Abstract
Ectomycorrhizal symbiosis results in profound morphological and physiological modifications in both plant and fungus. This in turn is the product of differential gene expression in both co-symbionts, giving rise to specialized cell types capable of performing novel functions. During the precolonization stage, chemical signals from root exudates are sensed by the ectomycorrhizal fungus, and vice versa, which are in principle responsible for the observed change in the symbionts developmental program. Little is known about the molecular mechanisms involved in the signaling and recognition between ectomycorrhizal fungi and their host plants. In the present work, we characterized a novel lactone, termed pinelactone, and identified a gene encoding for a histidine kinase in Pisolithus tictorius, the function of which is proposed to be the perception of the aforementioned metabolites. In this study, the use of closantel, a specific inhibitor of histidine kinase phosphorylation, affected the capacity for fungal colonization in the symbiosis between Pisolithus tinctorius and Pinus greggii, indicating that a 2-component system (TCS) may operate in the early events of plant-fungus interaction. Indeed, the metabolites induced the accumulation of Pisolithus tinctorius mRNA for a putative histidine kinase (termed Pthik1). Of note, Pthik1 was able to partially complement a S. cerevisiae histidine kinase mutant, demonstrating its role in the response to the presence of these metabolites. Our results indicate a role of a TCS pathway in the early stages of ectomycorrhizal symbiosis before colonization. Furthermore, a novel lactone from Pinus greggii root exudates may activate a signal transduction pathway that contributes to the establishment of the ectomycorrhizal symbiosis.
Keywords: Ectomycorrhiza, 2-Component System (TCS), Pinelactone
Introduction
Ectomycorrizal symbiosis results of the interaction between different basidiomycete fungi and roots of diverse plant species, the majority of them gymnosperms.1,2 While the fungus provides mineral nutrients to the plant, this in turn allocates fixed carbon to the fungus. The basidiomycete Pisolithus tinctorius is an almost ubiquitous ectomycorrhizal fungus, able to establish successful symbiosis with a wide range of plant species. Ectomycorrhized plants possess adaptive advantages under stress conditions such as marginal soils, drought, pathogen attack, extreme pH and temperatures, among others; thus its importance.3,4
The establishment of this symbiosis requires an exchange of diffusible chemical signals between the symbionts previous to root colonization. The perception of such signals, likely secondary metabolites, induces changes in gene expression in both plant and fungus; as a result, specialized cell types involved in nutrient exchange are developed.1,2 Thus, the fungus perceives specific compounds exuded from roots in the precolonization stage, although the receptors for such compounds have not been identified to date. Similarly, there is no information on the transduction pathway(s) that eventually activates the symbiotic program in the ectomycorrhizal fungus in response to such signals. In the Laccaria-Pinus symbiosis, a small-secreted protein from Laccaria, MiSSP7, is endocyted by root cells and is an effector for the establishment of the symbiosis. Indeed, this protein is secreted in response to the perception of diffusible signals by the fungus.5 On the other hand, a secreted protein, SP7, from Glomus intraradices suppresses the defense response in plant roots, increasing mycorrhization.6 Much less is known of the receptors for plant-derived signals and of the induced signaling pathway(s) in the fungal symbiont that results in the secretion of these proteins or, in general, of the activation of the symbiotic program.
In a parallel system, the arbuscular endomycorrhizal inducer is strigolactone (SL), which promotes the growth and branching of fungal hypha during the precolonization stage of Glomus intraradices and Gigaspora margarita.7-9 More recently it has been shown that strigolactone is a plant hormone that regulates root and shoot branching, so it has been speculated that this signaling system was hijacked from plants by mycorrhizal fungi.10-12 Additional Glomus intraradices signals that let the establishment of arbuscular mycorrhizal symbioses are a mix of sulphated and non-sulphated simple lipochitooligosaccharides (LCOs).13
In fungus-host interactions, it has been shown that 2-Component Systems (TCS), and hence histidine kinases (HK) are important regulators of pathogenicity in fungal pathogens of animals and plants.14 A functional parallel may be drawn between host recognition by pathogenic and symbiotic (mycorrhizal) fungi, in which a HK binds metabolites from the host, triggering a unique developmental program in the fungus. Consistent with the notion that a TCS is involved in the establishment of this symbiosis, a transcript for a putative HK is induced in the early symbiotic interaction of P. tinctorius with Eucalyptus globulus.15 The accumulation of a HK mRNA in P. tinctorius in response to plant metabolites lends additional support for the involvement of a TCS during early stages of the ectomycorrhizal symbiosis.16 TCS are signal transduction pathways involved in the response to a wide range of environmental cues in prokaryotes, and in fungi, and plants. TCS regulate cell growth, differentiation and development, and response to extracellular cues such as oxidative stress, osmotic shock, and secondary metabolites.17,18 It is also involved in the pathogenesis and virulence of important microbial pathogens (fungal and bacterial). In fungi and plants TCS activate a downstream mitogen-activated protein kinase (MAPK) pathway leading to the activation of several genes.14,17-19 The canonical TCS in prokaryotes usually consist of a membrane-bound (sensor) HK, involved in the perception of the stimulus, and a response regulator, which is phosphorylated by the HK and thus activated, leading to the response. The phosphate from the conserved histidine residue in the transmitter domain is rapidly transferred to an aspartate in the receiver domain of the response regulator.
The aim of this study was to analyze the effect of the root plant metabolites zeatin, acetosyringone, and a novel lactone characterized in the present work, termed pinelactone, on P. tinctorius growth and the identification and characterization of a gene encoding for a histidine kinase in P. tictorius, the function of which is proposed to be the perception of the aforementioned metabolites. The Pthik1 mRNA is accumulated when saprophytic mycelia are grown on media supplemented with these plant metabolites. The role of PtHIK1, and, in general, of HK, and hence TCS, in root colonization by P. tinctorius was determined through the use of closantel, an inhibitor of histidine kinase autophosphorylation and therefore of TCS. Indeed, this compound inhibited mycorrhization almost completely. An expression assay of PtHIK1 in a S. cerevisiae histidine kinase mutant suggests a partial functional complementation.
