ABSTRACT
A population of human immunodeficiency virus (HIV) within a host often descends from a single transmitted/founder virus. The high mutation rate of HIV, coupled with long delays between infection and diagnosis, make isolating and characterizing this strain a challenge. In theory, ancestral reconstruction could be used to recover this strain from sequences sampled in chronic infection; however, the accuracy of phylogenetic techniques in this context is unknown. To evaluate the accuracy of these methods, we applied ancestral reconstruction to a large panel of published longitudinal clonal and/or single-genome-amplification HIV sequence data sets with at least one intrapatient sequence set sampled within 6 months of infection or seroconversion (n = 19,486 sequences, median [interquartile range] = 49 [20 to 86] sequences/set). The consensus of the earliest sequences was used as the best possible estimate of the transmitted/founder. These sequences were compared to ancestral reconstructions from sequences sampled at later time points using both phylogenetic and phylogeny-naive methods. Overall, phylogenetic methods conferred a 16% improvement in reproducing the consensus of early sequences, compared to phylogeny-naive methods. This relative advantage increased with intrapatient sequence diversity (P < 10−5) and the time elapsed between the earliest and subsequent samples (P < 10−5). However, neither approach performed well for reconstructing ancestral indel variation, especially within indel-rich regions of the HIV genome. Although further improvements are needed, our results indicate that phylogenetic methods for ancestral reconstruction significantly outperform phylogeny-naive alternatives, and we identify experimental conditions and study designs that can enhance accuracy of transmitted/founder virus reconstruction.
IMPORTANCE When HIV is transmitted into a new host, most of the viruses fail to infect host cells. Consequently, an HIV infection tends to be descended from a single “founder” virus. A priority target for the vaccine research, these transmitted/founder viruses are difficult to isolate since newly infected individuals are often unaware of their status for months or years, by which time the virus population has evolved substantially. Here, we report on the potential use of evolutionary methods to reconstruct the genetic sequence of the transmitted/founder virus from its descendants at later stages of an infection. These methods can recover this ancestral sequence with an overall error rate of about 2.3%—about 15% more information than if we had ignored the evolutionary relationships among viruses. Although there is no substitute for sampling infections at earlier points in time, these methods can provide useful information about the genetic makeup of transmitted/founder HIV.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) is among the most genetically variable human pathogens. HIV-1 envelope diversity within a single host during chronic infection can exceed the global diversity of influenza during a given flu season (1); HIV's global diversity is more than an order of magnitude greater. This diversity, driven by HIV's rapid rate of evolution and frequent recombination (2), represents a major challenge to vaccine design. For example, vaccine-induced protection against one strain may not protect against another (3). Recently, however, there is growing evidence that transmitted/founder viruses—strain(s) that successfully establish a productive infection following the transmission bottleneck (4, 5)—may be substantially less diverse than the global viral population. Moreover, transmitted/founder viruses may possess certain genotypic or phenotypic characteristics (6–10), such as envelope sequences that display the CCR5 coreceptor usage phenotype (11), and enhanced sensitivity to antibody-meditated neutralization (12). If infecting strains indeed represent a particular subset of all circulating viruses, and if immunogens could be designed to specifically stimulate protective host responses against them, the prospect of a protective vaccine may be more hopeful. Characterizing transmitted/founder strains is therefore critically important, since this provides a more specific target for vaccine design.
The identification of transmitted/founder strains, however, remains challenging. Often, HIV is not diagnosed until the chronic phase of infection, at which point substantial intrahost evolution has already occurred. To date, identification of transmitted/founder strains has been accomplished through intensive monitoring of large prospective HIV cohorts of high-risk individuals, wherein new infections can be identified within days or weeks of exposure (see, for example, references 11, 13, and 14). Although highly valuable, such programs are resource intensive and require follow-up of a great number of individuals to capture a comparatively small number of founder viruses. In contrast, it is substantially less onerous to identify new HIV infections on a time scale of months or years; indeed, an abundance of published HIV sequence data is available from patients diagnosed after the acute stage. Whereas the sequence of a transmitted/founder strain can be inferred from the genotypes of its immediate descendants when these are captured in acute/early infection (11), the accuracy of reconstructing transmitted/founder strains from more distant descendants remains unknown. If it were possible to accurately reconstruct transmitted viral strains using later (or even chronic) intrapatient sequences, the wealth of existing sequence data would yield a much larger repository of transmitted/founder virus genotypes for study.
Recovering ancestral sequences from present-day descendants has been of interest to biologists since the middle of the 20th century (15). Extensive statistical and computational methods for ancestral sequence reconstruction, which rely on inferring the phylogeny that models patterns of common ancestry among the observed genetic sequences, have been developed and continue to be improved. Often, these techniques have been used by researchers interested in evolution on a macro scale (16). In these cases, the algorithms are untestable, since the ancestors in question are long extinct. In contrast, longitudinal intrapatient HIV sequence data provide a unique opportunity to apply these algorithms in a real-world setting and on a more immediate time scale. In an individual with untreated HIV infection, the equivalent of a million years of macroorganism evolution can be observed in a single year (17). Substantial published longitudinal intrapatient data exist on which to validate these algorithms. The best estimate of the transmitted/founder strain can be obtained by computing a consensus from intrapatient sequences sampled during acute infection, if these early samples are available. In other words, the consensus of the earliest sequences in a longitudinal data set are a “gold standard” estimate of the true transmitted/founder strain, which we can attempt to recover by performing ancestral reconstruction on the remainder of the data set. Although the output of an ancestral reconstruction procedure is the most recent common ancestor (MRCA) of the input sequences, HIV's documented transmission bottleneck (4, 5) makes the MRCA very likely to be identical to the transmitted/founder virus (with the notable exception of very early selection and rapid fixation of an immune escape mutation; see Discussion). If these techniques can be applied to accurately reconstruct transmitted/founder virus sequences using only longitudinal HIV sequences sampled after acute infection, we will have a more rapid and cost-effective method to examine and characterize many more infecting strains in the absence of early samples.
MATERIALS AND METHODS
Data collection.
We used the Los Alamos Intra-Patient search interface (www.hiv.lanl.gov/components/sequence/HIV/ipsearch/IPsearch.html) to identify longitudinal studies with two or more clonal or single-genome (SGS) sequences available at each time point, with a known timeline relative to one of several reference points: HIV infection, seroconversion, presentation of symptomatic seroconversion illness, or birth. Studies were selected where the earliest (“baseline”) sample was collected within 6 months (186 days) of this reference point, and where at least one of the subsequent (“follow-up”) time points occurred a minimum of 6 months after baseline. Any available follow-up time points less than 6 months after baseline were included in the analysis, as long as the total study duration (baseline to last follow-up) exceeded 6 months. As a critical step of ancestral sequence reconstruction is inferring the evolutionary relationships between sequences from different viruses in a within-host population, sequences derived from direct (“bulk”) sequencing of PCR products, and data sets with only one sequence per time point, were excluded. Known cases of superinfection (n = 19 patients) were also excluded. References for all published data sources are listed in the supplemental material.
We organized the data sets from these studies in a purpose-built SQLite database. When patients from two different studies shared one or more sequences (by GenBank accession number), they were treated as one individual with a single ID. The data were grouped by patient and by gene into data sets, defined as a collection of sequences from a single patient and a single HIV gene. HIV env and gag genes comprised the majority of data sets (77%, Table 1), but pol, nef, rev, tat, vif, vpr, and vpu were also represented. A patient may have been associated to more than one data set if more than one gene of that patient's viral population (for example, both env and gag) was sequenced. We assembled 335 unique data sets from 232 unique patients (Table 1), comprising a total of 19,486 sequences (if a sequence was used for two data sets because it contained two different genes, it was counted twice).
TABLE 1.
Data set characteristicsa
| Characteristic or parameter | Risk group |
||||||
|---|---|---|---|---|---|---|---|
| All | Heterosexual | MSM | MTCT | IDU | Transfusionb | Unknownc | |
| Patients (n) | 232 | 52 | 52 | 50 | 3 | 3 | 72 |
| Data sets (n) | 335 | 90 | 105 | 55 | 4 | 3 | 78 |
| env | 155 | 33 | 46 | 40 | 2 | 3 | 31 |
| gag | 102 | 39 | 13 | 8 | 1 | 41 | |
| pol | 28 | 11 | 9 | 5 | 1 | 2 | |
| nef | 11 | 5 | 3 | 3 | |||
| rev | 11 | 1 | 9 | 1 | |||
| tat | 15 | 1 | 12 | 2 | |||
| vif | 3 | 3 | |||||
| vpr | 3 | 3 | |||||
| vpu | 7 | 7 | |||||
| DNA | 201 | 81 | 42 | 36 | 2 | 40 | |
| RNA | 87 | 9 | 43 | 17 | 2 | 3 | 13 |
| DNA+RNA | 47 | 20 | 2 | 25 | |||
| Treated | 95 | 3 | 48 | 23 | 2 | 19 | |
| Untreated | 194 | 81 | 57 | 13 | 4 | 1 | 38 |
| Unknown treatment history | 46 | 6 | 19 | 21 | |||
| Parameter | |||||||
| Follow-up time points (n) | 3 (1–5) | 1 (1–2) | 5 (2–7) | 3 (2–3) | 3 (1–5) | 4 (4–4) | 4 (3–6) |
| Sequences/data set (n) | 49 (20–86) | 16 (12–38) | 90 (53–119) | 41 (20–60) | 88 (39–136) | 56 (48–70) | 52 (39–73) |
| Baseline (n) | 10 (6–20) | 6 (3–11) | 18 (10–32) | 10 (4–14) | 23 (22–24) | 8 (6–10) | 10 (7–13) |
| Follow-up (n) | 35 (12–61) | 11 (7–24) | 58 (36–93) | 29 (12–42) | 66 (17–113) | 48 (38–64) | 40 (29–58) |
| tfirst (days) | 32 (7–68) | 59 (23–93) | 25 (6–78) | 31 (14–62) | 142 (98–186) | 31 (16–31) | 16 (1–48) |
| tsecond (days) | 222 (72–769) | 1,262 (306–1,893) | 163 (37–571) | 186 (93–375) | 635 (216–1,054) | 62 (62–563) | 138 (70–224) |
| tlast (days) | 724 (408–1,253) | 1,262 (849–1,893) | 724 (341–1,166) | 504 (387–840) | 1,028 (1,001–1,054) | 341 (310–1,114) | 430 (357–645) |
Values are numbers (n) for patients and data sets or medians (interquartile ranges) for other data. Abbreviations: MSM, men who have sex with men; MTCT, mother-to-child transmission; IDU, injection drug user; tfirst, baseline time point; tsecond, first follow-up time point; tlast, final follow-up time point. “Days” refers to the number of days after the reference point.
