Abstract
We report here, the transcriptional regulation of 2 Calcium Dependent Protein Kinases in response to nutrient starvation of Chlamydomonas reinhardtii vegetative cells. The CDPK proteins, CDPK1 and CDPK3; share 53% identity among themselves, a maximum of 57% and 52% to higher plants respectively and 42% to apicomplexan protozoans. We expressed a CDPK1-GFP fusion protein in the C. reinhardtii vegetative cells and showed its distribution both in the cell body and the membrane-matrix fraction of the flagella. The fusion protein exhibits mobility shift in the presence of Ca2+, confirming its Ca2+-binding properties. To the best of our knowledge, this is the first report of transcriptional regulation of CDPKs from a unicellular chlorophyte in response to nutrient starvation namely acetate (A), phosphorus (P), and nitrogen (N).
Keywords: Chlamydomonas reinhardtii, CDPK1, CDPK3, Nutrient starvation, RT-PCR, CDPK1-GFP, Localization
Introduction
Plants lead a sedentary lifestyle and are exposed to high salt, extreme temperatures, drought, heavy metals, excessive light and radiation, and oxidative stress. They have developed response mechanisms that lead to their survival or death.1 For such purposes, plants use Ca2+ as a second messenger and harbor Ca2+-binding proteins as effector proteins leading to a signal transduction pathway.2 Plants possess 3 main families of Ca2+ sensors: the Calcium-dependent Protein Kinases (CDPKs), Calmodulin (CaM), and Calceneurin B-like (CBL-like) proteins.3 CaM and CBL-like proteins do not possess any enzymatic activities; however, CDPKs exhibit a protein kinase activity and calcium-sensing abilities present on the same polypeptide. CDPKs are specific to plants, algae, and some apicomplexan protozoans, but are absent in animals.4
Based on the homology they share with each other, the multigene family of CDPKs has now been classified into 4 subgroups. All of these differ by their pattern of expression in response to the stimuli used, sub-cellular localizations, substrate specificities, Ca2+ sensitivities, and, regulation by phosphorylation and lipids, all this resulting in functional specificity and redundancy.5 CDPKs consist of 4 domains: an N-terminal variable domain, a serine-threonine protein kinase domain, an auto-inhibitory junction domain, and a C-terminal calmodulin-like Ca2+-binding domain with EF hands, that bind Ca2+ to activate the serine-threonine kinase activity.6 Besides their involvement in the normal growth and development of plants, CDPKs have been associated with a response to biotic and abiotic stress agents and therefore may be involved in various signal transduction pathways. The array of abiotic stress agents that have shown CDPK regulation include abscisic acid, cold tolerance, drought, high salt, oxidative bursts, wounding, heavy metal, osmotic stress, etc.5,7-12 They are also known to play an important role in parasite motility, gamete formation, host cell attachment and invasion, exocytosis, and parasite transmission in apicomplexans.13
Calcium is known to exert a regulatory effect on the cellular activities of the unicellular, biflagellated green alga Chlamydomonas reinhardtii, in particular, with the flagellar related activities of motility, phototaxis, mating, flagellation/deflagellation, and flagellar length regulation.14-17 In the membrane-matrix fraction of the flagella there are kinases which get activated in the presence of calcium.18 A recent study, using in silico methods of analyses, recently reported the presence of 14 CDPKs in C. reinhardtii, of which CDPK3 (Mw 53 kDa) has been shown to be localized in the flagella and plays a role in flagellar biogenesis.19 CDPK1 has been annotated as FAP223 (Mw 67 kDa) in the flagellar proteome analysis and thus far, there have been no studies performed on this enzyme.20 The effect of some stresses on the physiology of C. reinhardtii shows that depending on the type of stress, the dose, and the length of exposure, the flagella can be paralyzed or lost and later, cells manifest “palmelloids” or apoptose or form actively motile gametes.21-27 As mentioned earlier, since Ca2+ is known to be involved in several stress induced phenomenons of Chlamydomonas, we set out to explore the regulation of the 2 CDPKs, CDPK1 and CDPK3, under varying stress conditions by studying their transcript profiles using RT-PCR.
Results and Discussion
In silico analyses of C. reinhardtii CDPK1 and CDPK3
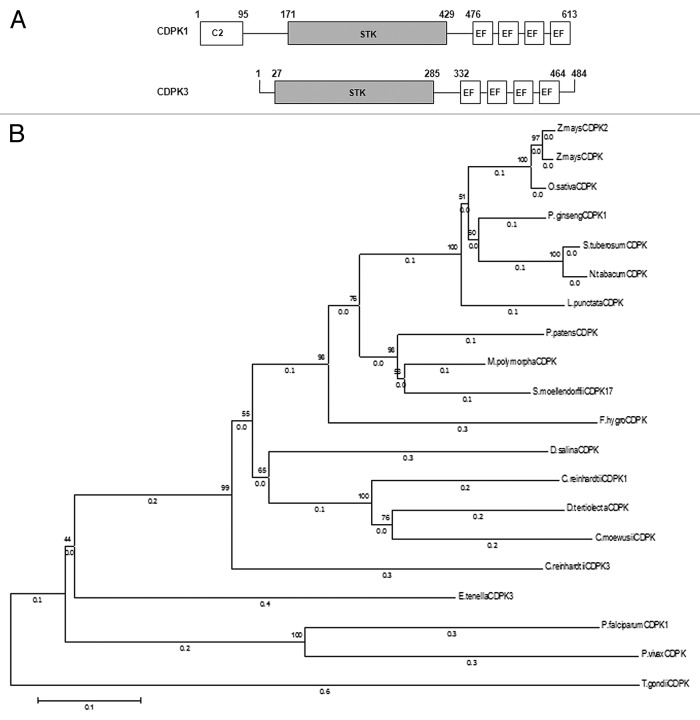
CDPK1 is a protein of 613 amino acids (4786 bp) while CDPK3 is a protein of 484 amino acids (3615 bp). CDPK1 and CDPK3, like other canonical CDPKs have a protein kinase domain, autoinhibitory-junction domain, and a C-terminal CaM-like binding domain and 4 EF domains (Fig. 1A). In addition to these domains, CDPK1 also exhibits an N-terminal C2 domain (aa 1 to 95; Fig. 1A) that is absent from any known CDPKs in higher plants. Our in silico analysis have revealed the presence of the C2 domain in CDPKs of at least 2 other unicellular algae such as Dunaliella tertiolecta and Volvox carteri f. nagariensis. It may be noted that the C2 domain was first identified as a calcium-binding motif in protein kinase C and has now been identified in a number of eukaryotic signaling proteins involved in membrane trafficking, protein phosphorylation, and activation of GTPases.28-32 Upon binding to calcium, C2 domain is known to bind to substrates such as phospholipids and intracellular proteins.33,34 In silico analysis using NCBI-BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) tool for multiple alignment of the entire CDPK1 and CDPK3 protein sequences with that of sequences obtained from NCBI’s Protein tool (http://www.ncbi.nlm.nih.gov/protein) showed that there is a 52–57% identity to higher plants and, both the CDPKs show a maximum of 42% identity to apicomplexan protozoans. Of the 500 sequences that were obtained in this blast, 20 have been chosen that showed > 30% identity with the CDPK1 and CDPK3 and used to generate a Phylogenetic tree using MEGA6: Molecular Evolutionary Genetics Analysis version 6.0 software (Fig. 1B).35-38 Care was administered to represent at least one sequence from the major phyla of higher plants, lower plants, algae, and apicomplexans. Also, the full-length protein sequences were accepted provided these harbored the consensus Ser/Thr protein kinase domain, conserved Asp and Lys residues with the active site (D[L/I/V]K motif), and the calmodulin domain comprising of the EF-hand motif. The tree showed 4 clusters each containing algal, higher plants, apicomplexans, and a mixed cluster containing lower plants (mosses, Lichens, Selaginella; Fig. 1B). Although the identity between CDPK1 and CDPK3 is 53%, they seem to have diverged. Algal and apicomplexan CDPKs seem to share a common node but have diverged or parallelly evolved, and that might be the reason that they share a lower homology.
