ABSTRACT
The adenovirus E4orf4 protein induces nonclassical apoptosis in mammalian cells through at least two complementing pathways regulated by the interactions of E4orf4 with protein phosphatase 2A (PP2A) and Src kinases. In Saccharomyces cerevisiae cells, which do not express Src, E4orf4 induces PP2A-dependent toxicity. The yeast Golgi apyrase Ynd1 was found to contribute to E4orf4-mediated toxicity and to interact with the PP2A-B55α regulatory subunit. In addition, a mammalian Ynd1 orthologue, the NTPDASE4 gene product Golgi UDPase, was shown to physically interact with E4orf4. Here we report that knockdown of NTPDASE4 suppressed E4orf4-induced cell death. Conversely, overexpression of the NTPDASE4 gene products Golgi UDPase and LALP70 enhanced E4orf4-induced cell killing. We found that similarly to results obtained in yeast, the apyrase activity of mammalian UDPase was not required for its contribution to E4orf4-induced toxicity. The interaction between E4orf4 and UDPase had two consequences: a PP2A-dependent one, resulting in increased UDPase levels, and a PP2A-independent outcome that led to dissociation of large UDPase-containing protein complexes. The present report extends our findings in yeast to E4orf4-mediated death of mammalian cells, and combined with previous results, it suggests that the E4orf4-NTPDase4 pathway, partly in association with PP2A, may provide an alternative mechanism for the E4orf4-Src pathway to contribute to the cytoplasmic death function of E4orf4.
IMPORTANCE The adenovirus E4orf4 protein contributes to regulation of the progression of virus infection from the early to the late phase, and when expressed alone, it induces a unique caspase-independent programmed cell death which is more efficient in cancer cells than in normal cells. The interactions of E4orf4 with cellular proteins that mediate its functions, such as PP2A and Src kinases, are highly conserved in evolution. The results presented here reveal that the NTPDASE4 gene product Golgi UDPase, first discovered to contribute to E4orf4 toxicity in Saccharomyces cerevisiae, associates with E4orf4 and plays a role in induction of cell death in mammalian cells. Details of the functional interaction between E4orf4, PP2A, and the UDPase are described. Identification of the evolutionarily conserved mechanisms underlying E4orf4 activity will increase our understanding of the interactions between the virus and the host cell and will contribute to our grasp of the unique mode of E4orf4-induced cell death.
INTRODUCTION
The adenovirus E4orf4 protein contributes to the progression of viral infection from the early to the late phase (1–7). When expressed outside the context of virus infection, E4orf4 induces caspase- and p53-independent cell death in transformed cells (34, 37, 39, 42). This nonclassical mode of programmed cell death is more efficient in oncogenically transformed cells than in normal cells (8), indicating that the study of E4orf4 may have implications for cancer therapy. Furthermore, a major part of the E4orf4 signaling network is highly conserved in evolution from Saccharomyces cerevisiae through Drosophila to mammalian cells (9–14), underscoring its importance to cell regulation.
E4orf4 associates with several cellular proteins (10, 11, 15–19), and one of its major partners is protein phosphatase 2A (PP2A). The interaction with PP2A is required for all E4orf4 functions known to date (1, 3, 7, 8, 16, 19). PP2A is composed of three subunits: the catalytic C subunit, a scaffolding A subunit, and one of several regulatory B subunits encoded by at least four unrelated gene families, PR55/B55/B, PR61/B56/B′, PR72/B″, and PR93/PR110/B‴ (20), which dictate substrate specificity of the PP2A holoenzyme. The interaction of E4orf4 with the B55α subunit of PP2A, but not the B56 subunits, contributes to E4orf4-induced cell death and cell cycle arrest in both yeast and mammalian cells (10, 12, 18, 19). Earlier reports indicated that PP2A phosphatase activity was required for various E4orf4 functions within the context of virus infection (4, 16, 21) and that E4orf4 recruited PP2A to novel substrates, such as the ACF chromatin remodeling complex, both in the context of virus infection and when overexpressed alone (15). Furthermore, overexpression of the PP2A-B55 subunit was reported to enhance E4orf4-induced cell death (18). In contrast, it was recently suggested that E4orf4 induces cell death by titrating out functional PP2A holoenzymes containing the B55 subunit, thus preventing dephosphorylation of substrates required for cell survival (22). It was also demonstrated that E4orf4 inhibited PP2A activity toward some substrates but not toward others (23). However, since physiological substrates of the E4orf4-PP2A complex have not been identified to date, it is not clear yet how E4orf4 may affect PP2A activity toward them and whether it prevents PP2A from dephosphorylating them.
In addition to its interaction with PP2A, E4orf4 associates with Src-family kinases, and this interaction produces a “cytoplasmic” death signal (24), which leads to remodeling of the actin cytoskeleton, alterations in recycling endosome trafficking, changes in Golgi membrane dynamics, and cell death (25, 26).
Based on the findings that at least part of the E4orf4 effector network was conserved from yeast to mammalian cells, a genetic screen was utilized in S. cerevisiae to identify novel E4orf4 effectors. This screen revealed that yeast nucleoside diphosphatase (Ynd1) contributed to E4orf4-induced toxicity and physically interacted with the viral protein (11). Ynd1 is a Golgi apyrase whose enzymatic activity is required for regulation of nucleotide-sugar import into the Golgi lumen (27, 28). We reported previously that Ynd1 interacted both physically and functionally with Cdc55, the yeast orthologue of the PP2A-B55 regulatory subunit. Deletions of Cdc55 and Ynd1 were shown to confer additive resistance to E4orf4, suggesting that these proteins participated in more than one pathway involved in mediating E4orf4 toxicity. On the other hand, overexpression of Cdc55 was more toxic to the cells in the absence of Ynd1 than in the presence of Ynd1, indicating that there may be functional interactions between the two proteins. Surprisingly, the Ynd1 apyrase activity was found to be dispensable for mediating E4orf4-induced toxicity in yeast (11), and the Ynd1 cytosolic tail was shown to be sufficient for this function (29).
The mammalian YND1 orthologue, NTPDASE4, encodes two proteins, Golgi UDPase and a lysosomal apyrase-like protein of 70 kDa (LALP70), translated from alternatively spliced NTPDASE4 mRNAs. The LALP70 mRNA contains an additional stretch of 24 bp, which encodes 8 amino acids inserted in-frame into the UDPase protein sequence (30–32). This additional 8-residue motif was reported to confer calcium sensitivity to LALP70 (33), and the apyrase products of the two splice variants were shown to possess somewhat different enzymatic properties (30). The two NTPDASE4-encoded proteins differ also in their reported cellular localization. UDPase localized mainly to the Golgi apparatus (32), whereas part of the LALP70 population was detected in lysosomal/autophagic vacuoles, in addition to Golgi localization (31). In mammalian cells, E4orf4 was shown to physically interact with the Golgi UDPase (11), suggesting the evolutionary conservation of this part of the E4orf4 effector network.
In the work presented here, we investigated the contribution of the mammalian Ynd1 orthologues to E4orf4-induced cell death and found that E4orf4 had both PP2A-dependent and -independent effects on UDPase that could contribute to induction of cell death.
MATERIALS AND METHODS
Cell lines, transfections, and plasmids.
HEK293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS). IRBα cells, constitutively expressing a PP2A-B55α-specific short hairpin RNA (shRNA), were grown in medium as described above, supplemented with 1 μg per ml puromycin (1).