That is, two blood and one factor VIII donations.
“Unknown” usually indicates that patients were participants in a large cohort comprising multiple risk groups.
Alignment.
Sequences were annotated in FASTA format using Biopython's SeqIO module (18) and aligned with Seaview (19), using a combination of the built-in MUSCLE interface (20) and manual adjustment. Alignments were trimmed to the interval where a minimum of 50% of sequences within the data set had sequence coverage over each codon. This interval tended to span the entire gene length for relatively short genes such as vif or nef (median alignment lengths 558 and 615 bp, respectively) or targeted specific regions such as the region encoding HIV protease and reverse transcriptase in pol (median length, 1,261 bp). Because codon models generally have no representation for stop codons (see below), columns containing any stop codons were removed. Terminal gaps due to incomplete sequences, which comprised 1.08% of sites across all sequences, were replaced with missing data characters (“?”). Finally, the alignments were split into two FASTA files: one containing the sequences from the baseline time point and the other containing all of the follow-up sequences on which ancestral reconstruction was to be performed.
Consensus sequences.
Ancestral reconstruction via consensus is appropriate when all taxa directly descend from a single common ancestor (a scenario represented by a “star” phylogeny) because the sequences represent independent outcomes. As has been shown previously (8, 11, 21, 22), a within-host population tends to briefly exhibit a star-like phylogeny in acute infection, so a consensus would be appropriate at this stage. However, as infection progresses, certain lineages in the phylogeny begin to proliferate over others as the host's immune system exerts selection pressure. Thus, we would expect to see the consensus diverge from the ancestor over time. Consensus sequences were therefore generated from the baseline sample, to be used as the best possible estimate of the “true” ancestor. We refer to these as “baseline consensus” sequences. The consensus nucleotide for each column of an alignment was taken as the most frequent non-missing nucleotide, whether or not it was present in more than 50% of the sequences (i.e., plurality rule consensus). A tie between two nucleotide frequencies resulted in an ambiguous DNA character in the consensus sequence; ties between a nucleotide and a gap were resolved to missing data (“?”). Consensus sequences were also generated in the same manner from the follow-up samples, to be used as a phylogeny-naive reconstruction of the ancestor, for comparison against the ancestral reconstruction approach. We refer to these as the “follow-up consensus” sequences.
Phylogenetic tree inference.
The software program Bayesian Evolutionary Analysis by Sampling Trees (BEAST) (23) was used to generate a random sample of rooted phylogenetic trees from the posterior distribution given each data set, under a molecular clock model of evolution. We specified a log-normal prior for the clock rate; this prior distribution has the mean and standard deviation as hyperparameters. The prior mean was computed as follows. Mansky and Temin (24) estimated an HIV mutation rate of 3.4 × 10−5 bp per replication cycle, and Perelson et al. (25) estimated the mean generation time of HIV to be ∼2.6 days. Thus, an estimate of the prior mean clock rate is calculated as follows: [(3.4 × 10−5 bp)/generation] × (1 generation/2.6 days) = (1.3 × 10−5 bp)/day.
We specified a prior standard deviation of 1 such that the prior distribution had a 95% confidence interval of 1.5 × 10−6 to 4.1 × 10−5. All other prior distributions were left at their default settings. We used an HKY85 model of nucleotide substitution rates (26), with stationary nucleotide frequencies estimated as model parameters, no variation in substitution rates across sites, and no partitioning of rate parameters by codon position. A prior distribution over trees was specified by a constant effective population size coalescent model with a random starting tree. These settings were applied across all 335 sequence alignments.
For each data set, we evaluated both strict and relaxed uncorrelated lognormal clock models. Two replicate 108-step MCMC chains were run for each clock model for a total of four chain samples, and trees were recorded to a log file every 10,000 steps. Convergence of the chains was assessed using the R package coda's implementation of the Gelman-Rubin diagnostic, which compares the variance within chains to the variance among chains. We took a Gelman-Rubin diagnostic value of ≤1.1 for the posterior traces as a criterion of convergence. If the replicate chains for a given clock model did not satisfy this criterion, we repeated the analysis with longer chain samples up to 109 steps until the criterion was passed. The criterion was not satisfied after 109 steps for 0 and 25 data sets for strict and relaxed molecular clock models, respectively. For these, the remaining clock model (which did pass the criterion) was carried forward. Otherwise, using the implementation of the smoothed marginal-likelihood estimator of Newton and Raftery (27) in Tracer (28), Bayes factors between the strict and relaxed clock models were calculated, and results from the favored model for each respective data set were carried forward. The first 10% of the sampled trees were disregarded as burn-in, and the remainder imported into Python and converted to the standard Newick format using Biopython's Phylo module (29).
For each MCMC run, the maximum clade credibility (MCC) tree (23) was computed using Treeannotator (30), and 10 other sample trees were chosen at regular intervals along the chain. At the end of this stage, a total of 11 trees were associated to each data set: the MCC tree, plus 10 additional trees randomly sampled from the posterior distribution (sample trees).
Codon reconstruction.
We used an implementation of the Muse-Gaut codon model (MG94) (31) in HyPhy (32) as described previously (33) to estimate the ancestral sequence at the root of the tree. This model takes into account the difference between synonymous and nonsynonymous codon substitutions. The reconstructed ancestral root sequence was estimated by joint maximum likelihood (ML sequence [34]), and 10 additional ancestral root sequences were sampled from the posterior distribution (“sample” sequences) for a total of 11 reconstructed ancestral sequences associated to each tree. Since there were 11 trees associated to each data set (1 MCC trees and 10 sample trees), this made for a total of 121 reconstructed ancestral sequences per data set that could be classified into four categories: a 1×MCC tree with an ML sequence (denoted as MCC/ML), a 10×MCC tree with a sample sequence (MCC/sample), a 10×sample tree with an ML sequence (sample/ML), and a 100×sample tree with a sample sequence (sample/sample). For each category except MCC/ML, we computed the consensus of the reconstructed ancestral sequences in that category, resulting in four reconstructed ancestral sequences per data set (one for each category).
Indel reconstruction.
A codon model such as the one we used in the previous step only considers one kind of mutational event: nucleotide substitutions. Other mutational events such as insertions and deletions (indels) are generally not accounted for by codon models. Consequently, we were required to reconstruct the evolution of indel polymorphisms in ancestral sequences separately. We evaluated three methods at this stage. The first two were the published software packages Indelign (35) and Ancestors (36). For Indelign, we assumed a geometric distribution for the lengths of the indels. The two distribution parameters, the average length of insertions and the average length of deletions, were estimated separately per data set. For Ancestors, we used the software's maximum-likelihood heuristic with the default parameters. Since both software packages lack a character representation for missing data, these were replaced with the consensus of the observed indel character states in their respective columns. That is, if the state of an indel polymorphism was unavailable at a given position, it was assumed to be in the most common state. This only occurred when the sequence in question was incomplete at either end; these sites represented 0.98% of all follow-up data at the nucleotide level. The programs were run on nucleotide sequence alignments. In addition, we evaluated a series of “gap character” models that were applied to dichotomized alignments in which indel polymorphisms were represented by “0” or “1” to indicate the absence or presence of a codon. Full details are provided in the supplemental material (see “Gap character models”).
We initially tested all three methods on a representative sample of data sets and found that Indelign and Ancestors performed comparably and significantly outperformed the binary character approach. However, both packages were designed to handle large regions from only a few taxa and were somewhat unstable with intrapatient phylogenies with many branches. We decided to use the Indelign package, since it was more stable with these data than Ancestors. Indelign produced one sequence per tree, totaling 11 reconstructed ancestral indel patterns per data set. We reduced this to two: a single pattern generated with the MCC tree and a consensus of the 10 patterns generated with sampled trees.
Finalization and comparison.
The codon reconstructions were overwritten with the indel patterns to obtain four ancestral sequences per data set. The reconstructed ancestors were then compared to the baseline consensus. These comparisons were undertaken at both the nucleotide and amino acid levels. We did not consider sites where the baseline consensus nucleotide or amino acid sequence had a missing data character (“?”) or was ambiguous, states that comprised only 0.7% of all sites across all data sets at the nucleotide level and 0.7% at the amino acid level. These sites occurred for one of two reasons: (i) either the majority of baseline sequences were missing (incomplete) at that site (ii) or, of the baseline data present, exactly half of the sequences had one character and half had another. In either case, we could not be sure of the true ancestral state at these sites, so comparison to the reconstruction would be meaningless.
At the nucleotide level, we considered all of the sites where both the reconstruction and baseline consensus had a residue (i.e., neither sequence had a gap at that site, and the baseline consensus did not have a missing data character). If the ancestral reconstruction did not match the baseline consensus residue, we called that site a “nucleotide error.” We then defined the nucleotide error rate as follows: nucleotide error rate = (the number of nucleotide errors/the number of sites) × 100%. The “amino acid error rate” was defined in the same way for the amino acid translations of the reconstructed and follow-up consensus sequences. For comparison, we also calculated these error rates when comparing the consensus of all follow-up sequences to the baseline consensus sequence. Given only sequences from late in infection, the follow-up consensus sequence represents a phylogeny-naive estimate of the transmitted/founder virus genotype. For example, this consensus sequence would not account for the effect of selection, which would cause certain lineages in the population to proliferate over others and skew the consensus away from the true ancestor. We do not intend to suggest this method as an “alternative” method of ancestral reconstruction but rather as a comparator to provide context for our results.
At each site, the reconstructed ancestor may either correctly match the baseline consensus, be ambiguous, or show one of three types of error: substitution (i.e., a nucleotide or amino acid error), insertion (a present nucleotide or amino acid where the baseline consensus is gapped), or deletion (a gap where the baseline consensus has a present nucleotide or amino acid). We define the “overall error rate,” on both the nucleotide and the amino acid levels, as the combined proportion of ambiguous characters and these three types of error, i.e., as [(the number of substitution errors + the number of insertion errors + the number of deletion errors + the number of ambiguous sites)/the number of sites] × 100%.
