Abstract
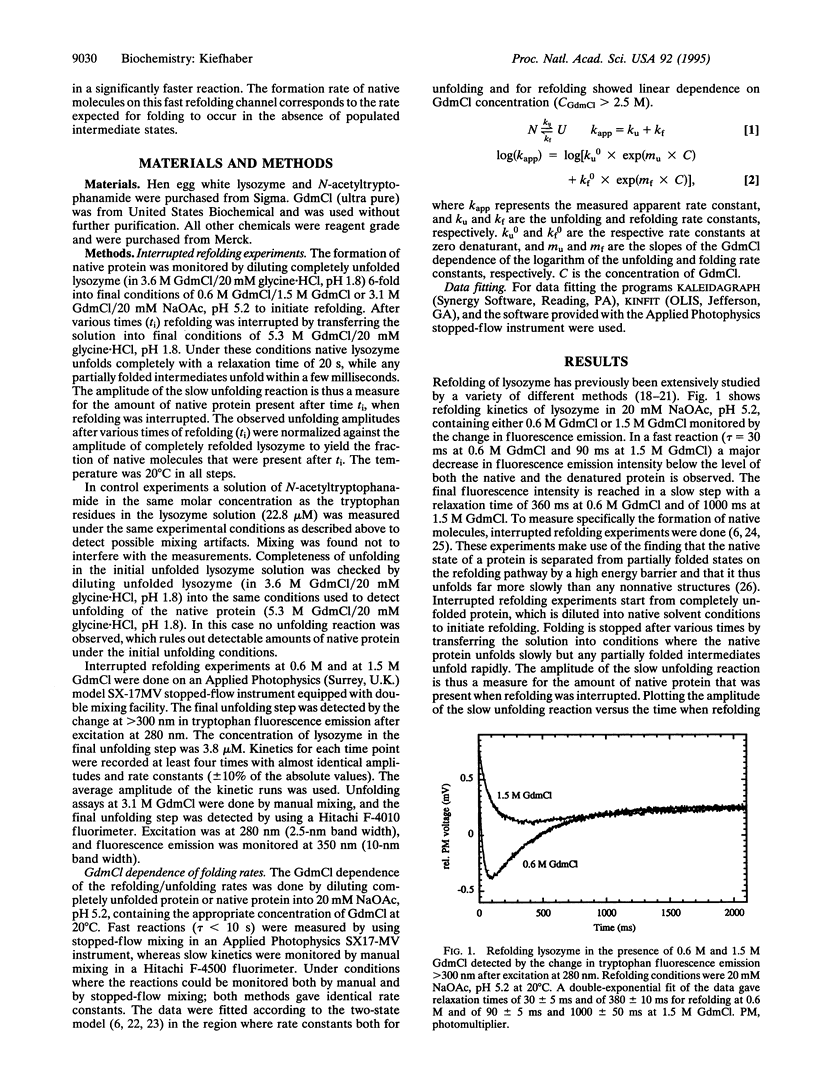
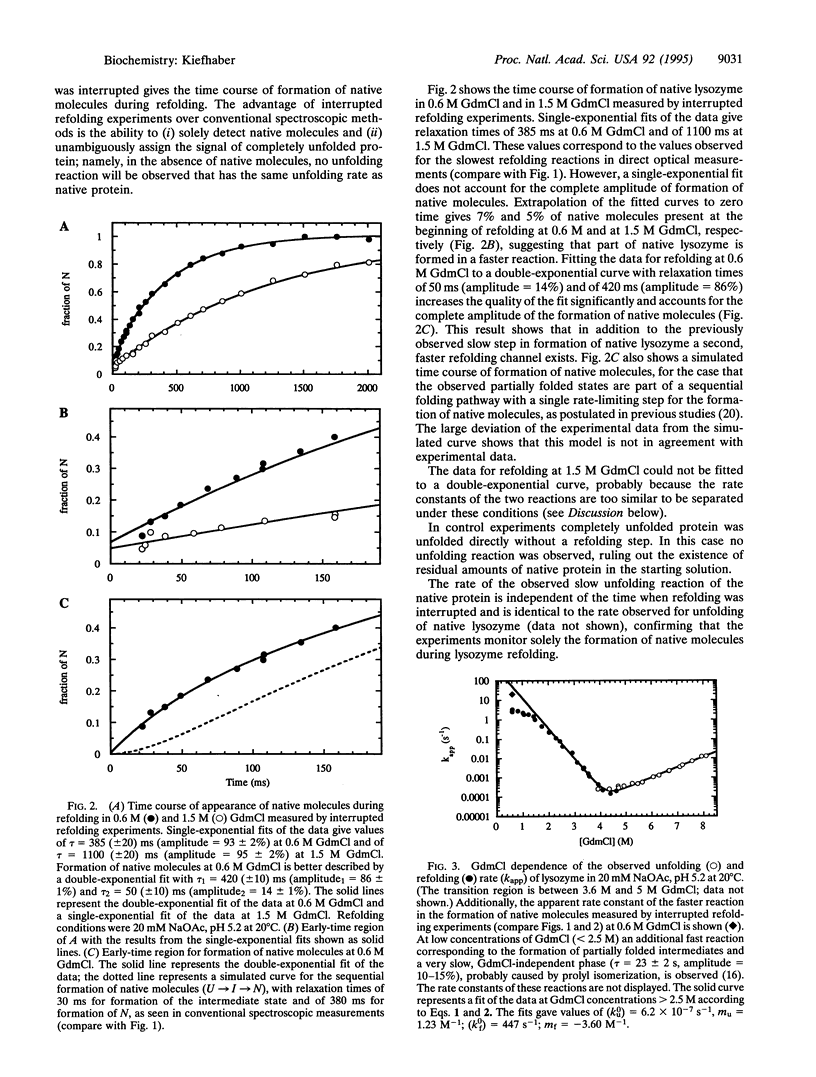
Folding of lysozyme from hen egg white was investigated by using interrupted refolding experiments. This method makes use of a high energy barrier between the native state and transient folding intermediates, and, in contrast to conventional optical techniques, it enables one to specifically monitor the amount of native molecules during protein folding. The results show that under strongly native conditions lysozyme can refold on parallel pathways. The major part of the lysozyme molecules (86%) refold on a slow kinetic pathway with well-populated partially folded states. Additionally, 14% of the molecules fold faster. The rate constant of formation of native molecules on the fast pathway corresponds well to the rate constant expected for folding to occur by a two-state process without any detectable intermediates. The results suggest that formation of the native state for the major fraction of lysozyme molecules is retarded compared with the direct folding process. Partially structured intermediates that transiently populate seem to be kinetically trapped in a conformation that can only slowly reach the native structure.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldwin R. L. The nature of protein folding pathways: the classical versus the new view. J Biomol NMR. 1995 Feb;5(2):103–109. doi: 10.1007/BF00208801. [DOI] [PubMed] [Google Scholar]
- Brandts J. F., Halvorson H. R., Brennan M. Consideration of the Possibility that the slow step in protein denaturation reactions is due to cis-trans isomerism of proline residues. Biochemistry. 1975 Nov 4;14(22):4953–4963. doi: 10.1021/bi00693a026. [DOI] [PubMed] [Google Scholar]
- Bryngelson J. D., Wolynes P. G. Spin glasses and the statistical mechanics of protein folding. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7524–7528. doi: 10.1073/pnas.84.21.7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C. J., Thirumalai D. Kinetics and thermodynamics of folding in model proteins. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6369–6372. doi: 10.1073/pnas.90.13.6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffotte A. F., Guillou Y., Goldberg M. E. Kinetic resolution of peptide bond and side chain far-UV circular dichroism during the folding of hen egg white lysozyme. Biochemistry. 1992 Oct 13;31(40):9694–9702. doi: 10.1021/bi00155a024. [DOI] [PubMed] [Google Scholar]