Figure 1. CDPKs from Chlamydomonas reinhardtii. (A) Schematic diagram of CDPK1 and CDPK3 showing the C2 domain from 1–95 aa in CDPK1, protein kinase catalytic domains (STK), and the 4 EF hands in both the CDPKs. (B) Phylogenetic relationship between C. reinhardtii CDPK1, CDPK3, and other CDPKs from algae, moss, higher plants, and apicomplexans. The phylogenetic tree was inferred using the Neighbor-Joining method. The optimal tree with the sum of branch length = 4.60159844 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths (next to the branches) in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. The analysis involved 20 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 371 positions in the final data set. Evolutionary analyses were conducted in MEGA6.
We too noted the non-clustering of the algal and plant CDPKs as observed in a recently reported study.39 In addition, algae such as D. tertiolecta and C. moewusii shared 63% and 58% identity to the CrCDPK1 while a 53% and 51% identity to CrCDPK3, respectively. As of now, these algal CDPKs have been characterized as Ca2+-binding and Ca2+-dependent Protein Kinases.40,41
Expression analysis of CDPK1 and CDPK3 transcripts under nutrient starvation
The vegetative cells of C. reinhardtii when exposed to acetate, phosphorus, and nitrogen starvation showed a significant change in their CDPK transcript expression profiles. In the case of acetate starvation, only CDPK3 showed a significant (11-fold) increase at 1 h which continued until 3h and then decreased to ~4-fold at 6 h after which it remained constant until 18 h. However, the transcript levels of CDPK1 did not show any significant changes (Fig. 2A). On the other hand, in cells starved of phosphorus, the transcripts of both CDPK1 and CDPK3 respectively showed an 8- and 6-fold increase at 3 h, after which it remained constant until 18 h (Fig. 2B). For cells starved of nitrogen, transcripts of CDPK1 showed a 14-fold increase at 1 h followed by a decrease and the same trend was observed for CDPK3 transcripts with ~11-fold increase at 1 h (Fig. 2C). Taken together, our results clearly demonstrate an almost equal regulation of CDPK1 and CDPK3 in response to media starved of P and N. CDPK1, on the other hand, is not significantly regulated under conditions of acetate starvation. In essence, there seems to be an important role for CDPKs when the vegetative cells of C. reinhardtii cells are nutritionally starved; whether Ca2+ is invoked as a second messenger needs to be addressed. Earlier reports of phosphate starvation leading to the accumulation of polyphosphate and Ca2+ in C. reinhardtii cells have been observed.42 These polyphosphate stores have been shown to be calcium stores and the subsequent potential of Ca2+ as a second messenger has been provided in these studies. When correlated with the current study, we suggest an accumulation of Ca2+ during P starvation thereby activating CDPKs (Fig. 2B). Ca2+ is released into the medium by cells that undergo mating and plays a vital role in this process.14 Therefore, there might be a rapid (within minutes) rise in the Ca2+ levels inside the cells post N starvation, leading to increase in the effector proteins and in the current scenario, both the CDPKs (Fig. 2C). As for the regulation of CDPK in other organisms is concerned, studies on Funaria and Arabidopsis have reported an increase in the expression of CDPK at the gene level in response to phosphate starvation using northern analysis and microarray, respectively.43,44 Also, the same study on Funaria has shown an upregulation of CDPK gene in response to nitrogen starvation using northern analysis.43 The findings on rice suggest that the OsCDPK1 is involved in the signal transduction pathway(s) in the low-nitrogen stress response.45 In addition, a recent study using in silico analyses predicted a few CDPKs as important regulators of phosphate deficiency-induced root hair remodeling in Arabidopsis.46 We see a substantial increase (~6- to 14-fold over the controls) in the transcript levels of the CDPKs, as against those reported for Funaria or Arabidopsis.
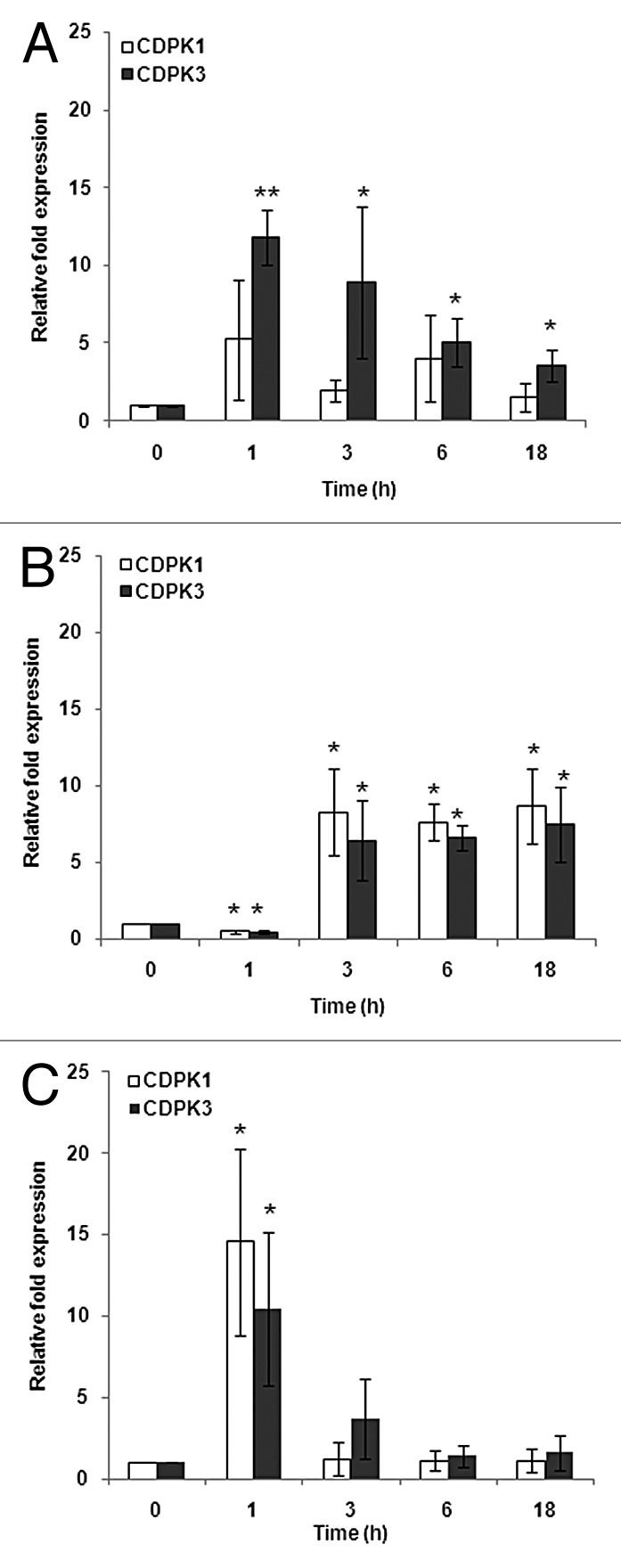
Figure 2. Expression patterns of CDPK1 and CDPK3 transcripts in response to nutrient starvation at 0, 1, 3, 6, and 18 h by real-time RT-PCR analysis. (A) Acetate starvation. (B) Phosphorus starvation. (C) Nitrogen starvation. The single and double asterisks indicate a statistically significant difference at P< 0.05, P< 0.005 respectively.