The following plasmids were used in this work: pEGFP-C1 (BD Bioscience), pCMV-E4orf4 (34), the pCMV/neo vector (35), pcDNA4/TO, pcDNA6/TR (Invitrogen), pcDNA4/To–E4orf4, pcDNA4/To, encoding the R81F84A E4orf4 mutant protein and a PP2A-B55α–HA mutant resistant to knockdown by the shRNA in the IRBα cells (1), pGEX-2TK (GE Healthcare), and pGEX-PP2A-B55α. The Golgi UDPase sequence was subcloned from GW1-UDPase-Myc (32) into a pCS2+ vector. LALP70 was subcloned from pEGFP-N3-LALP70 (31) into pCS2+, generating LALP70-Myc. Myc tagged catalytically inactive UDPase mutants with the mutations E223Q and S277A were generated using the QuikChange mutagenesis kit (Stratagene). A specific shRNA for Golgi UDPase was subcloned into pSuperior.neo-GFP (OligoEngine, Inc.) according to the manufacturer's protocol. The following primers were used: forward primer, 5′-GAT CCC CAG ACT ACA ATG CTG CTA AAT TCA AGA GAT TTA GCA GCA TTG TAG TCT TTT TTG GAA A-3′; reverse primer, 5′-AGC TTT TCC AAA AAA GAC TAC AAT GCT GCT AAA TCT CTT GAA TTT AGC AGC ATT GTA GTC TGG G-3′.
Transfections, immunoprecipitation, glutathione S-transferase (GST) pulldown assays, Western blot analysis, and antibodies.
Transfections were carried out using the JetPie reagent (Polyplus Transfection Inc., New York, NY). Cell extracts were prepared in CTAP buffer (50 mM HEPES-KOH [pH 8], 100 mM KCl, 2 mM EDTA, 10% glycerol, 0.1% Nonidet P-40, 2 mM dithiothreitol [DTT], 10 mM NaF, and 1/10 volume of Complete protease inhibitor cocktail [Roche]) by 3 freeze-thaw cycles and a 30-min incubation on ice and were denatured in SDS loading buffer at 65°C. Denatured proteins were analyzed by Western blotting using antibodies against the following proteins or protein tags: E4orf4 (34), PP2A-B55α (8), c-Myc (9E10 or C-33; Santa Cruz), NTPDase4 and alpha-tubulin (Sigma), hemagglutinin (HA) (Covance), and green fluorescent protein (GFP) (MBL, Woburn, MA).
For immunoprecipitation experiments, protein extracts were prepared as described above and incubated with antibodies to E4orf4 or the Myc tag. The immune complexes were heated to 65°C in SDS loading buffer and separated by SDS-PAGE. Immunoprecipitated proteins and input lysates were analyzed by Western blotting using specific antibodies.
GST fusion protein binding assays were carried out as described previously (16).
Apyrase assays.
Apyrase assays were performed using crude membrane preparations, as previously described (11, 32).
Cell death assays.
Cell death assays, including determination of nuclear aberrations and visualization of cell loss, have been described previously (15, 36). The average of two or three experiments, each containing duplicate plates, was calculated. Statistical significance of the results was determined by a one-tailed Student t test, unless otherwise indicated.
Protein stability assays.
Cells were transfected with plasmids expressing UDPase-Myc and/or E4orf4. Twenty hours later, cycloheximide suspended in ethanol was added to the cells at a concentration of 50 μg per ml, and one set of cells was treated with ethanol only. The cells were harvested one or two hours later and processed for Western blot analysis. Densitometry was used to quantify protein levels.
Size exclusion chromatography.
Protein extracts prepared as described above were chromatographed on a Superose 6 column (GE Healthcare). A 10% volume of each fraction was subjected to Western blot analysis. Molecular size markers (thyroglobulin and bovine serum albumin [BSA]; Sigma) were chromatographed under identical conditions.
Image acquisition and processing.
Plated cells were photographed by an Axiocam camera linked to a Zeiss Axioskop at a magnification of ×400. Gels were scanned with an Epson Photo 4990 scanner. Images were processed using Adobe Photoshop 5.0 or 7.0.
RESULTS
Golgi UDPase and LALP70 enhance E4orf4-induced cell death.
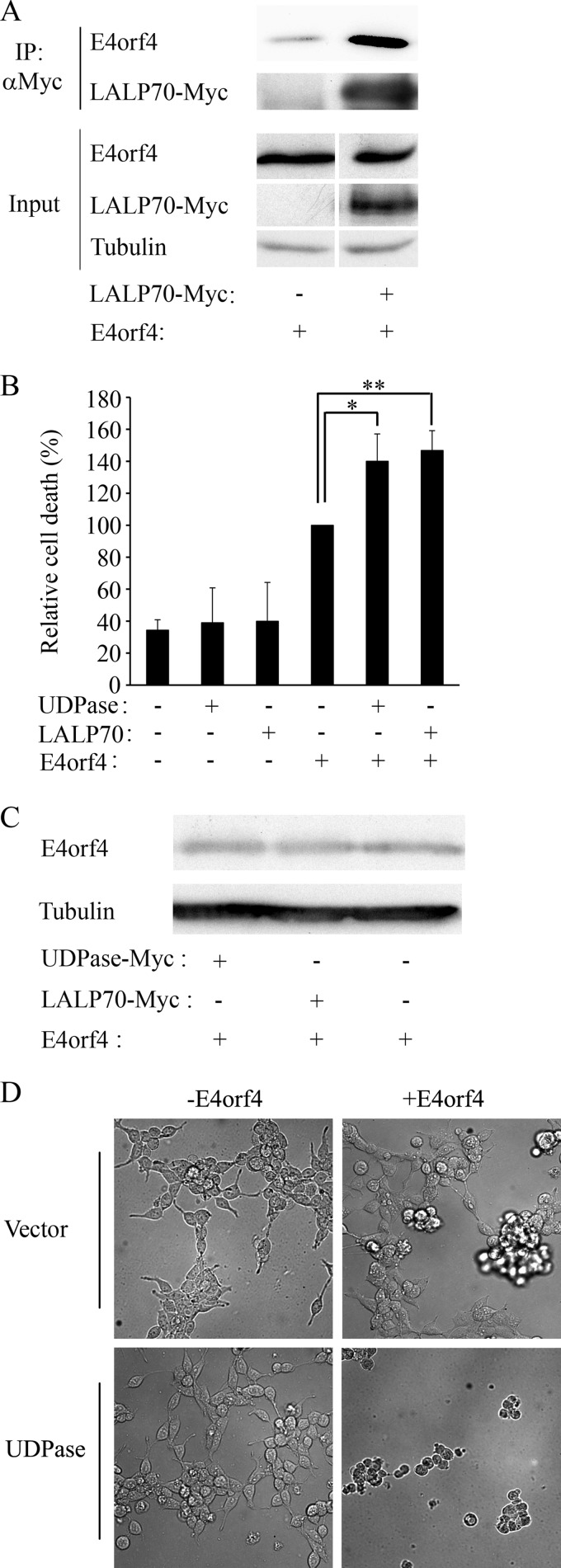
Since Ynd1 was shown to contribute to E4orf4-induced toxicity in yeast (11), we set out to determine whether the Ynd1 human orthologues Golgi UDPase and LALP70 participated in E4orf4-induced cell death in mammalian cells. We showed previously that E4orf4 physically associated with Golgi UDPase (11), and a coimmunoprecipitation experiment was carried out to test whether E4orf4 also associated with LALP70. E4orf4 was expressed alone or together with LALP70-Myc in 293T cells, and cell extracts were subjected to immunoprecipitation with antibodies to the Myc tag. Figure 1A demonstrates that E4orf4 was precipitated by the Myc-specific antibodies above background levels associated with the beads in the absence of LALP70-Myc. Thus, both UDPase (11) and LALP70 can associate with E4orf4.
FIG 1.
