Abstract
Hanwoo have been subjected over the last seventy years to intensive artificial selection with the aim of improving meat production traits such as marbling and carcass weight. In this study, we performed a signature of selection analysis to identify recent positive selected regions driven by a long-term artificial selection process called a breeding program using whole genome SNP data. In order to investigate homozygous regions across the genome, we estimated iES (integrated Extended Haplotype Homozygosity SNP) for the each SNPs. As a result, we identified two highly homozygous regions that seem to be strong and/or recent positive selection. Five genes (DPH5, OLFM3, S1PR1, LRRN1 and CRBN) were included in this region. To go further in the interpretation of the observed signatures of selection, we subsequently concentrated on the annotation of differentiated genes defined according to the iES value of SNPs localized close or within them. We also described the detection of the adaptive evolution at the molecular level for the genes of interest. As a result, this analysis also led to the identification of OLFM3 as having a strong signal of selection in bovine lineage. The results of this study indicate that artificial selection which might have targeted most of these genes was mainly oriented towards improvement of meat production.
Keywords: Hanwoo, Signatures of Selection, SNP
INTRODUCTION
Modern breeds of cattle were domesticated about 10,000 years ago to produce the distinct breed characteristics for milk or meat products from natural and human artificial selection (Bradley et al., 1999). During this history of artificial selection, mutations in genes that control important characteristics, such as high milk yield in modern dairy cows, have been selected to fixation. Hanwoo (Korean cattle) have become highly specialized for meat quality undergoing strong artificial selection (Yoon et al., 2008). Hanwoo have been intensively selected for marbling (intramuscular fat) through a progeny test in a breeding program since the 1930s. As a result of artificial selection, such as the Hanwoo progeny test, the breeding value for the marbling score could increase to 0.05 standard deviation (SD) in the Hanwoo population (Lee et al., 2011). Artificial selection might also affect genomic regions controlling Hanwoo marbling. Understanding the genetic mechanism leading to phenotypic differentiation requires identification of the genome regions that have been under long term artificial selection.
This strong artificial selection will increase the frequency of favorable alleles at the loci affecting meat quality traits in the meat production breeds. In this process a small region of the genome surrounding the mutations is also selected, resulting in a small genome region that shows reduced variation. This region of reduced variation is referred to as a signature of selection that is identified by distributions of nucleotides around favorable mutations that differ statistically from that expected purely by chance (Kim and Stephan, 2002). Many methods have been developed for detection of selection signatures from genome analyses. Most of methods are used to compare the distribution of allelic frequencies by calculating population genetics statistics such as FST (Weir et al., 2005), linkage disequilibrium (Kim and Nielsen, 2004), Tajima (Tajima, 1989), Wu’s H-test (Fay and Wu, 2000) and the integrated Haplotype Score (iHS) (Voight et al., 2006), which is a method based on extended haplotype homozygosity (EHH) statistics (Sabeti, et al., 2002). By identifying the signatures of past selection and then identifying the functional genes and mutations involved, it is possible to identify the major genetic and metabolic pathways that control important agricultural characteristics of our modern breeds. When data are available from a large number of populations by large scale SNP data, the analysis can distinguish the genetic variation between similar populations. Searches for signatures of selection have successfully revealed many genes that are important in livestock. For example, the International Bovine HapMap project described a range of breeds that has been historically selected for different phenotypic traits. Hayes also proposed to identify divergently selected regions of the genome between dairy cattle and beef cattle breeds within Bos taurus (Hayes et al., 2009). MacEachern et al. reported the results of comparison of allelic frequencies between Australian Angus and Holstein cattle (MacEachern et al., 2009).
The aim of this study is to perform a test for selection signatures within the Hanwoo population that share a similar phenotype and to detect divergently selected genomic regions. Analysis of within-population selection signatures indicate that at least some mutations, which have been differentially selected in Hanwoo, are still segregated within its population. Finally, positional candidate genes are determined in proximity to the genomic positions showing the most significant indication of selection.