Results
Identification of lactone and zeatin in P. greggii root exudates
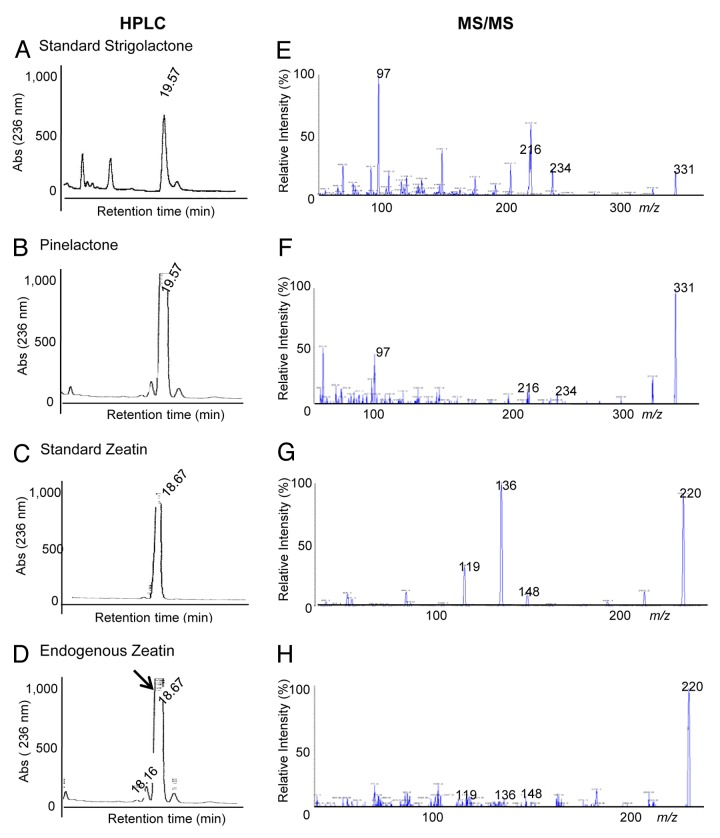
The identification of secondary plant metabolites was performed for P. greggii root exudates, based on the evidence of induced branching effects of lactones (strigolactone) in endomycorrizae,7 as well as the presence of zeatin, the positive effect of which on the growth of different mycorrhizal fungi has been described.22 Analogous molecules present in root exudates of Pinus greggii were identified, with retention times (tR) on HPLC (Fig. 1A, B, C, and D) and mass spectral data similar to lactone and zeatin standards (Fig. 1E, F, G, and H). The natural lactone identified was named pinelactone. The full-scan positive mass spectrum of pinelactone and zeatin (26 ng/ml and 30 ng/ml) are presented in Figure 1. Similar fragmentations were observed comparing the CE mass spectrum from strigolactone and zeatin standards and the mass spectrum of ionization peaks 331 and 220 from the activated charcoal fraction. A molecular precursor ion peak was observed at m/z 331, with characteristic product ion peaks at m/z 97 (a principal moiety for lactones), 234, and 216 (Fig. 1F). For zeatin, the CE showed a precursor ion peak at m/z 220 and characteristic weak product ions at m/z 148, 136, and 119, profiling zeatin (Fig. 1G). Thus, the presence of characteristic features of lactones and zeatin in root exudates allowed furthering testing their biological activity on mycorrhiza development. These data are consistent with previous studies showing the presence of cytokinin in root exudates of pine species.23
Figure 1. HPLC and MS/MS analysis of strigolactone and zeatin and natural homologous pinelactone and zeatin identified from root exudates. Pinus greggii root metabolites exudated to the hydroponic culture were trapped in an activated-charcoal filter as indicated in materials and methods. Fractions were resolved by HPLC (left panel) and selected peaks were then resolved by MS/MS with electrospray-positive scan mode (right panel). Standards and natural exudates are indicated. Note the precursor ion for lactone, at m/z = 331 and the characteristic product ions (97, 216, 234), common in the 2 upper panels. For zeatin (3rd and 4th panel, the precursor ion is at m/z = 220, with the characteristic product ions (119, 136, 148); common for the standard and natural zeatin.
Plant metabolites induce changes in P. tinctorius colonial morphology
The effect of acetosyringone (AS), strigolactone, and zeatin (a cytokinin, also a signaling molecule) on the radial growth of P. tinctorius colonies on solid MNM, was determined. This fungus displays a low growth rate, for which colony diameter was measured every 7 d for 3 weeks. In general, all compounds induced the formation of profuse aerial mycelium, compared with control fungus, grown on MNM. Despite that the colony diameter was similar, fungal cells treated with AS or lactone accumulated a black pigment, likely melanin, characteristic of this species when growing in soil (Fig S1). P. tinctorius actively secretes melanin, which is an indicative of an increase in metabolic rate. This was observed when the tested factors were added to the medium, consistent with previous observations of the effect of zeatin and hypaphorine on P. tinctorius growth.24 In contrast, zeatin did not induce the accumulation of black pigments in the fungus, although P. tinctorius growth was normal compared with the control.
Pthik1 encodes a 2-component hybrid histidine kinase from P. tinctorius
To clone the histidine kinase gene from P. tinctorius, degenerate primers were designed based on conserved regions of the histidine kinase domain containing the autophosphorylated histidine domain. First, P. tinctorius mycelia were treated with the phenolic compound AS, in which a 377-bp product was amplified using P. tinctorius cDNA from an AS-treated samples. The deduced amino acid sequence of the partial histidine kinase gene showed high similarity to a histidine kinase from Lentinula edodes, which is involved in the stress response and fruiting body development, a hypothetical protein from Coprinopsis cinerea, as well as with a TC-like hybrid sensor histidine kinase I from Cryptococcus neoformans. To obtain the full-length gene, GW and RACE were employed. An open reading frame of 2508 bases encoding 836 amino acids as well as a 3′ UTR region of 213 bp were identified (GenBank accession numbers: EU906855.1 GI: 219660833). The genomic clone includes 4 putative introns located in the open reading frame (Fig. S2). The protein contains a histidine kinase domain (HKD) and a C-terminal response regulator domain (RRD), which is characteristic of eukaryotic hybrid-type histidine kinases. Pthik1 contains the H box histidine residue that is probably autophosphorylated, the aspartic acid residue of the receiver D box, and a set of ATP-binding motifs. Four HAMP signatures are present in the N-terminus (Fig. S2). HAMP domains are ubiquitous in bacterial and archaeal TCS. Recently 2 distinct conformational states of the HAMP domain have been described, suggesting additional modulation of this signal transduction pathway through this domain (Fig. S2).25 While the deduced amino acid sequence is shorter than related HKs, this sequence is by no means unique in its length. Indeed, several other HKs, including one from Coprinopsis cinerea, show a similar size (~800–900 aa).
Pthik1 transcript accumulates in response to plant metabolites
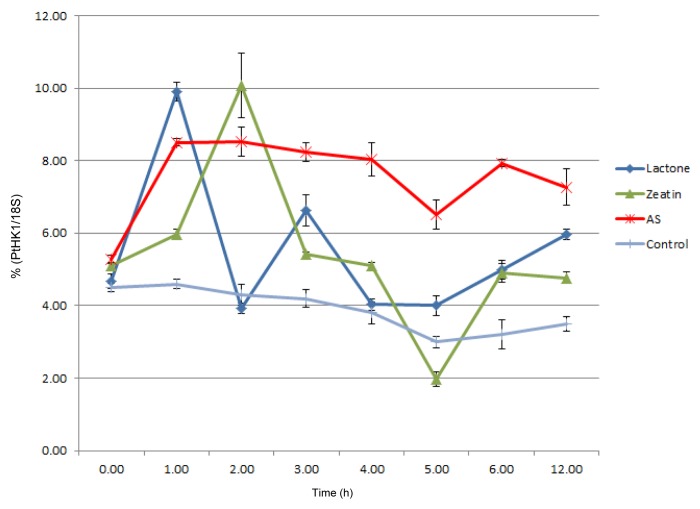
The accumulation of the Pthik1 transcript in the fungus in response to plant metabolites was estimated by quantitative RT-PCR. Total RNA from mycelium of P. tinctorius grown on MNM media supplemented either with AS, pinelactone, or zeatin was employed in this assay. A complex response to these compounds was observed. Despite the fungal low growth rate, the early accumulation of Pthik1 mRNA was registered within the first 4 hours of incubation with the metabolites as shown in Figure 2. In all the treatments, a basal mRNA accumulation was observed at time zero, suggesting the gene is expressed at low levels. However, the addition of lactone, zeatin or AS to the fungus increased Pthik1 mRNA accumulation. Lactone and AS induced its accumulation within one hour while lactone induced 2 accumulation peaks. Zeatin treatment induced the maximum accumulation at 2 hours, while AS caused a sustained accumulation of the transcript. Seven days after the different treatments, no induction of the Pthik1 transcript was observed (data not shown). This accumulation pattern of Pthik1 mRNA was consistently observed between triplicates.
Figure 2.Pthik1 mRNA accumulates when fungal cells are incubated with plant metabolites. Real-time Quantitative RT-PCR was assayed using total RNA extracted from previously incubated fungal cells with the indicated lactone, zeatin or acetosyringone (AS). Specific primers used for Pthik1 and 18S rRNA are indicated in material and methods. Pthik1 expression was normalized with the constitutively expressed gene 18S.