While, the enzymatic activity of CDPK1 remains to be tested, we outline a brief account of Ca2+-dependent protein kinase activity reported in other algal systems. The first report for the presence of a CDPK-like enzyme was performed in 1990 by Guo and Roux who partially purified and characterized the enzyme from D. salina. The enzyme, identified using an in-gel kinase assay and shown to be activated at free Ca2+ concentrations above 10−7 M, was not sensitive to phospholipids and was found to be inhibited by calmodulin antagonists.47 Almost a decade later, a 40 kDa Ca2+-dependent protein kinase was found to be activated in Dunaliella tertiolecta by heat shock, acidic stress, and H2O2 treatment.48Using an in-gel kinase assay, 3 CDPKs (62, 54, and 47 kDa) were identified in D. tertiolecta cell extracts.40 When the same technique was extended to homogenates of another green alga, Closterium ehrenbergii using histone H1, myelin basic protein, and casein as substrates, CDPKs ranging in relative molecular mass from 47–60 kDa were observed. Of these, the 55 kDa CDPK was found to be immunologically related to the 62 kDa D. tertiolecta CDPK.49 Another CDPK was found to share epitopes with the 62 kDa D. tertiolecta CDPK that was present in the internodal cells of the brackish water inhabiting Lamprothamnium succinctum charophyte. This enzyme of 53 kDa was found to be involved in processing turgor regulation in response to hypoosmotic treatment.50 The predicted Mw of CDPK1 and CDPK3 is 67 and 53 kDa, respectively, and seems to fall in the range of those observed for algal CDPKs, which belong to one cluster (Fig. 1B).
Extensive work on CDPK3 has already been reported and since the C2 domain appears to be novel to C. reinhardtii CDPK1, the current interest was pursued by the overexpression of a recombinant CDPK1-GFP protein for all further analyses (Fig. 3A).19
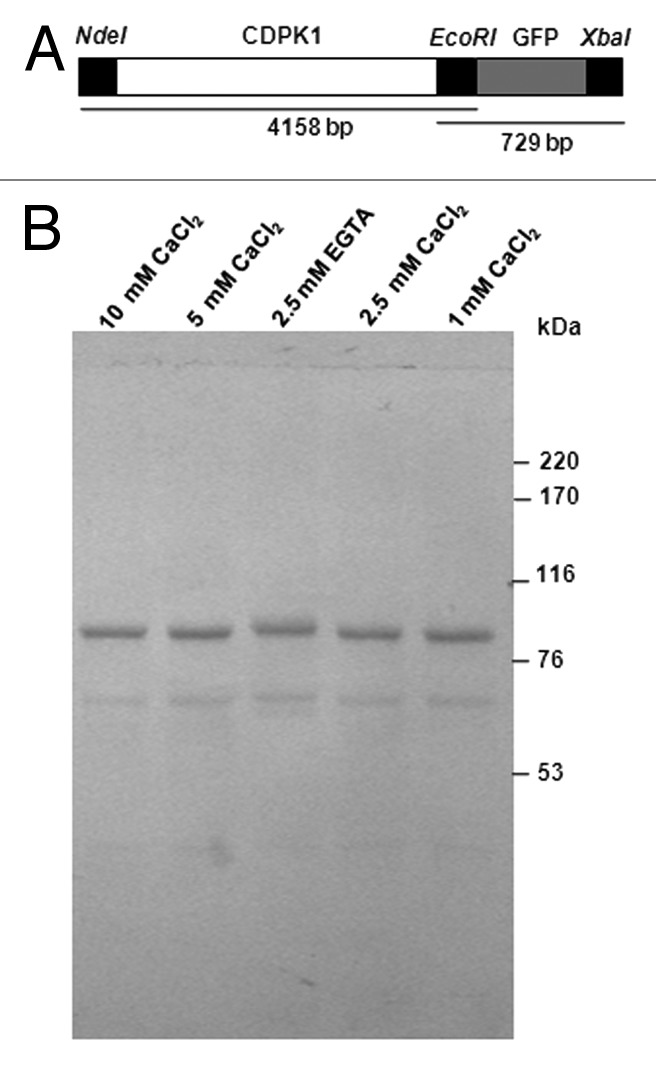
Figure 3. Ca2+ mobility of C. reinhardtii CDPK1-GFP. (A) CDPK1-GFP construct used to transform C. reinhardtii cells and (B) Ca2+ mobility shift of CDPK1-GFP fusion protein. Lanes 1, 2, 4, and 5 are treatments with different concentrations of CaCl2; lane 3 is the lysate treated with EGTA only. CDPK1-GFP cell lysate treated with CaCl2 (10 mM, 5 mM, 2.5 mM, and 1 mM) and EGTA (2.5 mM) were run on a 7% SDS -PAGE followed by Western Blotting using anti-GFP antibody. CDPK1 showed a mobility shift in presence of Ca2+.