MATERIALS AND METHODS
Animals and genotype assay
Carcass data and DNA samples for QTL analysis were obtained from 266 Hanwoo steers descending from 66 sires and unrelated dams (2 to 10 progeny per sire) from two NIAS experimental stations, Dae-Kwan-Ryoung and Nam-Won. Genomic DNA for genotyping assays was extracted from a blood sample and SeoLin Bioscience (Seoul, Korea) performed the SNP genotyping using the Affymetrix MegAllele GeneChip Bovine Mapping 10K SNP array (Affymetrix Inc., 2006). Three hundred steers were genotyped but 34 steers failed to genotype due to low DNA quality from phenol and chloroform contamination. Genotype data were received on 8,344 SNP and all those SNP were physically mapped to a chromosome (in bp) using the bovine genome sequence (Btau-3.1).
Analysis of SNP statistics
Genotypes were tested for Hardy-Weinberg equilibrium (HWE) to identify possible typing errors using a chi-square test in R/SNPassoc Package (R Development Core Team). SNP not in HWE (p<0.05), monomorphic SNPs and minor allele frequency (<1%) were removed in this study. Finally, a total of 8,344 SNPs, genotype data were received on 4,522 SNP.
Extended haplotype homozygosity (EHH)
The counting algorithm of Tang et al. (2007) was implemented for identifying differential extended haplotype homozygosity regions within Hanwoo (Korean cattle). A haplotype can be identified by patterns of SNPs. Haplotype maps can be used to determine complex genetic variations of inherited diseases or complex traits. For the proportion of homozygous individuals, EHHSi,j, at the ith and jth SNP were calculated in two steps. First, for each SNPi, EHHSi,j between SNPi and incrementally distant flanking SNPj were calculated until EHHSi, k<0; this was performed for both j>i and j<i.
Second, the extended haplotype homozygosity of SNPi was calculated: iESi = (EHHSi,j) for i_j_k for the region 3’ of i (or i_j_k for the region 5’ of i).
Extended haplotype homozygous regions were plotted based on the standardized log-ratio of iESi within Hanwoo. We calculated a standardized integrated extended haplotype homozygsity (iES(z)) value to identify significant regions of positive selection (p = 0.0001, z = 3.5).
Evolutionary analysis of genes in signatures of selection (SoS)
We identified genes within the signature of selection and obtained orthologous genes for the four species Homo sapiens, Mus musculus, Sus scrofa, Gallus gallus from the Ensembl Compara database (Flicek et al., 2008), which reports pairwise conserved synteny relations based on nucleotide alignments. Protein sequences of the orthologous genes were aligned with ClustalW. The protein sequence alignment and the corresponding coding sequences were converted into codon alignment using the pal2nal program (http://coot.embl.de/pal2nal). We obtained the orthologous sequences information for five genes in candidate regions.
If amino acid changes are selectively neutral (i.e., mutations that are neither advantageous or deleterious), they will be fixed at the same rate as synonymous mutations and ω ratio (dN/dS) = 1. ω values>1 are taken to indicate that amino acid changes are accumulating at a faster rate than is acceptable under a neutral mutation model. That is to say, the rate of amino acid changes (dN) significantly exceeds the rate of synonymous changes (dS) at the DNA level. The codon-based likelihood model proposed by Goldman and Yang (Goldman and Yang, 1994) is implemented in the program codeml of PAML package (Yang, 1997). The program is useful for estimating synonymous and non-synonymous substitution rates (dN/dS). To estimate the dN/dS values, model 0 (M0, null model) with a single dN/dS value for all branches of the tree and model 2 (M2, positive selection) with the branches of interest were assumed to have different dN/dS ratios. We obtained a log-likelihood value for each model. We tested for positive selection by comparing twice the log-likelihood difference among models (M0 vs. M2) from a chi-square distribution with n-1 df, where n is the number of branches of the phylogeny in the LRT (Yang and Bielawski, 2000). If a significant p-value is obtained, it can be concluded that the positive selection model (M2) is the favored model. Next, models of variable selective pressures among amino acid sites were used to test for the presence of sites under positive selection. The four models (M1a, M2a, M7, M8) in the CODEML program of the PAML package were tested (Lynn et al., 2005).