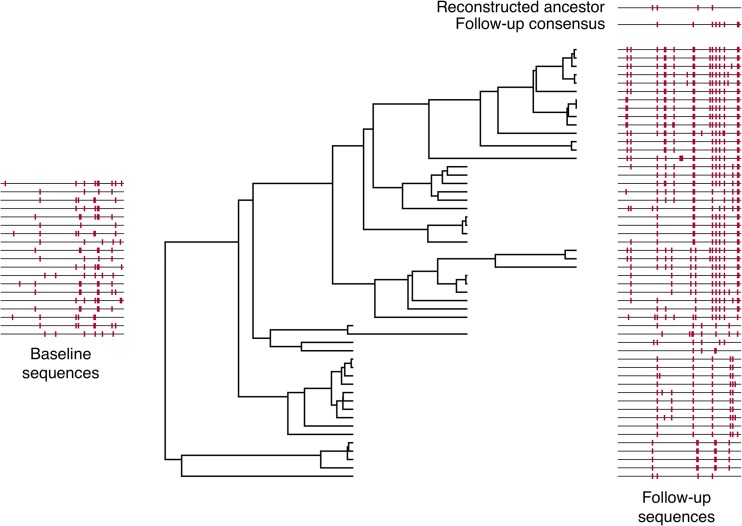
Since we are interested in all types of error, the overall error rate is a useful metric for evaluating reconstruction accuracy. Note that, in data sets with no indel polymorphism, the overall error rate is equal to the amino acid error rate. Figure 1 illustrates these definitions, and the components of ancestral reconstruction, for one data set on which reconstruction was performed with an overall error rate within 0.001% of the median. All sequences in the data set are displayed in the style of Highlighter plots (http://www.hiv.lanl.gov/) with respect to the baseline consensus. In the center is the maximum clade credibility phylogenetic tree. The baseline sequences, which we assume represent the ancestral sequence at the root of this tree, are on the left. On the right, the follow-up sequences are shown at the level of their corresponding tip node in the tree. The follow-up consensus and the reconstructed ancestral sequence are shown in comparison to the baseline consensus in the top right. Analogous figures for each data set studied can be found on our web server (link below).
FIG 1.
Illustration of ancestral reconstruction process. This figure summarizes the genetic composition and ancestral reconstruction for a representative data set (with the median reconstruction error rate). Baseline sequences are depicted on the left, with each line segment representing a sequence marked with mutations away from the baseline consensus, akin to a Highlighter plot (http://www.hiv.lanl.gov/). The phylogenetic tree was reconstructed from all follow-up sequences that were sampled at three time points after baseline. Follow-up sequences are depicted on the right with marks indicating mutations from the baseline consensus, and the reconstructed ancestral sequence using phylogenetic methods and the follow-up consensus sequence are depicted above.
Finally, since insertion and deletion events take place over whole sequence regions, rather than separately at individual sites, we defined a separate indel error rate to quantify the accuracy of indel reconstruction by regions rather than by sites. We defined an “indel region” as a contiguous group of alignment columns with a gap occurring at least once in either the follow-up sequences, or the baseline consensus, in every column. If at any site in the region the ancestral reconstruction showed a gap (“-”) and the baseline consensus showed a nucleotide or amino acid, or vice versa, we called the region an “indel error.” A total of 173 (51.6%) of the data sets had one or more indel regions; of these, the number of indel regions was, on average, low (median and interquartile range [IQR]: 3 [1 to 5], with a maximum of 15). The indel error rate was then defined in the same way as the other error rates, using counts of indel regions rather than individual sites, for data sets that had one or more indel regions: indel error rate = (the number of indel errors/the number of indel regions) × 100%.
All statistical analyses were conducted with R (37), and plots were made with the ggplot2 (38) package. The data files generated in all six steps of this project (alignment, trees, codon sequences, indel sequences, consensuses, and final reconstructions), our database, and analogues of Fig. 1 for all data sets, can be found at http://bioinfo.cfenet.ubc.ca/pub/ancre2/ (see the ReadMe in this directory for descriptions of all files, as well as the database schema).
RESULTS
Error rates.
Toward the goal of evaluating the accuracy of phylogenetic methods for reconstructing transmitted/founder viruses, we applied ancestral sequence reconstruction techniques to a large panel (n = 335 data sets, n = 19,486 sequences) of published intrapatient, longitudinal, HIV-1 clonal/SGS sequence data sets. The data originated from 55 studies published between 1991 and 2012 and varied widely with respect to geographic region, HIV subtype, transmission risk group, number of patients, and study design. The data sets were selected to contain at least two sample time points at least 6 months (186 days) apart, the first being within 6 months of HIV infection, seroconversion, onset of acute symptomatic seroconversion illness, or birth (in the case of mother-to-child transmission). The 6-month baseline cutoff was chosen as a liberal definition of “acute/early infection” in order to include as many and as varied data sets as possible (see also reference 11 for examples of prior investigations of HIV transmission using viruses sampled between 1 and 6 months postinfection). The published sequences had all been generated with either clonal or single-genome sequencing, so that multiple distinct sequences were associated with each time point.
Using the consensus of the earliest available (baseline) time point for each data set as the best estimate of the transmitted/founder sequence, we evaluated how well one could reconstruct this sequence using only data from subsequent (follow-up) samples. We evaluated several different procedures for extracting ancestral reconstructions from our phylogenetic analyses of these data sets. First, we used either a random sample of trees from the posterior distribution (this procedure is referred to as “sample”; [39]) or the maximum clade credibility (MCC) tree as the most representative case (see Materials and Methods). Second, we either used the joint maximum-likelihood (ML) reconstruction of the ancestral genotype at the root of the tree or a random sample from the posterior distribution of genotypes at the root (sample). Overall, there were four combinations of procedures evaluated for tree and sequence reconstructions, respectively, which we denoted as MCC/ML, sample/ML, MCC/sample, and sample/sample.
As a basis for comparison, we also evaluated the accuracy of the phylogenetically naive approach of taking a consensus sequence of follow-up samples (the “follow-up consensus”). The majority of differences between the follow-up and baseline consensuses are likely due to the selection of within-host mutations that increased in frequency or became fixed in the population between the baseline and follow-up time points. The frequency of these differences represents a phylogeny-naive error rate, corresponding to the degree of divergence of the host's viral population from the transmitted/founder sequence (see “Divergence and diversity” below).
The results are shown in Table 2. At both the nucleotide and the amino acid levels, all ancestral reconstruction methods yielded lower mean overall, substitution, and deletion error rates than the follow-up consensus approach. However, the mean rates of insertion errors were higher for all ancestral reconstruction methods than for the follow-up consensus. The difference in accuracy at the amino acid level between ancestral reconstruction and the follow-up consensus was highly significant when considered as an interaction effect with sequence diversity (+0.8 log odds, binomial GLM, P = 3.49 × 10−4). Controlling for this interaction effect and for the effect of entropy itself, ancestral reconstruction remained significantly more accurate than the follow-up consensus (+0.10 log odds, P = 0.04), and the magnitude of this improvement increased with the diversity of the data set (see “Divergence and diversity,” below).
TABLE 2.
Mean error rates of ancestral reconstruction and follow-up consensus at nucleotide and amino acid levelsa
| Error rate category | Mean error rate ×10−3 (range) |
||||
|---|---|---|---|---|---|
| Follow-up | MCC/ML | MCC/sample | Sample/ML | Sample/sample | |
| Nucleotide error rates | |||||
| Overall | 14.9 (0–177.9) | 13.1 (0–118.8) | 14.0 (0–118.8) | 13.9 (0–134.9) | 13.7 (0–123.0) |
| Substitution | 9.5 (0–135.2) | 8.6 (0–72.2) | 8.6 (0–75.0) | 8.1 (0–71.3) | 8.3 (0–76.9) |
| Insertion | 2.7 (0–66.9) | 3.7 (0–66.9) | 3.7 (0–65.9) | 4.8 (0–92.6) | 4.9 (0–92.6) |
| Deletion | 1.6 (0–55.6) | 0.69 (0–27.9) | 0.69 (0–27.9) | 0.39 (0–16.5) | 0.39 (0–16.5) |
| Missing or ambiguous | 1.1 (0–29.7) | 0 (0–0) | 1 (0–26.6) | 0.57 (0–21.5) | 0.12 (0–6.3) |
| By indel region | 240.9 (0–1,000.0) | 207.2 (0–1,000.0) | 207.2 (0–1,000.0) | 235.6 (0–1,000.0) | 235.6 (0–1,000.0) |
| Amino acid error rates | |||||
| Overall | 27.5 (0–237.4) | 23.0 (0–175.0) | 24.8 (0–200.0) | 23.7 (0–182.9) | 23.5 (0–182.9) |
| Substitution | 21.0 (0–187.0) | 18.6 (0–158.3) | 18.5 (0–161.1) | 17.4 (0–158.3) | 18.1 (0–169.4) |
| Insertion | 2.5 (0–66.9) | 3.7 (0–66.9) | 3.6 (0–64.1) | 4.7 (0–92.6) | 4.8 (0–92.6) |
| Deletion | 1.5 (0–48.8) | 0.67 (0–27.9) | 0.67 (0–27.9) | 0.38 (0–16.5) | 0.38 (0–16.5) |
| Missing or ambiguous | 2.5 (0–69.3) | 0.057 (0–15.1) | 2 (0–44.1) | 1.2 (0–53.4) | 0.23 (0–15.1) |
| By indel region | 237.8 (0–1,000.0) | 216.4 (0–1,000.0) | 216.4 (0–1000.0) | 245.7 (0–1,000.0) | 245.7 (0–1,000.0) |
Error rates of follow-up consensus and four ancestral reconstruction methods, broken down by error type (substitution, insertion, deletion, or ambiguous) at both nucleotide and amino acid levels are indicated. Error rates by indel region are also given.
Indel reconstruction was performed only for the n = 173 (51.6%) data sets with one or more indel polymorphisms in their follow-up sequences. When indel errors were counted as a proportion of indel regions, rather than as a proportion of individual sites, indel reconstruction was revealed to be much more challenging than the reconstruction of ancestral amino acids. At the amino acid level, the mean indel error rate, by regions, of the follow-up consensus was 23.8%. Since the Indelign package reconstructs only maximum-likelihood sequences, the same reconstructed indel pattern was used for both the ML and sample sequences for each tree, so that the indel error rate varied only with the tree used in the reconstruction. The mean indel error rates were 20.9% for indel reconstruction with the MCC tree, and 23.7% for indel reconstruction with the sample trees.
Since the MCC/ML method yielded the lowest overall error rate at both the nucleotide and amino acid levels, all results henceforth apply to MCC/ML sequences only. Furthermore, all subsequent references to “error rate” and “overall error rate” refer to the error rates at the amino acid, rather than nucleotide, level. The overall error rates stratified by gene are summarized in Fig. 2. The accessory and regulatory genes (nef, tat, rev, vif, vpr, and vpu) are grouped together, since they represented fewer than 20 alignments each (n = 51 total). HIV env was substantially more difficult to reconstruct than any other gene (Mann-Whitney test, P < 10−5), with gag having the second-highest error rate, followed by pol, and lastly the accessory and regulatory genes. Across all genes, the median error rates for ancestral reconstruction were lower than or equal to the median error rates for the follow-up consensus.