- Chen B. L., Baase W. A., Nicholson H., Schellman J. A. Folding kinetics of T4 lysozyme and nine mutants at 12 degrees C. Biochemistry. 1992 Feb 11;31(5):1464–1476. doi: 10.1021/bi00120a025. [DOI] [PubMed] [Google Scholar]
- Creighton T. E., Goldenberg D. P. Kinetic role of a meta-stable native-like two-disulphide species in the folding transition of bovine pancreatic trypsin inhibitor. J Mol Biol. 1984 Nov 5;179(3):497–526. doi: 10.1016/0022-2836(84)90077-9. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Protein folding. Biochem J. 1990 Aug 15;270(1):1–16. doi: 10.1042/bj2700001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton T. E. The energetic ups and downs of protein folding. Nat Struct Biol. 1994 Mar;1(3):135–138. doi: 10.1038/nsb0394-135. [DOI] [PubMed] [Google Scholar]
- Engel J., Prockop D. J. The zipper-like folding of collagen triple helices and the effects of mutations that disrupt the zipper. Annu Rev Biophys Biophys Chem. 1991;20:137–152. doi: 10.1146/annurev.bb.20.060191.001033. [DOI] [PubMed] [Google Scholar]
- Honeycutt J. D., Thirumalai D. Metastability of the folded states of globular proteins. Proc Natl Acad Sci U S A. 1990 May;87(9):3526–3529. doi: 10.1073/pnas.87.9.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzhaki L. S., Evans P. A., Dobson C. M., Radford S. E. Tertiary interactions in the folding pathway of hen lysozyme: kinetic studies using fluorescent probes. Biochemistry. 1994 May 3;33(17):5212–5220. doi: 10.1021/bi00183a026. [DOI] [PubMed] [Google Scholar]
- Jackson S. E., Fersht A. R. Folding of chymotrypsin inhibitor 2. 1. Evidence for a two-state transition. Biochemistry. 1991 Oct 29;30(43):10428–10435. doi: 10.1021/bi00107a010. [DOI] [PubMed] [Google Scholar]
- Jaenicke R. Folding and association of proteins. Prog Biophys Mol Biol. 1987;49(2-3):117–237. doi: 10.1016/0079-6107(87)90011-3. [DOI] [PubMed] [Google Scholar]