Calcium-dependent mobility shift of CDPK1
Studies have shown that the binding of Ca2+ to EF-hands of a CDPK results in the protein undergoing a conformational change. Due to this conformational change, a mobility shift is seen in the presence of calcium.49,51-54 The electrophoretic mobility of the CDPK1-GFP fusion protein was compared in the presence and absence (EGTA) of Ca2+. As expected, the mobility shift of the fusion protein was clearly seen in presence of Ca2+ when compared to that treated with EGTA, and a 4 kDa difference was observed (Fig. 3B), proving that CDPK1 is a calcium binding protein. Similar mobility shifts have been observed earlier, a difference of 3 kDa with soybean CDPK, 5 kDa with Vicia faba CDPK, ~5 kDa with Closterium ehrenbergii CDPK49,52,53
CDPK1 is a flagellar membrane-matrix protein
The CDPK1-GFP C. reinhardtii clones were used to study the sub-cellular localization of CDPK1. Although a strong promoter (PSAD) was used to express the CDPK1-GFP fusion protein, repeated attempts at directly observing GFP fluorescence using live or fixed cells were unsuccessful. Similar experience in the field prompted us to switch over to immunostaining experiments.55 Immunostaining of C. reinhardtii cells with anti-GFP antibody revealed the presence of CDPK1-GFP fusion protein in both the cell body as well as the flagella (Fig. 4A). For sub-flagellar localization experiments, flagellar isolation was performed. The flagella were further fractionated into the membrane-matrix and axonemal fractions; protein extracts were subjected to denaturing gel electrophoresis; followed by western analysis and immuno-probing using anti-GFP antibody. CDPK1 was present in the whole cells, cell body, flagella, and the membrane-matrix fraction, but was absent in the axonemal fraction (Fig. 4B). A recent study of CDPK3 being distributed in both the cell body and flagellar membrane-matrix fraction of C. reinhardtii cells has been shown.19 Moreover, whether the protein re-localizes upon an exposure to stress needs to be addressed. As far as the sub-cellular localization of CDPKs is concerned, they have been shown to occupy several parts of the plant cell, such as cytoplasm, peroxisomes, nucleus, endoplasmic reticulum, mitochondrial outer membrane, and plasma membrane.56-62 Studies on protozoans have shown that CDPKs are localized in the schizonts and pellicle in Plasmodium, in sporozoites in Eimeria, and also in the cilia and body of Paramecium.63-67 CDPK3 is localized in the C. reinhardtii membrane-matrix fraction of the flagella.19 Another 65 kDa CDPK has been shown to be localized in the C. moewusii flagella.41
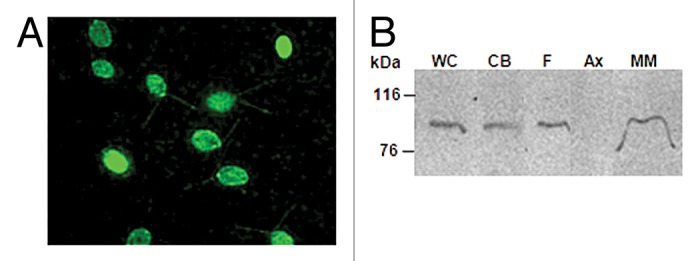
Figure 4. C. reinhardtii CDPK1 is localized in the cell body and flagella. (A) Immunostaining of cells expressing CDPK1-GFP with anti-GFP antibody. CDPK1-GFP fusion protein was localized in C. reinhardtii flagella and cell body. (B) Whole cell (WC), cell body (CB), isolated flagella (F), axoneme (Ax), and membrane-matrix (M-M) fractions of CDPK1-GFP clone were analyzed by immunoblotting using anti-GFP antibody. CDPK1- GFP fusion protein was present in the cell body and the membrane-matrix fraction of the flagella. The bar on (A) indicate 10 µm.
In conclusion, the 2 CDPKs from C. reinhardtii cells are regulated by nutrient starvation (P and N); however, it was only CDPK3 that was upregulated upon acetate starvation. This might suggest a plausible Ca2+-dependent pathway in response to nutrient stress.
Materials and Methods
Plant materials and abiotic stress treatment
The wild-type C. reinhardtii (strain cc124, mt-) cells were grown in Tris-Acetate Phosphate (TAP) medium at 24 °C in continuous light until the cell density reached 3x106 cells/ml.68 Acetate, phosphorus, and nitrogen deficient media was prepared as follows. Glacial acetic acid was omitted in the acetate-free medium and the pH was adjusted to 7.0 using HCl. K2HPO4 and KH2PO4 was replaced with 1.4 mM KCl in the phosphorus-free medium and NH4Cl was replaced with 7.4 mM KCl in the nitrogen-free medium.69 For the acetate, phosphorus, and nitrogen starvation experiments, cells were washed with TAP and divided into 2 sets (treated and control). The treated sets were washed with nutrient-starved medium (TAP-A, TAP-P, and TAP-N) and the control set with TAP. The cells were inoculated in the respective media to a cell density of 105 cells/ml. Cells were harvested at 0, 1, 3, 6, 18 h.
RNA isolation and cDNA synthesis
RNA was extracted as per standard protocol using Trizol reagent (Invitrogen, Cat. No. 15596–026). Nucleic acid was treated with DNaseI (Thermo Scientific, Cat. No. #EN0521) containing buffer to remove DNA. The total RNA was quantified using the Infinite® 200 PRO NanoQuant (TECAN). cDNA was synthesized using Revert Aid First Strand cDNA Synthesis kit (Thermo Scientific, Cat. No. K1621). The absence of DNA was confirmed by running a PCR with forward and reverse actin primers (Table 1) followed by a 2% Agarose Gel Electrophoresis.
Table 1. Primer sequences used in this study.