Golgi UDPase and LALP70 enhance E4orf4-induced cell death. Plasmids expressing UDPase-Myc, LALP70-Myc, or an empty vector were transfected into 293T cells with a plasmid expressing E4orf4 or with the corresponding empty vector. (A) Cell extracts were subjected to immunoprecipitation (IP) with antibodies to the Myc tag, and the presence of E4orf4 and LALP70-Myc proteins in the immune complexes and in input lysates was determined by a Western blot. Alpha-tubulin served as a loading control. The amount of proteins in the input represents 10% of the amount of proteins used for immunoprecipitation. (B) Cells in duplicate plates were stained 24 h after transfection with antibodies to E4orf4 and the Myc tag and with 4′,6-diamidino-2-phenylindole (DAPI). Nuclei with apoptotic morphology were counted in the double-transfected cell population expressing both E4orf4 and UDPase-Myc and in single-transfected cells expressing either E4orf4 or UDPase-Myc, and the percentage of cell death was determined. Cell death induced by E4orf4 alone was defined as 100%, and relative cell death was calculated for the other samples. Two independent experiments, each with duplicates, were carried out. Error bars represent the pooled standard deviation, and statistical significance was determined using a one-tailed t test (*, P = 0.043; **, P = 0.04). (C) Proteins extracted from a parallel set of E4orf4-transfected plates, as described for panel B, were analyzed by Western blotting using antibodies to E4orf4 and to alpha-tubulin. (D) Representative photographs of cells containing the indicated plasmids were taken 24 h after transfection.
Next, we examined whether overexpression of UDPase or LALP70 contributed to E4orf4-induced cell death. It was shown previously that the most typical morphologies associated with E4orf4-induced cell death include membrane blebbing, nuclear condensation or fragmentation, and cell detachment, whereas morphologies associated with classical apoptosis, such as caspase activation, DNA fragmentation, phosphatidylserine externalization, and mitochondrial changes, do not always accompany E4orf4-induced cell death (37–39). We thus measured E4orf4-induced cell death by assaying nuclear aberrations, or cell loss. UDPase and LALP70 were expressed in 293T cells alone or together with E4orf4, and cell death was measured by determination of the fraction of transfected cells exhibiting nuclear condensation or fragmentation. Figure 1B demonstrates that UDPase and LALP70 by themselves did not significantly enhance background levels of toxicity. However, when coexpressed with E4orf4, both proteins caused a significant increase in E4orf4-induced cell death (P < 0.044), without affecting E4orf4 protein levels (Fig. 1C). Similarly, when the cells were visualized by light microscopy (Fig. 1D), UDPase by itself did not significantly affect cell growth, and E4orf4 caused cell blebbing, cell detachment, and death. Cell death was further increased in the presence of UDPase. Taken together, these results suggest that UDPase or LALP70 overexpression synergized with E4orf4 to enhance cell death. Since UDPase and LALP70 appeared to have similar effects on E4orf4-induced cell killing, we continued the analysis of the E4orf4-NTPDASE4 interaction using only the Golgi UDPase.
UDPase catalytic activity is not required for the E4orf4-UDPase cooperation in induction of cell death.
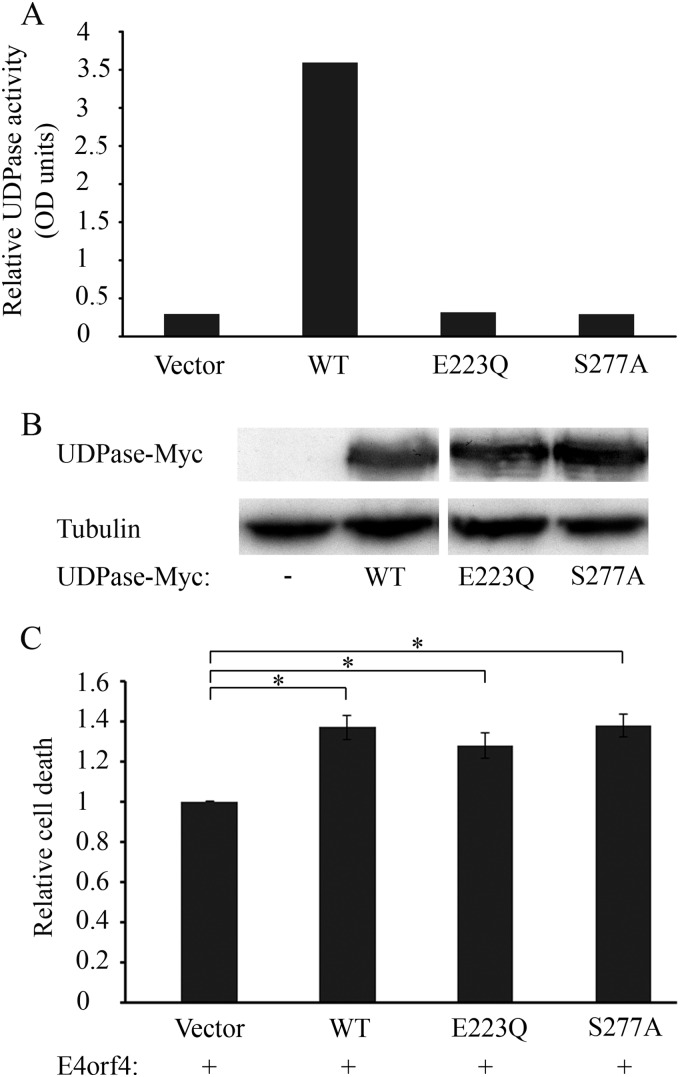
It was previously shown in yeast that Ynd1 apyrase activity is not required for E4orf4-induced toxicity. To investigate whether this holds true for a mammalian Ynd1 orthologue as well, we prepared two catalytically inactive UDPase mutants, with the mutations E223Q and S277A, which corresponded to the Ynd1 mutants E152Q and S189A used in yeast (11). We first verified that these mutants indeed lost their catalytic activity while maintaining wild-type (WT) levels of expression, as shown in Fig. 2A and B. We then compared the ability of WT UDPase and the mutants to enhance E4orf4-induced cell death. E4orf4 was expressed in 293T cells alone or together with WT UDPase or the mutants. The results shown in Fig. 2C indicate that WT UDPase and the two mutants enhanced E4orf4-induced cell death similarly, confirming that, as in yeast, the UDPase apyrase activity was dispensable for functional interaction with E4orf4.
FIG 2.
The UDPase catalytic activity is not required for E4orf4-induced cell death. (A) An empty vector and plasmids expressing WT UDPase-Myc and Myc tagged UDPase mutants (E223Q and S277A) were transfected into 293T cells. The cells were harvested 48 h later, and the UDPase catalytic activity was measured using an apyrase activity assay. (B) Protein extracts from the cells used for panel A were subjected to a Western blot analysis with antibodies to the Myc tag and alpha-tubulin. (C) Plasmids expressing WT UDPase-Myc and Myc tagged UDPase mutants (E223Q and S277A) or an empty vector were transfected into 293T cells together with a plasmid expressing E4orf4. The cells were fixed 24 h later and were stained with antibodies to E4orf4 and the Myc tag and with DAPI. The percentage of cell death in transfected cells was calculated as described in the legend to Fig. 1. Induction of cell death by E4orf4 in the presence of WT or mutant UDPase proteins was normalized to induction of cell death by E4orf4 in the absence of UDPase, defined as 1. Results of two independent experiments are shown. Error bars represent standard error. *, P < 0.002.
NTPDASE4 knockdown inhibits E4orf4-induced cell death.
