Abstract
Gene expression and consequent biological activity of adult tissue stem cells are regulated by signals emanating from the local microenvironment (niche). To gain insights into the molecular regulation of spermatogonial stem cells (SSCs), gene expression was characterized from SSCs isolated from their cognate niches of cryptorchid (stem cell-enriched), wild-type, and busulfan-treated (stem cell-depleted) mouse testes. Quantitative assessment of stem cell activity in each testis model was determined using an in vivo functional assay and correlated with gene expression using Affymetrix MGU74Av2 microarrays and the ChipStat algorithm optimized to detect gene expression from rare cells in complex tissues. We identified 389 stem/progenitor spermatogonia candidate genes, which exhibited significant overlap with genes expressed by embryonic, hematopoietic, and neural stem cells; enriched spermatogonia; and cultured SSCs identified in previous studies. Candidate cell surface markers identified by the microarray may facilitate the isolation and enrichment of stem and/or progenitor spermatogonia. Flow cytometric analyses confirmed the expression of chemokine receptor 2 (Ccr2) and Cd14 on a subpopulation cryptorchid testis cells (α6-integrin+, side scatterlo) enriched for SSCs. These cell surface molecules may mark progenitor spermatogonia but not SSCs because Ccr2+ and Cd14+ fractions failed to produce spermatogenesis upon transplantation to recipient testes. Functional annotation of candidate genes and subsequent immunohistochemistry revealed that proteins involved in post-transcriptional regulation are overrepresented in cryptorchid testes that are enriched for SSCs. Comparative analyses indicated that this is a recurrent biological theme among stem cells.
Keywords: Spermatogonial stem cell, Progenitor, Spermatogonia, Microarray, RNA-binding proteins, Post-transcriptional regulation
Introduction
Self-renewing systems in adult animals, such as epidermis, intestinal epithelium, hematopoiesis, and spermatogenesis, are maintained by resident stem cells. These tissues are in constant flux because cells that are lost because of terminal differentiation or death must be replaced continuously from a pool of tissue-specific progenitor cells. Homeostasis is achieved because of the unique capacity of stem cells to maintain their own population (self-renew) as well as replenish the pool of committed progenitors that give rise to the specified cell lineage. Concerted efforts are focused on harnessing the unique biological potential of stem cells to regenerate unhealthy or damaged tissues, and these efforts will be facilitated by the investigation of molecular mechanisms that control self-renewal and differentiation fate decisions.
Two pioneering studies, published in October 2002 [1, 2], were designed to identify a “stem cell molecular signature ” by comparing gene expression in hematopoietic stem cells (HSCs), embryonic stem cells (ESCs), and neural stem cells (NSCs). These studies were innovative but failed to converge on a consensus list of candidate genes [3–5]. However, these seminal studies provide an important framework for future studies. There is general consensus that common regulatory pathways exist among stem cells, which will be revealed by continued evaluation of stem cells from different tissues and different species [3].
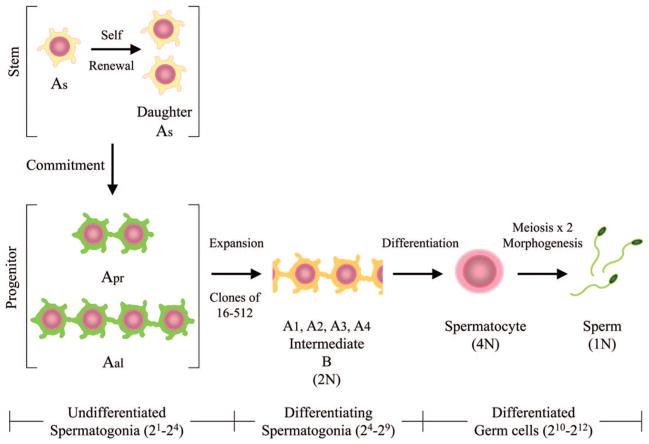
Spermatogonial stem cells lie at the foundation of spermatogenesis and are located on the basement membrane of seminiferous tubules in specific niches that are defined, in part, by close associations with Sertoli cells [6]. When a stem cell divides in mouse seminiferous tubules, the daughter cells either migrate to individual niches and become two new stem cells (Asingle spermatogonia [As]), or undergo incomplete cytokinesis, stay together (Apaired [Apr]), and enter the progenitor pool (which includes Apr and Aaligned [Aal] spermatogonia; Fig. 1). Subsequent divisions of Apr spermatogonia produce chains of 4, 8, and 16 undifferentiated progenitor spermatogonia (Aal) that will ultimately give rise to all cells of the spermatogenic lineage (reviewed in [7, 8]). Stem and progenitor spermatogonia (As, Apr, Aal) collectively make up the pool of classically defined undifferentiated spermatogonia. In the mouse, once a stem cell commits to differentiation, it undergoes 12 amplifying divisions (including 2 meiotic divisions) and morphological differentiation to generate a clone of mature spermatozoa (Fig. 1 integrates stem cell and spermatogenesis jargon). The developmental fate of dividing spermatogonial stem cells (self-renewal vs. commitment to differentiation) is subject to intrinsic and extrinsic controls [9] and likely influenced by the availability/quality of stem cell niches and the biological demand of the dependent system (spermatogenesis). Investigation of these early events in spermatogenesis will provide general insight into stem cell biology and specific information about spermatogonial stem cells (SSCs), which have the potential to restore fertility [10–14] and modify the germline of animals [15–20].
Figure 1.
Mouse spermatogenic lineage. When a spermatogonial stem cell (type As spermatogonia) divides, daughter cells can remain type As spermatogonia (self-renewal) or commit to differentiation, giving rise through 12 amplifying divisions (212) to a clone of spermatogenesis that could theoretically contain 4,096 spermatozoa. Once committed to differentiation, an As spermatogonial stem cell gives rise to a pair of undifferentiated spermatogonia (Apr) that are connected by a cytoplasmic bridge. Incomplete cytokinesis is a hallmark of germ cell proliferation. The first four population doubling divisions (21–24) generate chains of undifferentiated Apr and Aal spermatogonia; Apr, Aal4, Aal8, Aal16, which collectively make up the pool of progenitor spermatogonia. Differentiation becomes evident morphologically when a chain of Aal16 spermatogonia becomes a chain of type A116 spermatogonia. The next five population doubling divisions (25–29) generate types A2, A3, A4, intermediate, and B differentiating spermatogonia. The 10th division (210) produces primary spermatocytes (4N) and is followed by two meiotic divisions (211–212) and spermiogenesis to produce 1N, terminally differentiated spermatozoa. Abbreviations: Aal, Aaligned; Apr, Apaired; As, Asingle.
Biological investigation of these important cells is complicated because they are extremely rare, constituting approximately 1 in 3,000 cells of the adult mouse testis [21], and few distinguishing morphological or biochemical characteristics are known. The only way to definitively identify SSCs in adult testis cell populations is by observing their capacity to produce and maintain colonies of spermatogenesis in a functional assay. Therefore, the development of an SSC transplantation technique [22, 23] was an important advance that allowed functional assessment of male germline stem cells and facilitated dissection of their molecular and biological characteristics. Fluorescence-activated cell sorting followed by transplantation of selected populations has proven to be a particularly powerful tool for characterizing SSCs, which can now be described by the cell surface phenotype, α6-integrin+, β1-integrin+, Thy-1+, CD24+, CD9+, Cdh1+, MHC-1−, CD51−, Sca-1−, CD34−, and c-kit− [24–27]. However, these methods are slow, laborious, and primarily limited to the evaluation of cell surface characteristics.
Alternative strategies are necessary to gain a more comprehensive insight into the genetic and molecular mechanisms that regulate stem cell activity in the adult testis. High-density microarrays allow the simultaneous expression analysis of thousands of genes and are biased only by the number and quality of sequences on the array. Microarrays or other high throughput analyses (e.g., subtractive hybridization, serial analysis of gene expression, and differential display polymerase chain reaction) have now been used to identify genes expressed by stem/ progenitor cells in a variety of systems [1, 2], including the male germline [28–31]. These studies provide a tremendous informatics resource that, when coupled with additional comparative genetic analyses, will help identify unifying themes in stem cell biology.
In the current study, Affymetrix MGU74Av2 arrays (Affymetrix, Santa Clara, CA, http://www.affymetrix.com) were used to identify gene expression profiles that correlate with SSC activity in wild-type, busulfan-treated, and cryptorchid mouse testes. The current study confirms that these three testis models are highly divergent with respect to stem/progenitor cell content. The multipoint analysis (three testis models) of heterogeneous testis cell populations allowed molecular dissection of genes expressed by testicular somatic cells, differentiating germ cells, and stem/progenitor spermatogonia isolated directly from their cognate environments in the mouse testis. These studies were facilitated because the cryptorchid testis is enriched for SSCs. Although the cryptorchid procedure modifies the germ cell and somatic environment of the testis, the data generated can now be used to investigate the expression and function of candidate genes in normal mouse testes. Indeed, functional annotation of candidate genes in the current study suggests a role for post-transcriptional regulation, and we have begun confirming expression of specific regulators in stem/progenitor spermatogonia of normal mouse testes.
Materials and Methods
Animals
Testis cells for microarray experiments were obtained from adult cryptorchid, wild-type, and busulfan-treated C57BL/6 mice (Jackson Laboratory, Bar Harbor, ME, http://www.jax.org). For the transplantation experiments, ROSA26 mice (stock no. 2192; also on the C57BL/6 background) that express the Escherichia coli lacZ gene in all germ cells were used as donors. In vivo enrichment of spermatogonial stem cells was accomplished using the experimental cryptorchid procedure. Testes were attached inside the abdominal cavity, where exposure to core body temperature causes the loss of most germ cells. However, stem and progenitor spermatogonia remain [32] at the time of testis collection (2–3 months after surgery), and the stem cells are enriched 20–25-fold [33]. A germ cell-depleted testis model was generated by treatment with the chemotherapeutic agent busulfan (50 mg/kg, i.p.; Sigma-Aldrich, St. Louis, http://www.sigmaaldrich.com). The Animal Care and Use Committee of the University of Pennsylvania approved all experimental procedures in accordance with the Guide for Care and Use of Laboratory Animals of the National Academy of Sciences (Assurance no. A3079-01).