RESULTS AND DISCUSSION
Identification of candidate genes in the positively selected region
Figure 1 shows the plot of iES scores for one strong candidate region identified by our genome-wide scan. We extended core regions in both directions up to 1.5 cM from a core SNP (rs29012432, 41.75 cM) and annotated a subset of genes in the core region. The total distance between the first point to the left and to the right of the core SNP is from 40.93 cM to 43.87 cM of BTA3. We also detected the core SNPs that were 20.79 to 21.84 Mb of BTA22. There is a clear clustering of high values into the region where some SNPs show evidence of selection. As a result, five genes were determined as putative targets of recent artificial selection as follows: diphthine synthase 5 (DPH5), sphingosine-1-phosphate receptor 1 (S1PR1), olfactomedin 3 (OLFM3), leucine-rich repeat neuronal protein 1 (LRRN1) and protein cereblon (CRBN). A summary statistics for positively selected regions presenting the highest values of the iES analysis is shown in Table 1.
Figure 1.
Genome wide extended haplotype homozygosity (EHH) profiling to detect signature of selection in Hanwoo.
Table 1.
Summary statistics of the integrated EHHS (iES) values for selection signature in candidate genes
| Chromosome | Candidate region | Closet SNP name and position (bp) | iES value |
|---|---|---|---|
| BTA3 | DPH5 (DPH5 homolog (S. cerevisiae)) | rs29020061 (40,929,695) | 7.21 |
| S1PR1(sphingosine-1-phosphate receptor 1) | rs29018907 (41,249,954) | 7.46 | |
| OLFM3(olfactomedin 3) | rs29018230 (41,792,758) | 9.82 | |
| BTA12 | LRRN1(Leucine-rich repeat neuronal protein 1) | rs29015171 (20,796,087) | 12.55 |
| CRBN (Protein cereblon) | rs29017072 (21,841,651) | 11.71 |
DPH5 encodes a component of the diphthamide synthesis pathway. Diphthamide is a post-translationally modified histidine residue found only on translation elongation factor 2 (EF2). EF2 affects adipocyte differentiation in lipid and energy metabolism with differences in protein synthesis (Bluher et al., 2004). OLFM3 is an olfatomedin-related protein that interacts with myocilin (Torrado et al., 2002), which is a major cause of glaucoma and may play a role in cytoskeletal function (Stone et al., 1997). Sensory systems have undergone major evolutionary changes in mammalian lineages. Some studies suggest that a subset of olfactory genes have been positively selected (Sharon et al., 1999; Clark et al., 2003). Animals require great olfactory performances for social communication. Therefore, this result indicates that genes associated with the sensory system are determined by species specificity in terms of evolution. We also observed a signal for the selection targeting aspects of lipid metabolism. S1PR1 is one of five G protein-coupled receptors (S1PR1–5) of sphingosine-1-phosphate (S1P) (Hannun and Obeid, 2008), which is a potent lipid mediator produced from the metabolism of sphingolipid by the actions of sphingosine kinase. High density lipoprotein (HDL) is stimulated from the binding of S1P in HDL with its receptors EDG1/S1P1 and EDG3/S1P3 (Kimura et al., 2003). Regulation of lipid synthesis and degradation is important in meat animals. The human aims to decrease total cholesterol and increases the HDL fraction that is known as “good” cholesterol (Tang et al., 2001). Expression differences in EDG1 have been previously reported between a high-marbled steer group and a low-marbled steer group in musculus longissimus muscle across all ages (Sasaki et al., 2006). Recently, SNP in 5′ flanking region of EDG1 was associated with marbling in Japanese black cattle population (Yamada et al., 2009). LRRN encodes a type I transmembrane protein with unknown function and is associated with neural development (Andreae et al., 2007). CRBN directly interacts with the α1 subunit of AMP-activated protein kinase (AMPK) and reduces the activation of AMPK (Lee et al., 2011). AMPK is known to activate fatty acid oxidation in skeletal muscle by activating PPARα and PGC1 (Lee et al., 2006). We also found a gene (EDEM1, ER degradation enhancer, mannosidase alpha-like 1) related to the adipogenesis. It is located (BTA22:19.03–19.05 Mb) near selected regions of BTA22: 20.79 to 21.84 Mb. EDEM1 is one of the ER stress markers that strongly correlate with total adiposity. Recently, fatty acids also induced ER stress in some cell lines (Wei et al., 2007). It has been suggested that as a marker of obesity in humans it increases adipocyte expression (Sharma et al., 2008). Table 2 summarizes the functions of the candidate genes showing evidence for selection signatures using Gene Ontology (http://www.geneontology.org/) and KEGG pathway (http://www.genome.jp/kegg/). This observation probably reflects the signal of a partial selective sweep and may be an ongoing process in its flanking region known as “hitchhiking”.