| Name | Primer Sequence (5′-3′) |
|---|---|
| Primers for sequencing pCCDPK1GFP | |
| C1seqF1 | ACTCGTTGTGCATTCTAGGACC |
| C1seqR1 | AGCTTCAGCGTCTGGTCG |
| C1seqF2 | TGTGGCGTTGCACACAGAT |
| C1seqR2 | TTGTTGGTGGCGGAGTAAGT |
| C1seqF3 | GGACTGCTGGAAGGACTACG |
| C1seqR3 | TCCGACAGCAGGAAGTTCTC |
| C1seqF4 | CACAACATGGGCGTCATTC |
| C1seqR4 | CTGCACCGTACGTCTGCTG |
| C1seqF5 | CCGCAGAGTGTATGACATGG |
| C1seqR5 | ACGCGACGGAAACACATAG |
| C1seqF6 | CTGCGTCCACGCGTCTAC |
| C1seqR6 | TGCCGTTGATCTCGTCCA |
| C1seqF7 | CCTGCTGGTGTGTTTGCAAC |
| C1seqR7 | TCAATCACACGGCACATCG |
| C1seqF8 | GGAAGCGTCTGGTGAAGATG |
| C1seqR8 | GTCTTGTAGTTGCCGTCGTCC |
| C1seqF9 | AGCAGCACGACTTCTTCAAGTC |
| C1seqR9 | CACACCGACCCTGGTCAC |
| Primers for generating CDPK1-GFP | |
|---|---|
| CIGUF | GAGGCCTAGGAGACTGTGCATGTGAGGC |
| C1GR | CATATGGAGGCCTAGGAGACTGTGCA |
| C1GUFNdeI | CAACCCGCGCATCATTGCCACGAA |
| CIGREcorI | GAATTCCAACCCGCGCATCATTGCC |
| GFPfwdEcoRI | GAATTCATGGCCAAGGGCGAGGAGC |
| GFPrevXbaI | TCTAGATTACTTGTACAGCTCGTCCATGCCG |
| Primers for Actin PCR | |
|---|---|
| CreActin fwd | CGCTGGAGAAGACCTACGAG |
| CreActin rev | GGAGTTGAAGGTGGTGTCGT |
| Primers for Real Time PCR | |
|---|---|
| Cblp fwd | CTTCTCGCCCATGACCAC |
| Cblp rev | CCCACCAGGTTGTTCTTC |
| CDPK1RT1F | AGCCAACTTGGAATGAGGTG |
| CDPK1RT1R | CGTCTTGCCCTCTGTAAAGC |
| CDPK3RT1F | TTCTCGGTTGCAAACATCCT |
| CDPK3RT1R | ACGTCCTTCACGTCCTCCTT |
Real Time PCR
The gene expression analysis was performed using real-time-PCR. The reaction mixture (10 µl) contained 10 ng of prepared cDNA, 5 µl of 2 X Sso Fast Eva Green Supermix (BIORAD, Cat. No. 172–5201), and 0.5 µl of 5 picomoles/µl of forward and reverse primers each (See Table 1 for the list of primers). All reactions carried out in duplicates were performed in a CFX96TM Thermocycler (BIO-RAD) under the following conditions: 3 min at 95 °C, 40 cycles of 10 s at 95 °C, and 30 s at 60 °C and melt curve (65–95 °C) analysis. The C. reinhardtii β subunit-like polypeptide (Cblp) gene, which is constitutively expressed was used as the reference gene.70 Data was analyzed using the 2(-Delta Delta C(T)).71 The results of the RT-PCR experiments are represented as a fold increase over the controls, calculated from the Ct values. It may be noted that while sample duplicates were maintained for each Real Time PCR reaction, the entire experiment was conducted thrice.
Construction of CDPK1-GFP fusion protein
CDPK1 was amplified from C. reinhardtii gDNA using the KOD Xtreme kit (Calbiochem,Cat. No. 71975–3) and primers CIGUF and CIGR (Table 1). This was followed by a secondary amplification using primers with RE sites C1GUFNdeI and CIGREcoRI (Table 1). It was cloned in pUC19 and transformed in E. coli DH5α cells. The plasmid was isolated, purified, and RE digested with NdeI and EcoRI, ligated with pChlamiRNA3 (RE digested with NdeI and EcoRI), and transformed in E. coli DH5α cells. The GFP plasmid (pCRGFP) was obtained from Chlamy Resource Centre (http://www.chlamy.org). It was RE digested with BamHI and subsequently the 717 bp product of GFP gene was gel purified. The GFP gene was amplified using KOD Xtreme kit (Calbiochem,Cat. No. 71975–3) and primers GFPfwdEcoRI and GFPrevXbaI and cloned in pUC19 followed by transformation in E. coli DH5α cells (Table 1). The GFP amplicon was transformed into E. coli DH5α cells. The plasmid was isolated, purified, and RE digested with EcoRI and XbaI and ligated with pChlamiRNA3-CDPK1 plasmid (RE digested with EcoRI and XbaI). This resulting plasmid (henceforth called pCrCDPK1GFP) was transformed in E. coli DH5α cells (Fig. 3A). Sequencing of CDPK1-GFP plasmid was done using nested primers C1seqF1 to C1seqR9 (Table 1). After sequence confirmation, the vector was transformed into C. reinhardtii cells.
Transformation in C. reinhardtii cells and screening for CDPK1-GFP clones
The pCrCDPK1GFP plasmid was isolated, purified, and linearized with NotI. Transformation of pCrCDPK1GFP in C. reinhardtii was performed as per the protocol of Kindle.72 Selection for positive clones were done on (10 µg/ml) paromomycin in TAP agar plates. Clones were picked up from the plates and maintained on TAP agar plates. Transformants were screened for the presence of CDPK1-GFP fusion protein by PCR using primers GFPfwdEcoRI and C1seqR8 (Table 1), immunocytochemistry, and by western blotting using an anti-GFP antibody (Sigma, Cat. No. G 1544). Twelve positive clones were obtained.
Calcium-mobility gel shift assay
To determine if the protein binds to Ca2+, calcium mobility shift assay was performed. The cell pellet of CDPK1-GFP clone was resuspended in lysis buffer (50 mM Tris, 10% glycerol and 1 mM PMSF, pH 7.4) and sonicated. The cell lysate was centrifuged at 13,400 g for 5 min. The supernatant was incubated with 2.5 mM EGTA, 1 mM, 2.5 mM, 5 mM, and 10 mM CaCl2 on ice for 30 min followed by addition of 5 X SDS sample buffer and incubation for 15 min at 100 °C. Proteins were electrophoresed on 7% SDS-polyacrylamide gel by the method of Laemmli and electro-transferred to a nitrocellulose membrane.73 The membrane was blocked with 3% (w/v) BSA (Bovine Serum Albumin) in PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4) for 1 h and then incubated with anti-GFP antibody (1:1,000) (Sigma, Cat. No. G 1544) for 2 h at room temperature. The membrane was washed thrice (15 min each) with PBS containing 0.05% Tween 20 (PBST) and incubated with goat-anti rabbit IgG conjugated with HRP (horseradish peroxidase) (1:5,000) (Santa Cruz Biotechnology, INC, Cat. No. sc-2004) for 1h. The membrane was washed as above and developed using peroxide buffer and HRP substrate, DAB (3,3′-diaminobenzidine tetrahydrochloride) (Roche Applied Science, Cat. No.11718096001).
Immunofluorescence microscopy
To localize CDPK1-GFP protein, C. reinhardtii cells expressing the CDPK1-GFP fusion protein were grown until logarithmic phase. The transformed cells were resuspended in microtubule stabilizing buffer (MTSB-30 mM HEPES, 15 mM KCl, 5 mM MgSO4, 5 mM EGTA, 100 μM dithiothreitol, pH 7.0 with KOH) and allowed to adhere to polyethyleneimine-treated coverslip. They were washed with PBS and fixed in chilled methanol for 10 min. The cells fixed on the coverslip were washed with PBS and incubated with 3% (w/v) BSA in PBS blocking solution for 1 h. This was followed by a wash with PBS and incubation with primary antibody, anti-GFP (1:100) (Genscript, Cat No. A01388) for 1 h. The cells were washed briefly as before, followed by incubation with secondary antibody, Alexa Fluor 488 goat anti-rabbit IgG (1:100) (Invitrogen, Cat No. A-11008) for 1 h. The cells were finally washed with PBST and PBS and then mounted in ProLong® Gold Antifade Reagent (Invitrogen, Cat No. P36934). The cells were observed under a Nikon Eclipse 90i microscope with the Imaging Software NIS-Elements BR.