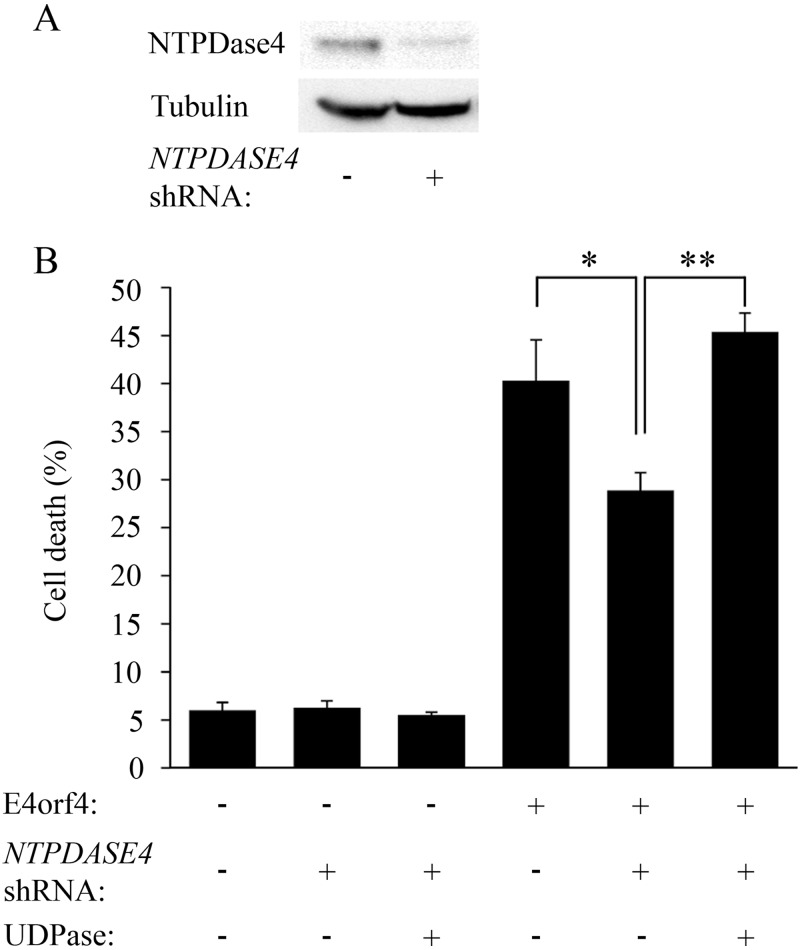
To determine whether the NTPDASE4 gene products were required for E4orf4-induced cell death, knockdown of NTPDASE4 expression was carried out. Western blot and densitometry analysis demonstrated that transient expression of a NTPDASE4-specific shRNA for 48 h in 293T cells reduced NTPDase4 protein levels 3-fold (Fig. 3A). The levels of E4orf4-induced cell death in the presence or absence of the NTPDASE4 shRNA were measured by determination of the fraction of transfected cells exhibiting nuclear condensation or fragmentation. As seen in Fig. 3B, E4orf4-induced cell death was inhibited significantly (P = 0.017) when NTPDASE4 expression was knocked down. Knockdown of NTPDASE4 in control cells did not alter cell death levels within the time frame of the experiment (Fig. 3B, left). To confirm that the decrease in cell death was indeed due to NTPDASE4 knockdown, a mutant UDPase containing silent mutations, which rendered it resistant to knockdown by the NTPDASE4 shRNA, was introduced into cells containing the shRNA by transient transfection. Restoration of UDPase to the cells led to a significant increase in E4orf4-induced cell death (P = 0.0001), reaching levels similar to or higher than those observed in cells that did not express the NTPDASE4 shRNA (Fig. 3B, right). These results confirm that NTPDASE4 contributes to E4orf4-induced cell death.
FIG 3.
NTPDASE4 contributes to E4orf4-induced cell death. (A) 293T cells were transfected with an empty vector or a vector expressing both GFP and a shRNA sequence targeting NTPDASE4. Protein extracts were prepared 48 h later and subjected to Western blot analysis using antibodies to NTPDase4 and alpha-tubulin. NTPDase4 protein levels were determined by densitometry and normalized to tubulin levels, demonstrating a 3-fold reduction in NTPDase4 expression upon knockdown. (B) 293T cells were transfected with an empty vector or a plasmid expressing a NTPDASE4-specific shRNA. These cells were further transfected 2 days later with a plasmid expressing a shRNA-resistant UDPase-Myc or a vector control together with a plasmid expressing E4orf4 or its corresponding empty vector control. The cells were fixed 24 h after the second transfection and stained with antibodies to E4orf4 and the Myc tag and with DAPI. Cells transfected with the shRNA were identified by the presence of GFP expressed from the same plasmid. The percentage of cell death in transfected cells was calculated. Two independent experiments, each with duplicates, were carried out. Error bars represent the standard error. *, P = 0.017; **, P = 0.0001.
UDPase only weakly enhances cell death induced by an E4orf4 mutant incapable of binding PP2A.
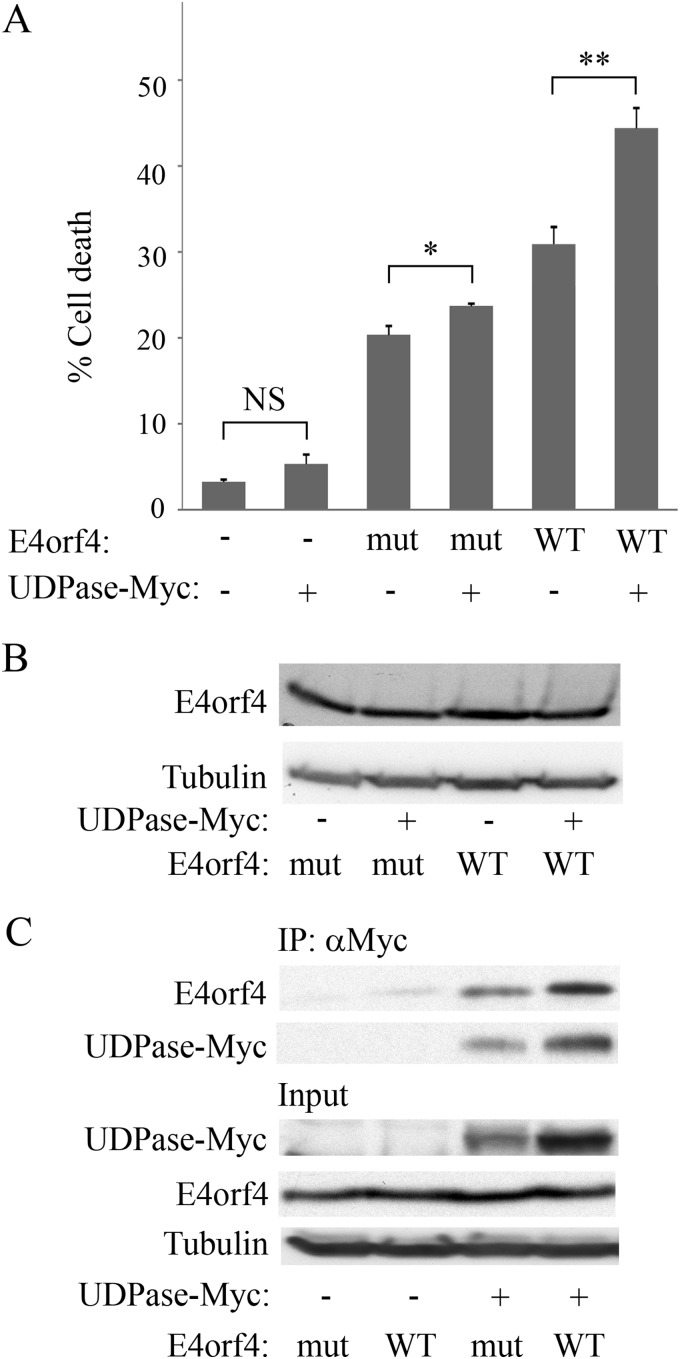
The PP2A- and Src-mediated pathways contribute additively to E4orf4-induced cell death. Therefore, an E4orf4 mutant which does not bind PP2A can still induce cell death through a Src-mediated pathway, albeit at lower levels than WT E4orf4 (24, 40). Since Ynd1 was identified as a partner of E4orf4 and PP2A in yeast cells, which do not express Src family kinases, we hypothesized that the mammalian Ynd1 orthologue, Golgi UDPase, functionally interacted with PP2A rather than with Src kinases in the E4orf4 signaling network. To test this hypothesis, we examined whether Golgi UDPase cooperated with the Src-dependent E4orf4 cell death pathway in mammalian cells. For this purpose we measured the ability of UDPase to enhance cell death induced by an E4orf4 mutant (R81F84A) that did not bind PP2A (1, 19) and induced cell death through the Src pathway (24). WT E4orf4 and the R81F84A mutant were expressed in 293T cells alone or together with a plasmid expressing UDPase-Myc. In addition, an empty vector or a plasmid expressing UDPase-Myc was introduced into the cells without E4orf4. Cell death levels were determined by measuring the percentage of transfected cells with nuclear aberrations 1 day posttransfection. Figure 4A confirms that the R81F84A mutant induced lower cell death levels than WT E4orf4. The results further demonstrate that UDPase did not significantly enhance cell death levels occurring in the absence of E4orf4 and slightly increased death induced by the R81F84A mutant (by an additional 3.4%; P = 0.009), whereas it synergistically increased WT E4orf4-induced cell death by an additional 13.5% (P = 0.002). The levels of WT and mutant E4orf4 proteins were similar and were not altered in the presence of UDPase (Fig. 4B). To understand the mechanistic reason for the reduced functional interaction between UDPase and the E4orf4 mutant, we set out to compare the abilities of E4orf4 and the R81F84A mutant to associate with UDPase. We found that UDPase levels were consistently higher in the presence of WT E4orf4 than in the presence of R81F84A, and coimmunoprecipitation of both E4orf4 proteins with UDPase was proportionate to UDPase levels (Fig. 4C). These results indicate that the reduced ability of UDPase to enhance cell death induced by the R81F84A mutant did not result from lack of an interaction between the two proteins but could possibly be explained by a different effect of WT E4orf4 and the R81F84A mutant on UDPase levels. The results further suggest that the E4orf4-PP2A-dependent pathway but not the E4orf4-Src pathway may cooperate with UDPase and that the interaction with PP2A may contribute to an effect of E4orf4 on UDPase levels.