Complementation of the TM182 yeast mutant with the Pthik1 gene
Given the possible role of zeatin and pinelactone as plant signals perceived by the fungus in the establishment of ectomycorrhizal symbiosis, we tested the putative role of PtHIK1 in the recognition of such signals. A complementation assay in a Saccharomyces cerevisiae strain deficient in the hybrid-sensor kinase SLN1, TM182,26 was performed to investigate the function of PtHIK1 in a heterologous system. For this purpose, the Pthik1 open reading frame was cloned in an expression vector under the direction of the GAL1 promoter, transformed in yeast and induced with galactose in the presence of the endogenous metabolite pinelactone (Fig. 3). A partial complementation was observed when the recombinant TM182 strain was incubated in the presence of galactose and the purified pinelactone. These results suggest the restoration of the endogenous signal transduction pathway involved in the response to osmotic stress in the TM182 strain transformed with Pthik1. Regarding complementation assays with AS and zeatin, no complementation was observed at the tested concentration.
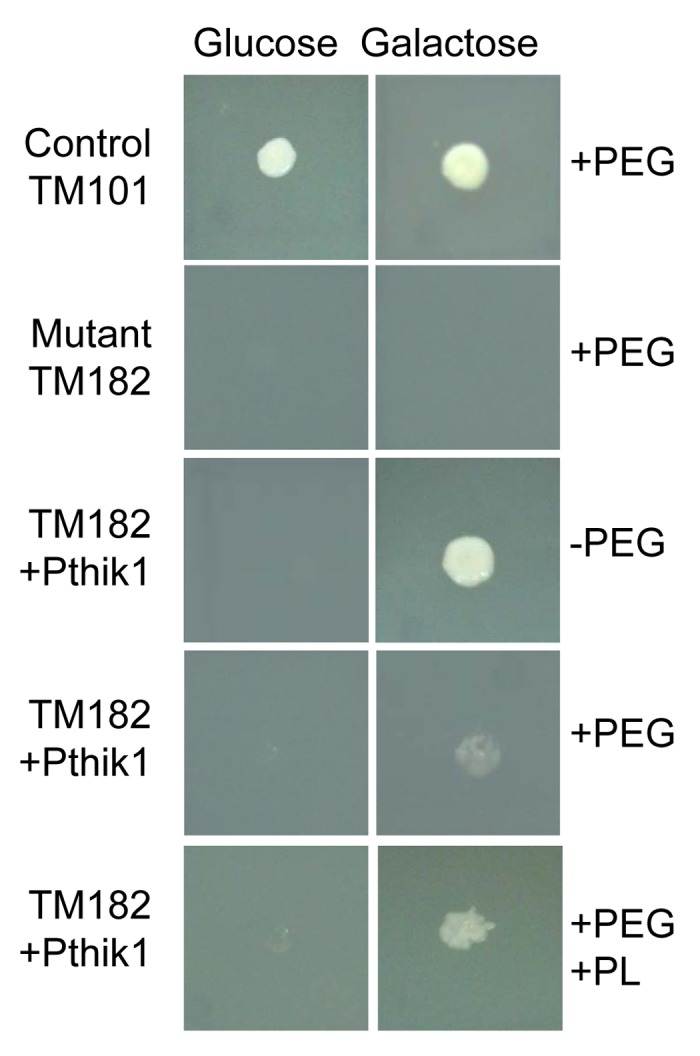
Figure 3.Pthik1 gene partially complements TM182 HKyeast mutant. Strain TM101 is wild type yeast, TM182 is histidine kinase deficient strain, transformed with Ptkih1 gene. PEG, polyethylenglycol, PL, Pinelactone.
Effect of the histidine kinase-inhibitor closantel on fungal colonization of roots
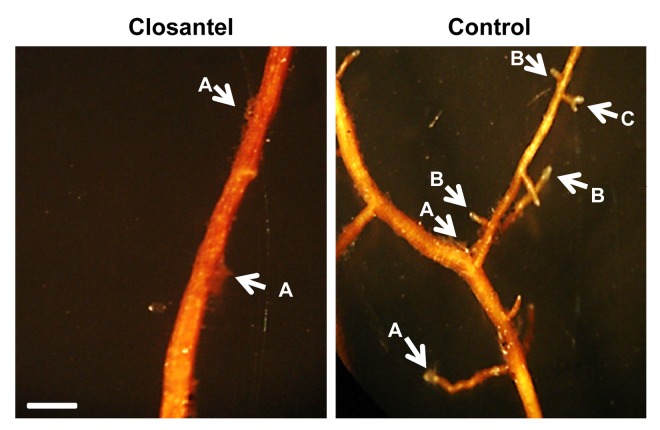
Pinus greggii plantlets were inoculated with P. tinctorius mycelia previously incubated with sublethal concentration of closantel, a competitive inhibitor of the histidine kinase activity, broadly used for its antimycotic and antihelmintic activity in animals. Indeed, this compound inhibits in vitro the autophosphorylation of HK, although it also binds and inhibits helmintic chitinases.27,28 Root branching was quantified as indicative of mycorrhization (Table 1). In roots inoculated with closantel-treated P. tinctorius, branching was dramatically reduced to 0.4 secondary branches/cm (sb/cm); in contrast, 2 sb/cm were obtained in control assays. X2 test was calculated at < 0.05, indicating the statistical difference is significant. To discard that the inability of colonization was due to toxicity in the fungus, a vital stain (Live/Dead, Molecular Probes Invitrogen) was applied to assess cell viability. In the same way, root plantlets that interacted with closantel-treated fungi did not show morphological differences when compared with controls. Figure 4 shows the lack of branching capacity in roots incubated with closantel-treated mycelia. Histological assays on closantel-treated roots confirmed the absence of mantle and Hartig net, features of a successful mycorrhization (data not shown). Our results indicate that mycelia treated with closantel were metabolically active but presumably unable to respond to the inductive plant signals in the precolonization stage. As a result, the fungus would not produce the chemical signals that trigger root branching, which in turn leads to the establishment of ectomycorrhizal symbiosis.
Table 1. Effect of the histidine kinase-inhibitor closantel on the establishment of symbiosis.
| Sample | % Colonization ± SD |
|---|---|
| Control | 2.2 ± 1.47 |
| Closantel-treated roots | 0.4 ± 0.08 |
SD was calculated from 3 independent experiments. Thirty plants were used in each experiment.
Figure 4. Closantel-treated P. tinctorius mycelia are unable to colonize plant roots. Morphology of P. greggii roots inoculated with P. tinctorius, which were previously incubated with the histidine kinase inhibitor closantel; where no ramifications were observed. Arrow A: P. tinctorius mycelium growing on the root surface. Arrow B: Root ramifications in control roots. Arrow C: P. greggii roots with the typical bifurcations structure in ectomycorrhized plants.