Flagella isolation and fractionation
Flagella were isolated from CDPK1-GFP clone as previously described.74 An aliquot of flagella was stored separately at –20 °C. The remaining flagella were extracted in 1% Igepal for 10 min on ice. After centrifugation (31,000 g for 20 min at 4 °C), the membrane-matrix (M-M) was obtained in the soluble phase while the insoluble phase (axoneme) was washed with HMDEK buffer (10 mM HEPES, pH 7.2, 5 mM MgSO4, 1 mM dithiothreitol, 1 mM EDTA, and 25 mM KCl) followed by resuspension in the same buffer. The M-M and axonemal fractions were stored at –20 °C until further use.
Sub-flagellar localization of CDPK1
Whole cells, cell body, flagella, axonemal proteins, and membrane-matrix fraction of the CDPK1-GFP clone were separated on an 8% SDS-polyacrylamide gel and electro-transferred to a nitrocellulose membrane. The membrane was blocked with 3% (w/v) BSA in PBS for 1 h and then incubated with anti-GFP antibody (1:1,000) (Sigma, Cat. No. G 1544) for 2 h at room temperature. The membrane was washed thrice (15 min each) with PBST and incubated with goat-anti rabbit IgG conjugated with HRP (1:7,500) (Calbiochem, Cat. No. DCO3L) for 1h. This was followed by washes with PBST and development of the blot using peroxide buffer and HRP substrate, DAB.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was funded by the Department of Atomic Energy, India.
References
- 1.Knight H, Knight MR. Abiotic stress signalling pathways: specificity and cross-talk. Trends Plant Sci. 2001;6:262–7. doi: 10.1093/molbev/msh197. [DOI] [PubMed] [Google Scholar]
- 2.Nagata T, Iizumi S, Satoh K, Ooka H, Kawai J, Carninci P, Hayashizaki Y, Otomo Y, Murakami K, Matsubara K, et al. Comparative analysis of plant and animal calcium signal transduction element using plant full-length cDNA data. Mol Biol Evol. 2004;21:1855–70. doi: 10.1093/molbev/msh197. [DOI] [PubMed] [Google Scholar]
- 3.Luan S, Kudla J, Rodriguez-Concepcion M, Yalovsky S, Gruissem W. Calmodulins and calcineurin B-like proteins: calcium sensors for specific signal response coupling in plants. Plant Cell. 2002;14(Suppl):S389–400. doi: 10.1105/tpc.001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hrabak EM, Chan CW, Gribskov M, Harper JF, Choi JH, Halford N, Kudla J, Luan S, Nimmo HG, Sussman MR, et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003;132:666–80. doi: 10.1104/pp.102.011999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boudsocq M, Sheen J. CDPKs in immune and stress signaling. Trends Plant Sci. 2013;18:30–40. doi: 10.1016/j.tplants.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harmon AC, Gribskov M, Harper JF. CDPKs - a kinase for every Ca2+ signal? Trends Plant Sci. 2000;5:154–9. doi: 10.1016/S1360-1385(00)01577-6. [DOI] [PubMed] [Google Scholar]
- 7.Asano T, Hayashi N, Kikuchi S, Ohsugi R. CDPK-mediated abiotic stress signaling. Plant Signal Behav. 2012;7:817–21. doi: 10.4161/psb.20351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubrovina AS, Kiselev KV, Khristenko VS. Expression of calcium-dependent protein kinase (CDPK) genes under abiotic stress conditions in wild-growing grapevine Vitis amurensis. J Plant Physiol. 2013;170:1491–500. doi: 10.1016/j.jplph.2013.06.014. [DOI] [PubMed] [Google Scholar]
- 9.Chmielowska-Bąk J, Lefèvre I, Lutts S, Deckert J. Short term signaling responses in roots of young soybean seedlings exposed to cadmium stress. J Plant Physiol. 2013;170:1585–94. doi: 10.1016/j.jplph.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 10.Jiang S, Zhang D, Wang L, Pan J, Liu Y, Kong X, Zhou Y, Li D. A maize calcium-dependent protein kinase gene, ZmCPK4, positively regulated abscisic acid signaling and enhanced drought stress tolerance in transgenic Arabidopsis. Plant Physiol Biochem. 2013;71:112–20. doi: 10.1016/j.plaphy.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 11.Ding Y, Cao J, Ni L, Zhu Y, Zhang A, Tan M, Jiang M. ZmCPK11 is involved in abscisic acid-induced antioxidant defence and functions upstream of ZmMPK5 in abscisic acid signalling in maize. J Exp Bot. 2013;64:871–84. doi: 10.1093/jxb/ers366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J, Xue B, Xia X, Yin W. A novel calcium-dependent protein kinase gene from Populus euphratica, confers both drought and cold stress tolerance. Biochem Biophys Res Commun. 2013;441:630–6. doi: 10.1016/j.bbrc.2013.10.103. [DOI] [PubMed] [Google Scholar]
- 13.Moreno SNJ, Ayong L, Pace DA. Calcium storage and function in apicomplexan parasites. Essays Biochem. 2011;51:97–110. doi: 10.1042/bse0510097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodenough UW, Shames B, Small L, Saito T, Crain RC, Sanders MA, Salisbury JL. The role of calcium in the Chlamydomonas reinhardtii mating reaction. J Cell Biol. 1993;121:365–74. doi: 10.1083/jcb.121.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheshire JL, Evans JH, Keller LR. Ca2+ signaling in the Chlamydomonas flagellar regeneration system: cellular and molecular responses. J Cell Sci. 1994;107:2491–8. doi: 10.1242/jcs.107.9.2491. [DOI] [PubMed] [Google Scholar]
- 16.Smith EF. Regulation of flagellar dynein by calcium and a role for an axonemal calmodulin and calmodulin-dependent kinase. Mol Biol Cell. 2002;13:3303–13. doi: 10.1091/mbc.E02-04-0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wakabayashi K, Ide T, Kamiya R. Calcium-dependent flagellar motility activation in Chlamydomonas reinhardtii in response to mechanical agitation. Cell Motil Cytoskeleton. 2009;66:736–42. doi: 10.1002/cm.20402. [DOI] [PubMed] [Google Scholar]
- 18.Bloodgood RA. Calcium-regulated phosphorylation of proteins in the membrane-matrix compartment of the Chlamydomonas flagellum. Exp Cell Res. 1992;198:228–36. doi: 10.1016/0014-4827(92)90375-I. [DOI] [PubMed] [Google Scholar]