FIG 2.

Error rates stratified by gene. Box-and-whisker plots show that ancestral reconstruction has a lower median error rate than the follow-up consensus across all genes. Error rates of env data are elevated compared to other genes. “Follow-up consensus” denotes the error rate in phylogeny-naive reconstructions from follow-up samples. “Reconstructed ancestor” denotes the error rate in phylogenetic ancestral reconstructions from the same follow-up samples.
Determinants of error rate.
The fact that the data we analyzed originate from diverse study types introduces many possible sources of reconstruction error. We expected both the design of the study and the clinical characteristics of patients involved to have an impact on the error rate. To identify which variables had an impact on error rates and to what extent, we identified several study design parameters and data features and evaluated their association with error rates using a generalized linear model (GLM). For a data set, we define “Δt” as the number of days between the baseline and first follow-up samples. In other words, Δt is the amount of time evolution has progressed unobserved within the patient after the initial sampling and thus quantifies the amount of diversification we must “undo” to recover the founder strain. Other study design factors that we examined were as follows: the mean number of days between follow-up samples (intrasample time), the mean number of sequences per time point, and the number of samples per year of additional follow-up (0 for only one follow-up time point). The data features we considered were risk group/mode of transmission (MSM, heterosexual, IDU, MTCT, or transfusion), gene (env, gag, pol, or accessory/regulatory), molecule type (DNA, RNA, or mixed), and treatment history (treated or untreated). Molecule type was taken from the “mol_type” field of each GenBank entry, with RNA generally originating from free virus in plasma and DNA originating from peripheral blood mononuclear cells. mRNA molecules were grouped with RNA. If patients ever received antiretroviral therapy (as detailed in the original publication), they were labeled as treated, regardless of the duration or efficacy of the treatment.
To identify which of these factors were predictors of reconstruction error rates, we fit a GLM with a logit link function to the overall error rate expressed as a binomial outcome (substitution, insertion, or deletion error, or missing data = failure, no error = success). Risk group was unavailable for n = 78 (23%) of the data sets, since these data originated from cohort studies comprising multiple risk groups and the clinical characteristics of individual patients were not specified. Similarly, for n = 46 (14%) it was not specified whether the patients were treated or untreated. These cases were excluded from the GLM but are included in all reported univariate relationships. n = 21 (6%) data sets were missing both pieces of information, so the total number of excluded data sets was n = 103 (31%) We did not consider interaction effects in the GLM because the significant colinearity among some variables would have made it difficult to interpret the interaction terms of the resulting model (see “Sensitivity to route of infection, sampling timeline, and viral genome type,” below). The profile of the resulting GLM is shown in Table 3. Coefficients are relative to the most frequent data features, which were MSM risk group, env gene, DNA-derived sequences, and no treatment.
TABLE 3.
GLM profilea
| Parameter | Coefficient | 95% CI (minimum, maximum) | P |
|---|---|---|---|
| Intercept (MSM, env, DNA, untreated) | 2.75 | 2.53, 3.0 | <10−5 |
| Heterosexual | –0.21 | −0.37, −0.04 | 0.01 |
| MTCT | –0.07 | −0.27, 0.1 | 0.54 |
| IDU | 0.98 | 0.64, 1.4 | <10−5 |
| Transfusion | –2.05 | −2.47, −1.6 | <10−5 |
| Baseline time (years) | –1.03 | −1.44, −0.6 | <10−5 |
| Δt (years) | –0.08 | −0.12, −0.04 | 1.20 × 10−4 |
| Follow-up samples/year (n) | 0.34 | 0.28, 0.4 | <10−5 |
| Mean sequences/time point (n) | 0.03 | 0.02, 0.04 | <10−5 |
| gag | 1.37 | 1.21, 1.5 | <10−5 |
| pol | 1.84 | 1.52, 2.2 | <10−5 |
| Accessory/regulatory | 0.56 | 0.19, 1.0 | 4.57 × 10−3 |
| RNA | –0.03 | −0.20, 0.1 | 0.72 |
| DNA+RNA | 0.03 | −0.19, 0.3 | 0.78 |
| Treated | 0.26 | 0.13, 0.4 | 1.06 × 10−4 |
Data were determined using a generalized linear model. Regression coefficients (log odds), 95% confidence intervals (95% CI), and P values for the fitted GLM are shown. The parameters used in the GLM are risk group (heterosexual, MTCT, transfusion, and IDU, all relative to MSM), gene (gag, pol, and accessory/regulatory, all relative to env), molecule type (RNA, RNA+DNA, all relative to DNA), Δt, follow-up samples/year, sequences/time point, and baseline sample time.
Increased time between critical time points (t) made reconstruction significantly more difficult (−0.08 log odds per year, P = 1.15 × 10−4; Table 2). This intuitive result was confirmed by examination of the univariate relationship between error rates and t (Spearman's ρ = 0.5, P < 10−5; Fig. 3) and with the time between infection/seroconversion and baseline. The more HIV evolves unobserved, the further its pattern of evolution deviates from the predictive models used in ancestral reconstruction. On the other hand, increasing the number of follow-up time points per year was associated with a higher rate of successful reconstruction (+0.3 log odds per follow-up sample per year, P < 10−5), as was increasing the number of sequences per time point, though the effect was more modest (+0.03 log odds per additional sequence, P < 10−5). Reconstruction on HIV RNA-derived data sets was slightly less accurate than on HIV DNA-derived data sets, but this was not significant (−0.03 log odds, P = 0.7). Finally, treatment had a positive effect on reconstruction accuracy (+0.3 log odds, P < 10−4).
FIG 3.
Relationship of reconstruction error to Δt and to entropy, env, and non-env, stratified by risk group. The scatter plots depict the univariate associations between overall error rates and amino acid entropy (left) and the time elapsed between the baseline and first follow-up sample (Δt, right), for env (top) and HIV genes other than env (bottom). Trend lines were derived from linear regressions stratified by risk group as indicated in the inset figure legend.
Closer examination of the 17 data sets comprising the highest 5% of error rates did not reveal a single unifying cause for the poor reconstructions. The majority of the data sets were env (14 env, 2 accessory/regulatory, and 1 gag), and the mean alignment length was shorter than the average (247 versus 309 codons). The mean t of these data was more than double the average (1,272 versus 493 days), while the number of follow-up sequences was just over half the average (27 versus 44). Two of three patients in the transfusion risk group were present among the 17, which suggests that these two patients may have been coinfected with more than one founder virus. The other risk groups were represented roughly in proportion to the total (10 heterosexual, 4 MSM, and 1 MTCT), as were molecule types (14 DNA and 3 RNA) and treatment histories (12 untreated, 4 treated, and 1 unknown). The diversity of the high error rate sequences was over three times the average (mean entropies, 0.3 versus 0.10).
Divergence and diversity.
Regardless of study design, ancestral reconstruction is generally impeded by two features of the available data: divergence and diversity. Divergence represents the accumulation and fixation of mutations in the virus population, taking the consensus genotype further away from the transmitted/founder genotype (40). Diversity refers to how different the follow-up sequences are from each other or how “spread out” viral evolution has been within the patient (40). We first examined the relationship between diversity and reconstruction accuracy, quantifying diversity with the entropy of the data set. The entropy of an alignment with respect to a set of allowable characters is a measure of diversity, where a higher value indicates a more diverse alignment. The amino acid entropy of an alignment, which is the entropy of the alignment with respect to the amino acid alphabet, was defined by Henikoff and Henikoff (41). We also consider indel entropy, which is analogously defined with respect to the binary alphabet representing presence and absence. A high amino acid entropy indicates a high rate of nonsynonymous polymorphism; high indel entropy indicates more variable sequence length. The “overall entropy” is the sum of amino acid and indel entropies.
An important measure of ancestral reconstruction's utility is how well the techniques scale with diversity. As shown in Fig. 3, the positive correlation between entropy and error rate is pronounced. The more diverse a population, the more difficult reconstruction becomes for any method, phylogenetic or otherwise. However, Fig. 4 shows that, as entropy increases, the disparity between methods increases. At very high entropy, both methods are disadvantaged, but the relative improvement in accuracy of phylogenetics over the follow-up consensus increases. We note also that both entropy and our definition of error rate are normalized by sequence length. The median length of the alignments was 233 codons, and we found no significant difference in reconstruction error rates between sequences above and below this value (Wilcoxon rank-sum test, P = 0.4).
FIG 4.
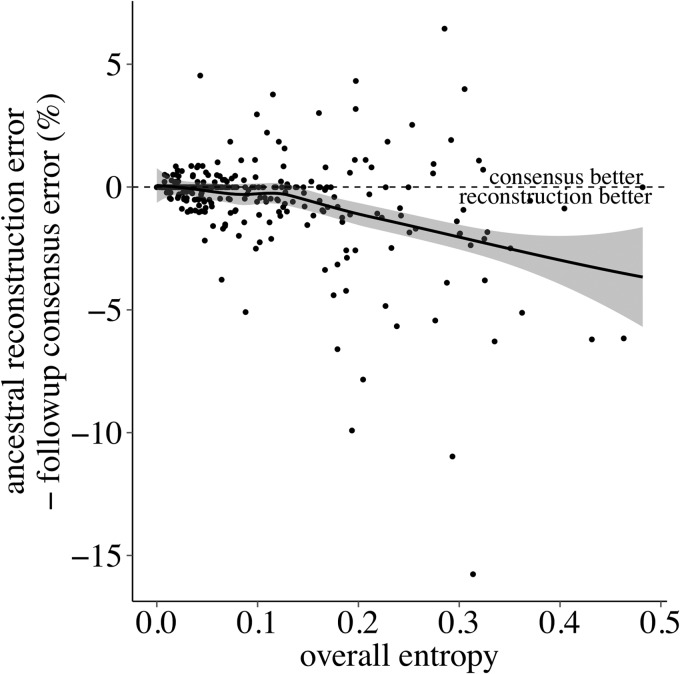
Advantage of phylogenetics over naive method increases with diversity. As entropy of data set increases, the difference between the error rates of the phylogenetic and naive methods increases, in the direction of the phylogenetic method having a lower error rate. Points below the vertical line are data sets where phylogenetics outperforms consensus. Trend line is a local regression (Loess) with 95% confidence interval (shaded area).