A 2-component system groups fungi in symbiotic, saprobe or pathogenic clades
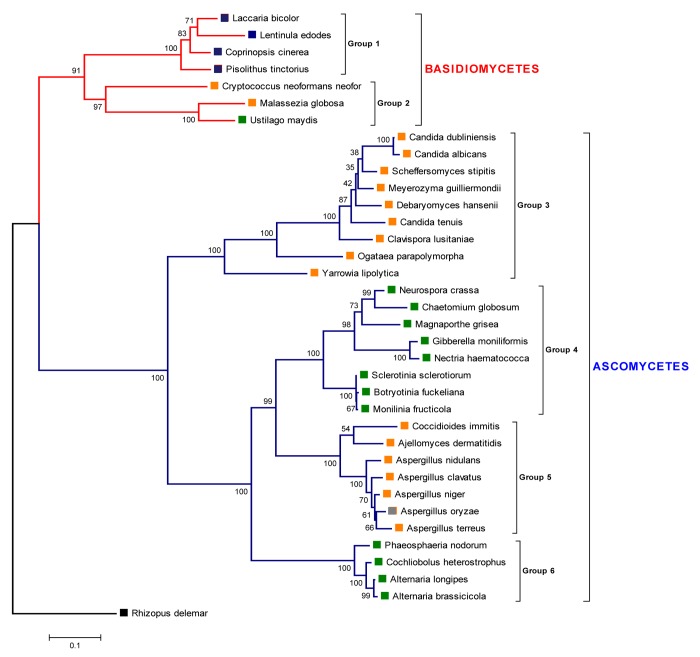
The deduced Pthik 2-C amino acid sequence was employed to compare it with related sequences from the extant databases. The phylogenetic tree of the histidine kinase family in true fungi (Fig. 5) showed a defined division between the taxa basidiomycetes and ascomycetes. Mention must be made to the fact that no homologous sequences were found in the extant databases corresponding to histidine kinases from ectomycorrhizae present in ascomycetes. Thus, it is not possible at this time to assess the role of a TCS in the establishment of symbiosis in ascomycetes. In contrast, despite a limited number of available sequences of histidine kinases from ectomycorrhizal basidiomycetes, a calculated phylogeny was obtained, consistent with both the inferred lineage of this group and the nature of the host and its interaction (i.e., saprobe, parasitic or symbiotic). An early branching gave rise to 2 clades, the first one containing saprobes and ectomycorrhizal fungi and the second containing both plant and animal pathogens.
Figure 5. Phylogenetic tree of the histidine kinase family in true fungi. The evolutionary history was inferred using the Minimum Evolution method. Protein sequences were aligned using ClustalX. Evolutionary distances were computed using the Dayhoff matrix based method. The Neighbor-joining algorithm was used to generate the initial tree. All ambiguous positions were removed for each sequence pair. The zygomycete Rhizopus delemar was used as outgroup. Bootstrap values are shown (1000 replicates). The evolutionary analyses were conducted in MEGA5. Red lines show basidiomycetes, while blue lines show the radiation of ascomycetes. The blue square labels show ectomycorrhyzal symbionts. Green and orange squares show plant and animal pathogens, respectively. Grey squares are considered as non-pathogenic. Sequence accession numbers are listed in Table 2.
A similar distribution is shown in ascomycetes: plant and animal pathogens are clustered in independent clades, likely indicating that the grouping on the basis of 2-C homology reflects common mechanisms of interaction with the respective hosts within each clade. Within basidiomycetes, the compact group 1 (Fig. 5) comprises the ectomycorrhyzal fungi Laccaria bicolor, Lentinula edodes, Coprinopsis cinerea, and Pisolithus tinctorius, widely distributed in symbiotic association to diverse plant taxa. The group 2 includes opportunistic pathogenic fungi such as Cryptococcus neoformans and Malassezia globosa both animal and human pathogens and Ustilago maydis, a biotroph maize pathogen. The ascomycete taxa formed 4 large clades, including in group 3 animal pathogens (Candida dubliniensis, C. albicans, Meyerozyma (Candida) guilliermondii, C. tenuis, and commensals in insects (Clavispora lusitaniae, Scheffersomyces (Pichia) stipites, Ogataea parapolymorpha). Although considered as non-pathogenic, some of the following taxa: Neurospora crassa, Yarrowia lipolytica, Debaryomyces hansenii, and Chaetomium globosum have recently proven to be weak pathogens or to possess at least some genes involved in pathogenesis.29 Candida lipolytica, also known as Yarrowia lipolytica, is a weakly pathogenic fungus known to be an opportunistic pathogen found recurrently in hospitalized patients with catheter related fungemia.29 Candida famata also known as Debaryomyces hansenii has previously been described in human infections in patients with peritoneal dialysis.30 Group 4 includes Neurospora crassa which harbors genes similar to those require for plant pathogenesis, including all the signal transduction genes identified in ascomycete pathogenesis.31 In addition plant-associated fungi are also grouped here, such as Chaetomium globosum that induces root necrosis in carrot cell cultures and in hydroponically grown barley.32,33 C. globosum has been described epiphytically colonizing the root epidermis and even a few hyphae are able to penetrate and grow endophytically in axenic growth. Although these results do not reflect the natural growth conditions and do not intend to prove that C. globosum is a plant pathogen, they show that C. globosum holds genes involved in plant invasive behavior.33 Important plant pathogens such as Magnaporthe grisea, Giberella moniliformis, Nectria hematococca, Sclerotinia sclerotiorum, Botryotinia fuckeliana and Monilia fructicola belongs to this clade. The group 5 comprises important animal pathogens (Coccidiodes immitis, Ajellomyces dermatitidis, Aspergillus nidulans, A. clavatus, A. niger, and A terreus) with the exception of A. oryzae, a saprobe fungus. While the last clade identified as group 6, comprises also the plant pathogens Phaeosphaeria nodorum, Cochliobolus heterostrophus, Alternaria longipes, and A. brassicicola, all producing important losses in agriculture worldwide.
Discussion
A number of natural metabolites exuded by plant roots have been described to act as signaling molecules in the ectomycorrhizal symbiosis. Among these metabolites, the cytokinin zeatin promotes the establishment of the arbuscular endomycorrhizal symbiosis (AM),34 while strigolactone is a plant signaling molecule that negatively regulates root and shoot branching and is required for parasitic plant colonization,10-12 but is also involved in the induction of hyphal branching in AM (and is therefore the first step in the establishment of endomycorrhiza.7 On the other hand, the synthetic phenolic compound AS24,35 binds to receptor histidine kinases found in some TCS in prokaryotes such as Agrobacterium tumefaciens;36 previous results in our group suggested that this compound positively regulated the germination of spores of P. tinctorius. Indeed, we identified a novel lactone, termed pinelactone in the present work, and zeatin, in Pinus greggii root exudates. Along with AS, we tested their capacity to induce molecular and physiological changes in Pisolithus tinctorius. The presence of lactones in root exudates from pines and their role in ectomycorrhizal symbiosis signaling have not been reported previously.
Ectomycorrhizal fungi perceive chemical signals from its host in the presymbiotic stage. This leads to morphological changes making feasible a symbiotic relationship with the compatible plant root.37 Since an mRNA for a HK is induced in extraradical mycelium of P. tinctorius,16 we hypothesized that a TCS could be involved in the transduction of host signals. This HK, which we termed Pthik1, was further characterized. Its expression is likely controlled at both transcriptional and posttranslational level, as can be inferred by the accumulation of the transcript when mycelia were exposed to the aforementioned compounds. We observed that strigolactone and AS induced phenotypic changes in P. tinctorius, such as melanin secretion, although the overall diameter of the colonies was not affected. Since these compounds alter processes that potentially occur early during mycorrhization, we tested their effect on the accumulation of the Pthik1 transcript; it is possible that this HK that may be the receptor of plant signals that start the progression of this symbiosis.
The Pthik1 transcript was detected within one hour when the in vitro propagated mycelium was incubated with pinelactone, 2 hours when treated with zeatin, and a sustained accumulation after 5 hours with AS treatment. This early and temporary induction by plant metabolites is consistent with a role for Pthik1 in the early steps of the symbiosis. In fungi, TCS also mediate responses to osmotic stress; however, this transcript did not accumulate when tested on hypertonic media, suggesting that other HKs may be involved in the response to such stimulus. Indeed, a genomic approach is necessary to identify additional HK genes, which could be conceivably involved in the response to other stimuli.