- 19.Liang Y, Pan J. Regulation of flagellar biogenesis by a calcium dependent protein kinase in Chlamydomonas reinhardtii. PLoS One. 2013;8:e69902. doi: 10.1371/journal.pone.0069902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pazour GJ, Agrin N, Leszyk J, Witman GB. Proteomic analysis of a eukaryotic cilium. J Cell Biol. 2005;170:103–13. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hessen DO, Donk VE, Anderson T. Growth responses, P-uptake and loss of flagellae in Chlamydomonas reinhardtii exposed to UV-B. J Plankton Res. 1995;17:17–27. doi: 10.1093/plankt/17.1.17. [DOI] [Google Scholar]
- 22.Dharmadhikari JA, D'Souza JS, Gudipati M, Dharmadhikari AK, Rao BJ, Mathur D. Sensitive, real-time monitoring of UV-induced stress in a single, live plant cell using an optical trap. J Sens & Act-B. 2006;115:439–43. doi: 10.1016/j.snb.2005.10.006. [DOI] [Google Scholar]
- 23.Jamers A, De Coen W. Effect assessment of the herbicide paraquat on a green alga using differential gene expression and biochemical biomarkers. Environ Toxicol Chem. 2010;29:893–901. doi: 10.1002/etc.102. [DOI] [PubMed] [Google Scholar]
- 24.Moharikar S, D'Souza J, Kulkarni AB. Rao BJ. UV-C induced apoptotic-like cell death process in the unicellular chlorophyte Chlamydomonas reinhardtii. J Phycol. 2006;42:423–33. doi: 10.1111/j.1529-8817.2006.00207.x. [DOI] [Google Scholar]
- 25.Moharikar S, D’Souza JS, Rao BJ. A homologue of the defender against the apoptotic death gene (dad1 )in UV-exposed Chlamydomonas cells is downregulated with the onset of programmed cell death. J Biosci. 2007;32:261–70. doi: 10.1007/s12038-007-0026-z. [DOI] [PubMed] [Google Scholar]
- 26.Yordanova ZP, Iakimova ET, Cristescu SM, Harren FJ, Kapchina-Toteva VM, Woltering EJ. Involvement of ethylene and nitric oxide in cell death in mastoparan-treated unicellular alga Chlamydomonas reinhardtii. Cell Biol Int. 2010;34:301–8. doi: 10.1042/CBI20090138. [DOI] [PubMed] [Google Scholar]
- 27.Sager R, Granick S. Nutritional control of sexuality in Chlamydomonas reinhardi. J Gen Physiol. 1954;37:729–42. doi: 10.1085/jgp.37.6.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rizo J, Südhof TC. C2-domains, structure and function of a universal Ca2+-binding domain. J Biol Chem. 1998;273:15879–82. doi: 10.1074/jbc.273.26.15879. [DOI] [PubMed] [Google Scholar]
- 29.Cho W. Membrane targeting by C1 and C2 domains. J Biol Chem. 2001;276:32407–10. doi: 10.1074/jbc.R100007200. [DOI] [PubMed] [Google Scholar]
- 30.Parker PJ, Coussens L, Totty N, Rhee L, Young S, Chen E, Stabel S, Waterfield MD, Ullrich A. The complete primary structure of protein kinase C--the major phorbol ester receptor. Science. 1986;233:853–9. doi: 10.1126/science.3755547. [DOI] [PubMed] [Google Scholar]
- 31.Coussens L, Parker PJ, Rhee L, Yang-Feng TL, Chen E, Waterfield MD, Francke U, Ullrich A. Multiple, distinct forms of bovine and human protein kinase C suggest diversity in cellular signaling pathways. Science. 1986;233:859–66. doi: 10.1126/science.3755548. [DOI] [PubMed] [Google Scholar]
- 32.Knopf JL, Lee MH, Sultzman LA, Kriz RW, Loomis CR, Hewick RM, Bell RM. Cloning and expression of multiple protein kinase C cDNAs. Cell. 1986;46:491–502. doi: 10.1016/0092-8674(86)90874-3. [DOI] [PubMed] [Google Scholar]
- 33.Nalefski EA, Falke JJ. The C2 domain calcium-binding motif: structural and functional diversity. Protein Sci. 1996;5:2375–90. doi: 10.1002/pro.5560051201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yáñez M, Gil-Longo J, Campos-Toimil M. Calcium binding proteins. Adv Exp Med Biol. 2012;740:461–82. doi: 10.1007/978-94-007-2888-2_19. [DOI] [PubMed] [Google Scholar]
- 35.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 36.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–91. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 37.Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. In: Evolving Genes and Proteins. Bryson V and Vogel HJ, eds. New York: Academic Press, 1965; 97-166. [Google Scholar]
- 38.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hamel LP, Sheen J, Séguin A. Ancient signals: comparative genomics of green plant CDPKs. Trends Plant Sci. 2013;S1:360–85. doi: 10.1016/j.tplants.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pinontoan R, Yuasa T, Anderca MI, Matsuoka T, Uozumi N, Mori H, Muto S. Cloning of a cDNA encoding a 66-kDa Ca2+-dependent protein kinase (CDPK) from Dunaliella tertiolecta (Chlorophyta) J Phycol. 2000;36:545–52. doi: 10.1046/j.1529-8817.2000.99185.x. [DOI] [PubMed] [Google Scholar]
- 41.Siderius M, Henskens H, Porto-leBlanche A, van Himbergen J, Musgrave A, Haring M. Characterisation and cloning of a calmodulin-like domain protein kinase from Chlamydomonas moewusii (Gerloff) Planta. 1997;202:76–84. doi: 10.1007/s004250050105. [DOI] [PubMed] [Google Scholar]
- 42.Siderius M, Musgrave A, van den Ende H, Koerten H, Cambier P, van der Meer P. Chlamydomonas eugamentos (chlorophyta) stores phosphate in polyphosphate bodies together with calcium. J Phycol. 1996;32:402–9. doi: 10.1111/j.0022-3646.1996.00402.x. [DOI] [Google Scholar]
- 43.Mitra D, Johri MM. Enhanced expression of a calcium-dependent protein kinase from the moss Funaria hygrometrica under nutritional starvation. J Biosci. 2000;25:331–8. doi: 10.1007/BF02703786. [DOI] [PubMed] [Google Scholar]
- 44.Wu P, Ma L, Hou X, Wang M, Wu Y, Liu F, Deng XW. Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol. 2003;132:1260–71. doi: 10.1104/pp.103.021022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Asano T, Wakayama M, Aoki N, Komatsu S, Ichikawa H, Hirochika H, Ohsugi R. Overexpression of a calcium-dependent protein kinase gene enhances growth of rice under low-nitrogen conditions. Plant Biotechnol. 2010;27:369–73. doi: 10.5511/plantbiotechnology.27.369. [DOI] [Google Scholar]
- 46.Lan P, Li W, Schmidt W. Genome-wide co-expression analysis predicts protein kinases as important regulators of phosphate deficiency-induced root hair remodeling in Arabidopsis. BMC Genomics. 2013;14:210–21. doi: 10.1186/1471-2164-14-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo Y-L, Roux SJ. Partial purification and characterization of a Ca(2+)-dependent protein kinase from the green alga, Dunaliella salina. Plant Physiol. 1990;94:143–50. doi: 10.1104/pp.94.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuasa T. Identification of a 40 kDa Protein Kinase activated by stress in a halotolerant green alga Dunaliella tertiolecta. Microbes Environ. 2002;17:39–47. doi: 10.1264/jsme2.2002.39. [DOI] [Google Scholar]