- Kato S., Shimamoto N., Utiyama H. Identification and characterization of the direct folding process of hen egg-white lysozyme. Biochemistry. 1982 Jan 5;21(1):38–43. doi: 10.1021/bi00530a007. [DOI] [PubMed] [Google Scholar]
- Kiefhaber T., Grunert H. P., Hahn U., Schmid F. X. Folding of RNase T1 is decelerated by a specific tertiary contact in a folding intermediate. Proteins. 1992 Feb;12(2):171–179. doi: 10.1002/prot.340120210. [DOI] [PubMed] [Google Scholar]
- Kim P. S., Baldwin R. L. Intermediates in the folding reactions of small proteins. Annu Rev Biochem. 1990;59:631–660. doi: 10.1146/annurev.bi.59.070190.003215. [DOI] [PubMed] [Google Scholar]
- Kim P. S., Baldwin R. L. Specific intermediates in the folding reactions of small proteins and the mechanism of protein folding. Annu Rev Biochem. 1982;51:459–489. doi: 10.1146/annurev.bi.51.070182.002331. [DOI] [PubMed] [Google Scholar]
- Kuwajima K., Hiraoka Y., Ikeguchi M., Sugai S. Comparison of the transient folding intermediates in lysozyme and alpha-lactalbumin. Biochemistry. 1985 Feb 12;24(4):874–881. doi: 10.1021/bi00325a010. [DOI] [PubMed] [Google Scholar]
- Matthews C. R. Effect of point mutations on the folding of globular proteins. Methods Enzymol. 1987;154:498–511. doi: 10.1016/0076-6879(87)54092-7. [DOI] [PubMed] [Google Scholar]
- Miranker A., Radford S. E., Karplus M., Dobson C. M. Demonstration by NMR of folding domains in lysozyme. Nature. 1991 Feb 14;349(6310):633–636. doi: 10.1038/349633a0. [DOI] [PubMed] [Google Scholar]
- Miranker A., Robinson C. V., Radford S. E., Aplin R. T., Dobson C. M. Detection of transient protein folding populations by mass spectrometry. Science. 1993 Nov 5;262(5135):896–900. doi: 10.1126/science.8235611. [DOI] [PubMed] [Google Scholar]
- Nall B. T., Garel J. R., Baldwin R. L. Test of the extended two-state model for the kinetic intermediates observed in the folding transition of ribonuclease A. J Mol Biol. 1978 Jan 25;118(3):317–330. doi: 10.1016/0022-2836(78)90231-0. [DOI] [PubMed] [Google Scholar]
- Odefey C., Mayr L. M., Schmid F. X. Non-prolyl cis-trans peptide bond isomerization as a rate-determining step in protein unfolding and refolding. J Mol Biol. 1995 Jan 6;245(1):69–78. doi: 10.1016/s0022-2836(95)80039-5. [DOI] [PubMed] [Google Scholar]
