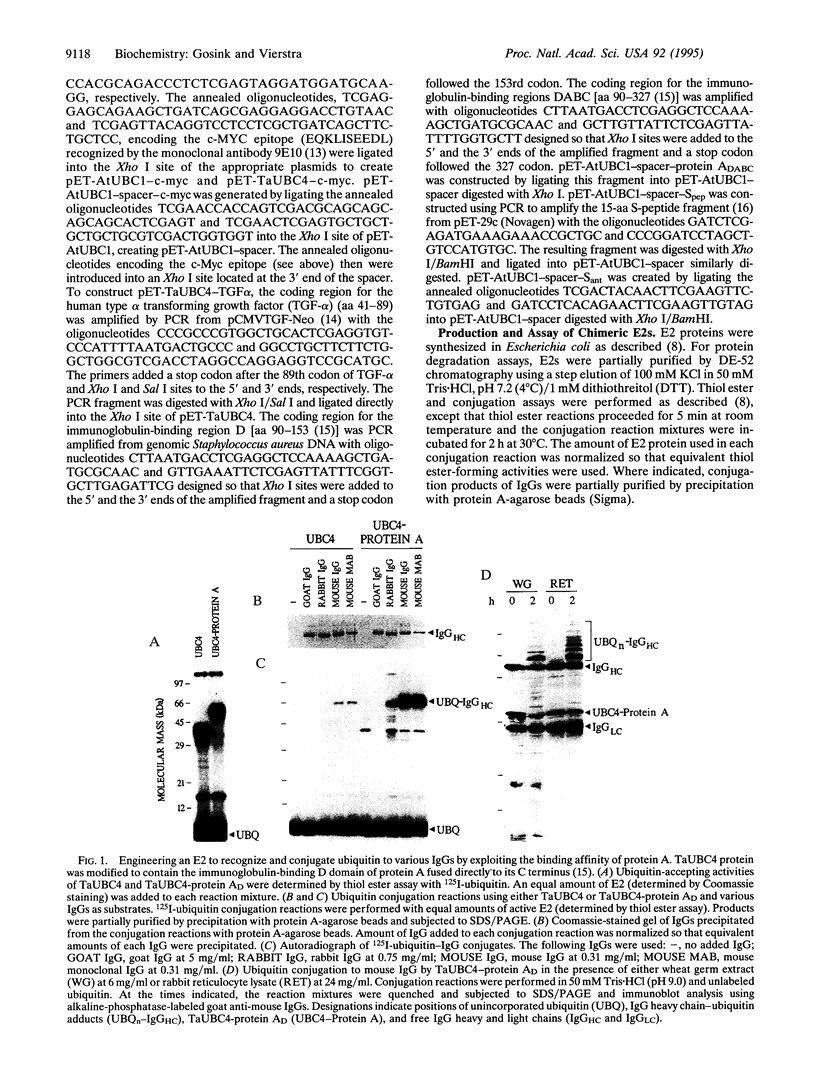
Abstract
Depletion of specific cellular proteins is a powerful tool in biological research and has many medical and agricultural benefits. In contrast to genetic methods currently available to attenuate protein levels, we describe an alternative approach that redirects the ubiquitin-dependent proteolytic pathway to facilitate specific proteolytic removal. Degradation via the ubiquitin pathway requires the prior attachment of multiple ubiquitins to the target protein. This attachment is accomplished, in part, by a family of enzymes designated E2s (or ubiquitin-conjugating enzymes), some of which use domains near their C termini for target recognition. Here, we demonstrate that E2 target recognition can be redefined by engineering E2s to contain appropriate protein-binding peptides fused to their C termini. In five dissimilar examples, chimeric E2s were created that recognized and ubiquitinated their respective binding partners with high specificity. We also show that ubiquitination of one protein targeted by this method led to its ATP-dependent degradation in vitro. Thus, by exploiting interacting domains derived from natural and synthetic ligands, it may be possible to design E2s capable of directing the selective removal of many intracellular proteins.