Population divergence is more intuitively quantified than diversity, simply by counting the differences between the baseline and follow-up consensus sequences. In other words, the divergence of the population can be directly quantified by the error rate of the follow-up consensus. As the ultimate goal of ancestral reconstruction is an error-free recovery of the transmitted/founder virus sequence, we examined the cases where perfect reconstruction was accomplished by either method. Cases where the baseline and follow-up consensuses achieved an exact match—data sets with zero divergence—provided a sanity check on the ancestral reconstruction methods. Examination of cases where ancestral reconstruction was error-free informed us about how much divergence one could reasonably expect to “undo” with computational methods. Finally, the cases where neither method was perfect indicated which technique was more accurate on the more difficult data sets.
There were 79 cases where both methods performed a perfect reconstruction. In 14 cases, the follow-up consensus was perfect but ancestral reconstruction had errors. On the other hand, there were 27 cases of perfect reconstruction by phylogenetics but not by the follow-up consensus. Unsurprisingly, the overall error rates of the follow-up consensus on these data sets were lower than the average (mean [range] error rates 1.1% [0.1 to 6.3%]; see “Error rates,” above). All risk groups except IDU and transfusion were represented in these data sets (10 MSM, 9 unknown, 4 MTCT, and 4 heterosexual). The majority were DNA derived (n = 14); 5 were RNA derived, and 8 were mixed DNA and RNA data sets. Non-env genes were over-represented in this sample (63%). Finally, when neither method was perfect, phylogenetics outperformed the follow-up consensus by ca. 0.6% of sites (mean error rates = 3.5% [0.2 to 17.5%] for ancestral reconstruction and 4.1% [0.1 to 23.7%] for follow-up consensus).
Errors within immunologically relevant positions.
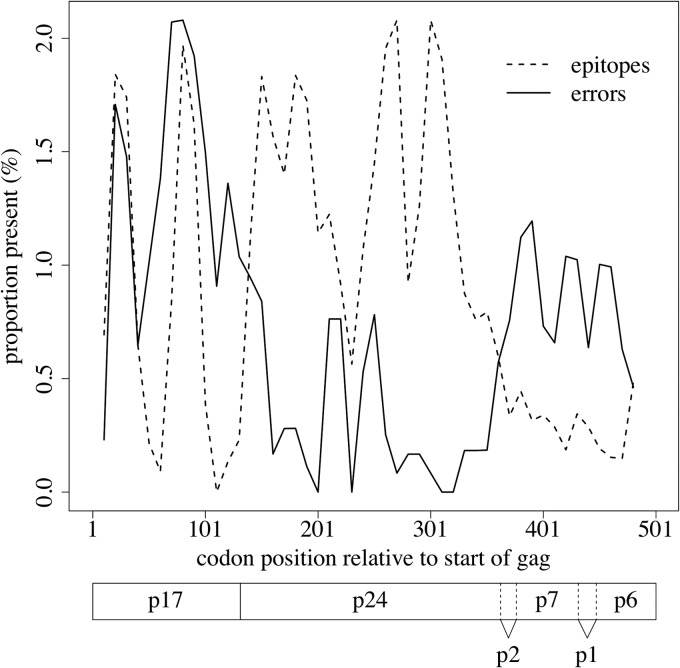
Errors occurring in immunologically important positions, such as CD8+ T-lymphocyte (CTL) epitopes in gag, may also be of particular relevance to vaccine design. We observed a weak, but statistically significant, inverse correlation between overall error rate and density of best-defined epitopes (42) (Spearman's ρ = −0.1, P = 0.02), a trend particularly noticeable in p24 onward (Fig. 5). However, it is possible that this was due simply to those regions of gag which are most epitope dense being most conserved and therefore easier to reconstruct (see Discussion). HIV gag overall was easier to reconstruct that env (see “Error rates,” above) but more difficult than pol or the accessory/regulatory genes, although sample size may have been a factor in this result.
FIG 5.
Proportions of errors and CTL epitopes within HIV gag. Inverse correlation between epitope density and error density. Lines represent sliding window weighted averages of error rates (solid) and epitope densities (dashed) with a window size of 20 codons. Locations of the respective gene products of the Gag precursor, including the matrix (p17), capsid (p24), and nucleocapsid (p7) proteins, are indicated below the plot (based on the HXB2 reference sequence, GenBank accession number K03455).
Rapid divergence from founder.
In the preceding analyses, we have measured the accuracy of ancestral reconstruction by comparing a phylogenetically imputed ancestor to the consensus of samples taken in acute infection. For this comparison to be meaningful, the baseline consensus must actually represent the true transmitted/founder strain. However, there may be cases where the baseline consensus is different from the transmitted/founder strain such as, for example, if one or more mutations occurred very early in infection and proliferated to a high frequency by the time of the first (baseline) sample. This possibility introduces another source of uncertainty into our reported error rates. Unfortunately, since the baseline sequences are the earliest available for each data set, any differences between the baseline consensus and transmitted/founder strain are not directly measurable.
To estimate the impact of this source of uncertainty, we performed a similar analysis on n = 11 longitudinal data sets from a study of simian immunodeficiency virus (SIV) env evolution in macaques experimentally infected by a known inoculum of SIVmac239 (43). If sequences diverged rapidly from the inoculum genotype by baseline, then one should expect higher error rates when comparing reconstructed genotypes directly to the inoculum than against the baseline consensus that will more likely resemble genetic variants from follow-up samples. In these data, the consensus of SIV env sequences sampled at baseline (63 to 168 days postinfection) differed from the inoculum genotype at 0.1% of amino acids on average (range, 0 to 0.35%); sequences ranged from 857 to 878 amino acids in length. This level of rapid divergence was similar to what has been reported in longitudinal studies of acute HIV infections (21, 44, 45) (see Discussion). The error rates of the follow-up consensus method were slightly higher when evaluated against the inoculum (mean, 0.33% [0.12 to 0.69%]) than against the baseline consensus (mean, 0.3% [0.12 to 0.58%]). On the other hand, we found no measurable difference in error rates using phylogenetic methods, with means of 0.19% (range, 0 to 0.58%) and 0.19% (range, 0 to 0.47%), respectively, relative to the inoculum and baseline consensuses. These results suggest that rapid divergence of HIV from the transmitted/founder virus prior to baseline is not a primary source of error in ancestral reconstruction by phylogenetic methods. Since rapid divergence before the baseline sample tends to be limited to a small number of sites, however, these error rates are close to the level of stochastic error, making it difficult to measure differences between methods with a high degree of confidence.
DISCUSSION
Previous work.
The concept of ancestral reconstruction was first theorized by Zuckerkandl and Pauling (15), whose goal was to “extract evolutionary history from molecules.” Since then, advances in computational techniques and sequencing technologies have allowed researchers to apply ancestral reconstruction to a wide variety of organisms and time scales. In particular, a few studies have made use of ancestral sequence reconstruction to infer properties of unavailable HIV strains. These studies share the limitation that their methods are unverifiable, as the true ancestor could not be sequenced. For example, Rolland et al. (46) developed a “center of tree” reconstruction method and applied it to gag, tat, and nef sequences from several different hosts. The reconstructed sequences were synthesized and found to be both functional and immunogenic. However, the center of tree sequence was not meant to represent the (unrecoverable) common ancestor to all of the input sequences but rather a hypothetical sequence which minimized evolutionary distance to the extant strains. Ancestral env sequences, reconstructed with maximum-likelihood methods, have likewise been shown to be both functional and immunogenic (47, 48). In (33), a reconstruction pipeline from which this work was derived in part was used to examine the pattern of the HIV coreceptor switch within a given host by estimating ancestral sequences at all nodes of several intrapatient phylogenies. Although the reconstructed sequences were theorized to have existed in the respective patients at some point, this could not be confirmed by sequencing as the study population comprised chronic HIV infections and no early samples were available.
On the other hand, several validations of phylogenetic tree reconstruction, a critical step of ancestral reconstruction, have been done with in vivo HIV data. Hillis et al. (49) correctly inferred a between-host HIV phylogeny with Δt between 2 and 3 years. Phylogenetic methods described previously (50) were used to estimate the time elapsed since the most recent common ancestor (tMRCA) of the viral population of several patients, roughly corresponding to date of infection. The estimates corresponded well to clinical measures of infection date. In an earlier study (51), a known HIV phylogeny comprising 13 patients was accurately inferred through a variety of phylogenetic methods.
A natural next step is to apply the idea of testing on real data from the second body of work to the methodological framework of the first—that is, use ancestral reconstruction on groups of extant sequences where the ancestor is already known or can be accurately estimated. This has been applied in a few limited cases, but most often in a controlled laboratory setting. Hillis et al. (52) successfully recovered the branching order (but not the branch lengths) of an experimentally created bacteriophage T7 phylogeny by five different reconstruction methods, and parsimony was used to estimate the ancestral character states with 98.6% accuracy. However, the mutation rate of bacteriophage T7 is substantially lower than HIV. Researchers using experiments with synthesized ancestral env sequences (48) evaluated their reconstruction methods on a group of macaques inoculated with SIVmacBK28 and were able to recover the inoculum genotype with 99.8% accuracy.
The recent availability of longitudinal, intrapatient HIV sequence data sets provides a large number of test cases for evaluation of ancestral reconstruction on real data, with phylogenies having evolved on their own outside a laboratory setting. To our knowledge, ancestral reconstruction has only been applied to one such data set (53). In that case, the objective of the study was not to validate ancestral reconstruction but to examine the relationship of extant sequences to the most recent common ancestor (MRCA). In the present work, we evaluated the performance of phylogenetic methods to reconstruct transmitted HIV variants on a much larger number of cases. We found that, overall, phylogenetic methods tend to provide a better reconstruction of the amino acid composition of this ancestral sequence than a consensus method. The relative advantage of the phylogenetic method increased significantly with sequence diversity, where reconstruction is generally more challenging for either approach. On the other hand, neither method could reliably reconstruct ancestral indel variation, with mean rates of indel reconstruction error exceeding 21.6%. Finally, reconstruction error rates were significantly reduced in CTL epitope-dense regions of the HIV genome; this association may be due to the fact that these regions tend to be evolutionarily conserved.
Sensitivity to phylogenetic methods.