FIG 4.
UDPase does not synergistically activate cell death induced by an E4orf4 mutant that binds Src but not PP2A. (A) The E4orf4 R81F84A mutant (mut) and WT E4orf4 were expressed in 293T cells alone or together with UDPase-Myc. Cells in duplicate plates were fixed 24 h after transfection and stained with antibodies to E4orf4 and the Myc tag and with DAPI. The percentage of cell death in transfected cells was calculated. Two independent experiments, each with duplicates, were carried out. Error bars represent standard errors. NS, not significant. *, P = 0.009; **, P = 0.002. (B) An additional set of cells that were transfected similarly was harvested 24 h after transfection, and a Western blot analysis was performed with E4orf4- and alpha-tubulin-specific antibodies. (C) Cell extracts as described for panel B were subjected to immunoprecipitation with antibodies to the Myc tag. A Western blot containing both immune complexes and input lysates was stained with antibodies to E4orf4 and the Myc tag. Alpha-tubulin served as a loading control. The blots were subjected to densitometry, and the results indicated that UDPase levels were 2-fold higher in the presence of WT E4orf4 than in the presence of the R81F84A mutant and the levels of WT and mutant E4orf4 proteins found in the immune complexes mirrored this difference (1.75:1).
E4orf4 increases UDPase levels in a PP2A-dependent manner.
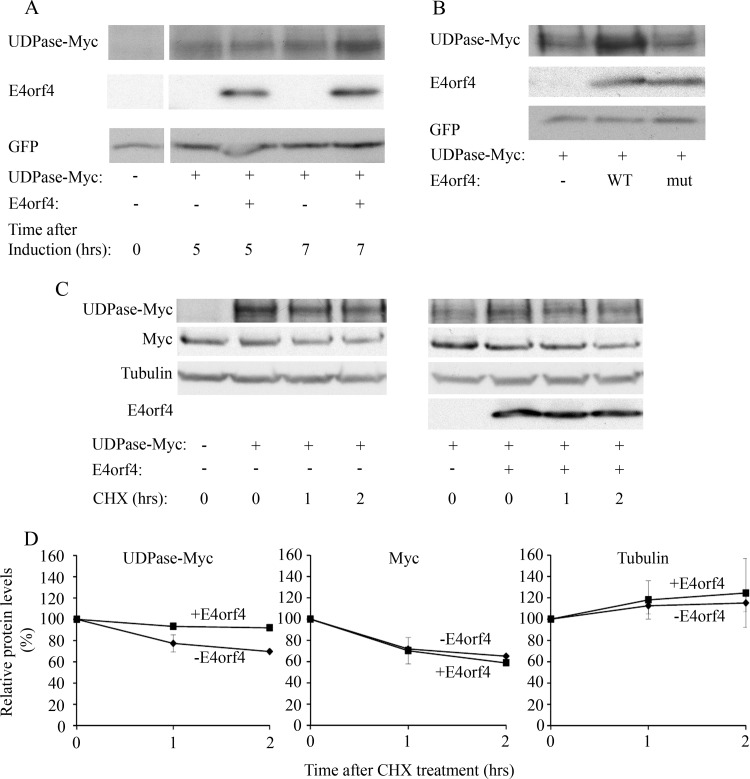
Based on the results shown in Fig. 4C, we continued to explore how E4orf4 expression led to changes in UDPase protein levels. First we tested how soon after the onset of E4orf4 expression UDPase levels were affected. A plasmid expressing E4orf4 from a doxycycline-inducible promoter was introduced into 293T cells together with plasmids expressing the Tet repressor, UDPase-Myc, and GFP. E4orf4 expression was induced for various lengths of time by the addition of doxycycline, and protein levels were determined by Western blotting. As seen in Fig. 5A, induction of E4orf4 expression for 5 h was not sufficient to increase UDPase protein levels, but a 7-h expression led to a 40% rise in UDPase quantities. GFP levels, serving as a transfection and loading control, were not altered during the same time period. To confirm that an interaction with PP2A was required for the E4orf4-induced effect on UDPase expression, plasmids expressing UDPase-Myc and GFP together with an empty vector or a vector expressing WT E4orf4 or the R81F84A mutant, which does not bind PP2A, were introduced into 293T cells. Figure 5B demonstrates that UDPase-Myc levels were increased in the presence of WT but not mutant E4orf4 protein, and control GFP levels were not altered at all. These results suggest that an interaction with PP2A is required for an E4orf4-induced increase in UDPase levels.
FIG 5.
E4orf4 increases UDPase protein levels. (A) UDPase-Myc was expressed in 293T cells together with inducibly expressed E4orf4 and with GFP. The transfection was carried out in duplicate. One day after transfection, E4orf4 expression was induced by the addition of 5 μg/ml doxycycline to the cells (+E4orf4), and a duplicate plate was treated with ethanol (-E4orf4). The cells were harvested 5 and 7 h later, and equal amounts of proteins were separated by SDS-PAGE. A Western blot was stained with the indicated antibodies. Densitometry of the blot and normalization to the levels of the GFP transfection control revealed that UDPase levels increased by 40% in the presence of E4orf4 after 7 h induction. (B) Plasmids expressing UDPase-Myc and GFP were transfected into 293T cells together with an empty vector or a vector expressing WT E4orf4 or the R81F84A mutant, which does not bind PP2A-B55α (mut). The cells were harvested 1 day after transfection, and protein levels of UDPase-Myc, E4orf4, and GFP were determined by Western blot analysis using specific antibodies. (C) UDPase-Myc was expressed in 293T cells with or without E4orf4. Cycloheximide (CHX; 50 μg/ml) or an equal volume of ethanol was added to the medium 1 day after the transfection, and cells were harvested at 0, 1, and 2 h after addition of the drug. The levels of UDPase-Myc, endogenous Myc, alpha-tubulin, and E4orf4 were visualized by Western blotting. (D) Protein levels were determined by densitometry of the Western blots. Protein levels at time zero were defined as 100%, and relative protein levels are shown in the graphs. Diamonds, samples with empty vector; squares, samples with E4orf4. Error bars represent the standard errors from two independent experiments. Errors smaller than 0.03 are not depicted.