Testis Cell Isolation
To ascertain the effect of cryptorchid and busulfan treatment procedures, testes were weighed; cryptorchid testes weighing more than 28 mg and busulfan-treated testes weighing more than 25.5 mg were excluded from analysis. Single-cell suspensions from the testes of cryptorchid, wild-type, and busulfan-treated mice were generated by two-step enzymatic digestion, as previously described [34]. Cells for SSC transplantation were suspended in Minimum Essential Medium α (Invitrogen, Carlsbad, CA, http://www.invitrogen.com) containing 10% fetal bovine serum (FBS; Hy-Clone, Logan, UT, http://www.hyclone.com). For RNA isolation, cells were suspended in lysis buffer (RNeasy RNA isolation kit; Qiagen, Hilden, Germany, http://www1.qiagen.com).
Spermatogonial Stem Cell Transplantation
To determine relative SSC concentrations, testis cell suspensions from cryptorchid, wild-type, and busulfan-treated mice were introduced by efferent duct injection into the testes of busulfan-treated 129svCP × C57BL/6 (129/C57) hybrid mice, as previously described [34]. Recipient mice were treated with busulfan (50 mg/kg, i.p.) to remove endogenous germ cells 6 weeks prior to transplantation. Testes of recipient mice were collected 2 months after transplantation and stained with the β-galactosidase substrate, 5-bromo-4-chloro-3-indolyl-β-D-galactoside, to visualize donor-derived spermatogenesis. Transgenic (ROSA26) spermatogonial stem cells are defined by the ability to produce and maintain blue colonies of spermatogenesis in recipient testes, and each colony is clonally derived from a single transplanted SSC [35, 36]. Differentiated germ cells do not produce and sustain colonies of spermatogenesis, and endogenous germ cells do not express the lacZ trans-gene. Approximately 10 μl of donor testis cell suspension was injected per recipient testis. Because donor testis cell concentrations varied, colony number was normalized to 105 cells injected per testis.
RNA Isolation and Microarray Analysis
RNA was isolated from cell suspensions of cryptorchid, wild-type, and busulfan-treated testes using an RNeasy RNA isolation kit (Qiagen). At least three animals per group were used for each replicate to normalize biological variation, and three replicate experiments were performed using a total of nine Affymetrix Mouse Genome U74Av2 arrays (three groups, with three replicates per group). RNA quality was confirmed by spectrophotometer (samples with 260/280 nm ratio ≥1.8 were used) and by visual inspection of 18S and 28S ribosomal RNA bands on agarose gels. For each sample, 5 μg of total RNA was reverse-transcribed to produce double-stranded cDNA. Biotinylated cRNA was generated by in vitro transcription and fragmented for hybridization to microarrays. Raw file processing was performed and presence/absence determination was made using Affymetrix GeneChip software. Differentially expressed genes were identified using ChipStat [37] with a pairwise p value of .05 and an overall false discovery rate (FDR) expectation of 10 or fewer genes per list (FDR, 10 of 12,844), which corresponds to a target p value of 8 × 10−4. Empirical calibration of the ChipStat expectation curve was performed as previously described [37]. Composite lists (Boolean combinations) were generated from initial pairwise comparisons using in-house software (S.M. and L.C.). Assignment to Unigene clusters and chromosomes was also performed using in-house software written in PERL.
Fluorescence-Activated Cell Sorting
Fluorescence-activated cell sorting (FACS) was used to verify candidate genes encoding cell surface markers in a subpopulation of cryptorchid testis cells designated α6-integrinhi and exhibiting low side scatter of incident light that is enriched for SSCs [24]. This SSC-enriched cryptorchid testis cell fraction was evaluated for the expression of the candidate cell surface markers, chemokine receptor 2 (Ccr2), Ccr5, and Cd14. Cells were sorted into marker+ and marker− fractions and transplanted into the testes of recipient mice to determine relative SSC content. The Ccr2 primary antibody was previously described [38]. The Ccr5 and Cd14 primary antibodies and secondary reagents were from BD Biosciences (San Jose, CA, http://www.bdbiosciences.com).
Immunohistochemistry
Testes of adult C57BL/6 mice were fixed in 4% paraformaldehyde, paraffin-embedded, and sectioned (5 μm). Following high-temperature citrate buffer antigen retrieval, sections were stained with rabbit anti-Rbmy IgG (1:200 dilution; gift from Dr. David Elliott, University of Newcastle upon Tyne, Newcastle upon Tyne, U.K.) and Armenian hamster anti-PLZF IgG (1:800 dilution; gift from R. Hobbs and P.P. Pandolfi, Sloan-Kettering Institute, New York). Primary antibodies were detected with goat anti-rabbit IgG Alexa-Fluor555 (1:200; Invitrogen), biotin-conjugated anti-Armenian hamster IgG fluorescein isothiocyanate (1:50; Jackson Immunoresearch Laboratories, West Grove, PA, http://www.jacksonimmuno.com), and streptavidin-488 (1:100; Invitrogen). Samples were mounted with VectaShield mounting medium containing 4,6-di-amidino-2-phenylindole (Vector Laboratories, Burlingame, CA, http://www.vectorlabs.com).
Statistical Analyses
General estimating equations were used to test for differences among groups in the spermatogonial transplantation experiments. Association to Gene Ontology annotation data was performed using the EASE software package (NIAID, NIH, Frederick, MD, http://david.abcc.ncifcrf.gov/content.jsp?file=ease/ease.htm&type=1) [39], and a Monte Carlo estimate of the within-system false discovery rate was also estimated using the software. FDR <0.05 were reported as significant associations. Because EASE does not report significantly underrepresented categories, we used the cumulative hypergeometric distribution to calculate the probability of spermatogenesis underrepresentation in somatic cells. Chromosomal associations were performed using a Bonferroni-corrected cumulative hypergeometric distribution. Pairwise estimates of the statistical significance of associations between our lists and previously described stem cell lists were also derived using the cumulative hypergeometric distribution.
Results
Each organ system and species presents unique challenges and opportunities for dissecting the molecular mechanisms controlling stem cell activity. In the current study, gene expression was examined in three mouse testis models with different proportions of somatic cells, germ cells, and stem/progenitor spermatogonia (cryptorchid, wild-type, and busulfan-treated). RNA was isolated from triplicate experimental testis samples, and RNA probes were generated to hybridize with Affymetrix microarrays (three groups with three replicates per group, for a total of nine mouse genome U74Av2 arrays; Affymetrix). Hypothetical expression profiles or clusters were established (corresponding with somatic cell, male germ cell or stem and progenitor spermatogonia content) to facilitate the interpretation of gene expression results (Fig. 2; discussed below).
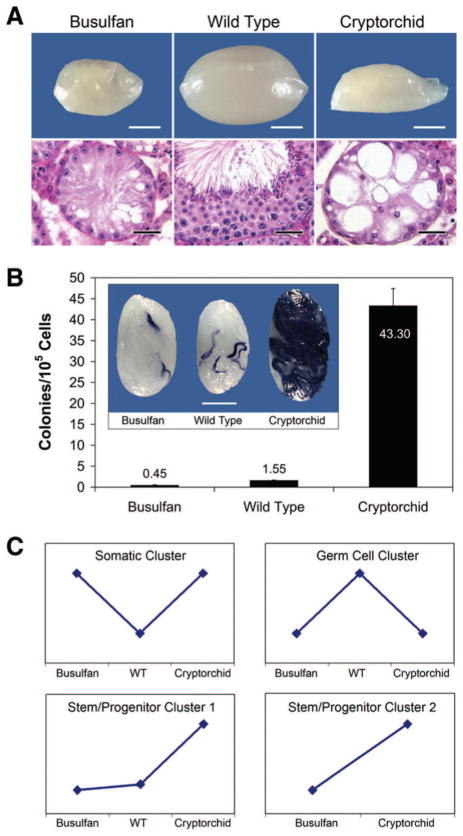
Figure 2.
Busulfan-treated (Bu), WT, and cryptorchid (Cr) testis models were evaluated for stem cell activity and gene expression. (A): Macroscopic (top panels; scale bar = 2 mm) and histological (bottom panels; scale bar = 50 μm) examination of Bu (left panels), WT (center panels), and Cr (right panels) testes. Compared to WT testes (92.26 ± 6.33 mg; 35.26 ± 4.1 × 106 cells per testis), Bu and Cr testes were smaller (20.6 ± 0.53 and 23.54 ± 0.62 mg, respectively) and contained fewer cells (1.37 ± 0.17 × 106 and 1.38 ± 0.16 × 106 cells per testis, respectively) because they were nearly devoid of germ cells. Histological sections were stained with hematoxylin and eosin. (B): The spermatogonial stem cell transplantation technique was used to determine the stem cell activity (mean ± SEM) in Bu (0.45 ± 0.13 colonies per 105 cells transplanted), WT (1.55 ± 0.13 colonies per 105 cells transplanted), and Cr (43.30 ± 4.17 colonies per 105 cells transplanted) donor testes. The spermatogonial stem cell transplantation technique has an efficiency of 5%. Therefore, the stem cell concentrations in Bu, WT, and Cr testes were 1 in 11,111 (105 cells transplanted per 0.45 colonies × 5%), 1 in 3,226, and 1 in 115, respectively. Inset, relative stem cell activity in the three testis models is demonstrated by the extent of donor spermatogenesis (blue) produced following transplantation of 500,000 transgenic (lacZ) donor testis cells. Scale bar = 2 mm. (C): Four hypothetical gene expression clusters were predicted on the basis of the morphological and functional (stem cell) analyses in (A) and (B). Gene expression data were organized into clusters predicted to represent somatic cells (WT < Cr, Bu), germ cells (WT > Cr, Bu), stem/ progenitor cluster 1 (Cr > WT, Bu; most specific), and stem/progenitor cluster 2 (Cr > Bu; less restrictive; descriptions given in text). Abbreviation: WT, wild-type.