Full text
PDF
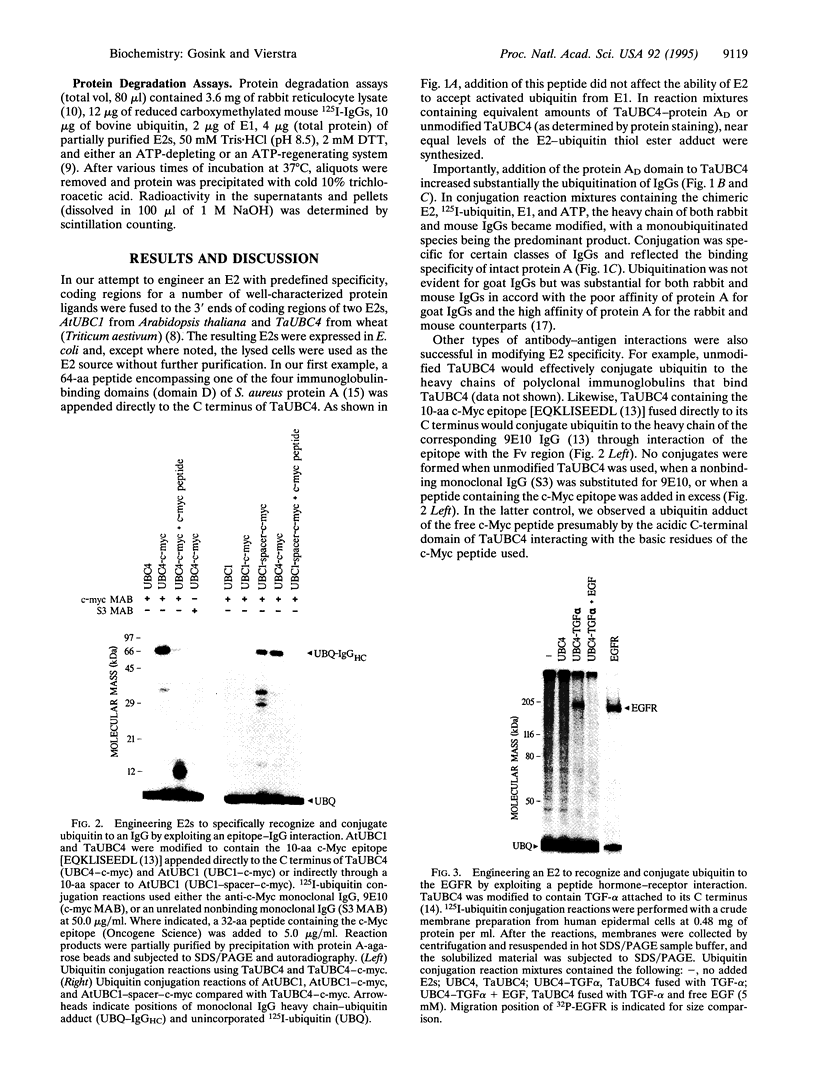
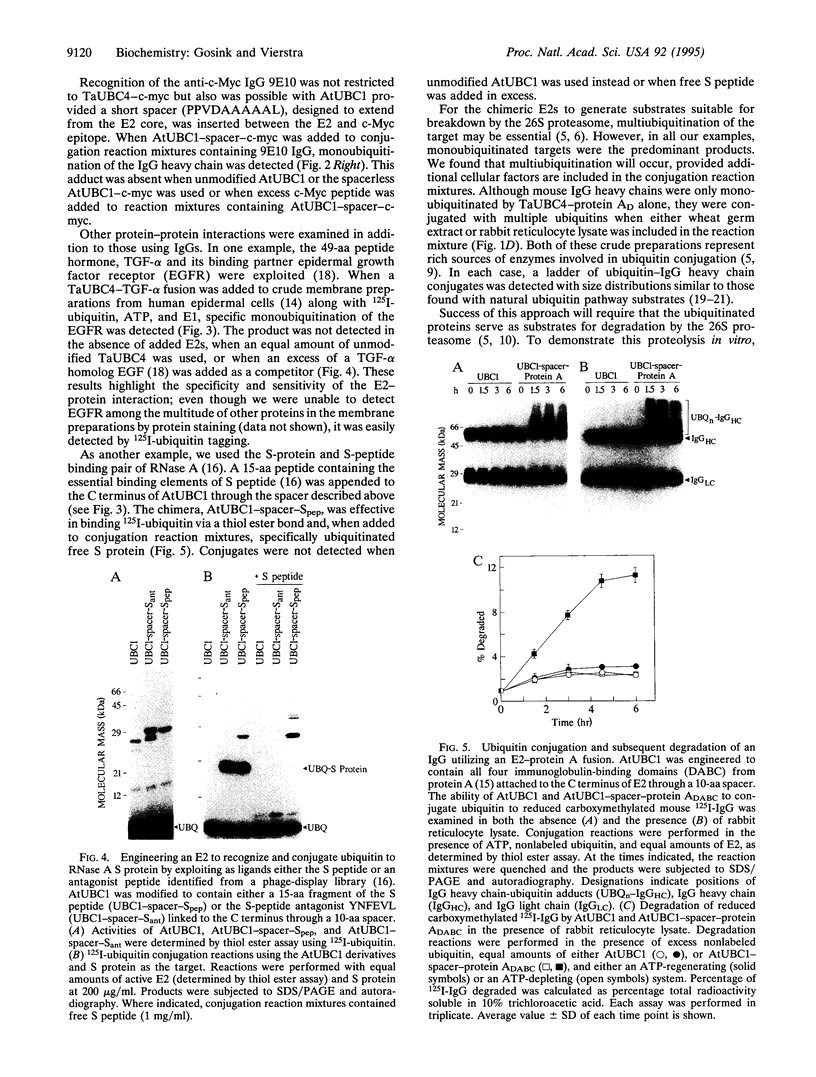

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brondyk W. H., Kujoth G. C., Fahl W. E. Transforming growth factor-alpha expression produces only morphological transformants of diploid human fibroblasts. Cancer Res. 1993 May 1;53(9):2162–2167. [PubMed] [Google Scholar]
- Dohmen R. J., Wu P., Varshavsky A. Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science. 1994 Mar 4;263(5151):1273–1276. doi: 10.1126/science.8122109. [DOI] [PubMed] [Google Scholar]
- Dropulić B., Jeang K. T. Gene therapy for human immunodeficiency virus infection: genetic antiviral strategies and targets for intervention. Hum Gene Ther. 1994 Aug;5(8):927–939. doi: 10.1089/hum.1994.5.8-927. [DOI] [PubMed] [Google Scholar]
- Evans A. C., Jr, Wilkinson K. D. Ubiquitin-dependent proteolysis of native and alkylated bovine serum albumin: effects of protein structure and ATP concentration on selectivity. Biochemistry. 1985 Jun 4;24(12):2915–2923. doi: 10.1021/bi00333a015. [DOI] [PubMed] [Google Scholar]
- Glotzer M., Murray A. W., Kirschner M. W. Cyclin is degraded by the ubiquitin pathway. Nature. 1991 Jan 10;349(6305):132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- Hoffman L., Pratt G., Rechsteiner M. Multiple forms of the 20 S multicatalytic and the 26 S ubiquitin/ATP-dependent proteases from rabbit reticulocyte lysate. J Biol Chem. 1992 Nov 5;267(31):22362–22368. [PubMed] [Google Scholar]
- Jabben M., Shanklin J., Vierstra R. D. Ubiquitin-phytochrome conjugates. Pool dynamics during in vivo phytochrome degradation. J Biol Chem. 1989 Mar 25;264(9):4998–5005. [PubMed] [Google Scholar]
- Jentsch S. The ubiquitin-conjugation system. Annu Rev Genet. 1992;26:179–207. doi: 10.1146/annurev.ge.26.120192.001143. [DOI] [PubMed] [Google Scholar]
- Kolodziej P. A., Young R. A. Epitope tagging and protein surveillance. Methods Enzymol. 1991;194:508–519. doi: 10.1016/0076-6879(91)94038-e. [DOI] [PubMed] [Google Scholar]