- 49.Yuasa T, Hashimoto H. A Calcium-dependent Protein Kinase (CDPK) in the Unicellular Green Alga Closterium ehrenbergii. Microbes Environ. 2006;21:278–83. doi: 10.1264/jsme2.21.278. [DOI] [Google Scholar]
- 50.Yuasa T, Okazaki Y, Iwasaki N, Muto S. Involvement of a Calcium-Dependent Protein Kinase in Hypoosmotic Turgor Regulation in a Brackish Water Characeae Lamprothamnium succinctum. Plant Cell Physiol. 1997;38:586–94. doi: 10.1093/oxfordjournals.pcp.a029208. [DOI] [Google Scholar]
- 51.Gifford JL, Walsh MP, Vogel HJ. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem J. 2007;405:199–221. doi: 10.1042/BJ20070255. [DOI] [PubMed] [Google Scholar]
- 52.Harmon AC, Putnam-Evans C, Cormier MJ. A calcium-dependent but calmodulin-independent protein kinase from soybean. Plant Physiol. 1987;83:830–7. doi: 10.1104/pp.83.4.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li J, Lee YR, Assmann SM. Guard cells possess a calcium-dependent protein kinase that phosphorylates the KAT1 potassium channel. Plant Physiol. 1998;116:785–95. doi: 10.1104/pp.116.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuasa T, Takahashi K, Muto S. Purification and Characterization of a Ca2+-Dependent Protein Kinase from the Halotolerant Green Alga Dunaliella tertiolecta. Plant Cell Physiol. 1995;36:699–708. [Google Scholar]
- 55.Dymek EE, Smith EF. PF19 encodes the p60 catalytic subunit of katanin and is required for assembly of the flagellar central apparatus in Chlamydomonas. J Cell Sci. 2012;125:3357–66. doi: 10.1242/jcs.096941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pical C, Fredlund KM, Petit PX, Sommarin M, Møller IM. The outer membrane of plant mitochondria contains a calcium-dependent protein kinase and multiple phosphoproteins. FEBS Lett. 1993;336:347–51. doi: 10.1016/0014-5793(93)80835-I. [DOI] [PubMed] [Google Scholar]
- 57.Patharkar OR, Cushman JC. A stress-induced calcium-dependent protein kinase from Mesembryanthemum crystallinum phosphorylates a two-component pseudo-response regulator. Plant J. 2000;24:679–91. doi: 10.1046/j.1365-313x.2000.00912.x. [DOI] [PubMed] [Google Scholar]
- 58.Anil VS, Harmon AC, Rao KS. Spatio-temporal accumulation and activity of calcium-dependent protein kinases during embryogenesis, seed development, and germination in sandalwood. Plant Physiol. 2000;122:1035–43. doi: 10.1104/pp.122.4.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu SX, Hrabak EM. An Arabidopsis calcium-dependent protein kinase is associated with the endoplasmic reticulum. Plant Physiol. 2002;128:1008–21. doi: 10.1104/pp.010770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dammann C, Ichida A, Hong B, Romanowsky SM, Hrabak EM, Harmon AC, Pickard BG, Harper JF. Subcellular targeting of nine calcium-dependent protein kinase isoforms from Arabidopsis. Plant Physiol. 2003;132:1840–8. doi: 10.1104/pp.103.020008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ueoka-Nakanishi H, Sazuka T, Nakanishi Y, Maeshima M, Mori H, Hisabori T. Thioredoxin h regulates calcium dependent protein kinases in plasma membranes. FEBS J. 2013;280:3220–31. doi: 10.1111/febs.12301. [DOI] [PubMed] [Google Scholar]
- 62.Lu SX, Hrabak EM. The myristoylated amino-terminus of an Arabidopsis calcium-dependent protein kinase mediates plasma membrane localization. Plant Mol Biol. 2013;82:267–78. doi: 10.1007/s11103-013-0061-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gundersen RE, Nelson DL. A novel Ca2+-dependent protein kinase from Paramecium tetraurelia. J Biol Chem. 1987;262:4602–9. [PubMed] [Google Scholar]
- 64.Son M, Gundersen RE, Nelson DL. A second member of the novel Ca(2+)-dependent protein kinase family from Paramecium tetraurelia. Purification and characterization. J Biol Chem. 1993;268:5940–8. [PubMed] [Google Scholar]
- 65.Choi KM, Kim JY, Moon SU, Lee HW, Sattabongkot J, Na BK, Kim DW, Suh EJ, Kim YJ, Cho SH, et al. Molecular cloning of Plasmodium vivax calcium-dependent protein kinase 4. Korean J Parasitol. 2010;48:319–24. doi: 10.3347/kjp.2010.48.4.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Holder AA, Mohd Ridzuan MA, Green JL. Calcium dependent protein kinase 1 and calcium fluxes in the malaria parasite. Microbes Infect. 2012;14:825–30. doi: 10.1016/j.micinf.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 67.Han HY, Zhu SH, Jiang LL, Li Y, Dong H, Zhao QP, Kong CL, Huang B. Molecular characterization and analysis of a novel calcium-dependent protein kinase from Eimeria tenella. Parasitology. 2013;140:746–55. doi: 10.1017/S0031182012002107. [DOI] [PubMed] [Google Scholar]
- 68.Harris E. The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use San Diego: Academic Press, 1989 [DOI] [PubMed] [Google Scholar]
- 69.Quisel JD, Wykoff DD, Grossman AR. Biochemical characterization of the extracellular phosphatases produced by phosphorus-deprived Chlamydomonas reinhardtii. Plant Physiol. 1996;111:839–48. doi: 10.1104/pp.111.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mus F, Dubini A, Seibert M, Posewitz MC, Grossman AR. Anaerobic acclimation in Chlamydomonas reinhardtii: anoxic gene expression, hydrogenase induction, and metabolic pathways. J Biol Chem. 2007;282:25475–86. doi: 10.1074/jbc.M701415200. [DOI] [PubMed] [Google Scholar]
- 71.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 72.Kindle KL. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 1990;87:1228–32. doi: 10.1073/pnas.87.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 74.Witman GB. Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 1986;134:280–90. doi: 10.1016/0076-6879(86)34096-5. [DOI] [PubMed] [Google Scholar]