Germ cells make up ~95% of testicular cell content, and when these cells are removed by ablative therapies (i.e., cryptorchid, busulfan; Fig. 2A, right and left panels, respectively), the remaining cell populations (e.g., somatic cells: interstitial, myoid, and Sertoli) are effectively enriched approximately 20-fold. Therefore, a gene expression profile that is higher in cryptorchid and busulfan-treated testes than in wild-type testes should correlate with somatic cell content (Fig. 2C). Conversely, genes that are expressed at a higher level in wild-type testes than cryptorchid or busulfan-treated testes should correlate with germ cell content. Finally, although cryptorchid testes are nearly depleted of germ cells (Fig. 2A, right panels), stem and progenitor cells remain [32, 33, 40], and the transplant data in Figure 2B demonstrate that cryptorchid testes have a significantly higher concentration of stem cells than wild-type (p < .001) or busulfan-treated (p < .001) testes (28-fold and 97-fold, respectively). Therefore, we hypothesize that genes expressed at a higher level in cryptorchid testes than in wild-type and busulfan-treated testes correspond to stem and progenitor spermatogonia.
A typical strategy for analyzing microarray data and limiting false-positive rates is to incorporate fold-change criteria (e.g., twofold) in the search for differentially regulated genes. This strategy is adequate for studying pure target cell populations where large changes in gene expression are likely to reflect true biological phenomena. However, in complex tissues where the cells of interest are rare, such as SSCs in the testis, greater sensitivity is required. Small but reproducible changes in gene expression at the whole-organ level may reflect significant changes of a subpopulation of cells within the organ. On Affymetrix microarrays, each probe set (transcript) comprises 16 oligonucleotide probe pairs (perfect match and mismatch). Expression values for each probe pair are typically combined with the values for the other probe pairs of the same transcript to obtain a robust estimate of transcript abundance. However, one drawback of this approach is that the individual probe pair information is discarded, thus decreasing statistical power. We (S.M. and L.C.) recently described a novel algorithm (ChipStat) for sensitive detection of gene expression changes in a complex tissue (mammary gland) using all probe pair-level information from Affymetrix microarrays [37]. In the current study, Chip-Stat analysis was used to evaluate Affymetrix gene expression data from cryptorchid, wild-type, and busulfan-treated testes, in which SSC concentrations were determined to be 1 of 115, 1 of 3,226, and 1 of 11,111, respectively (calculations given in Fig. 2B legend). Gene expression data were then organized into expression clusters predicted to represent germ cells, somatic cells, and stem/progenitor spermatogonia (Fig. 2C).
Among 12,422 probe sets (excluding controls) on the Affymetrix MGU74Av2 array, 5,957 were called present on one or more of the three testis cell populations. Analysis of the expression data was performed using ChipStat with statistical thresholds set for an expected FDR of less than 10 of 12,422 (FDR = 8 × 10−4) for each pairwise comparison. This analysis revealed that 1,311 probe sets fell into the germ cell expression cluster (wild-type [Wt] > cryptorchid [Cr], busulfan-treated [Bu]; supplemental online Table 1), 2,222 probe sets exhibited the profile of the somatic cell expression cluster (Wt < Cr, Bu; supplemental online Table 2), 88 genes were in stem/progenitor spermatogonia cluster 1 (Cr > Wt, Bu; Table 1), and 389 genes were in stem/progenitor cluster 2 (Cr > Bu; supplemental online Table 3). The rationale for generating two stem/progenitor candidate gene lists is that it is likely that many genes essential for stem/progenitor spermatogonia cell function are also expressed by more differentiated germ cells. These genes would be excluded from the more specific stem/progenitor cluster 1 (Cr > Wt, Bu). Therefore, we modified the criteria by excluding the wild-type data to generate the more relaxed stem/progenitor cluster 2 (Cr > Bu). Dazl and Ddx4 (mouse vasa homolog) are two examples of genes that were present on stem/progenitor spermatogonia cluster 2 but excluded from cluster 1. Expression of both genes has been demonstrated in undifferentiated spermatogonia [41, 42], and both are essential for the establishment and/or maintenance of spermatogenesis [43–46].
Table 1.
Stem/progenitor cluster 1 (cryptorchid > wild-type, busulfan-treated; 88 probe sets)
| Row | Probe set | Accession no. | Gene | Description |
|---|---|---|---|---|
| 1 | 98783_at | Y15131 | Rbmy1a1 | RNA binding motif protein, Y chromosome |
| 2 | 102627_at | AF061569 | Igf2bp1 | Insulin-like growth factor 2 mRNA binding protein |
| 3 | 95063_at | AI606257 | Cdca7 | Cell division cycle associated 7 |
| 4 | 98108_at | X15789 | Crabp1 | Cellular retinoic acid binding protein I |
| 5 | 92290_at | Z19580 | Siah1b | Seven in absentia 1B |
| 6 | 101883_s_at | L22977 | Xlr3a /// Xlr3b | X-linked lymphocyte-regulated 3A /// X-linked |
| 7 | 92607_at | AF017994 | Mest | Mesoderm specific transcript |
| 8 | 161787_f_at | AV363418 | Cdt1 | Chromatin licensing and DNA replication factor |
| 9 | 93397_at | U56819 | Ccr2 | Chemokine (C-C motif) receptor 2 |
| 10 | 94743_f_at | M29009 | LOC214403 | Complement factor H-related protein |
| 11 | 100479_at | AA756653 | Dnmt3a | DNA methyltransferase 3A |
| 12 | 96169_at | AA683788 | Sall4 | Sal-like 4 (Drosophila) |
| 13 | 92208_at | AI152966 | C1qdc1 | C1q domain containing 1 |
| 14 | 160702_at | AA791885 | Rbm35a | RNA binding motif protein 35A |
| 15 | 103301_i_at | AA866668 | Sox3 | SRY-box containing gene 3 |
| 16 | 101853_f_at | M12660 | Cfh | Complement component factor h |
| 17 | 94761_at | X70058 | Ccl7 | Chemokine (C-C motif) ligand 7 |
| 18 | 161968_f_at | AV370035 | Ccr5 | Chemokine (C-C motif) receptor 5 |
| 19 | 104116_at | AW124049 | D5Ertd593e | DNA segment, Chr 5, ERATO Doi 593, exp |
| 20 | 93112_at | D86725 | Mcm2 | Minichromosome maintenance deficient 2 rr |
| 21 | 104576_at | AW212397 | Ski | Sloan-Kettering viral oncogene homolog |
| 22 | 96047_at | U63146 | Rbp4 | Retinol binding protein 4, plasma |
| 23 | 101160_at | X53798 | Cxcl2 | Chemokine (C-X-C motif) ligand 2 |
| 24 | 100325_at | M65027 | Gp49a | Glycoprotein 49 A |
| 25 | 92291_f_at | M29008 | Cfhr1 | Complement factor H-related 1 |
| 26 | 97444_at | AI844520 | Ifi30 | Interferon gamma inducible protein 30 |
| 27 | 93028_at | X58196 | H19 | H19 fetal liver mRNA |
| 28 | 102710_at | AF020313 | Apbb1ip | Amyloid beta (A4) precursor protein-binding |
| 29 | 97356_at | AI839653 | 1810008O21F | RIKEN cDNA 1810008O21 gene |
| 30 | 104680_at | AJ250489 | Ramp1 | Receptor (calcitonin) activity modifying protein |
| 31 | 161984_f_at | AV234303 | Col3a1 | Procollagen, type III, alpha 1 |
| 32 | 161698_f_at | AV232762 | Rprc1 | Arginine/proline rich coiled-coil 1 |
| 33 | 100397_at | AF024637 | Tyrobp | TYRO protein tyrosine kinase binding protein |
| 34 | 98817_at | Z29532 | Fst | Follistatin |
| 35 | 94982_f_at | AI852470 | Nme3 | Expressed in non-metastatic cells 3 |
| 36 | 103033_at | X06454 | C4b /// C4a /// | Complement component 4B (Childo blood g |
| 37 | 103075_at | M34381 | Pou5f1 | POU domain, class 5, transcription factor 1 |
| 38 | 162483_f_at | AV112006 | Col18a1 | Procollagen, type XVIII, alpha 1 |
| 39 | 101019_at | U74683 | Ctsc | Cathepsin C |
| 40 | 103981_at | AI842281 | U2af1-rs2 | U2 small nuclear ribonucleoprotein auxiliary |
| 41 | 97487_at | X70296 | Serpine2 | Serine (or cysteine) peptidase inhibitor, clad |
| 42 | 104237_at | AI838071 | Pcbd2 | Pterin 4 alpha carbinolamine dehydratase/di |
| 43 | 93538_at | AW228036 | Ttrap | Traf and Tnf receptor associated protein |
| 44 | 104337_f_at | AW208938 | Pkp2 | Plakophilin 2 |
| 45 | 104156_r_at | U19118 | Atf3 | Activating transcription factor 3 |
| 46 | 93041_at | D26089 | Mcm4 | Minichromosome maintenance deficient 4 h |
| 47 | 103534_at | V00722 | Hbb-b2 | Hemoglobin, beta adult minor chain |
| 48 | 103081_at | AA863928 | Baz1b | Bromodomain adjacent to zinc finger domain |