There are a myriad of programs available for each step of the reconstruction process, and no widely accepted standard for which to use. The packages in our pipeline were chosen based on a combination of literature recommendations and prior experience (50) and limits on computational resources. A great many packages exist for the first step, multiple sequence alignment (see Table 3.1 in reference 54) for a list). After all steps of processing, a very poor reconstruction could often be traced back to an inaccurate alignment. Predictably, the hypervariable loops of env were very difficult to align owing to high indel rates in these regions and were likely at least partly responsible for the high error rates seen in env reconstructions compared to other genes. Many methods also exist for phylogenetic inference, but the necessity of rooting the tree left us with only two broad options—rooting with an outgroup or using a molecular clock model—and we opted for the latter. As discussed previously (55), outgroup rooting works well only when the outgroup is fortuitously chosen close to the ancestor.
To reconstruct sequences at the root of the tree, we chose the Muse-Gaut model of codon substitution (31), which is aware of both nucleotide and amino acid space (in particular, it makes the distinction between a synonymous and nonsynonymous substitution). Mostly, this choice was based on prior experience (50) and having an implementation of the model already available. On the other hand, reconstructing the ancestral indel pattern was a particularly challenging step. To our knowledge, only two published programs, Indelign (35) and Ancestors (36), were available at the time of our study for ancestral indel reconstruction. Moreover, these programs are tailored for whole-genome, macroorganism evolution, where insertions and deletions tend to be large and infrequent. In HIV, indel polymorphisms tend to occur most often in highly variable regions of the HIV genome, such as the region encoding the HIV envelope glycoprotein gp120 variable loops V1 and V2. These regions are inherently difficult to align due to a lack of sequence homology and can undergo extensive change between time points. Finally, our merging of the indel and codon reconstructions by simply “overwriting” one with the other implicitly assumes that codons and indels evolve independently, which is unlikely to be the case. On our data, the best phylogenetic methods in the public domain for reconstructing ancestral indel variation were unable to outperform a rudimentary method of taking the consensus of follow-up sequences. This is a significant concern in the context of HIV, where indel variation can play an important role, for example, in mediating escape from the neutralizing antibody response (56). Reconstructing ancestral indels is a major challenge because, by definition, the sequence homology that conventional phylogenetic methods rely on to reconstruct ancestral variation lacks insertions and deletions. As such, this remains an important open area for future work with immediate applications to understanding the role of indel variation in transmitted/founder variants of HIV.
Although it complicates the construction of an ancestral reconstruction pipeline, the variety of available methods also provides opportunities for further exploration. In particular, the models of evolution used in phylogenetics do not account for recombination as a possible evolutionary event. As such, when recombination does occur, it has the potential to greatly confound both phylogenetic inference (57, 58) and the reconstruction of ancestral sequences (59). Although recombination rates in HIV are high (60), it has recently been suggested that HIV's effective recombination rate may be 1 to 2 orders of magnitude lower than previously thought (61). In addition, in cases of infection by a single transmitted/founder virus sequence, the only possible types of recombination events are within, as opposed to between, divergent lineages (62). Although such events might potentially distort the shape of the estimated phylogeny, the ancestral sequence at the root should be markedly less affected than if interlineage recombinants were introduced. Regardless of magnitude, recombination is almost certainly a contributing factor to the error rate of ancestral reconstruction. Phylogenetic techniques for ancestral reconstruction which take recombination into account, which could help offset this source of error, are an important area for future research.
Finally, we considered whether sampling trees or sequences conferred an advantage compared to maximum clade credibility phylogenies and maximum-likelihood reconstructions. Hanson-Smith et al. (63) suggest that the Bayesian approach of sampling from the posterior distribution is unnecessary and will not improve reconstruction accuracy. Our analysis of longitudinal HIV sequence data agrees with this result, although the apparent loss of accuracy resulting from sampling was very small. This suggests a fairly high degree of uncertainty for our data about both the phylogeny (perhaps owing to the relatively high homology of intrapatient data sets) and the underlying evolutionary process. Indeed, although the heights of the reconstructed phylogenies (which represent the time to most recent common ancestor, i.e., tMRCA) should have been in agreement with the known timelines of longitudinal sampling, this was not always the case, since many of the tree heights were much further in the past than the known reference points (see Fig. S1, left, in the supplemental material). We do not believe this to be entirely attributable to coinfection, since it was observed across all risk groups and even in the SIV data sets we examined, which were known to have only one transmitted/founder. We investigated the possibility that the assumption of a constant population size was having a deleterious effect on tree heights by using a Bayesian skyline model to marginalize out uncertainty about population dynamics; however, the Bayes factors indicated strong support for the constant population size model in most data sets (data not shown). We also observed a significant correlation between the variance in tree heights sampled from the posterior and reconstruction error (see Fig. S1, right, in the supplemental material), further contributing to the evidence for a high degree of phylogenetic uncertainty in these data.
Sensitivity to route of infection, sampling timeline, and viral genome type.
A core assumption of applying phylogenetic methods to the problem of reconstructing the sequence of the transmitted/founder virus is the existence of a population bottleneck at transmission, resulting in a single productive viral lineage (64). Multiple studies have reported that, although the majority of new infections stem from a single variant, coinfection with two or more variants does occur (7, 22, 62, 65). Using the methods of Keele et al. (11), we isolated n = 135 data sets where the estimated tMRCA fell within 3 months on either side of the known reference point. Given the potential margin for error in the reference point itself (which is often reported as the midpoint between the last seronegative and first seropositive HIV test), we could be reasonably certain that these data sets satisfied the hypothesis of a single transmitted/founder virus. Error rates for these data sets were lower than average (mean error rates [range] = 1.6% [0 to 14.6%] for ancestral reconstruction and 1.9% [0 to 16.0%] for follow-up consensus), but the relative advantage of phylogenetics over the naive approach (18.5%) was roughly consistent with the rest of the data. A histogram of discrepancies between tMRCA estimates and known reference points is provided in Fig. S2 in the supplemental material.
On the other hand, modes of HIV transmission that bypass the recipient's mucosal barriers (e.g., via injecting drugs or blood transfusion) can result in a greater number of infecting strains (66). Since our methods rely on the assumption of a single founder virus, we would expect the reconstructions to be of greatly diminished accuracy in these cases. Although we only had data from six patients in these categories, we observed elevated rates of overall reconstruction error (relative to the MSM risk group) that were consistent with this hypothesis. In the transfusion risk group (n = 3) the error rates were well above average (13.3, 14.6, and 6.6%); the error rates of the data sets in the IDU risk group (n = 4) were 0, 0, 1.9, and 3.4%.
We also observed significantly greater reconstruction errors among individuals in the heterosexual risk category relative to MSM (mean error rates [range] = 3.7% [0 to 17.5%] for heterosexuals versus 1.8% [0 to 11.1%] for MSM). However, it is very likely that this result was due to confounding with other study design variables. Values of Δt were much higher for the heterosexual data, averaging several years for the heterosexual transmission group but only months for the MSM group (median days [IQR] = 1,202 [252 to 1,852] for heterosexuals versus 89 [30 to 489] for MSM). The heterosexual group also had fewer sequences per time point than the MSM group (median number [IQR] = 10 [7 to 14] for heterosexuals versus 15 [10 to 20] for MSM).
To compare risk groups while controlling for Δt, we performed a matched test by selecting pairs of data sets, one from each risk group, with values of Δt within 15 days of each other. We then compared the error rates between matched pairs of data sets using a paired Wilcoxon signed-rank test. This controls for the effect of Δt on error rates; however, due to the wide variation of t values, matched pairs comprised only a small subset of the data sets. Using this procedure, we found no significant differences between the heterosexual and MSM risk groups (n = 26, P = 0.7), the MSM and MTCT risk groups (n = 31, P = 0.2), or the heterosexual and MTCT risk groups (n = 20, P = 0.09).
We hypothesize that the effect of HIV molecular type (proviral DNA versus RNA) on reconstruction error was due to the presence of “archived” proviral HIV sequences in the reservoir of latently infected cells (67), which has been previously observed to affect the accuracy of phylogenetic inference (68). Moreover, data sets where both DNA and RNA were sequenced tended to have higher error rates than data sets derived from only one molecular type. This outcome suggests that the evolutionary processes within cellular reservoirs represented by the samples of HIV DNA are very different from the processes shaping variation among free virions in plasma. A single evolutionary model may not adequately capture this heterogeneity. As with risk group, the HIV molecular type was correlated with other study design variables. In particular, RNA data sets tended to have lower Δt, a shorter interval between the reference point and baseline sample, and more follow-up time points per year than DNA data sets (all Wilcoxon rank-sum test, P < 10−5). Although confounding between study parameters makes it difficult to attribute the impact of each parameter on reconstruction error rates, our model analysis indicates that a combination of RNA data, low Δt, high numbers of sequences per time point, and frequent follow-up provides the best chance of an accurate ancestral reconstruction. In addition, the techniques may be inappropriate for modes of transmission where the probability of coinfection is high, such as via blood transfusion or the use of injection drugs.
Finally, there is no guarantee that the consensus of the baseline sequences is an accurate representation of the true transmitted/founder virus. Discrepancies may occur if, for example, a mutation in the HIV genome appearing very early in infection reaches a high frequency in the virus population before the baseline sample is taken. In this case, neither a naive consensus nor phylogenetic approaches would be able to accurately reconstruct the individual transmitted/founder virus. Instead, estimates of this sequence will erroneously include substitutions that became fixed in the population between transmission and the earliest date of sampling. To assess the impact of rapid divergence from the founder genotype, we analyzed longitudinal data sets of SIV env in 11 macaques experimentally infected with a clonal SIVmac239 inoculum, such that the transmitted/founder virus genotype was known without ambiguity. In these data, there was limited amino acid divergence (mean, 0.1%) from the inoculum genotype by the baseline sample. As expected, we observed slightly higher mean error rates when the follow-up consensus sequences were evaluated against the inoculum instead of the baseline consensus. In contrast, we observed no difference in mean ancestral reconstruction error rates with respect to the inoculum or baseline consensus, suggesting that phylogenetic methods were less sensitive to rapid sequence divergence before baseline.