- Radford S. E., Dobson C. M., Evans P. A. The folding of hen lysozyme involves partially structured intermediates and multiple pathways. Nature. 1992 Jul 23;358(6384):302–307. doi: 10.1038/358302a0. [DOI] [PubMed] [Google Scholar]
- Sali A., Shakhnovich E., Karplus M. How does a protein fold? Nature. 1994 May 19;369(6477):248–251. doi: 10.1038/369248a0. [DOI] [PubMed] [Google Scholar]
- Schmid F. X. Fast-folding and slow-folding forms of unfolded proteins. Methods Enzymol. 1986;131:70–82. doi: 10.1016/0076-6879(86)31035-8. [DOI] [PubMed] [Google Scholar]
- Schmid F. X. Mechanism of folding of ribonuclease A. Slow refolding is a sequential reaction via structural intermediates. Biochemistry. 1983 Sep 27;22(20):4690–4696. doi: 10.1021/bi00289a013. [DOI] [PubMed] [Google Scholar]
- Segawa S., Sugihara M. Characterization of the transition state of lysozyme unfolding. I. Effect of protein-solvent interactions on the transition state. Biopolymers. 1984 Nov;23(11 Pt 2):2473–2488. doi: 10.1002/bip.360231122. [DOI] [PubMed] [Google Scholar]
- Shakhnovich E. I., Finkelstein A. V. Theory of cooperative transitions in protein molecules. I. Why denaturation of globular protein is a first-order phase transition. Biopolymers. 1989 Oct;28(10):1667–1680. doi: 10.1002/bip.360281003. [DOI] [PubMed] [Google Scholar]
- Shakhnovich E, Farztdinov G, Gutin AM, Karplus M. Protein folding bottlenecks: A lattice Monte Carlo simulation. Phys Rev Lett. 1991 Sep 16;67(12):1665–1668. doi: 10.1103/PhysRevLett.67.1665. [DOI] [PubMed] [Google Scholar]
- Sosnick T. R., Mayne L., Hiller R., Englander S. W. The barriers in protein folding. Nat Struct Biol. 1994 Mar;1(3):149–156. doi: 10.1038/nsb0394-149. [DOI] [PubMed] [Google Scholar]
- Weissman J. S., Kim P. S. Reexamination of the folding of BPTI: predominance of native intermediates. Science. 1991 Sep 20;253(5026):1386–1393. doi: 10.1126/science.1716783. [DOI] [PubMed] [Google Scholar]
- Zhang J. X., Goldenberg D. P. Amino acid replacement that eliminates kinetic traps in the folding pathway of pancreatic trypsin inhibitor. Biochemistry. 1993 Dec 28;32(51):14075–14081. doi: 10.1021/bi00214a001. [DOI] [PubMed] [Google Scholar]