| 49 | 96083_s_at | AI226368 | Hnrpdl | Heterogeneous nuclear ribonucleoprotein D− |
| 50 | 103449_at | AA795923 | Lsm14b | LSM14 homolog B (SCD6, Saccharomyces cerevisiae) |
| 51 | 98092_at | AA790307 | Plac8 | Placenta-specific 8 |
| 52 | 96886_at | AW060556 | Stab1 | Stabilin 1 |
| 53 | 160458_at | AI853261 | Mcam | Melanoma cell adhesion molecule |
| 54 | 95627_at | AW046038 | Wwtr1 | WW domain containing transcription regulat |
| 55 | 96891_at | AI842771 | Anp32b /// LO | Acidic nuclear phosphoprotein 32 family, me |
| 56 | 97750_at | X06406 | Rpsa | Ribosomal protein SA |
| 57 | 94954_at | AI846628 | Anapc4 | Anaphase promoting complex subunit 4 |
| 58 | 103016_s_at | X68273 | Cd68 | CD68 antigen |
| 59 | 96138_at | AA682129 | Mars | Methionine-tRNA synthetase |
| 60 | 97199_at | AI854767 | Rbm12 /// Cpn | RNA binding motif protein 12 /// copine I |
| 61 | 95641_at | AI853899 | Plac9 | Placenta specific 9 |
| 62 | 98624_at | X75316 | Rnpc1 | RNA-binding region (RNP1, RRM) containin |
| 63 | 99475_at | U88327 | Socs2 | Suppressor of cytokine signaling 2 |
| 64 | 98059_s_at | D49733 | Lmna | Lamin A |
| 65 | 98088_at | X13333 | Cd14 | CD14 antigen |
| 66 | 95791_s_at | U14648 | Sfrs2 | Splicing factor, arginine/serine-rich 2 (SC-35 |
| 67 | 104524_at | AI842825 | Gltp | Glycolipid transfer protein |
| 68 | 92794_f_at | M35970 | Nme1 | Expressed in non-metastatic cells 1, protein |
| 69 | 103916_at | AI850713 | 8430420C20F | RIKEN cDNA 8430420C20 gene |
| 70 | 95944_at | AV299153 | Dhx36 | DEAH (Asp-Glu-Ala-His) box polypeptide 36 |
| 71 | 100468_g_at | X57687 | Lyl1 | Lymphoblastomic leukemia |
| 72 | 94839_at | M96823 | Nucb1 | Nucleobindin 1 |
| 73 | 98346_at | AI593759 | Lrrc54 | Leucine-rich repeat containing 54 |
| 74 | 100957_at | AA881160 | Ssbp1 | Single-stranded DNA binding protein 1 |
| 75 | 94024_at | AA671429 | Cdt1 | Chromatin licensing and DNA replication factor |
| 76 | 160182_at | AI846595 | Sfrs6 | Splicing factor, arginine/serine-rich 6 |
| 77 | 100732_at | X73829 | Rps8 | Ribosomal protein S8 |
| 78 | 98934_at | AI847513 | 0610007P06R | RIKEN cDNA 0610007P06 gene |
| 79 | 93089_at | X12507 | Eif4a2 | Eukaryotic translation initiation factor 4A2 |
| 80 | 103312_f_at | AW047929 | 2610101J03R | RIKEN cDNA 2610101J03 gene |
| 81 | 95657_f_at | AW125164 | D13Wsu177e | DNA segment, Chr 13, Wayne State University |
| 82 | 96710_at | AI854262 | H2afv | H2A histone family, member V |
| 83 | 104573_at | AA921069 | Bola2 | BolA-like 2 (E. coli) |
| 84 | 99336_at | U78085 | Rps5 | Ribosomal protein S5 |
| 85 | 99093_at | AI852864 | Rps10 /// LOC | Ribosomal protein S10 /// similar to ribosomal |
| 86 | 94767_at | U93864 | Rps11 /// LOC | Ribosomal protein S11 /// similar to ribosomal |
| 87 | 101129_at | X83590 | Rpl5 | Ribosomal protein L5 |
| 88 | 93092_at | U35323 | H2-DMa | Histocompatibility 2, class II, locus DMa |
Supplemental online Table 3 indicates overlap between stem/progenitor cluster 1 candidate genes (this table), stem/progenitor cluster 2, hematopoietic, neural, and embryonic stem cells and undifferentiated spermatogonia.
Each expression cluster contained candidate genes that were predictable on the basis of the morphological and functional analysis. The germ cell cluster contained known spermatogenesis genes, including glyceraldehyde-3-phosphate dehydrogenase, spermatogenic (Gapdhs); preproacrosin (Acr); transition protein 2 (Tpn2); germ cell-specific gene 1 (Gsg1); meiosis expressed gene 1 (Meig1); protamines 1 and 3 (Prm1, Prm3); acrosomal vesicle protein 1 (Acrv1); tektins 1 and 2 (Tekt1, Tekt2); proacrosin-binding protein (Acrbp); A kinase anchor protein 4 (Akap4); and glial cell line-derived neurotrophic factor family receptor α2 (Gfrα2). Furthermore, functional annotation using Gene Ontology categories [47] revealed that spermatogenesis genes were significantly overrepresented in the germ cell cluster (Bonferroni-corrected p = 9.5 × 10−8; Fig. 3). In contrast, spermatogenesis genes were significantly underrepresented in the somatic cell cluster (p = 6.7 × 10−4; Fig. 3), which included known products of Sertoli and Leydig cells of the testis, such as inhibin α (Inha), kit ligand (Kitl), 17β hydroxysteroid dehydrogenase (Hsd17b12), steroidogenic acute regulatory protein (Star), doublesex and mab-3 related transcription factor (Dmrt), Desert hedgehog (Dhh), and glial cell line-derived neurotrophic factor (Gdnf). As expected from the experimental design, stem/progenitor clusters 1 and 2 had less overlap with the Spermatogenesis Gene Ontology (p > .2). Thus, germ cell and testicular somatic cell gene expression profiles were accurately dissected from heterogeneous testis cell populations, providing a high degree of confidence in the power of the experimental design to reveal molecular mechanisms of stem/progenitor cell function.
Figure 3.
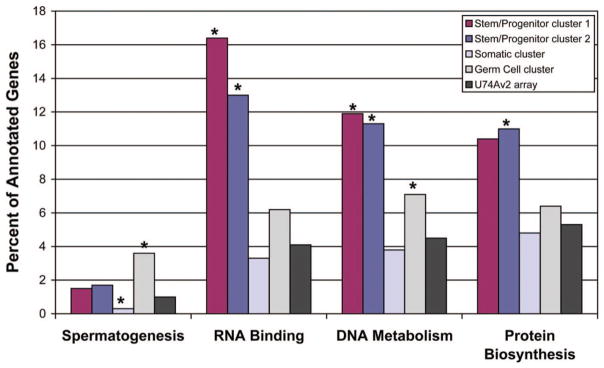
Functional annotation of candidate gene lists. Stem/progenitor cluster 1 (maroon columns), stem/progenitor cluster 2 (dark blue columns), somatic cell (light blue columns), and germ cell (light gray columns) candidate gene clusters were annotated on the basis of all functional categories in Gene Ontology and compared with the functional distribution of genes on the U74Av2 array (dark gray columns). Results for spermatogenesis, RNA binding, DNA metabolism, and protein biosynthesis functional categories are shown. The percentages of genes in each cluster that fall in the indicated functional categories are indicated on the y-axis. Significant differences (false discovery rate, <0.05) compared with the percentage of functionally annotated genes on the U74Av2 array in each functional category are indicated with an asterisk (*).
To further evaluate the two stem/progenitor spermatogonia gene expression lists (cluster 1 and cluster 2), we compared our results from the analysis of freshly isolated testis cells to previous reports on gene expression in partially purified or cultured mouse spermatogonia. Shima et al. used Affymetrix Mouse Genome U74Av2 arrays to identify 6,102 genes expressed by type A spermatogonia isolated using the Staput method ([30]; personal communication with M. Griswold). Oatley et al. used Affymetrix Mouse Genome 430 2.0 arrays to identify 16,453 genes expressed in cultures of mouse spermatogonial stem cells [31], 4,889 of which have a “best match” on the MGU74Av2 array (Affymetrix MG-U74v2 to mouse 430 best match array comparison spreadsheet). The degree of overlap between stem/ progenitor cluster 2 genes in the current study and the candidate gene lists generated in these two previous studies is highly significant and is summarized in Table 2. In addition, Wang et al. used cDNA subtraction to identify candidate genes in a spermatogonia-enriched fraction of testis cells isolated using Staput [28]. Among the 36 “spermatogonially expressed” genes identified in that study, 14 were on the Affymetrix MGU74Av2 array used in the current study, and 9 of these were also identified in stem/progenitor cluster 2 (Cr > Bu; supplemental online Table 3). The degree of overlap among these studies is striking considering that the starting cell populations and the experimental and analytical methods were different. A gene-by-gene comparison of overlap between stem/progenitor/spermatogonia genes identified in the current study and previous studies is included in supplemental online Table 3.
Table 2.
Comparative gene expression among stem cells and spermatogonia
| Stem/progenitor cluster 2 (Cr > Bu) SSC (389)a | Cultured SSCs (4,889)b | Type A spermatogonia (6,102)c | HSC (2,598)d | NSC (3,036)d | ESC (2,176)d | |
|---|---|---|---|---|---|---|
| No. of probe setse | 389 | 275 | 352 | 110 | 147 | 147 |
| Expectedf | – | 184 | 191 | 81 | 95 | 68 |
| Probability (p)g | – | <1.0 × 10−15 | <1.0 × 10−16 | <0.0003 | 1.9 × 10−9 | 3.3 × 10−16 |
In the current study, Affymetrix mouse genome U74Av2 arrays containing 12,422 probe sets were used to evaluate gene expression in Cr, wild-type, and Bu testis.
Supplemental online Table 3 gives a complete list of genes in stem/progenitor cluster 2, indicating where each gene overlaps with gene lists for HSCs, NSCs, ESCs, SSCs, and type A spermatogonia. Overlap with stem/progenitor cluster 1 is also indicated in supplemental online Table 3.