- Marquardt H., Hunkapiller M. W., Hood L. E., Twardzik D. R., De Larco J. E., Stephenson J. R., Todaro G. J. Transforming growth factors produced by retrovirus-transformed rodent fibroblasts and human melanoma cells: amino acid sequence homology with epidermal growth factor. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4684–4688. doi: 10.1073/pnas.80.15.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman D. D., Cleveland P. H., Oxman M. N., Johnson K. M. The binding of staphylococcal protein A by the sera of different animal species. J Immunol. 1982 May;128(5):2300–2305. [PubMed] [Google Scholar]
- Scheffner M., Münger K., Huibregtse J. M., Howley P. M. Targeted degradation of the retinoblastoma protein by human papillomavirus E7-E6 fusion proteins. EMBO J. 1992 Jul;11(7):2425–2431. doi: 10.1002/j.1460-2075.1992.tb05307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuttleworth H. L., Duggleby C. J., Jones S. A., Atkinson T., Minton N. P. Nucleotide sequence analysis of the gene for protein A from Staphylococcus aureus Cowan 1 (NCTC8530) and its enhanced expression in Escherichia coli. Gene. 1987;58(2-3):283–295. doi: 10.1016/0378-1119(87)90383-0. [DOI] [PubMed] [Google Scholar]
- Smith G. P., Schultz D. A., Ladbury J. E. A ribonuclease S-peptide antagonist discovered with a bacteriophage display library. Gene. 1993 Jun 15;128(1):37–42. doi: 10.1016/0378-1119(93)90150-2. [DOI] [PubMed] [Google Scholar]
- Sullivan M. L., Vierstra R. D. Cloning of a 16-kDa ubiquitin carrier protein from wheat and Arabidopsis thaliana. Identification of functional domains by in vitro mutagenesis. J Biol Chem. 1991 Dec 15;266(35):23878–23885. [PubMed] [Google Scholar]
- Treier M., Staszewski L. M., Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994 Sep 9;78(5):787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- Weber W., Bertics P. J., Gill G. N. Immunoaffinity purification of the epidermal growth factor receptor. Stoichiometry of binding and kinetics of self-phosphorylation. J Biol Chem. 1984 Dec 10;259(23):14631–14636. [PubMed] [Google Scholar]