Table 2.
Gene Ontology and KEGG pathway of the candidate genes showing evidence for selection signatures
| Candidate gene | GO term | KEGG pathway |
|---|---|---|
| DPH5 | Peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) | - |
| S1PR1 | Angiogenesis (GO:0001525), cell adhesion (GO:0007155), G-protein coupled receptor protein signaling pathway (GO:0007186), inhibition of adenylate cyclase activity by G-protein signaling pathway (GO:0007193), brain development (GO:0007420) | Neuroactive ligand-receptor interaction |
| OLFM3 | Eye photoreceptor cell development (GO:0042462) | - |
| LRRN1 | Integral to membrane (GO:0016021) | - |
| CRBN | Negative regulation of protein homooligomerization (GO:0032463), negative regulation of ion transmembrane transport (GO:0034766) | - |
Evidence of positive selection between species
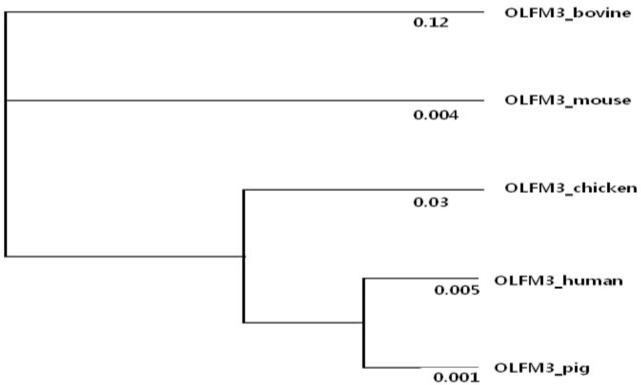
We next implemented a further test to study interspecific divergence between bovine and the other species against the five candidate genes. It is important to know how these genes have been positively selected along bovine lineage with different selective pressures. The evolutionary forces operating on particular genes use the ratio of non-synonymous (dN) to synonymous (dS) substitution. To study differences in selection pressures, we conducted likelihood ratio tests comparing a one-ratio model to a two-ratio alternative model. Table 3 shows the results of the likelihood ratio test using different evolutionary models. Only one gene showed significant acceleration in the ω-ratio on the bovine lineage. For OLFM3, the two-ratio (ω = 0.12) models detected significant positive selection in bovine lineage (Figure 2). It suggests that OLFM3 is an accelerated protein evolution driven by positive selection or a relaxation of constraints. The branch shows evidence of positive selection. To identify particular codon sites subjected to positive selection in the gene, recommended site-specific models (NSsites = 1, 2, 7, 8) implemented in the PALM program were applied. We compared the lnL values from M1a, M2a, M7 and M8. M1a and M2a are neural models with ω fixed = 1 and selection model with ω fixed = 1, respectively. Model 7 uses a β-distribution of sites between the intervals ω = 0 and ω = 1. M8 adds an extra class of sites to the M7 model, allowing for sites with ω>1. If the ω -ratios for some sites are >1, sites with ith posterior probabilities for those sites are likely to be under positive selection. However, neither model (M1a vs M2a and M7 vs M8) detected site classes as significantly favored (data not shown). In other words, no particular codon (amino acid) sites are subjected to adaptive evolution. Among candidate genes, OLFM3 is also likely to have undergone adaptive evolution in the bovine lineage. Its role could be more essential in bovine lineage because this species maintained the complete functions.
Table 3.
Likelihood estimates of different evolutionary models (Model0 vs Model2)
| Gene Name | dN/dS | Degree of freedom | x2 | p-value |
|---|---|---|---|---|
| S1PR1 | 0.05 | 1 | 1.93 | NS |
| DPH5 | 0.09 | 1 | 0.18 | NS |
| OLFM3 | 0.12 | 1 | 31.47 | <0.001* |
| LRRN1 | 0.03 | 1 | 5.34 | NS |
| CRBN | 0.05 | 1 | 1.21 | NS |
Degree of freedom is the difference in the number of parameters between evolutionary models.
x2 is twice the difference of log likelihood between models.
p-value is the probability that two models should differ in log likelihood given the degree of freedom.
Figure 2.
Phylogeny of OLFM3. Branch lengths were estimated by maximum likelihood under the free-ratio model that assumes an independent ω-value for each branch. ω-values are shown for each branch.