Oatley et al. [31] used Affymetrix MG340 2.0 arrays to evaluate gene expression in cultured SSCs. MGU74Av2 contains 8975 best matches with MG340 2.0, and 4,889 of these were expressed by cultured SSCs.
Shima et al. [30] used Affymetrix MGU74Av2 arrays (same arrays used in the current study) to identify 6,102 genes expressed by isolated type A spermatogonia.
Ivanova et al. [1] and Ramalho-Santo et al. [2] characterized and compared gene expression in three different stem cell populations. The stem cell candidate gene lists from these two studies were merged to create composite gene lists for HSCs, NSCs, and ESCs. Both studies used Affymetrix MGU74Av2 arrays.
Number of probe sets that overlap with stem/progenitor cluster 2 (Cr > Bu) candidate SSC gene list generated in the current study.
Hypergeometric distribution was used to determine the level of overlap between candidate gene lists expected by chance. For example, the type A spermatogonia list contains 6,102 candidate probe sets, which comprise 49% (6,102 of 12,422) of probe sets on the mouse MGU74Av2 array. Therefore, 49% (191) of the 389 stem/progenitor cluster 2 probe sets are expected to overlap with the Shima et al. [30] list by chance.
The probability (p) that the level of overlap observed is different from that expected by chance.
Abbreviations: Bu, busulfan-treated; Cr, cryptorchid; ESC, embryonic stem cell; HSC, hematopoietic stem cell; NSC, neural stem cell; SSC, spermatogonial stem cell.
The quality and specificity of stem/progenitor clusters 1 and 2 (Cr > Wt, Bu; Cr > Bu) was confirmed, in part, because many of the candidate genes have known expression by undifferentiated spermatogonia, including RNA-binding motif protein (Rbmy1A1) [48], cellular retinoic acid-binding protein (Crabp1) [49], Sal-like 4 (Sall4) [28], Sry-box containing gene 3 (Sox3) [50, 51], Oct 4 (Pou5f1) [52], and Plzf (Zbtb16) [53, 54] (complete lists given in Table 1 and supplemental Table 3). Each of these genes was also identified in previous studies of isolated and cultured spermatogonia (supplemental online Table 3; [30, 31]). The c-Kit gene was expressed by cryptorchid testis cells, but the expression profile did not meet the criteria for assignment to the stem/progenitor clusters in this study. Interestingly, both Ret and GFRα1 received absent calls on three replicate arrays of cryptorchid testis cells in the current study. These coreceptors for Gdnf also received absent or marginal calls in the previous analysis of isolated spermatogonia [30] but were expressed by cultured SSCs [31]. These differences may be explained by the observation that the SSC pool in mice is heterogeneous with respect to GFRα1 expression [55]. Genetic manipulation of Gdnf signaling in mouse models demonstrated the importance of this growth factor for maintenance of spermatogenesis [56]. Although that in vivo study did not assay stem cells, Gdnf is clearly critical for maintaining SSCs in culture [57, 58]. Perhaps culture conditions select SSCs with high Gfrα1 expression that can respond to the mitogenic signal, whereas the cryptorchid condition selects for quiescent SSCs that do not express Gfrα1.
Experiments were designed to confirm the expression of other candidate genes from stem/progenitor cluster 1 on spermatogonial stem cells. These studies are complicated because SSCs are rare and have few defining characteristics. However, we have previously demonstrated that FACS followed by transplantation of selected fractions is a powerful tool for identifying cell surface characteristics of SSCs [24, 25, 59]. Expression of three cell surface molecules, Ccr2, Ccr5, and Cd14 antigen, was evaluated in a subpopulation of testis cells from cryptorchid mice characterized as α6-integrin (CD49f)+ and SSClo (side scatter of incident light) and known to be significantly enriched for SSCs [24].
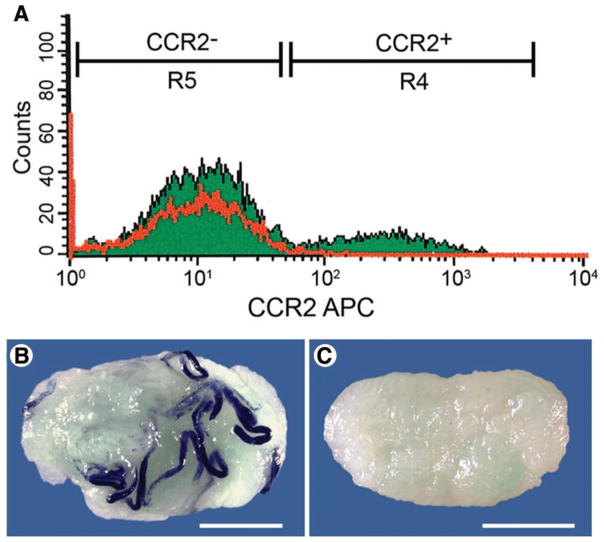
The FACS analysis in Figure 4 confirms the results of the microarray by demonstrating that a subpopulation of cryptorchid, α6-integrin+, SSClo testis cells expresses Ccr2 (Fig. 4A). Similar results were obtained for Cd14, but the results for Ccr5 were more equivocal (data not shown). However, functional evaluation of Ccr2− and Ccr2+ fractions by transplantation into the testes of busulfan-treated recipient mice revealed that all stem cell activity was in the Ccr2− fraction (Fig. 4B). Cells from the Ccr2− and Ccr2+ fractions produced 67.49 ± 15.85 and 0 ± 0 colonies per 105 cells transplanted, respectively. These results highlight the importance of confirming microarray data with additional experimentation. Although it appears that Ccr2 is expressed by a subpopulation of cells in cryptorchid mouse testes, which likely includes undifferentiated germ cells, either this surface marker is not expressed by SSCs or the CCR2 antigen was destroyed during trypsin digestion for cell processing.
Figure 4.
Flow cytometric analysis of cryptorchid testis cells for expression of the cell surface marker, CCR2. (A): The α6-integrin+, side scatterlo subpopulation cryptorchid testis cells, which is enriched for SSCs [24], was evaluated for CCR2 expression. CCR2− and CCR2+ gates were established on the basis of analysis of cryptorchid testis cells stained with an IgG2b isotype control antibody (red line). Distribution of cryptorchid testis cells was 7.26% in fraction R5 (α6-integrin+, SSClo, CCR2−) and 1.37% in fraction R4 (α6-inte-grin+, SSClo, CCR2+). The CCR2− and CCR2+ fractions were isolated by fluorescence-activated cell sorting (FACS) and transplanted to the testes of busulfan-treated mice to determine the relative SSC activity. Testes of recipient mice were analyzed 2 months after transplantation for colonies of donor-derived (blue) spermatogenesis (B, C). Colony number was normalized to the number of blue spermatogenic colonies produced per 105 cells transplanted: unsorted cryptorchid testis cells, 20.41 ± 4.83; fraction R5 (CCR2−), 67.49 ± 15.85; and fraction R4 (CCR2+), 0 ±0 colonies per 105 cells transplanted (mean ± SEM). Representative recipient testes transplanted with CCR2− (B) and CCR2+ (C) cells are shown. Scale bars =2 mm (B, C). Abbreviations: APC, allophycocyanin; CCR2, chemokine receptor 2.
Clustering of stem cell genes in specific chromosomal regions may allow for coregulation of expression for specific cellular functions. In an examination of genes expressed by embryonic, hematopoietic, and neural stem cells, Ramalho-Santos et al. found a greater abundance of chromosome 17 genes than expected by chance [2]. Furthermore, several of these stem cell genes mapped to the t-complex of chromosome 17, a region containing genes involved in embryonic development and male fertility [60]. In another study, Wang et al. found an abundance of X-linked genes expressed by spermatogonia [28]. In contrast, we found that the 88 candidate genes in stem/progenitor spermatogonia cluster 1 were evenly distributed among chromosomes (supplemental online Fig. 1a). Although X chromosome genes were not differentially expressed by stem/progenitor spermatogonia, we did observe over- and underrepresentation of X chromosome genes in the somatic cell and germ cell candidate gene lists, respectively (supplementary online Fig. 1b; additional results and discussion are given in supplementary online material).
Functional annotation of stem/progenitor spermatogonia candidates was conducted to gain insight into the cellular mechanisms controlling spermatogonial stem cell biological activity. We determined the distribution of our stem/progenitor cluster 1 and cluster 2 genes among all functional categories in Gene Ontology [47] and compared the results with the distribution of functions of all annotated genes on the MGU74Av2 array. The functional categories DNA metabolism, RNA-binding proteins, and protein biosynthesis were significantly overrepresented by stem/progenitor cluster 1 and/or cluster 2 (FDR <0.01; Fig. 3). Overrepresentation of the latter two categories suggests that post-transcriptional mechanisms play a role in regulating the function of stem/progenitor spermatogonia in cryptorchid testes. Indeed, a role for RNA-binding proteins in germline development is well established and evolutionarily conserved [61–64]. RNA-binding proteins regulate diverse cellular processes in the testis, including transcript splicing [65] and storage [66]. Consistent with our results, Wang et al. observed an abundance of RNA-binding proteins in mouse spermatogonia [28]. Finally, genes involved in transcriptional and post-transcriptional processes are conserved in the transcriptomes of mouse ESCs, HSCs, and NSCs [2].