Next we set out to determine whether the E4orf4-induced increase in UDPase levels resulted from alterations in UDPase protein stability. UDPase-Myc was introduced into 293T cells in the presence or absence of E4orf4. Cycloheximide (CHX) was added to the cells 24 h posttransfection, and protein extracts were prepared at 0, 1, and 2 h after CHX addition. The levels of UDPase-Myc, endogenous Myc, and alpha-tubulin were determined by Western blotting (Fig. 5C), and the blots were subjected to densitometry to quantify the changes in protein levels (Fig. 5D). As seen in Fig. 5C and D, the levels of the short-lived endogenous Myc protein were reduced similarly in the presence and absence of E4orf4, indicating that E4orf4, which was previously shown to reduce Myc transcription (1), did not affect Myc protein stability. The levels of the more stable alpha-tubulin were also not affected by E4orf4. However, E4orf4 did affect UDPase levels. During the first hour of CHX addition and in the absence of E4orf4, UDPase-Myc levels decreased by 23%, whereas UDPase-Myc levels in the presence of E4orf4 decreased by only 7%. Between 1 and 2 h of CHX treatment, the rate of decrease in UDPase-Myc levels was reduced both in the absence and in the presence of E4orf4. These results suggest that the UDPase protein population consisted of at least two subpopulations, one of which was less stable than the other. The stability of the more labile UDPase proteins was increased in the presence of E4orf4.
UDPase interacts with PP2A-B55α.
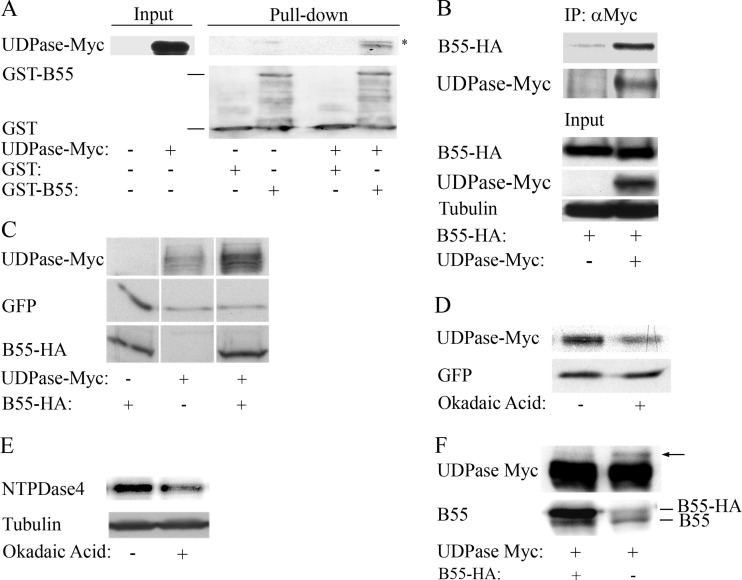
Previous reports demonstrated that E4orf4 interacted with PP2A through its regulatory B55α subunit and that in both mammalian cells and yeast, this interaction was required for E4orf4 toxicity (8, 10, 12, 16, 19). Furthermore, we showed in yeast that Ynd1 associated both physically and functionally with Cdc55, the yeast PP2A-B55 subunit (11). To examine whether the mammalian Ynd1 orthologue, UDPase, interacted with PP2A-B55α, we carried out GST pulldown and coimmunoprecipitation experiments. GST alone and GST–PP2A-B55α were purified on glutathione beads and incubated with protein extracts from 293T cells expressing UDPase-Myc or containing an empty vector. As seen in Fig. 6A, UDPase-Myc bound to GST–PP2A-B55α but not to GST alone, although the interaction levels were low compared with those of input lysates. To confirm that this interaction occurred in cells as well, HA-tagged PP2A-B55α was expressed alone in 293T cells or together with UDPase-Myc. Protein extracts were prepared 1 day posttransfection and subjected to immunoprecipitation with antibodies to the Myc tag. Figure 6B shows results of a representative experiment which demonstrate that PP2A-B55α associated with UDPase, at a level that was higher than background. Thus, the results indicate that the UDPase-PP2A-B55α interaction is conserved from yeast to mammals.
FIG 6.
PP2A-B55α interacts with UDPase and increases UDPase protein levels. (A) UDPase-Myc was expressed in 293T cells and protein extracts were prepared 24 h later. These extracts were incubated with bacterially produced PP2A-B55α fused to GST or with GST alone for 2 h at 4°C. A Western blot analysis was performed with Myc tag- and GST-specific antibodies. The amount of proteins in the input extracts from 293T cells represents 10% of the amount of proteins used for incubation with GST proteins. The asterisk marks a nonspecific band. (B) HA-tagged PP2A-B55α was expressed in 293T cells alone or together with UDPase-Myc. One day after transfection, the cells were harvested and protein extracts were subjected to immunoprecipitation (IP) with a Myc tag antibody. A Western blot shows the presence of PP2A-B55α–HA and UDPase-Myc in the immune complexes (IP) and in input lysates. Alpha-tubulin served as a loading control. The amount of proteins in the input represents 10% of the amount of proteins used for IP. Results of one representative experiment out of three are shown. (C) IRBα cells that constitutively express a PP2A-B55α-specific shRNA were transfected with a plasmid expressing UDPase-Myc and a plasmid expressing GFP (serving as a transfection efficiency control) with or without a plasmid expressing mutant PP2A-B55α–HA, which was resistant to the PP2A-B55α-specific shRNA. The cells were harvested 24 h later, and Western blot analysis was performed with antibodies to the HA and Myc tags and to GFP. (D) Two plates of IRBα cells were transfected with plasmids expressing UDPase-Myc, GFP, and shRNA-resistant PP2A-B55α–HA. After 24 h, 5 nM okadaic acid was added to one plate and ethanol was added to the other plate. The cells were harvested 4 h later, and protein extracts were subjected to Western blot analysis with antibodies to the Myc tag and GFP. Densitometry of the blot and normalization to the GFP control revealed that okadaic acid treatment led to a 2-fold reduction in UDPase levels. Similar results were obtained in a second independent experiment. (E) 293T cells were treated with 5 nM okadaic acid or with ethanol in three independent experiments and protein extracts were prepared 4 h later. Western blots were stained with antibodies to NTPDase4 proteins and alpha-tubulin. Densitometry of the blots and normalization to alpha-tubulin levels revealed an okadaic acid-induced drop of 33% to 75% in NTPDase4 protein levels in the three experiments. One representative blot is shown. (F) IRBα cells were transfected with a plasmid expressing UDPase-Myc and with a plasmid expressing PP2A-B55α–HA or an empty vector. The cells were harvested 1 day later, and protein extracts were prepared and loaded on a large SDS gel. Western blot analysis was performed with antibodies to the Myc tag and to PP2A-B55α. An arrow marks a slower-migrating UDPase-Myc isoform. The figure is representative of three independent experiments.
PP2A increases UDPase protein levels.