It may be difficult to generalize results from SIV to the rapid divergence of HIV in vivo. However, we found comparable levels of divergence within similar time frames (within 6 months of infection) have been reported in studies of acute HIV infections with exceptionally early samples. Herbeck et al. (21) analyzed whole HIV genome sequences (∼9,100 bp) sampled longitudinally from three seroconverters, including samples as early as 3 days after the onset of symptoms, and found that between 9 and 18 mutations (0.1 to 0.2% of the genome) had accumulated under positive selection by 6 to 7 months. These researchers also noted that the transition from a star- to lineage-based phylogeny may occur as early as 50 days into infection, which would shift the baseline consensus away from the transmitted variant. Similarly, Henn et al. (44) performed whole-genome “deep” sequencing on longitudinal samples of HIV from one subject, including a sample taken within 15 to 20 days of infection, and identified 2 mutations (∼ 0.02%) that attained >50% prevalence by 6 months after presentation. Finally, Goonetilleke et al. (45) performed longitudinal single-genome amplification sequencing on three patients who were at Fiebig stage II (18 to 34 days postinfection) at screening and identified between 4 and 12 mutations (0.05 to 0.15% of the genome) which had become dominant by 6 months after screening. Although our analysis of SIV data sets suggests that phylogenetic methods are less sensitive to this rapid divergence, we can incorporate the upper limit of reported HIV sequence divergence by 6 months postinfection (0.2%) to yield conservative estimates of the overall error rates, averaging from 2.1 to 2.5% for ancestral reconstruction and from 2.6 to 3.0% for the follow-up consensus.
Despite these limitations, the best possible method to reconstruct the transmitted HIV genotype is to examine the genetic composition of samples from an acute or early stage of infection. Obtaining such samples in substantial numbers, however, unavoidably requires implementing and maintaining a large-scale prospective cohort of individuals at risk of HIV infection. When early samples cannot be obtained, the best that one can do is to extrapolate the ancestral sequence from HIV sequences derived from the samples that are available, while interpreting such results in light of limitations discussed above. Our ability to reconstruct ancestors wanes as the HIV sequences inevitably become more diverse and complex over time. Beyond this certainty, we have lacked a quantitative understanding of how accurately we can reconstruct transmitted HIV variants from available sequence data. In the present study, we have shown that a significant quantity of ancestral genotypic information is recoverable through phylogenetic analysis, but not by a phylogeny-naive examination of extant sequences, and that the amount of new information revealed through phylogenetics increases with sequence diversity. However, the advantage of phylogenetic methods does not carry over to the reconstruction of ancestral indels. This reveals a significant gap in the state-of-the-art of phylogenetics and a critical area for future work because of its key implications for our understanding of HIV, where indel variation can play a central role in mediating escape from the neutralizing antibody response.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Alejandro Gonzalez-Serna and Mark Brockman for their insightful suggestions on the effects of molecule type on reconstruction accuracy.
This study was supported in part by Operating Grants from the Canadian Institutes of Health Research (CIHR) to Z.L.B. (grant HOP115700) and to A.F.Y.P. (grant HOP111406). Z.L.B. is the recipient of a CIHR New Investigator Award and a Scholar Award from the Michael Smith Foundation for Health Research. A.F.Y.P. is the recipient of a Scholar Award from the Michael Smith Foundation for Health Research/St. Paul's Hospital Foundation–Providence Health Care Research Institute Career Investigator program and a CIHR New Investigator Award supported by the Canadian HIV Vaccine Initiative (Vaccine Discovery and Social Research). R.M.M. is the recipient of a Vice President Research Undergraduate Student Research Award (VPR USRA) from Simon Fraser University. P.R.H. is supported by a CIHR/GSK research chair in Clinical Virology.
Footnotes
Published ahead of print 19 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00483-14.
REFERENCES
- 1.Korber B, Gaschen B, Yusim K, Thakallapally R, Kesmir C, Detours V. 2001. Evolutionary and immunological implications of contemporary HIV-1 variation. Br. Med. Bull. 58:19–42. 10.1093/bmb/58.1.19 [DOI] [PubMed] [Google Scholar]
- 2.Srinivasan A, York D, Jannoun-Nasr R, Kalyanaraman S, Swan D, Benson J, Bohan C, Luciw P, Schnoll S, Robinson R. 1989. Generation of hybrid human immunodeficiency virus by homologous recombination. Proc. Natl. Acad. Sci. U. S. A. 86:6388–6392. 10.1073/pnas.86.16.6388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaschen B, Taylor J, Yusim K, Foley B, Gao F, Lang D, Novitsky V, Haynes B, Hahn BH, Bhattacharya T, Korber B. 2002. Diversity considerations in HIV-1 vaccine selection. Science 296:2354–2360. 10.1126/science.1070441 [DOI] [PubMed] [Google Scholar]
- 4.Wolinsky SM, Wike CM, Korber BT, Hutto C, Parks WP, Rosenblum LL, Kunstman KJ, Furtado MR, Muñoz JL. 1992. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science 255:1134–1137. 10.1126/science.1546316 [DOI] [PubMed] [Google Scholar]
- 5.Zhu T, Mo H, Wang N, Nam DS, Cao Y, Koup RA, Ho DD. 1993. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science 261:1179–1181. 10.1126/science.8356453 [DOI] [PubMed] [Google Scholar]
- 6.Redd AD, Collinson-Streng AN, Chatziandreou N, Mullis CE, Laeyendecker O, Martens C, Ricklefs S, Kiwanuka N, Nyein PH, Lutalo T, Grabowski MK, Kong X, Manucci J, Sewankambo N, Wawer MJ, Gray RH, Porcella SF, Fauci AS, Sagar M, Serwadda D, Quinn TC. 2012. Previously transmitted HIV-1 strains are preferentially selected during subsequent sexual transmissions. J. Infect. Dis. 206:1433–1442. 10.1093/infdis/jis503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haaland RE, Hawkins PA, Salazar-Gonzalez J, Johnson A, Tichacek A, Karita E, Manigart O, Mulenga J, Keele BF, Shaw GM, Hahn BH, Allen SA, Derdeyn CA, Hunter E. 2009. Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype A and C HIV-1. PLoS Pathog. 5:e1000274. 10.1371/journal.ppat.1000274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, Li H, Decker JM, Wang S, Baalwa J, Kraus MH, et al. 2009. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J. Exp. Med. 206:1273–1289. 10.1084/jem.20090378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Go EP, Hewawasam G, Liao HX, Chen H, Ping LH, Anderson JA, Hua DC, Haynes BF, Desaire H. 2011. Characterization of glycosylation profiles of HIV-1 transmitted/founder envelopes by mass spectrometry. J. Virol. 85:8270–8284. 10.1128/JVI.05053-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilen CB, Parrish NF, Pfaff JM, Decker JM, Henning EA, Haim H, Petersen JE, Wojcechowskyj JA, Sodroski J, Haynes BF, et al. 2011. Phenotypic and immunologic comparison of clade b transmitted/founder and chronic HIV-1 envelope glycoproteins. J. Virol. 85:8514–8527. 10.1128/JVI.00736-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105:7552–7557. 10.1073/pnas.0802203105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derdeyn CA, Decker JM, Bibollet-Ruche F, Mokili JL, Muldoon M, Denham SA, Heil ML, Kasolo F, Musonda R, Hahn BH, Shaw GM, Korber BT, Allen S, Hunter E. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303:2019–2022. 10.1126/science.1093137 [DOI] [PubMed] [Google Scholar]
- 13.Drumright LN, Gorbach PM, Little SJ, Strathdee SA. 2009. Associations between substance use, erectile dysfunction medication and recent HIV infection among men who have sex with men. AIDS Behavior 13:328–336. 10.1007/s10461-007-9330-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Routy JP, Vanhems P, Rouleau D, Tsoukas C, Lefèbvre E, Côté P, LeBlanc R, Conway B, Alary M, Bruneau J, et al. 2000. Comparison of clinical features of acute HIV-1 infection in patients infected sexually or through injection drug use. J. Acquir. Immune Defic. Syndr. 24:425–432. 10.1097/00126334-200008150-00004 [DOI] [PubMed] [Google Scholar]
- 15.Zuckerkandl E, Pauling L. 1965. Molecules as documents of evolutionary history. J. Theor. Biol. 8:357–366. 10.1016/0022-5193(65)90083-4 [DOI] [PubMed] [Google Scholar]
- 16.Thornton J. 2004. Resurrecting ancient genes: experimental analysis of extinct molecules. Nat. Rev. Genet. 5:366–375. 10.1038/nrg1324 [DOI] [PubMed] [Google Scholar]
- 17.Holland J, Spindler K, Horodyski F, Grabau E, Nichol S, VandePol S. 1982. Rapid evolution of RNA genomes. Science 215:1577–1585. 10.1126/science.7041255 [DOI] [PubMed] [Google Scholar]
- 18.Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJL. 2009. Biopython: freely available python tools for computational molecular biology and bioinformatics. Bioinformatics 25:1422–1423. 10.1093/bioinformatics/btp163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gouy M, Guindon S, Gascuel O. 2010. Seaview version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27:221–224. 10.1093/molbev/msp259 [DOI] [PubMed] [Google Scholar]
- 20.Edgar RC. 2004. Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herbeck JT, Rolland M, Liu Y, McLaughlin S, McNevin J, Zhao H, Wong K, Stoddard JN, Raugi D, Sorensen S, Genowati I, Birditt B, McKay A, Diem K, Maust BS, Deng W, Collier AC, Stekler JD, McElrath MJ, Mullins JI. 2011. Demographic processes affect HIV-1 evolution in primary infection before the onset of selective processes. J. Virol. 85:7523–7534. 10.1128/JVI.02697-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abrahams MR, Anderson JA, Giorgi EE, Seoighe C, Mlisana K, Ping LH, Athreya GS, Treurnicht FK, Keele BF, Wood N, Salazar-Gonzalez JF, Bhattacharya T, Chu H, Hoffman I, Galvin S, Mapanje C, Kazembe P, Thebus R, Fiscus S, Hide W, Cohen MS, Karim SA, Haynes BF, Shaw GM, Hahn BH, Korber BT, Swanstrom R, Williamson C, CAPRISA Acute Infection Study Team, Center for HIV-AIDS Vaccine Immunology Consortium 2009. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-Poisson distribution of transmitted variants. J. Virol. 83:3556–3567. 10.1128/JVI.02132-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29:1969–1973. 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mansky LM, Temin HM. 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 69:5087–5094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. 1996. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271:1582–1586. 10.1126/science.271.5255.1582 [DOI] [PubMed] [Google Scholar]