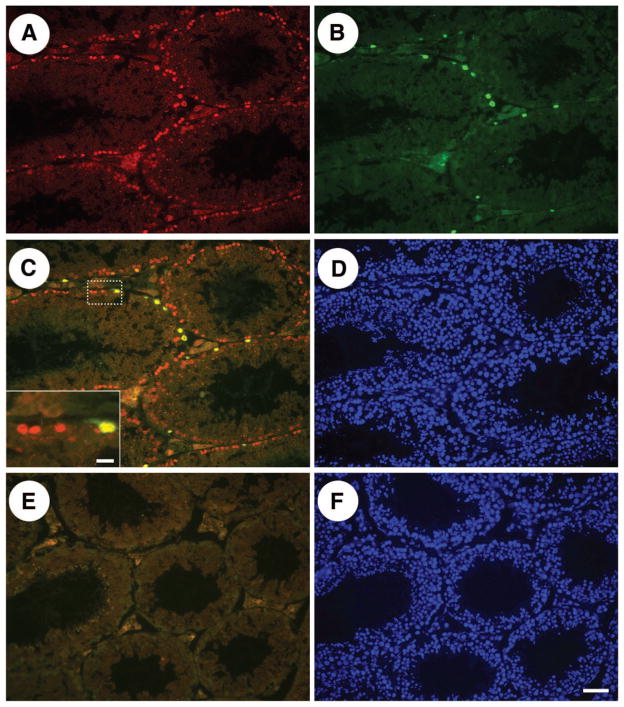
Our immunohistochemistry results (Fig. 5) confirm previously published reports that the RNA-binding protein Rbmy (identified in the current study) is expressed by spermatogonia on the basement membrane of normal adult mouse seminiferous tubules [48, 67]. Furthermore, costaining with Plzf (Fig. 5C) demonstrates that Rbmy is expressed by stem and/or progenitor spermatogonia. Plzf is a transcriptional regulator required for SSC self-renewal and maintenance [53, 54] that was identified in the current study (stem/progenitor cluster 2), as well as previous studies of isolated and cultured spermatogonia [30, 31]. This result confirms the microarray and validates the potential of the experimental design to provide additional insights into the molecular characteristics of stem/progenitor spermatogonia.
Figure 5.
The RNA-binding protein Rbmy colocalizes with Plzf in normal adult mouse seminiferous tubules. (A): Rbmy immunoreactivity (red) was restricted to cells on the basement membrane of adult mouse seminiferous tubules. (B): Immunofluorescent staining for Plzf (green), a consensus marker of stem/progenitor spermatogonia that is expressed by rare cells on the basement membrane of mouse seminiferous tubules. (C): Overlay of Plzf and Rbmy fluorescent signals. Inset displays a higher magnification of cells in the dashed white box. (D): 4,6-Di-amidino-2-phenylindole (DAPI) staining of the section depicted in (A–C) demonstrates that all seminiferous tubules contain multiple germ cell layers. (E): Negative control section in which primary antibodies were replaced with an isotype control. (F): DAPI staining of section in (E). Scale bars = 50 μm (F) and 10 μm ([C], inset).
All stem cells are defined by their dual functional capacities to self-renew and differentiate, and these activities must be balanced to exactly meet the biological demand of the dependent tissue. The genetic and molecular determinants of these cell fate decisions are not well understood, but some features are likely to be shared among stem cells. Two previous studies [1, 2] comparing the transcriptomes of ESCs, HSCs, and NSCs did not converge on a consensus stem cell molecular signature, but it is likely that each study independently identified genes essential for stem cell function. Furthermore, both groups made their gene expression data publicly available and have provided advice and additional information for the current study. Therefore, we combined the data from these two studies to produce grand lists of HSC-, NSC-, and ESC-expressed genes, containing 2,598, 3,036, and 2,176 probe sets, respectively (Table 2). Examination of these gene expression lists also identified 1,979 probe sets that were expressed by two of the three stem cell types and 303 probe sets that were expressed in all three stem cell populations (HSCs, NSCs, and ESCs).
We compared stem/progenitor cluster 2 (389 probe sets, which also includes all genes in stem/progenitor cluster 1) with the HSC, NSC, and ESC transcriptomes described above. The degree of overlap between stem/progenitor cluster 2 and each of the other stem cell populations was significantly greater than expected by chance (Table 2; HSC, p < .0003; NSC, p = 1.9 × 10−9; ESC, p = 3.3 × 10−16). Furthermore, 139 of the 389 probe sets in stem/progenitor cluster 2 were expressed by two (p = 3.8 × 10−13) or three (p = .0006) of the three other stem cell populations, including several encoding RNA-binding proteins (Rbmy1a1, U2af1-rs2, Snrpg, Sfrs2, Fbl, Cstf2t, Lsm8, Sfrs6, and Nip7), again suggesting a role for post-transcriptional regulation of stem cell function (overlap between gene lists is given in supplemental online Table 3). Finally, 21 probe sets were conserved among stem/progenitor spermatogonia cluster 2 and the merge lists for HSCs, NSCs, and ESCs (p = .0006; supplemental online Table 3). Among the 21 conserved genes, 4 were also on the more specific stem/progenitor cluster 1, including suppressor of cytokine signaling 2 (Socs2), leucine-rich repeat containing 54 (Lrrc54), splicing factor, arginine/serine-rich 6 (Sfrs6), and an expressed sequence tag (accession no. AW047929). Socs family proteins are intracellular inhibitors of cytokine and growth factor-induced Jak-Stat signaling. Of note, Socs2 is expressed in neural progenitor cells and required for differentiation into neurons [68]. A role for Socs2 in mouse germline development has not been described, but Socs2−/− mice are fertile ([69]; W.S. Alexander, personal communication).
Supplemental online Table 3 is the master table for this study. The table lists the 389 probe sets in stem/progenitor cluster 2 of the current study and indicates overlap with stem/ progenitor cluster 1, as well as gene expression lists for hematopoietic stem cells, neural stem cells, embryonic stem cells [1, 2], and spermatogonia [28, 30, 31]. It also identifies the 39 candidate genes encoding RNA-binding proteins.
Discussion
The experimental design and cluster analysis enabled us to generate candidate gene lists for stem/progenitor spermatogonia of cryptorchid testes (Table 1; supplemental online Table 3) as well as differentiating germ cells (supplemental online Table 1) and testicular somatic cells (supplemental online Tables 2). Functional annotation and comparison with the spermatogenesis Gene Ontology category [47] helped validate the experimental design by confirming the presence of known spermatogenesis genes on the germ cell candidate gene cluster and a lack of spermatogenesis genes on the somatic cell cluster (Fig. 3). Therefore, it is likely that the stem/progenitor clusters contain important expression data that will provide a foundation for understanding the molecular mechanisms controlling spermatogonial stem cell biology.
Stem/progenitor cluster 1 (88 genes) and cluster 2 (389 genes), generated from differential expression analysis of cryptorchid, wild-type, and busulfan-treated adult mouse testes in the current study, exhibited substantial overlap with the transcriptomes of cultured SSCs (4,889 genes on the U74Av2 array [31]) and isolated spermatogonia (6,102 genes [30]) that were previously described. Furthermore, the concordant gene expression between SSCs in vivo (current study) and in vitro [31] may validate the power of the SSC culture to dissect regulatory pathways controlling stem cell function.
The biological activity of stem cells is controlled intrinsically by the genes that they express and extrinsically by the local microenvironment (niche). However, adult tissue stem cells are generally difficult to identify and study in their resident tissues because they are rare, have few defining characteristics, and cannot be quantified without a robust biological assay. In some previous studies, genetic analyses were facilitated by the availability of enriched target populations [28, 30, 31]. However, isolation, purification, and/or culture procedures resulted in the disassociation of stem/ progenitor cells from their niches for hours or days prior to analysis. The level to which these disruptions altered gene expression compared with the in vivo situation is not known. Conversely, attempts to characterize SSC gene expression in the testes of jsd and W mutant mice [70] were confounded because the relative stem cell activity in the two models was not determined.
All gene expression studies have strengths and weaknesses, and results must be interpreted with caution and confirmed through additional experimentation. The strengths of the current study are that spermatogonial stem cells were isolated directly from their cognate microenvironments in three different testis models (cryptorchid, wild-type, and busulfan-treated). Spermatogonial stem cell transplantation provided a quantitative functional assessment of stem cell activity in each testis cell population. Although stem cells are rare in these models (range, 1 in 115 in cryptorchid to 1 in 11,111 in busulfan-treated), we were able to exploit the statistical power of the nearly 200,000 probe pairs on Affymetrix MGU74Av2 microarrays, using the ChipStat algorithm to identify gene expression that correlated with spermatogonial stem cell activity. One of the limitations of this experimental design is that it might also identify genes that are activated by the cryptorchid procedure (which exposes the testis to core body temperature) or busulfan treatment, which are not necessarily expressed by stem/progenitor spermatogonia. Indeed, several candidate genes encode heat shock proteins, and immunohistochemical analysis demonstrated that one of these proteins (Serpinh1, HSP47) was expressed by testicular interstitial cells, not spermatogonia (data not shown).
In contrast, our histochemical analyses confirmed that the candidate gene encoding the RNA-binding protein, Rbmy, is expressed by cells located on the basement membrane of normal adult mouse seminiferous tubules that also express Plzf, a consensus SSC marker [53, 54]. Roles for RNA-binding proteins in germ cell development, self-renewal, and differentiation are evolutionarily conserved [71, 72]. Rbmy and Dazl (deleted in azoospermia-like) are two examples identified in the current study that have been implicated in male infertility and play roles in RNA splicing and protein translation, respectively [71, 73]. Alternative splicing is particularly prevalent in the testis [65], as well as in hematopoietic and embryonic stem cells [74]. The extent and functional significance of alternative splicing in stem cells is under-studied, and better tools are needed for global expression analysis of alternative transcripts [74]. Dazl-null mice are infertile, and males are characterized by the presence of undifferentiated spermatogonia that proliferate but fail to differentiate from Aal to A1 spermatogonia [45]. Daz family proteins regulate protein translation, but the specific mRNA targets of Dazl have not been elucidated [75].
Conclusion
Identification of RNA or protein targets of candidate RNA-binding proteins may provide future inroads to understanding the cellular mechanisms controlling SSC biological activity. Post-transcriptional regulation may enable SSCs to respond rapidly (without new transcription) to environmental cues for self-renewal, differentiation, or pathological/toxic conditions. However, whether these or other candidate RNA-binding proteins are essential for SSCs must be confirmed by additional mechanistic and functional experimentation.