The approach undertaken in the present paper will allow signatures of selection to be identified for the unique high intramuscular fat (marbling) of the Hanwoo breed under very intensive artificial selection pressure in the process of breeding programs. In the past 30 years, the body weight at 18 months of age increased from 331 to 574 kg. The average annual genetic gain for carcass traits and marbling was also 4.05 kg and 0.37 grade (1 to 7 grades). The annual genetic gain was also 0.02 to 0.82 kg/yr. As a result, it was assumed that the Hanwoo breed might achieve dramatically increased genetic improvement. This suggests that although Hanwoo have experienced recent selective pressure with a short divergence time, signatures of selection have been observed with a fitness advantage during the process of an artificial breeding program. However, this study has a limitation in that low density SNP data were used for identifying highly homozygous regions. In addition, we observed the putative signatures of selection with only the Hanwoo breed. Therefore, additional biological studies are necessary to identify putative selection signatures and differences between Hanwoo and other breeds. Robust results can be clearly observed and obtained by application of this method.
REFERENCES
- Andreae LC, Peukert D, Lumsden A, Gilthorpe JD. Analysis of Lrrn1 expression and its relationship to neuromeric boundaries during chick neural development. Neural Dev. 2007;2:22. doi: 10.1186/1749-8104-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluher M, Wilson-Fritch L, Leszyk J, Laustsen PG, Corvera S, Kahn CR. Role of insulin action and cell size on protein expression patterns in adipocytes. J Biol Chem. 2004;279:31902–31909. doi: 10.1074/jbc.M404570200. [DOI] [PubMed] [Google Scholar]
- Bradley DG, Cunningham EP. Genetic aspects of domestication. In: Fries R, Ruvinsky A, editors. The genetics of cattle. CABI Publishing; Wallingford: 1999. pp. 15–31. [Google Scholar]
- Clark AG, Glanowski S, Nielsen R, Thomas PD, Kejariwal A, Todd MA, Tanenbaum DM, Civello D, Lu F, Murphy B, et al. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science. 2003;302:1960–1963. doi: 10.1126/science.1088821. [DOI] [PubMed] [Google Scholar]
- Fay JC, Wu CI. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Aken B, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T. Ensembl 2008. Nucleic Acids Res. 2008;36(Suppl 1):D707. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- Hayes BJ, Chamberlain AJ, Maceachern S, Savin K, McPartlan H, MacLeod I, Sethuraman L, Goddard ME. A genome map of divergent artificial selection between Bos taurus dairy cattle and Bos taurus beef cattle. Anim Genet. 2009;40:176–184. doi: 10.1111/j.1365-2052.2008.01815.x. [DOI] [PubMed] [Google Scholar]
- Kim Y, Nielsen R. Linkage disequilibrium as a signature of selective sweeps. Genetics. 2004;167:1513–1524. doi: 10.1534/genetics.103.025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Stephan W. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics. 2002;160:765–777. doi: 10.1093/genetics/160.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Sato K, Malchinkhuu E, Tomura H, Tamama K, Kuwabara A, Murakami M, Okajima F. High-density lipoprotein stimulates endothelial cell migration and survival through sphingosine 1-phosphate and its receptors. Arterioscler Thromb Vasc Biol. 2003;23:1283–1288. doi: 10.1161/01.ATV.0000079011.67194.5A. [DOI] [PubMed] [Google Scholar]
- Lee KM, Jo S, Kim H, Lee J, Park CS. Functional modulation of AMP-activated protein kinase by cereblon. Biochim Biophys Acta. 2011;1813:448–455. doi: 10.1016/j.bbamcr.2011.01.005. [DOI] [PubMed] [Google Scholar]
- Lee SH, van der Werf JH, Kim NK, Lee SH, Gondro C, Park EW, Oh SJ, Gibson JP, Thompson JM. QTL and gene expression analyses identify genes affecting carcass weight and marbling on BTA14 in Hanwoo (Korean Cattle) Mamm. Genome. 2011;22:589–601. doi: 10.1007/s00335-011-9331-9. [DOI] [PubMed] [Google Scholar]