- 26.Hasegawa M, Kishino H, Yano T. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22:160–74. 10.1007/BF02101694 [DOI] [PubMed] [Google Scholar]
- 27.Newton MA, Raftery AE. 1994. Approximate Bayesian inference with the weighted likelihood bootstrap. J. R. Stat. Soc. Ser. B (Methodological) 56:3–48 [Google Scholar]
- 28.Rambaut A, Drummond A. 2007. Tracer v1.4. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh, Scotland: http://beast.bio.ed.ac.uk/software/ [Google Scholar]
- 29.Talevich E, Invergo B, Cock P, Chapman B. 2012. Bio.phylo: a unified toolkit for processing, analyzing and visualizing phylogenetic trees in biopython. BMC Bioinformatics 13:209. 10.1186/1471-2105-13-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rambaut A, Drummond A. 2007. TreeAnnotator v1.6.1. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh, Scotland: http://beast.bio.ed.ac.uk/software/ [Google Scholar]
- 31.Muse SV, Gaut BS. 1994. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Mol. Biol. Evol. 11:715–724 [DOI] [PubMed] [Google Scholar]
- 32.Pond S, Muse S. 2005. Hyphy: hypothesis testing using phylogenies. Stat. Methods Mol. Evol. 2005:125–181. 10.1007/0-387-27733-1_6 [DOI] [PubMed] [Google Scholar]
- 33.Poon AFY, Swenson LC, Bunnik EM, Edo-Matas D, Schuitemaker H, van't Wout AB, Harrigan PR. 2012. Reconstructing the dynamics of HIV evolution within hosts from serial deep sequence data. PLoS Comput. Biol. 8:e1002753. 10.1371/journal.pcbi.1002753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pupko T, Pe I, Shamir R, Graur D. 2000. A fast algorithm for joint reconstruction of ancestral amino acid sequences. Mol. Biol. Evol. 17:890–896. 10.1093/oxfordjournals.molbev.a026369 [DOI] [PubMed] [Google Scholar]
- 35.Kim J, Sinha S. 2007. Indelign: a probabilistic framework for annotation of insertions and deletions in a multiple alignment. Bioinformatics 23:289–297. 10.1093/bioinformatics/btl578 [DOI] [PubMed] [Google Scholar]
- 36.Diallo A, Makarenkov V, Blanchette M. 2010. Ancestors 1.0: a web server for ancestral sequence reconstruction. Bioinformatics 26:130–131. 10.1093/bioinformatics/btp600 [DOI] [PubMed] [Google Scholar]
- 37.R Core Team. 2012. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 38.Wickham H. 2009. ggplot2: elegant graphics for data analysis. Springer, New York, NY [Google Scholar]
- 39.Huelsenbeck JP, Bollback JP. 2001. Empirical and hierarchical Bayesian estimation of ancestral states. Syst. Biol. 50:351–366. 10.1080/106351501300317978 [DOI] [PubMed] [Google Scholar]
- 40.Shankarappa R, Margolick JB, Gange SJ, Rodrigo AG, Upchurch D, Farzadegan H, Gupta P, Rinaldo CR, Learn GH, He X, Huang XL, Mullins JI. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489–10502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henikoff S, Henikoff JG. 1992. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. U. S. A. 89:10915–10919. 10.1073/pnas.89.22.10915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Llano A, Frahm N, Brander C. 2009. How to optimally define optimal cytotoxic T lymphocyte epitopes in HIV infection, p 3–24 In Yusim K, Korber BTM, Brander C, Haynes BF, Koup R, Moore JP, Walker BD, Watkins DI. (ed), HIV molecular immunology 2009. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM [Google Scholar]
- 43.Şahin GÖ, Bowles EJ, Parker J, Uchtenhagen H, Sheik-Khalil E, Taylor S, Pybus OG, Mäkitalo B, Walther-Jallow L, Spångberg M, et al. 2010. Generation of neutralizing antibodies and divergence of SIVmac239 in cynomolgus macaques following short-term early antiretroviral therapy. PLoS Pathog. 6:e1001084. 10.1371/journal.ppat.1001084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henn MR, Boutwell CL, Charlebois P, Lennon NJ, Power KA, Macalalad AR, Berlin AM, Malboeuf CM, Ryan EM, Gnerre S, et al. 2012. Whole genome deep sequencing of HIV-1 reveals the impact of early minor variants upon immune recognition during acute infection. PLoS Pathog. 8:e1002529. 10.1371/journal.ppat.1002529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goonetilleke N, Liu MK, Salazar-Gonzalez JF, Ferrari G, Giorgi E, Ganusov VV, Keele BF, Learn GH, Turnbull EL, Salazar MG, et al. 2009. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J. Exp. Med. 206:1253–1272. 10.1084/jem.20090365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rolland M, Jensen MA, Nickle DC, Yan J, Learn GH, Heath L, Weiner D, Mullins JI. 2007. Reconstruction and function of ancestral center-of-tree human immunodeficiency virus type 1 proteins. J. Virol. 81:8507–8514. 10.1128/JVI.02683-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kothe DL, Li Y, Decker JM, Bibollet-Ruche F, Zammit KP, Salazar MG, Chen Y, Weng Z, Weaver EA, Gao F, Haynes BF, Shaw GM, Korber BT, Hahn BH. 2006. Ancestral and consensus envelope immunogens for HIV-1 subtype C. Virology 352:438–449. 10.1016/j.virol.2006.05.011 [DOI] [PubMed] [Google Scholar]
- 48.Doria-Rose NA, Learn GH, Rodrigo AG, Nickle DC, Li F, Mahalanabis M, Hensel MT, McLaughlin S, Edmonson PF, Montefiori D, Barnett SW, Haigwood NL, Mullins JI. 2005. Human immunodeficiency virus type 1 subtype B ancestral envelope protein is functional and elicits neutralizing antibodies in rabbits similar to those elicited by a circulating subtype B envelope. J. Virol. 79:11214–11224. 10.1128/JVI.79.17.11214-11224.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hillis D, Huelsenbeck J, Cunningham C. 1994. Application and accuracy of molecular phylogenies. Science 264:671–677. 10.1126/science.8171318 [DOI] [PubMed] [Google Scholar]
- 50.Poon A, McGovern R, Mo T, Knapp D, Brenner B, Routy J, Wainberg M, Harrigan P. 2011. Dates of HIV infection can be estimated for seroprevalent patients by coalescent analysis of serial next-generation sequencing data. AIDS 25:2019. 10.1097/QAD.0b013e32834b643c [DOI] [PubMed] [Google Scholar]
- 51.Leitner T, Escanilla D, Franzén C, Uhlén M, Albert J. 1996. Accurate reconstruction of a known HIV-1 transmission history by phylogenetic tree analysis. Proc. Natl. Acad. Sci. U. S. A. 93:10864–10869. 10.1073/pnas.93.20.10864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hills DM, Bull JJ, White ME, Badgett MR, Molineux IJ. 1992. Experimental phylogenetics: generation of a known phylogeny. Science 255:589–592. 10.1126/science.1736360 [DOI] [PubMed] [Google Scholar]
- 53.Herbeck JT, Nickle DC, Learn GH, Gottlieb GS, Curlin ME, Heath L, Mullins JI. 2006. Human immunodeficiency virus type 1 env evolves toward ancestral states upon transmission to a new host. J. Virol. 80:1637–1644. 10.1128/JVI.80.4.1637-1644.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Higgins D, Lemey P. 2009. Multiple sequence alignment, p 68–96 In Lemey P, Salemi M, Vandamme AM. (ed), The phylogenetic handbook. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 55.Huelsenbeck JP, Bollback JP, Levine AM. 2002. Inferring the root of a phylogenetic tree. Syst. Biol. 51:32–43. 10.1080/106351502753475862 [DOI] [PubMed] [Google Scholar]
- 56.Frost SD, Wrin T, Smith DM, Kosakovsky Pond SL, Liu Y, Paxinos E, Chappey C, Galovich J, Beauchaine J, Petropoulos CJ, Little SJ, Richman DD. 2005. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc. Natl. Acad. Sci. U. S. A. 102:18514–18519. 10.1073/pnas.0504658102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Posada D, Crandall KA. 2002. The effect of recombination on the accuracy of phylogeny estimation. J. Mol. Evol. 54:396–402. 10.1007/s00239-001-0034-9 [DOI] [PubMed] [Google Scholar]
- 58.Schierup MH, Hein J. 2000. Recombination and the molecular clock. Mol. Biol. Evol. 17:1578–1579. 10.1093/oxfordjournals.molbev.a026256 [DOI] [PubMed] [Google Scholar]
- 59.Arenas M, Posada D. 2010. The effect of recombination on the reconstruction of ancestral sequences. Genetics 184:1133–1139. 10.1534/genetics.109.113423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jetzt AE, Yu H, Klarmann GJ, Ron Y, Preston BD, Dougherty JP. 2000. High rate of recombination throughout the human immunodeficiency virus type 1 genome. J. Virol. 74:1234–1240. 10.1128/JVI.74.3.1234-1240.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Batorsky R, Kearney MF, Palmer SE, Maldarelli F, Rouzine IM, Coffin JM. 2011. Estimate of effective recombination rate and average selection coefficient for HIV in chronic infection. Proc. Natl. Acad. Sci. U. S. A. 108:5661–5666. 10.1073/pnas.1102036108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H, Bar KJ, Wang S, Decker JM, Chen Y, Sun C, Salazar-Gonzalez JF, Salazar MG, Learn GH, Morgan CJ, et al. 2010. High multiplicity infection by HIV-1 in men who have sex with men. PLoS Pathog. 6:e1000890. 10.1371/journal.ppat.1000890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hanson-Smith V, Kolaczkowski B, Thornton JW. 2010. Robustness of ancestral sequence reconstruction to phylogenetic uncertainty. Mol. Biol. Evol. 27:1988–1999. 10.1093/molbev/msq081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Learn GH, Muthui D, Brodie SJ, Zhu T, Diem K, Mullins JI, Corey L. 2002. Virus population homogenization following acute human immunodeficiency virus type 1 infection. J. Virol. 76:11953–11959. 10.1128/JVI.76.23.11953-11959.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Delwart E, Magierowska M, Royz M, Foley B, Peddada L, Smith R, Heldebrant C, Conrad A, Busch M. 2002. Homogeneous quasispecies in 16 out of 17 individuals during very early HIV-1 primary infection. AIDS 16:189–195. 10.1097/00002030-200201250-00007 [DOI] [PubMed] [Google Scholar]
- 66.Bar KJ, Li H, Chamberland A, Tremblay C, Routy JP, Grayson T, Sun C, Wang S, Learn GH, Morgan CJ, et al. 2010. Wide variation in the multiplicity of HIV-1 infection among injection drug users. J. Virol. 84:6241–6247. 10.1128/JVI.00077-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, et al. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300. 10.1126/science.278.5341.1295 [DOI] [PubMed] [Google Scholar]
- 68.Rodrigo AG, Shpaer EG, Delwart EL, Iversen AK, Gallo MV, Brojatsch J, Hirsch MS, Walker BD, Mullins JI. 1999. Coalescent estimates of HIV-1 generation time in vivo. Proc. Natl. Acad. Sci. U. S. A. 96:2187–2191. 10.1073/pnas.96.5.2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.