Supplementary Material
Acknowledgments
We thank C. Freeman and R. Naroznowski for assistance with animal maintenance and experimentation. J. Hayden provided help with photography. Bintu Sherif helped with statistical analyses. Gene expression data for embryonic, hematopoietic, neural, and spermatogonial stem cells and undifferentiated spermatogonia was posted on the online (http://www.sciencemag.org/cgi/content/full/sci;1072530/DC1, http://www.sciencemag.org/cgi/content/full/sci;1073823/DC1, and http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GSM52694), and additional data were provided by N. Ivanova (Princeton University). The Rbmy antibody was a gift from D. Elliott (University of Newcastle upon Tyne), and the Plzf antibody was a gift from R. Hobbs and P.P. Pandolfi (Sloan-Kettering Institute, New York, NY). K.E.O. was supported by NIH Grant RR18500 and the Magee-Women’s Research Institute and Foundation; R.L.B. was supported by NIH Grant HD044445 and HD052728, the Commonwealth and General Assembly of Pennsylvania, and the Robert J. Kleberg, Jr., and Helen C. Kleberg Foundation. B.-Y.R. is currently affiliated with the Department of Animal Sciences and Technology, Chung-Ang University, Ansung-Si, Gyungki-Do, Korea.
Footnotes
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
References
- 1.Ivanova NB, Dimos JT, Schaniel C, et al. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- 2.Ramalho-Santos M, Yoon S, Matsuzaki Y, et al. “Stemness”: Transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 3.Burns CE, Zon LI. Portrait of a stem cell. Dev Cell. 2002;3:612–613. doi: 10.1016/s1534-5807(02)00329-5. [DOI] [PubMed] [Google Scholar]
- 4.Fortunel NO, Otu HH, Ng HH, et al. Comment on “Stemness”: Transcriptional profiling of embryonic and adult stem cells and a stem cell molecular signature (I) Science. 2003;302:393. doi: 10.1126/science.1086384. [DOI] [PubMed] [Google Scholar]
- 5.Evsikov AV, Solter D. Comment on “Stemness”: Transcriptional profiling of embryonic and adult stem cells and a stem cell molecular signature (II) Science. 2003;302:393. doi: 10.1126/science.1082380. [DOI] [PubMed] [Google Scholar]
- 6.Shinohara T, Orwig KE, Avarbock MR, et al. Restoration of spermatogenesis in infertile mice by Sertoli cell transplantation. Biol Reprod. 2003;68:1064–1071. doi: 10.1095/biolreprod.102.009977. [DOI] [PubMed] [Google Scholar]
- 7.Russell LD, Ettlin RA, SinhaHikim AP, et al. Mammalian spermatogenesis. In: Russell LD, Ettlin RA, SinhaHikim AP, et al., editors. Histological and Histopathological Evaluation of the Testis. Clearwater, FL: Cache River Press; 1990. pp. 1–40. [Google Scholar]
- 8.de Rooij DG, Grootegoed JA. Spermatogonial stem cells. Curr Opin Cell Biol. 1998;10:694–701. doi: 10.1016/s0955-0674(98)80109-9. [DOI] [PubMed] [Google Scholar]
- 9.Eddy EM. Male germ cell gene expression. Recent Prog Horm Res. 2002;57:103–128. doi: 10.1210/rp.57.1.103. [DOI] [PubMed] [Google Scholar]
- 10.Ogawa T, Dobrinski I, Avarbock MR, et al. Transplantation of male germ line stem cells restores fertility in infertile mice. Nat Med. 2000;6:29–34. doi: 10.1038/71496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shinohara T, Orwig KE, Avarbock MR, et al. Remodeling of the post-natal mouse testis is accompanied by dramatic changes in stem cell number and niche accessibility. Proc Natl Acad Sci U S A. 2001;98:6186–6191. doi: 10.1073/pnas.111158198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z, Renfree MB, Short RV. Successful intra- and interspecific male germ cell transplantation in the rat. Biol Reprod. 2003;68:961–967. doi: 10.1095/biolreprod.102.009480. [DOI] [PubMed] [Google Scholar]
- 13.Brinster CJ, Ryu BY, Avarbock MR, et al. Restoration of fertility by germ cell transplantation requires effective recipient preparation. Biol Reprod. 2003;69:412–420. doi: 10.1095/biolreprod.103.016519. [DOI] [PubMed] [Google Scholar]
- 14.Orwig KE, Schlatt S. Cryopreservation and transplantation of spermatogonia and testicular tissue for preservation of male fertility. J Natl Cancer Inst Monogr. 2005:51–56. doi: 10.1093/jncimonographs/lgi029. [DOI] [PubMed] [Google Scholar]
- 15.Nagano M, Shinohara T, Avarbock MR, et al. Retrovirus-mediated gene delivery into male germ line stem cells. FEBS Lett. 2000;475:7–10. doi: 10.1016/s0014-5793(00)01606-9. [DOI] [PubMed] [Google Scholar]
- 16.Nagano M, Brinster CJ, Orwig KE, et al. Transgenic mice produced by retroviral transduction of male germ-line stem cells. Proc Natl Acad Sci U S A. 2001;98:13090–13095. doi: 10.1073/pnas.231473498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagano M, Watson DJ, Ryu BY, et al. Lentiviral vector transduction of male germ line stem cells in mice. FEBS Lett. 2002;524:111–115. doi: 10.1016/s0014-5793(02)03010-7. [DOI] [PubMed] [Google Scholar]
- 18.Orwig KE, Avarbock MR, Brinster RL. Retrovirus-mediated modification of male germline stem cells in rats. Biol Reprod. 2002;67:874–879. doi: 10.1095/biolreprod.102.005538. [DOI] [PubMed] [Google Scholar]
- 19.Hamra FK, Gatlin J, Chapman KM, et al. Production of transgenic rats by lentiviral transduction of male germ-line stem cells. Proc Natl Acad Sci U S A. 2002;99:14931–14936. doi: 10.1073/pnas.222561399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryu BY, Orwig KE, Oatley JM, et al. Efficient generation of transgenic rats through the male germline using lentiviral transduction and transplantation of spermatogonial stem cells. J Androl. 2007;28:353–360. doi: 10.2164/jandrol.106.001511. [DOI] [PubMed] [Google Scholar]
- 21.Tegelenbosch RAJ, de Rooij DG. A quantitative study of spermatogonial multiplication and stem cell renewal in the C3H/101 F1 hybrid mouse. Mutat Res. 1993;290:193–200. doi: 10.1016/0027-5107(93)90159-d. [DOI] [PubMed] [Google Scholar]
- 22.Brinster RL, Zimmermann JW. Spermatogenesis following male germ-cell transplantation. Proc Natl Acad Sci U S A. 1994;91:11298–11302. doi: 10.1073/pnas.91.24.11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brinster RL, Avarbock MR. Germline transmission of donor haplotype following spermatogonial transplantation. Proc Natl Acad Sci U S A. 1994;91:11303–11307. doi: 10.1073/pnas.91.24.11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shinohara T, Orwig KE, Avarbock MR, et al. Spermatogonial stem cell enrichment by multiparameter selection of mouse testis cells. Proc Natl Acad Sci U S A. 2000;97:8346–8351. doi: 10.1073/pnas.97.15.8346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kubota H, Avarbock MR, Brinster RL. Spermatogonial stem cells share some, but not all, phenotypic and functional characteristics with other stem cells. Proc Natl Acad Sci U S A. 2003;100:6487–6492. doi: 10.1073/pnas.0631767100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanatsu-Shinohara M, Toyokuni S, Shinohara T. CD9 is a surface marker on mouse and rat male germline stem cells. Biol Reprod. 2004;70:70–75. doi: 10.1095/biolreprod.103.020867. [DOI] [PubMed] [Google Scholar]
- 27.Tokuda M, Kadokawa Y, Kurahashi H, et al. CDH1 is a specific marker for undifferentiated spermatogonia in mouse testes. Biol Reprod. 2007;76:130–141. doi: 10.1095/biolreprod.106.053181. [DOI] [PubMed] [Google Scholar]
- 28.Wang PJ, McCarrey JR, Yang F, et al. An abundance of X-linked genes expressed in spermatogonia. Nat Genet. 2001;27:422–426. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- 29.Hamra FK, Schultz N, Chapman KM, et al. Defining the spermatogonial stem cell. Dev Biol. 2004;269:393–410. doi: 10.1016/j.ydbio.2004.01.027. [DOI] [PubMed] [Google Scholar]
- 30.Shima JE, McLean DJ, McCarrey JR, et al. The murine testicular transcriptome: Characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod. 2004;71:319–330. doi: 10.1095/biolreprod.103.026880. [DOI] [PubMed] [Google Scholar]
- 31.Oatley JM, Avarbock MR, Telaranta AI, et al. Identifying genes important for spermatogonial stem cell self-renewal and survival. Proc Natl Acad Sci U S A. 2006;103:9524–9529. doi: 10.1073/pnas.0603332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishimune Y, Aizawa S, Komatsu T. Testicular germ cell differentiation in vivo. Fertil Steril. 1978;29:95–102. doi: 10.1016/s0015-0282(16)43045-1. [DOI] [PubMed] [Google Scholar]
- 33.Shinohara T, Avarbock MR, Brinster RL. Functional analysis of spermatogonial stem cells in steel and cryptorchid infertile mouse models. Dev Biol. 2000;220:401–411. doi: 10.1006/dbio.2000.9655. [DOI] [PubMed] [Google Scholar]
- 34.Ogawa T, Aréchaga JM, Avarbock MR, et al. Transplantation of testis germinal cells into mouse seminiferous tubules. Int J Dev Biol. 1997;41:111–122. [PubMed] [Google Scholar]