Since PP2A-B55α physically associated with the Golgi UDPase (Fig. 6A and B), and because the E4orf4 R81F84A mutant that bound Golgi UDPase (Fig. 4C) but lost the ability to bind PP2A (19) did not increase UDPase levels, we examined whether PP2A-B55α had an effect on UDPase levels. For this purpose we used IRBα cells, which constitutively express a PP2A-B55α-specific shRNA and consequently contain low PP2A-B55α levels (1). A plasmid expressing UDPase-Myc was introduced into IRBα cells together with an empty vector or with a plasmid expressing a HA-tagged mutant PP2A-B55α subunit containing silent mutations which rendered it resistant to knockdown by the B55α shRNA. A plasmid expressing GFP was added to the transfection mixture and served as a transfection control. UDPase-Myc, PP2A-B55α–HA, and GFP protein levels were determined by Western blotting. Figure 6C demonstrates that UDPase-Myc levels increased when PP2A-B55α was overexpressed in the IRBα cells. The levels of the control GFP protein, expressed from an identical promoter, were not altered. These results demonstrate that the presence of PP2A-B55α resulted in enhanced UDPase protein levels. To determine whether phosphatase activity was required for the increase in UDPase levels, UDPase-Myc, B55α, and GFP were expressed in IRBα cells, and 5 nM okadaic acid, a PP2A inhibitor, was added to the cells 4 h prior to harvest. As seen in Fig. 6D, UDPase-Myc levels were reduced 2-fold in the presence of okadaic acid, as determined by densitometry of the Western blots, whereas GFP levels did not change. Similarly, we tested whether okadaic acid affected protein levels of the endogenous NTPDASE4-encoded proteins in 293T cells. In three independent experiments, endogenous NTPDase4 protein levels were decreased by 33% to 75% in the presence of the PP2A inhibitor (Fig. 6E). These results suggest that phosphatase activity was required for the PP2A-B55α-mediated increase in UDPase levels.
When protein extracts containing UDPase-Myc were separated on a long SDS-polyacrylamide gel, a slower-migrating UDPase band was regularly observed in extracts from IRBα cells expressing UDPase-Myc alone but not in extracts from cells coexpressing exogenously added PP2A-B55α (Fig. 6F). This observation is consistent with the possibility that PP2A complexes containing the B55α subunit are required for dephosphorylation of the UDPase. UDPase dephosphorylation might then lead to an increase in UDPase levels, whereas a population of highly phosphorylated UDPase polypeptides may be unstable and would disappear fast.
E4orf4 expression leads to a PP2A-independent dissociation of high-molecular-weight protein complexes containing UDPase.
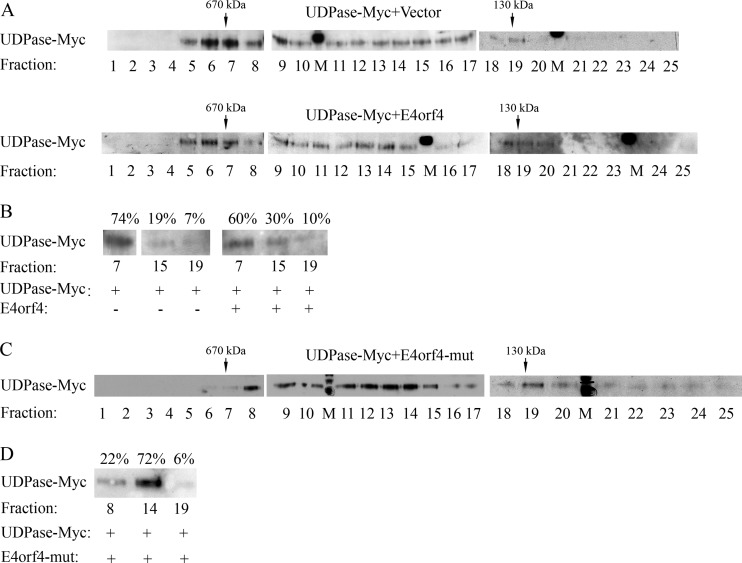
To further investigate the functional consequences of the E4orf4-UDPase interaction, we used gel filtration chromatography to examine the effects of E4orf4 on protein complexes containing UDPase. Whole-cell protein extracts were prepared from 293T cells expressing UDPase-Myc in the absence or presence of E4orf4 and were chromatographed on a Superose 6 column. As seen in Fig. 7A, in the absence of E4orf4, most of the UDPase-Myc population was concentrated in fractions 6 and 7, which contained protein complexes of around 670 kDa. In contrast, UDPase-Myc from cells expressing E4orf4 appeared to be more spread out both in fractions 5 to 7 and in fractions 11 to 20. To confirm that the differences did not result from altered transfer efficiency of the blots, representative fractions from each column run (numbers 7, 15, and 19) were separated on a single SDS-polyacrylamide gel. The intensity of the UDPase bands was measured by densitometry, and the percentage of intensity of the UDPase band in each fraction was calculated. Figure 7B demonstrates that the UDPase in the high-molecular-weight fraction was 74% of UDPase present in the three fractions in the absence of E4orf4, whereas it was 60% in the presence of E4orf4. UDPase levels in the lower-molecular-weight fractions increased from 26% in the absence of E4orf4 to 40% in its presence. These results suggest that E4orf4 expression led to partial dissociation of high-molecular-weight protein complexes containing UDPase. To determine whether an interaction with PP2A was required for E4orf4-induced dissociation of UDPase-containing complexes, we tested the ability of the R81F84A mutant, which does not bind PP2A, to dissociate UDPase complexes. Plasmids expressing UDPase-Myc and the R81F84A mutant were introduced into 293T cells, and protein extracts were separated on a Superose 6 column as described above. The presence of UDPase-Myc in the various fractions was determined by Western blotting of all fractions (Fig. 7C) or of representative fractions (Fig. 7D). The results demonstrate that expression of the R81F84A mutant protein led to an even more efficient dissociation of UDPase-containing complexes than the WT E4orf4 protein, resulting in the presence of only 22% of the UDPase protein in the high-molecular-weight fraction compared with 78% in the two lower-molecular-weight fractions. It appears therefore that PP2A is not required for E4orf4-induced dissociation of UDPase complexes and may even be inhibitory to it.
FIG 7.
A high-molecular-weight UDPase-containing complex is partially dissociated in the presence of E4orf4, regardless of the ability of E4orf4 to bind PP2A. (A) Protein extracts from 293T cells expressing UDPase-Myc in the presence or absence of E4orf4 were separated independently on a Superose 6 column. Twenty-five fractions were collected from each run, and 10% of each fraction were loaded onto SDS gels and subjected to Western blot analysis with antibodies to the Myc tag. In addition, thyroglobulin and BSA dimers were chromatographed separately on the column as molecular mass markers (670 and 130 kDa, respectively). M denotes a marker lane. (B) Fractions 7, 15, and 19 from the two column runs whose results are shown in panel A were loaded onto one SDS gel, and a blot was stained with Myc tag-specific antibodies. The intensities of UDPase-Myc protein bands were quantified by densitometry and the sum of UDPase-Myc levels in the 3 fractions of each column run was defined as 100%. The percentage of UDPase-Myc in each fraction is shown above the blot. (C and D) A column run of protein extracts expressing UDPase-Myc in the presence of the R81F84A E4orf4 mutant (mut) was analyzed as described for panels A and B. The results are representative of 3 independent experiments.
DISCUSSION
Two important conclusions can be drawn from this work. First, a significant part of the E4orf4 network required for induction of a unique mode of cell death is highly conserved from yeast to mammals. Thus, Ynd1 was required for E4orf4-induced toxicity in yeast in an apyrase-independent manner (11) and the Ynd1 mammalian orthologue UDPase contributed to E4orf4-induced cell death in mammalian cells, while its apyrase activity was not required for this function (Fig. 1 to 3). Furthermore, both Ynd1 and UDPase interacted physically with the major E4orf4 partner PP2A (11) (Fig. 6). The second conclusion from this work posits that the interaction between E4orf4 and UDPase had at least two consequences, one that was PP2A dependent and led to an increase in UDPase levels (Fig. 4 to 6) and another that was PP2A independent and led to dissociation of large UDPase-containing protein complexes (Fig. 7).
