants. Development. 1999;126:4405–19. doi: 10.1242/dev.126.20.4405. [DOI] [PubMed] [Google Scholar]
- 39.Camas A, Cárdenas L, Quinto C, Lara M. Expression of different calmodulin genes in bean (Phaseolus vulgaris L.): role of nod factor on calmodulin gene regulation. Mol Plant Microbe Interact. 2002;15:428–36. doi: 10.1094/MPMI.2002.15.5.428. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.