Pretreatment of in vitro-grown P. tinctorius with closantel, an inhibitor of histidine kinases autophosphorylation, blocked the colonization of plant roots by P. tinctorius. Closantel is a benzamidate extensively used to for inhibition of HK activity in fungi.27,38,39 These results suggest that HK autophosphorylation, and therefore a TCS, plays an essential role in the early colonization between P. tinctorius and P. greggii roots. Non-mycorrhized plants displayed a similar phenotype to controls, indicating that closantel did not have a deleterious effect on neither the fungus nor the plant at the tested concentration. A similar result was obtained with the human pathogen Anaplasma phagocytophilum; here, closantel treatment blocked fungal infection completely through inhibition of TCS.27 However, in higher doses closantel can be toxic; for this reason 2 controls were included to ensure that the cells were viable, using a vital stain, and testing whether these were metabolically active, detecting respiratory activity. Taken together, both assays suggest that the concentration of closantel used is sublethal and does not cause respiratory damage in P. tinctorius. It must be mentioned that closantel can also inhibit filarial chitinases, so an effect on plant or fungal chitinases cannot be discarded. However, the effect seems to be specific for this type of enzymes. Additionally, there is no information regarding the inhibition of non-animal chitinases.28
Interestingly, Pthik1 expression in an SLN1 yeast strain partially restored its growth when pinelactone was added to the medium. It is not surprising that the complementation was comparable to positive controls, given that the interaction of the expressed protein with its downstream targets occurred in a heterologous system, Indeed, the presence of determinants of specificity have been described in TCS signal transduction40 that could explain this partial complementation. This reinforces the notion that PtHIK1 may actually interact with this compound, and that it may mediate the earliest steps of P. greggii root colonization by P. tinctorius.
There are several HK sequences in fungi that interact with plants either as symbionts or as pathogens; interestingly, in basidiomycetes, Pthik1 shows a high degree of similarity with HK genes from other ectomycorrhizal fungi, suggesting a similar role in the ectomycorrhizal symbiosis. On a speculative note and according to the calculated phylogeny based on HK amino acid sequences, the 2-component system could be mediating fungus-host interaction, allowing plant pathogens to radiate from 2 unknown ancestors, while animal pathogens diversified from 2 different yet unknown nodes. In contrast, symbiotic fungi, based on the analysis of the extant sequences, likely emerged from one common ancestor. As an outgroup, HK amino acid sequence of the saprobe Rhizopus delemar was employed, which was distantly located in an early branching clade, thus confirming the hypothesis of the 2-C fungal evolution in association to diverse hosts.
The resulting phylogeny is also suggestive of an early divergence of pathogenic and symbiotic fungi, occurring after the radiation of both basidiomycete and ascomycete taxa.
Experimental evidence on TCS determining host-fungus interaction in saprobes, symbiotic or parasitic ecological relationships will help to understand these complex interactions in nature. If TCS mediate the establishment of such interactions, this could be proposed as a key target in order to control diseases produced by pathogenic fungi in both plants and animals, including the human.
In the present study, plant metabolites obtained from P. greggii root exudates were characterized. The accumulation of a HK mRNA in P. tinctorius in response to these compounds, the complementation of an HK-deficient strain of yeast by expression of this gene, and the almost complete inhibition of mycorrhization by closantel, an inhibitor of HK autophosphorylation, supports the notion that a TCS operates in the early stages of the ectomycorrhizal symbiosis. Additional work will help to elucidate in a more precise way the role of Pthik in the early ectomycorrhizal symbiosis.
Materials and Methods
Strains and culture conditions
A P. tinctorius strain isolated from Querétaro, Central México, was used in this study.41,42 This fungus was propagated using MNM medium43 by placing a small piece of agar-containing mycelia onto the center of the petri dish containing the medium. The MNM medium was supplemented with one of the following compounds: 200 μM acetosyringone (AS) (Sigma-Aldrich, St Louis MO), 10 μM zeatin (Sigma-Aldrich), 10 μM pinelactone or 3.5 pM 5-deoxy-strigol (strigolactone) per petri dish (kindly provided by Prof. K. Akiyama, Osaka University, Japan). Colony growth was measured as the diameter (in mm) the fungal cells reached every week up to 5 wk. Escherichia coli strain DH5α (Invitrogen, Carlsbad CA) was used for plasmid propagation and was grown in TB liquid medium.44 Saccharomyces cerevisiae TM182 sln1 (MATα leu2 ura3 his3 sln1::hisG) containing the plasmid pSSP25 (PGAL1- PTP2 URA3 ADE3) and TM101 (MAT leu2 ura3 his3) harboring pSSP25 were grown in YNB medium supplemented with dropout amino acid mixture (DO), leucine and galactose or glucose. S. cerevisiae was grown at 28 °C for 5 d.
Cloning and sequencing of Pthik1 gene
Genomic DNA of P. tinctorius was extracted from freeze-dried mycelia using the DNeasy plant total DNA isolation kit (Qiagen, Santa Clarita CA). Fungal HK were aligned and the following degenerate primers were designed based on conserved amino acid regions. HKF1 forward 5′-CAYGARATNM GNACNCCNAT NAAYGGNAT-3′ and HKR1 reverse 5′-GTRAAYTTNA CNGCRTTNCC NACNARRTT-3′. PCR was performed with ExTaq polymerase following the recommendations of the supplier (Takara BIO Inc. Japan), using a Mastercycler personal thermal cycler (Eppendorf, Hamburg, Germany) with the following settings: first cycle at 94 °C for 5 min, followed by 30 cycles at 94 °C, 30 s; 58 °C, 30 s; 72 °C, 1 min, and a final extension step at 72 °C for 4 min. The PCR product (of approximately 370 bp) was gel-purified and cloned into the pCRII TOPO vector (Invitrogen, Carlsbad, CA). DNA sequence analysis of the cloned fragment showed that the fragment encoded a histidine kinase domain. The gene was obtained by Genome Walker (GW) and PCR-RACE techniques. For GW, genomic DNA of P. tinctorius was digested with one of the following restriction enzymes: EcoRV, SstI or RsaI; the GW libraries were ligated to adaptors and the primary and secondary PCRs were performed as indicated by the supplier (Clontech Biosciences, Mountain View CA). For the RACE procedure, total RNA was isolated from P. tinctorius grown for 2 weeks, using the Qiagen RNeasy plant kit. The Pthik1 cDNA was amplified by RACE.45 The 5′ and 3′ cDNA ends for P. tinctorius Pthik1 were amplified by PCR using homologous primers designed from the conserved, previously amplified fragment.