- 35.Zhang X, Ebata KT, Nagano MC. Genetic analysis of the clonal origin of regenerating mouse spermatogenesis following transplantation. Biol Reprod. 2003;69:1872–1878. doi: 10.1095/biolreprod.103.019273. [DOI] [PubMed] [Google Scholar]
- 36.Kanatsu-Shinohara M, Inoue K, Miki H, et al. Clonal origin of germ cell colonies after spermatogonial transplantation in mice. Biol Reprod. 2006;75:68–74. doi: 10.1095/biolreprod.106.051193. [DOI] [PubMed] [Google Scholar]
- 37.Master S, Stoddard AJ, Bailey LC, et al. Genomic analysis of early murine mammary gland development using novel probe-level algorithms. Genome Biol. 2005;6:R20.1–R20.16. doi: 10.1186/gb-2005-6-2-r20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mack M, Cihak J, Simonis C, et al. Expression and characterization of the chemokine receptors CCR2 and CCR5 in mice. J Immunol. 2001;166:4697–4704. doi: 10.4049/jimmunol.166.7.4697. [DOI] [PubMed] [Google Scholar]
- 39.Hosack DA, Dennis G, Jr, Sherman BT, et al. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Rooij DG, Okabe M, Nishimune Y. Arrest of spermatogonial differentiation in jsd/jsd, Sl17H/Sl17H, and cryptorchid mice. Biol Reprod. 1999;61:842–847. doi: 10.1095/biolreprod61.3.842. [DOI] [PubMed] [Google Scholar]
- 41.Reijo RA, Dorfman DM, Slee R, et al. DAZ family proteins exist throughout male germ cell development and transit from nucleus to cytoplasm at meiosis in humans and mice. Biol Reprod. 2000;63:1490–1496. doi: 10.1095/biolreprod63.5.1490. [DOI] [PubMed] [Google Scholar]
- 42.Toyooka Y, Tsunekawa N, Takahashi Y, et al. Expression and intracellular localization of mouse Vasa-homologue protein during germ cell development. Mech Dev. 2000;93:139–149. doi: 10.1016/s0925-4773(00)00283-5. [DOI] [PubMed] [Google Scholar]
- 43.Ruggiu M, Speed R, Taggart M, et al. The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature. 1997;389:73–77. doi: 10.1038/37987. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka SS, Toyooka Y, Akasu R, et al. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 2000;14:841–853. [PMC free article] [PubMed] [Google Scholar]
- 45.Schrans-Stassen BHGJ, Saunders PTK, Cooke HJ, et al. Nature of the spermatogenic arrest in dazl−/− mice. Biol Reprod. 2001;65:771–776. doi: 10.1095/biolreprod65.3.771. [DOI] [PubMed] [Google Scholar]
- 46.Lin Y, Page DC. Dazl deficiency leads to embryonic arrest of germ cell development in XY C57BL/6 mice. Dev Biol. 2005;288:309–316. doi: 10.1016/j.ydbio.2005.06.032. [DOI] [PubMed] [Google Scholar]
- 47.Ashburner M, Ball CA, Blake JA, et al. Gene Ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahadevaiah SK, Odorisio T, Elliott DJ, et al. Mouse homologues of the human AZF candidate gene RBM are expressed in spermatogonia and spermatids, and map to a Y chromosome deletion interval associated with a high incidence of sperm abnormalities. Hum Mol Genet. 1998;7:715–727. doi: 10.1093/hmg/7.4.715. [DOI] [PubMed] [Google Scholar]
- 49.Zheng WL, Bucco RA, Schmitt MC, et al. Localization of cellular retinoic acid-binding protein (CRABP) II and CRABP in developing rat testis. Endocrinology. 1996;137:5028–5035. doi: 10.1210/endo.137.11.8895377. [DOI] [PubMed] [Google Scholar]
- 50.Weiss J, Meeks JJ, Hurley L, et al. Sox3 is required for gonadal function, but not sex determination, in males and females. Mol Cell Biol. 2003;23:8084–8091. doi: 10.1128/MCB.23.22.8084-8091.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rizzoti K, Brunelli S, Carmignac D, et al. SOX3 is required during the formation of the hypothalamo-pituitary axis. Nat Genet. 2004;36:247–255. doi: 10.1038/ng1309. [DOI] [PubMed] [Google Scholar]
- 52.Pesce M, Wang X, Wolgemuth DJ, et al. Differential expression of the Oct-4 transcription factor during mouse germ cell differentiation. Mech Dev. 1998;71:89–98. doi: 10.1016/s0925-4773(98)00002-1. [DOI] [PubMed] [Google Scholar]
- 53.Costoya JA, Hobbs RM, Barna M, et al. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet. 2004;36:653–659. doi: 10.1038/ng1367. [DOI] [PubMed] [Google Scholar]
- 54.Buaas FW, Kirsh AL, Sharma M, et al. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet. 2004;36:647–652. doi: 10.1038/ng1366. [DOI] [PubMed] [Google Scholar]
- 55.Buageaw A, Sukhwani M, Ben-Yehudah A, et al. GDNF family receptor alpha1 phenotype of spermatogonial stem cells in immature mouse testes. Biol Reprod. 2005;73:1011–1016. doi: 10.1095/biolreprod.105.043810. [DOI] [PubMed] [Google Scholar]
- 56.Meng X, Lindahl M, Hyvonen ME, et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 2000;287:1489–1493. doi: 10.1126/science.287.5457.1489. [DOI] [PubMed] [Google Scholar]
- 57.Kanatsu-Shinohara M, Ogonuki N, Inoue K, et al. Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol Reprod. 2003;69:612–616. doi: 10.1095/biolreprod.103.017012. [DOI] [PubMed] [Google Scholar]
- 58.Kubota H, Avarbock MR, Brinster RL. Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proc Natl Acad Sci U S A. 2004;101:16489–16494. doi: 10.1073/pnas.0407063101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ryu BY, Orwig KE, Kubota H, et al. Phenotypic and functional characteristics of spermatogonial stem cells in rats. Dev Biol. 2004;274:158–170. doi: 10.1016/j.ydbio.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 60.Silver LM. The peculiar journey of a selfish chromosome: Mouse t haplotypes and meiotic drive. Trends Genet. 1993;9:250–254. doi: 10.1016/0168-9525(93)90090-5. [DOI] [PubMed] [Google Scholar]
- 61.Jaruzelska J, Kotecki M, Kusz K, et al. Conservation of a Pumilio-Nanos complex from Drosophila germ plasm to human germ cells. Dev Genes Evol. 2003;213:120–126. doi: 10.1007/s00427-003-0303-2. [DOI] [PubMed] [Google Scholar]
- 62.Lamont LB, Crittenden SL, Bernstein D, et al. FBF-1 and FBF-2 regulate the size of the mitotic region in the C. elegans germline. Dev Cell. 2004;7:697–707. doi: 10.1016/j.devcel.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 63.Bachorik JL, Kimble J. Redundant control of the Caenorhabditis elegans sperm/oocyte switch by PUF-8 and FBF-1, two distinct PUF RNA-binding proteins. Proc Natl Acad Sci U S A. 2005;102:10893–10897. doi: 10.1073/pnas.0504593102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fox M, Urano J, Reijo Pera RA. Identification and characterization of RNA sequences to which human PUMILIO-2 (PUM2) and deleted in Azoospermia-like (DAZL) bind. Genomics. 2005;85:92–105. doi: 10.1016/j.ygeno.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 65.Elliott DJ, Grellscheid SN. Alternative RNA splicing regulation in the testis. Reproduction. 2006;132:811–819. doi: 10.1530/REP-06-0147. [DOI] [PubMed] [Google Scholar]
- 66.Steger K. Transcriptional and translational regulation of gene expression in haploid spermatids. Anat Embryol. 1999;199:471–487. doi: 10.1007/s004290050245. [DOI] [PubMed] [Google Scholar]
- 67.Saunders PT, Turner JM, Ruggiu M, et al. Absence of mDazl produces a final block on germ cell development at meiosis. Reproduction. 2003;126:589–597. doi: 10.1530/rep.0.1260589. [DOI] [PubMed] [Google Scholar]
- 68.Turnley AM, Faux CH, Rietze RL, et al. Suppressor of cytokine signaling 2 regulates neuronal differentiation by inhibiting growth hormone signaling. Nat Neurosci. 2002;5:1155–1162. doi: 10.1038/nn954. [DOI] [PubMed] [Google Scholar]
- 69.Metcalf D, Greenhalgh CJ, Viney E, et al. Gigantism in mice lacking suppressor of cytokine signalling-2. Nature. 2000;405:1069–1073. doi: 10.1038/35016611. [DOI] [PubMed] [Google Scholar]
- 70.Tanaka K, Tamura H, Tanaka H, et al. Spermatogonia-dependent expression of testicular genes in mice. Dev Biol. 2002;246:466–479. doi: 10.1006/dbio.2002.0671. [DOI] [PubMed] [Google Scholar]
- 71.Elliott DJ. The role of potential splicing factors including RBMY, RBMX, hnRNPG-T and STAR proteins in spermatogenesis. Int J Androl. 2004;27:328–334. doi: 10.1111/j.1365-2605.2004.00496.x. [DOI] [PubMed] [Google Scholar]
- 72.Xu EY, Moore FL, Pera RAR. A gene family required for human germ cell development evolved from an ancient meiotic gene conserved in metazoans. Proc Natl Acad Sci U S A. 2001;98:7414–7419. doi: 10.1073/pnas.131090498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tung J, Rosen M, Nelson L, et al. Novel missense mutations of the Deleted-in-AZoospermia-Like (DAZL) gene in infertile women and men. Reproductive Biology and Endocrinology. 2006;4:40. doi: 10.1186/1477-7827-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pritsker M, Doniger TT, Kramer LC, et al. Diversification of stem cell molecular repertoire by alternative splicing. Proc Natl Acad Sci U S A. 2005;102:14290–14295. doi: 10.1073/pnas.0502132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yen PH. Putative biological functions of the DAZ family. Int J Androl. 2004;27:125–129. doi: 10.1111/j.1365-2605.2004.00469.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.