- Lee WJ, Kim M, Park HS, Kim HS, Jeon MJ, Oh KS, Koh EH, Won JC, Kim MS, Oh GT. AMPK activation increases fatty acid oxidation in skeletal muscle by activating PPARα and PGC-1. Biochem Biophys Res Commun. 2006;340:291–295. doi: 10.1016/j.bbrc.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Lynn DJ, Freeman AR, Murray C, Bradley DG. A genomics approach to the detection of positive selection in cattle: adaptive evolution of the T-cell and natural killer cell-surface protein CD2. Genetics. 2005;170:1189–1196. doi: 10.1534/genetics.104.039040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacEachern S, Hayes B, McEwan J, Goddard M. An examination of positive selection and changing effective population size in Angus and Holstein cattle populations (Bos taurus) using a high density SNP genotyping platform and the contribution of ancient polymorphism to genomic diversity in Domestic cattle. BMC Genomics. 2009;10:181. doi: 10.1186/1471-2164-10-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, Schaffner SF, Gabriel SB, Platko JV, Patterson NJ, McDonald GJ, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Nagai K, Nagata Y, Doronbekov K, Nishimura S, Yoshioka S, Fujita T, Shiga K, Miyake T, Taniguchi Y, Yamada T. Exploration of genes showing intramuscular fat deposition-associated expression changes in musculus longissimus muscle. Anim Genet. 2006;37:40–46. doi: 10.1111/j.1365-2052.2005.01380.x. [DOI] [PubMed] [Google Scholar]
- Sharma NK, Das SK, Mondal AK, Hackney OG, Chu WS, Kern PA, Rasouli N, Spencer HJ, Yao-Borengasser A, Elbein SC. Endoplasmic reticulum stress markers are associated with obesity in nondiabetic subjects. J Clin Endocrinol Metab. 2008;93:4532–4541. doi: 10.1210/jc.2008-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon D, Glusman G, Pilpel Y, Khen M, Gruetzner F, Haaf T, Lancet D. Primate evolution of an olfactory receptor cluster: diversification by gene conversion and recent emergence of pseudogenes. Genomics. 1999;61:24–36. doi: 10.1006/geno.1999.5900. [DOI] [PubMed] [Google Scholar]
- Stone EM, Fingert JH, Alward WL, Nguyen TD, Polansky JR, Sunden SL, Nishimura D, Clark AF, Nystuen A, Nichols BE. Identification of a gene that causes primary open angle glaucoma. Science. 1997;275:668–670. doi: 10.1126/science.275.5300.668. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang K, Thornton KR, Stoneking M. A new approach for using genome scans to detect recent positive selection in the human genome. PLoS Biol. 2007;5:e171. doi: 10.1371/journal.pbio.0050171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Kerry JP, Sheehan D, Buckley DJ, Morrissey PA. Antioxidative effect of added tea catechins on susceptibility of cooked red meat, poultry and fish patties to lipid oxidation. Food Res Int. 2001;34:651–657. [Google Scholar]
- Torrado M, Trivedi R, Zinovieva R, Karavanova I, Tomarev SI. Optimedin: a novel olfactomedin-related protein that interacts with myocilin. Hum Mol Genet. 2002;11:1291–1301. doi: 10.1093/hmg/11.11.1291. [DOI] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Wang D, Pagliassotti MJ. Saturated fatty acid-mediated endoplasmic reticulum stress and apoptosis are augmented by trans-10, cis-12-conjugated linoleic acid in liver cells. Mol Cell Biochem. 2007;303:105–113. doi: 10.1007/s11010-007-9461-2. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cardon LR, Anderson AD, Nielsen DM, Hill WG. Measures of human population structure show heterogeneity among genomic regions. Genome Res. 2005;15:1468–1476. doi: 10.1101/gr.4398405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Sasaki S, Sukegawa S, Miyake T, Fujita T, Kose H, Morita M, Takahagi Y, Murakami H, Morimatsu F, et al. Novel SNP in 5′ flanking region of EDG1 associated with marbling in Japanese Black beef cattle. Anim Sci J. 2010;80:486–489. doi: 10.1111/j.1740-0929.2009.00665.x. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z, Bielawski JP. Statistical methods for detecting molecular adaptation. Trends Ecol Evol. 2000;15:496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon D, Kwon YS, Lee KY, Jung WY, Sasazaki S, Mannen H, Jeon JT, Lee JH. Discrimination of Korean cattle (Hanwoo) using DNA markers derived from SNPs in bovine mitochondrial and SRY genes. Asian Australas J Anim Sci. 2008;21:25–28. [Google Scholar]