Identification of lactone and zeatin in root exudates from Pinus greggii
The identification of lactone and zeatin from P. greggii root exudates was performed as described with modifications.7 Seeds of P. greggii were disinfected and germinated as described above. In sterile conditions, 750 germinated seeds were transferred to hydroponic conditions in a sterile acrylic vessel with an adapted pump containing an activated-charcoal cartridge. Sterile modified Hoagland nutrient solution without phosphorus (per distilled water liter: Ca(NO3)2 4H2O 656.4 mg, KNO3 606.6 mg, MgSO4 7H20 240.8 mg, NH4H2PO4 110 mg, H3Bo3 2.86 mg, MnCl 4H2O 1.81 mg, ZnSO4 7H20 0.22 mg, CuSO4 5H2O 0.08 mg, H2MoO4 0.02 mg, iron 5.0 mg) was used for the hydroponic P. greggii culture. NH4H2PO4 was omitted to improve root exudates signals involved in the ectomycorrhization process. Approximately 14L of the hydroponic culture were collected; pH adjusted to 2.0 and filtrated across membranes 0.45 µm pore size (Millipore, Temecula, CA) for the extraction process with ethyl acetate (3:1 v/v hydroponic solution-ethyl acetate). On the other hand, metabolites adsorbed to the activated charcoal were eluted with acetone (150 ml), and then extracted with ethyl acetate. The extracts (hydroponic solution and acetone) were partitioned separately with 1 volume of K2HPO4 0.2 M pH 9, both ethyl acetate phases were dried over anhydrous Na2SO4 and concentrated in vacuo to obtain 2 neutral fractions. The lactone and zeatin purification was performed by Solid Phase Extraction (SPE) using Superclean LC-SI SPE columns (6 ml, 1g silica gel) (Supelco, Bellafonte, PA) and eluted stepwise with solvents of increasing polarity from n-hexane to ethyl acetate, with 10% increments. The fractions obtained were collected and concentrated in vacuum conditions. Hydroponic solution fraction as well as acetone fraction were eluted with 40% ethyl acetate (where strigolactone elutes) and then analyzed by HPLC in a LDC Analytical constaMetric 3500, Solvent delivery system (Midland, ON, Canada) using an ODS Hypersil (C18) column (100 x 2.1 mm, 5 µm, Thermo Fisher Scientific Inc., Waltham, MA, USA) at 236 nm absorbance. The mobile phase used was 80% acetonitrile in water at 0.5 ml/min flow rate. According to the retention time (tR) of standards (synthetic (± ) 5-deoxy-strigol) and zeatin in acetronitrile:water (80:10), 2 peaks were collected from activated charcoal fraction at different tR; for pinelactone at 19.41 min, and an adjacent peak for zeatin at 18.16 min. The 2 peaks were collected from both, hydroponic solution fraction and activated charcoal fraction. The water and the acetonitrile were evaporated from both fractions by vacuo and then the small pellets obtained were suspended in acetonitrile (500 µl) for further Mass Spectrometry analysis.
Mass Spectrometry
Mass Spectrometry was performed on an Applied Biosystems MDS SCIEX, 3200 Q TRAP LC/MS/MS System (Applied Biosystems, Foster City, CA) equipped with an electrospray ionization source in the positive ion mode. Air was used as drying and nebulizer gas; 15 psi, curtain gas; 100 °C, drying temperature; 5300 V capillary voltage and 150 °C, interface temperature. MS/MS assays were performed using nitrogen as collision gas. Fragmentation was performed by collision energy (CE) of 15V for strigolactone standard and 20V for trans-zeatin standard. Declustering potential was 60V for strigolactone and 55V for zeatin. Data acquisition and analysis were performed with Analyst v1.4.2 software.
The full-scan positive mass spectrum of strigolactone and zeatin was obtained (26 ng/ml for lactone and 30 ng/ml for zeatin in 80% acetonotrile in water, delivered with an infusion pump at 1 µl/min). For the analysis of strigolactone and zeatin with Multiple Reaction Monitoring (MRM), transition m/z 331 > 234 and transition 331 > 97 were selected for strigolactone and transition m/z 220 > 148 and transition 220 > 136 for zeatin. The optimal Collision Energy (CE) was different by each transition, for transition m/z 331 > 234 the CE was 15 V, the CE for transition 331 > 97 CE was 30 V. For zeatin the transition m/z 220 > 148 the CE was 20 and for transition 220 > 136 the CE was 25 V.
Detection of the Pthik1 transcript by real-time RT-PCR
RQ1 RNase-free DNase (Promega, Madison WI) treated-RNA, obtained from P. tinctorius grown on MNM, was reverse-transcribed (RT) in a 20 µl reaction containing SuperScript III (Invitrogen) with 10 μM oligo(dT) 5′- (GA)10CTAGTCTCGA GT(T)18-3′ following the manufacturer’s instructions. Control reactions, without the addition of reverse transcriptase, were included in every experiment, in order to detect amplification from contaminating genomic DNA. 100 ng of cDNA was then employed to amplify Pthik1 using the primers forward RT-HK5 5′-AGCTTGTTAC TCGTGCACTC CTTGGC-3′ and reverse RT-HK3 5′-GTGAGTGAGG TCTAGGTTGT TCTGCGATG-3′. As control, 18S rDNA was amplified with the primers forward 18S5 5′-ATCTTTCCCT CACGGGACTT GTTC-3′ and reverse 18S3 5′-GGTAGGAACA CCCGCTGAAC TTA-3′, according to the 18S rDNA sequence of P. tinctorius (AY739178). A One-Step RT-PCR commercial system was employed and labeling with Syber-Green (Invitrogen), utilizing the primers: GFP 5′ CATGGCAAGT AAAGGAGAAG AACTTTT 3′; GFP3′ 5′ CTTCATATGA TCTGGGTATC TTG 3′. Rotorgene RG-3000 from Corbett Research was employed in this quantification. C(t) values were obtained by triplicate and average employed to calculate mass, related to standard curves. A ratio between Pthik1 and 18S Ct values was calculated to graph the relative Pthik1 mRNA accumulation.
Cloning of Pthik1 in yeast expression vector and complementation assay
For complementation of S. cerevisiae mutant with P. tinctorius Pthik1, the open reading frame (ORF, 2508 bp) was amplified by RT-PCR using SuperScript III (Invitrogen) and TaKaRa Taq polymerase (TaKaRa, Japan) with primers ORF5′ 5′-TCTAGAATGA CAGTGAACTC CATGGTCTCC C-3′ and ORF3′ 5′-CTCGAGCTAC AAATCGGCAG GCACGACGTG C-3′ containing the restriction sites XbaI-XhoI. cDNA was synthesized and then cloned into the pCR2.1-TOPO vector, and sequenced. The ORF was then subcloned into p4Pthik1 (PGAL1-Pthik1 URA3 ADE3, under the control of GAL1 promoter and CYC1 terminator.46 The resulting recombinant plasmid p415-HK was transformed to S. cerevisiae TM182 sln1 (MATα leu2 ura3 his3 sln1::hisG pSSP25) by electroporation and recovered in YNB medium containing glucose or galactose added without leucine. As a control, S. cerevisiae TM101 + pSSP25 [MATα leu2 ura3 his3 pSSP25] (pSSP25 [PGAL1-PTP2 URA3 ADE3]) was employed.46 S. cerevisiae TM102 expressing Pthik1 gene was grown in selective medium and Pthik1 expression was induced with galactose in minimum medium supplemented with glucose, galactose and/or pinelactone. To provide osmotic stress, plates were supplemented with 2M PEG 8000.
Root colonization with ectomycorrhizal fungus P. tinctorius
Seeds of P. greggii collected from Sierra Gorda, Querétaro México were disinfected with sodium hypochlorite (5% v/v) for 30 min and rinsed with sterile distilled water. The seeds were aseptically placed over nutrient agar contained in petri dishes and maintained in the dark at room temperature until the seeds germinated. The seedlings obtained were grown in an autoclaved vermiculite: peat moss: nutrient solution (840:60:600 ml) substrate.45 The tubes were maintained in a controlled environment chamber (Conviron, Manitoba, Canada) programmed with the following conditions: 16 h light period at 27 °C and 18 °C during the dark period. Two weeks after seeding, Pinus greggii plantlets were inoculated with P. tinctorius mycelia grown in MNM liquid media with or without 200 μM closantel (Sigma-Aldrich), a histidine kinase inhibitor. The fungus was grown under the presence of the closantel one week before being in contact with P. greggii plantlets. Mycelium was washed upon harvesting, before inoculating the plantlets with it. Fungal colonization was quantified as the number of first- and second-order lateral roots present in each primary root from colonized and non-colonized roots. X2 statistical tool was employed to calculate significance at < 0.05. Cross-sections of fresh tissue, excised from colonized roots, were visualized to confirm the presence of fungal sheath.
Bioinformatic analyses for HK phylogeny
Deduced amino acid sequences from fungal HK system were retrieved from the GenBank, using BLAST and P. tinctorius HK as bait (Table 2). .Amino acid sequences were then aligned using CLUSTALX2,46 and manually edited with SEAVIEW4 software.47 Maximum-likelihood and Neighbor-Joining phylogenies were calculated form the alignments with MEGA5.48 The substitution matrix P-distance49 was employed for the Neighbor-joining and Dayhoff algorithm for Maximum-likelihood reconstruction.50 Pairwise deletion Gaps/missing data treatment was chosen for Neighbor-joining method and partial deletion with a site coverage cut off of 95% for the Maximum-likelihood method. Replicates consisting in 1000 bootstraps were used for the statistical support of the phylogenetic trees.
Table 2. Histidine kinase sequences used to construct the phylogenetic tree.
| Basidiomycetes: Pisolithus tinctorius, ACL30962.1; Coprinopsis cinerea okayama, EAU80294.2; Laccaria bicolor, EDR03396.1; Lentinula edodes, ABP65341.1; Cryptococcus neoformans var neoformans BAF47077.1; Ustilago maydis 521, XP_758886.1; Malassezia globosa, XP_001730099.1. |
| Ascomycetes: Ogataea parapolymorpha, EFW96014.1; Candida tenius, EGV65371.1; Candida dubliniensis, XP_002421462.1; Yarrowia lipolytica, XP_503487.1; Botryotinia fuckeliana, AAL37947.1; Sclerotinia sclerotiorum, XP_001586119.1; Scheffersomyces stipites, XP_001385881.2; Meyerozymaguilliermondii, XP_001486691.1; Nectria hematococca, AAD09491.1, Monilinia fructicola, ABF60145.1; Candida albicans, AAC23929.1; Debaryomyces hansenii, XP_462511.2; Clavispora lusitaniae, ABO84860.1; Neurospora crassa, XP_964471.1; Chaetomium globosum, EAQ93286.1; Gibberellamoniliformis, AAR30126.1; Magnaporthe grisea BAB40947.1; Coccidioides immitis RS, XP_001245071.1; Aspergillus clavatus, EAW11400.1; Alternaria longipes, ACN62989.1; Alternaria brassicicola, AAU10313.1; Aspergillus niger, XP_001391885.2; Aspergillus nidulans, CBF77424.1; Cochliobolus heterostrophus, BAC78679.1; Phaeosphaeria nodorum SN15, XP_001801869.1; Ajellomyces dermatitidis, ATCC 18188, EGE84246.1; Aspergillus oryzae, RIB40, XP_001823967.2; Aspergillus terreus NIH2624, EAU29725. |
| Zygomycete: Rhizopus delemar RA99–880, EIE86498.1 |
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank to the members of the lab for helpful discussions, to Dr. Kohki Akiyama from Osaka University for provided us with strigolactone, to Dr. Cecilia Silva-Sánchez and Elvira Ríos-Leal from Centro de Investigación y Estudios Avanzados from Mexico DF for the technical assistance in the HPLC and MS/MS assays. Aseneth Herrera-Martinez was a CONACyT doctoral fellow. The present work was supported by Consejo Nacional de Ciencia y Tecnología, México (Grants no. 19885, 0192242 (BXC) and 156162 (RRM)).
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/psb/article/28604
References
- 1.Wiemken V, Boller T. Ectomycorrhiza: gene expression, metabolism and the wood-wide web. Curr Opin Plant Biol. 2002;5:355–61. doi: 10.1016/S1369-5266(02)00269-8. [DOI] [PubMed] [Google Scholar]
- 2.Nehls U. Mastering ectomycorrhizal symbiosis: the impact of carbohydrates. J Exp Bot. 2008;59:1097–108. doi: 10.1093/jxb/erm334. [DOI] [PubMed] [Google Scholar]
- 3.Duchesne LC, Peterson RL, Ellis BE. The time course of disease suppression and antibiosis by the ectomycorrhizal fungus Paxillus involutus. New Phytol. 1989;111:693–8. doi: 10.1111/j.1469-8137.1989.tb02364.x. [DOI] [PubMed] [Google Scholar]
- 4.Smith SE, Read DJ. 1997. Mycorrhizal Symbiosis, 2nd ed. London UK: Academic Press, 175-203. [Google Scholar]
- 5.Plett JM, Kemppainen M, Kale SD, Kohler A, Legué V, Brun A, Tyler BM, Pardo AG, Martin F. A secreted effector protein of Laccaria bicolor is required for symbiosis development. Curr Biol. 2011;21:1197–203. doi: 10.1016/j.cub.2011.05.033. [DOI] [PubMed] [Google Scholar]
- 6.Kloppholz S, Kuhn H, Requena N. A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr Biol. 2011;21:1204–9. doi: 10.1016/j.cub.2011.06.044. [DOI] [PubMed] [Google Scholar]
- 7.Akiyama K, Matsuzaki K, Hayashi H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature. 2005;435:824–7. doi: 10.1038/nature03608. [DOI] [PubMed] [Google Scholar]
- 8.Akiyama K, Hayashi H. Strigolactones: chemical signals for fungal symbionts and parasitic weeds in plant roots. Ann Bot. 2006;97:925–31. doi: 10.1093/aob/mcl063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Besserer A, Puech-Pagès V, Kiefer P, Gomez-Roldan V, Jauneau A, Roy S, Portais J-C, Roux C, Bécard G, Séjalon-Delmas N. Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol. 2006;4:e226. doi: 10.1371/journal.pbio.0040226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng X, Ruyter-Spira C, Bouwmeester H. The interaction between strigolactones and other plant hormones in the regulation of plant development. Front Plant Sci. 2013;4:199. doi: 10.3389/fpls.2013.00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rasmussen A, Depuydt S, Goormachtig S, Geelen D. Strigolactones fine-tune the root system. Planta. 2013;238:615–26. doi: 10.1007/s00425-013-1911-3. [DOI] [PubMed] [Google Scholar]
- 12.Ruyter-Spira C, Al-Babili S, van der Krol S, Bouwmeester H. The biology of strigolactones. Trends Plant Sci. 2013;18:72–83. doi: 10.1016/j.tplants.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 13.Maillet F, Poinsot V, André O, Puech-Pagès V, Haouy A, Gueunier M, Cromer L, Giraudet D, Formey D, Niebel A, et al. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature. 2011;469:58–63. doi: 10.1038/nature09622. [DOI] [PubMed] [Google Scholar]
- 14.Fassler JS, West AH. Histidine phosphotransfer proteins in fungal two-component signal transduction pathways. Eukaryot Cell. 2013;12:1052–60. doi: 10.1128/EC.00083-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voiblet C, Duplessis S, Encelot N, Martin F. Identification of symbiosis-regulated genes in Eucalyptus globulus-Pisolithus tinctorius ectomycorrhiza by differential hybridization of arrayed cDNAs. Plant J. 2001;25:181–91. doi: 10.1046/j.1365-313x.2001.00953.x. [DOI] [PubMed] [Google Scholar]
- 16.Herrera-Martínez A, Ruiz-Medrano R, Valdés M, Xoconostle-Cázares B. Detection of a histidine kinase mRNA in extraradical mycelium of Pisolithus tinctorius induced by the plant metabolites. Pak J Biol Sci. 2009;12:189–91. doi: 10.3923/pjbs.2009.189.191. [DOI] [PubMed] [Google Scholar]
- 17.Capra EJ, Laub MT. Evolution of two-component signal transduction systems. Annu Rev Microbiol. 2012;66:325–47. doi: 10.1146/annurev-micro-092611-150039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung K, Fried L, Behr S, Heermann R. Histidine kinases and response regulators in networks. Curr Opin Microbiol. 2012;15:118–24. doi: 10.1016/j.mib.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 19.Li S, Dean S, Li Z, Horecka J, Deschenes RJ, Fassler JS. The eukaryotic two-component histidine kinase Sln1p regulates OCH1 via the transcription factor, Skn7p. Mol Biol Cell. 2002;13:412–24. doi: 10.1091/mbc.01-09-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaller GE, Shiu SH, Armitage JP. Two-component systems and their co-option for eukaryotic signal transduction. Curr Biol. 2011;21:R320–30. doi: 10.1016/j.cub.2011.02.045. [DOI] [PubMed] [Google Scholar]
- 21.Nongpiur R, Soni P, Karan R, Singla-Pareek SL, Pareek A. Histidine kinases in plants: cross talk between hormone and stress responses. Plant Signal Behav. 2012;7:1230–7. doi: 10.4161/psb.21516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun YP, Fries N. The effect of tree-root exudates on the growth rate of ectomycorrhizal and saprotrophic fungi. Mycorrhiza. 1992;1:63–9. doi: 10.1007/BF00206138. [DOI] [Google Scholar]
- 23.Gogola N. Regulation of mycorrhizal infection by hormonal factors produced by host and fungi. Experientia. 1991;47:331–40. doi: 10.1007/BF01972074. [DOI] [Google Scholar]
- 24.Lagrange H, Jay-Allgand C, Lapeyrie F. Rutin, the phenolglycoside from eucalyptus root exudates, stimulates Pisolithus hyphal growth at picomolar concentrations. New Phytol. 2001;149:349–55. doi: 10.1046/j.1469-8137.2001.00027.x. [DOI] [PubMed] [Google Scholar]
- 25.Gushchin I, Gordeliy V, Grudinin S. Two distinct states of the HAMP domain from sensory rhodopsin transducer observed in unbiased molecular dynamics simulations. PLoS One. 2013;8:e66917. doi: 10.1371/journal.pone.0066917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maeda T, Wurgler-Murphy SM, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–5. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- 27.Cheng Z, Kumagai Y, Lin M, Zhang C, Rikihisa Y. Intra-leukocyte expression of two-component systems in Ehrlichia chaffeensis and Anaplasma phagocytophilum and effects of the histidine kinase inhibitor closantel. Cell Microbiol. 2006;8:1241–52. doi: 10.1111/j.1462-5822.2006.00704.x. [DOI] [PubMed] [Google Scholar]
- 28.Segura-Cabrera A, Bocanegra-García V, Lizarazo-Ortega C, Guo X, Correa-Basurto J, Rodríguez-Pérez MA. A computational analysis of the binding mode of closantel as inhibitor of the Onchocerca volvulus chitinase: insights on macrofilaricidal drug design. J Comput Aided Mol Des. 2011;25:1107–19. doi: 10.1007/s10822-011-9489-y. [DOI] [PubMed] [Google Scholar]
- 29.Chang CL, Park TH, Lee EY, Lim YT, Son HC. Recurrent self-limited fungemia caused by Yarrowia lipolytica in a patient with acute myelogenous leukemia. J Clin Microbiol. 2001;39:1200–1. doi: 10.1128/JCM.39.3.1200-1201.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grupta A, Mi H, Wroe C, Jaques B, Talbot D. Fatal Candida famata peritonitis in a peritoneal dialysis patient. Nephrol Dial Transplant. 2007;21:2036–7. doi: 10.1093/ndt/gfl040. [DOI] [PubMed] [Google Scholar]
- 31.Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, FitzHugh W, Ma LJ, Smirnov S, Purcell S, et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–68. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- 32.Kurosaki F, Nishi A. Elicitation of phytoalexin production in cultured carrot cells. Physiol Plant Pathol. 1984;24:169–76. doi: 10.1016/0048-4059(84)90025-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reissinger A, Winter S, Steckelbroeck S, Hartung W, Sikora RA. Infection of barley roots by Chaetomium globosum: evidence for a protective role of the exodermis. Mycol Res. 2003;107:1094–102. doi: 10.1017/S0953756203008189. [DOI] [PubMed] [Google Scholar]
- 34.van Rhijn P, Fang Y, Galili S, Shaul O, Atzmon N, Wininger S, Eshed Y, Lum M, Li Y, To V, et al. Expression of early nodulin genes in alfalfa mycorrhizae indicates that signal transduction pathways used in forming arbuscular mycorrhizae and Rhizobium-induced nodules may be conserved. Proc Natl Acad Sci U S A. 1997;94:5467–72. doi: 10.1073/pnas.94.10.5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tagu D, Delp G, Delp G, Jane Barker S Regulation of root and fungal morphogenesis in mycorrhizal symbioses. Plant Physiol. 1998;116:1201–7. doi: 10.1104/pp.116.4.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brencic A, Winans SC. Detection of and response to signals involved in host-microbe interactions by plant-associated bacteria. Microbiol Mol Biol Rev. 2005;69:155–94. doi: 10.1128/MMBR.69.1.155-194.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duplessis S, Courty PE, Tagu D, Martin F. Transcript patterns associated with ectomycorrhiza development in Eucalyptus globulus and Pisolithus microcarpus. New Phytol. 2005;165:599–611. doi: 10.1111/j.1469-8137.2004.01248.x. [DOI] [PubMed] [Google Scholar]
- 38.Kumagai Y, Cheng Z, Lin M, Rikihisa Y. Biochemical activities of three pairs of Ehrlichia chaffeensis two-component regulatory system proteins involved in inhibition of lysosomal fusion. Infect Immun. 2006;74:5014–22. doi: 10.1128/IAI.00735-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fleischer R, Heermann R, Jung K, Hunke S. Purification, reconstitution, and characterization of the CpxRAP envelope stress system of Escherichia coli. J Biol Chem. 2007;282:8583–93. doi: 10.1074/jbc.M605785200. [DOI] [PubMed] [Google Scholar]
- 40.Podgornaia AI, Laub MT. Determinants of specificity in two-component signal transduction. Curr Opin Microbiol. 2013;16:156–62. doi: 10.1016/j.mib.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 41.Valdés M. Survival and growth of pines with specific ectomycorrhizae after three years on a highly eroded site. Can J Bot. 1985;64:885–8. doi: 10.1139/b86-115. [DOI] [Google Scholar]
- 42.Rodríguez-Tovar AV, Ruiz-Medrano R, Herrera-Martínez A, Barrera-Figueroa BE, Hidalgo-Lara ME, Reyes-Márquez BE, Cabrera-Ponce JL, Valdés M, Xoconostle-Cázares B. Stable genetic transformation of the ectomycorrhizal fungus Pisolithus tinctorius. J Microbiol Methods. 2005;63:45–54. doi: 10.1016/j.mimet.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 43.Marx DH. Tree host range and world distribution of the extomycorrhizal fungus Pisolithus tinctorius. Can J Microbiol. 1977;23:217–23. doi: 10.1139/m77-033. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, Russell DW. Molecular Cloning a Laboratory Manual, 3rd ed. US: Cold Spring Harbor Laboratory Press, 2001 [Google Scholar]
- 45.Marx DH, Bryan WC. Growth and ectomycorrhizal development of loblolly pine seedlings in fumigated soil and infested with fungal symbiont Pisolithus tinctorius. For Sci. 1975;21:245–254. doi: 10.1126/science.188.4185.245. [DOI] [Google Scholar]
- 46.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 47.Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–4. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 48.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York, NY: Oxford University Press, 2000 [Google Scholar]
- 50.Schwarz R, Dayhoff M. Matrices for detecting distant relationships. In: Atlas of Protein Sequences. Dayhoff M, National Biomedical Research Foundation, ed. 1979:353-358. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.