A PP2A holoenzyme containing the B55α subunit regulated UDPase protein levels, possibly by dephosphorylating a subpopulation of the apyrase, leading to its stabilization (Fig. 5 and 6). The slower-migrating UDPase band, observed when B55 levels were low, may represent a hyperphosphorylated form of the protein (Fig. 6F). This band is much weaker than the majority of the UDPase population, likely because highly phosphorylated forms of UDPase may be unstable and could be degraded fast. A variety of UDPase bands is sometimes observed in Western blots (for example, see Fig. 6C), depending on the PAGE conditions utilized, and they conceivably represent diverse phosphorylated and glycosylated UDPase forms that may not always be separated efficiently. When E4orf4 was introduced into the system, it increased UDPase levels in a PP2A-dependent manner by enhancing the stability of a subpopulation of UDPase polypeptides (Fig. 5). The nature of the UDPase subpopulation influenced by E4orf4 is not known, but it may include phosphorylated UDPase molecules with or without additional modifications, such as glycosylation. The results demonstrating that E4orf4 must bind PP2A to regulate UDPase stability by a process requiring PP2A phosphatase activity raise at least two possibilities: E4orf4 may increase PP2A activity toward the UDPase, or it could augment UDPase accessibility to the phosphatase. An increase in UDPase protein levels contributed to E4orf4-induced cell death but was not sufficient, as UDPase overexpression alone did not result in cell death within the time frame of our experiments (Fig. 1). An E4orf4 mutant (R81F84A) that was unable to bind PP2A but could still induce cell killing in a Src-dependent manner (Fig. 4) could not increase UDPase levels (Fig. 5B), and its ability to induce cell death was not much enhanced by addition of UDPase (Fig. 4). These results indicate that UDPase cooperates with components of the E4orf4-PP2A pathway, but not the E4orf4-Src pathway, to contribute to cell death. Cooperation with the PP2A pathway, which leads to increased UDPase levels, may also include interactions with other E4orf4-PP2A effectors or a requirement for dephosphorylation of UDPase by PP2A, which would affect UDPase function. To date our results suggest that PP2A is required to dephosphorylate UDPase, leading to its stabilization (Fig. 5 and 6), but there is no indication whether it may also affect UDPase function.
We previously demonstrated in yeast that the Ynd1 cytosolic tail mediated the interaction with E4orf4 and was sufficient for providing the Ynd1 contribution to induction of toxicity by the viral protein (29). This Ynd1 domain was shown to bind several membrane proteins involved in protein trafficking in the cell (41). The E4orf4-Src pathway in mammalian cells was reported to affect cellular protein trafficking through regulation of Src family kinases, Cdc42, actin dynamics, and Rab11a, leading to modulation of Golgi membranes and cell death (25). Therefore, we suggested that E4orf4, in addition to its interaction with Src, may utilize an alternative mechanism involving Ynd1, which would allow E4orf4 to interact directly with components of the secretory pathway to affect protein trafficking, thus further transducing its toxic signal (29). Consequently, the E4orf4-Ynd1/UDPase and E4orf4-Src pathways may be parallel pathways that achieve similar results, with the UDPase pathway being at least partially dependent on PP2A. In yeast cells, which do not express Src, the Ynd1 pathway may be the sole contributor of this function, whereas in mammalian cells, the Src pathway provides a major contribution. The finding that the NTPDASE4-specific shRNA did not reduce E4orf4-induced cell death by more than 25% (Fig. 3) may be consistent with the existence of an additional E4orf4-induced cell death mechanism, which partially overlaps the NTPDASE4-PP2A-E4orf4 cell death pathway. However, the NTPDASE4- and Src-dependent pathways are not completely redundant, since NTPDASE4 knockdown decreased E4orf4-induced cell death despite the ability of E4orf4 to function via the Src-dependent pathway (Fig. 3).
In addition to affecting the UDPase by a PP2A-dependent mechanism, E4orf4 appears to manipulate the UDPase by a PP2A-independent pathway as well (Fig. 8). As observed in Fig. 7, both WT E4orf4 and the R81F84A mutant could dissociate high-molecular-weight UDPase-containing complexes. The finding that the mutant dissociated these complexes more efficiently than the WT protein raises the possibility that recruitment of PP2A to the UDPase may inhibit excess dissociation. This inhibition could be an indirect result of the increase in UDPase levels, which leads to the formation of more complexes with its partners. The nature of the high-molecular-weight complexes associating with the UDPase is not yet known. However, the previous results in yeast (29) suggested that the Ynd1 cytosolic tail may have a scaffolding function, associating with at least 10 proteins involved in the secretory pathway and with E4orf4. The Saccharomyces genome database (http://www.yeastgenome.org/) reports that six of these 10 proteins interact among themselves. Thus, it is possible that a multiprotein complex containing proteins involved in trafficking associates with the Ynd1 or UDPase cytosolic tails, and this complex is targeted for dissociation by E4orf4. Future research will be carried out to identify components of the UDPase-containing complexes before and after treatment with E4orf4.
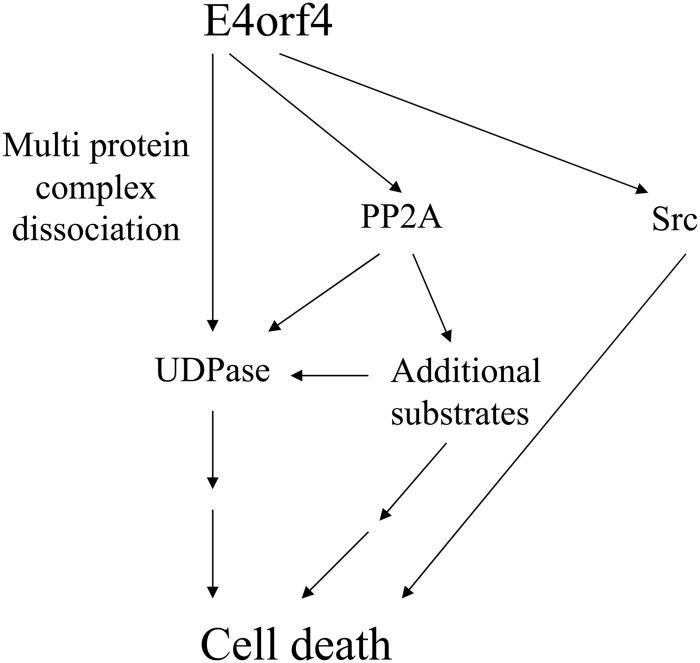
FIG 8.
A model of the interaction between UDPase, PP2A-B55α, and E4orf4, and their contribution to induction of cell death by E4orf4. E4orf4 affected UDPase in at least two ways. First, E4orf4 enhanced UDPase levels in a PP2A-dependent manner, and the increased UDPase levels enhanced E4orf4-induced cell death. Since high levels of UDPase alone did not cause cell death, we suggest that other substrates of PP2A were required for enhancing E4orf4 toxicity in parallel with UDPase or in cooperation with it. The finding that UDPase levels were not much influenced by an E4orf4 mutant that bound Src but not PP2A suggests that UDPase cooperated with the E4orf4-PP2A but not the E4orf4-Src pathway. Second, independently of PP2A, E4orf4 dissociated high-molecular-weight complexes that included UDPase. Because the Ynd1 cytoplasmic tail in yeast appears to act as a scaffold that binds E4orf4 and several proteins of the secretory pathway and mediates E4orf4 toxicity (29), we hypothesize that dissociation of a complex tethered to the UDPase cytosolic tail may contribute to induction of cell death. The Src- and UDPase-dependent pathways may provide partially overlapping contributions to E4orf4-induced cell death as they both interact, physically or functionally, with the protein trafficking machinery.
ACKNOWLEDGMENTS
We are grateful to T. F. Wang and H. P. Elsasser for gifts of plasmids. We thank Odelia Siboni for technical assistance and Antonina Pechkovsky for help with statistical analysis.
This work was supported (in part) by the Israel Science Foundation (grant 769/07), by the Deutsche Forschungsgemeinschaft (DFG) within the framework of the German-Israeli Project Cooperation (DIP), and by the Rappaport Faculty of Medicine and Research Institute, Technion—Israel Institute of Technology.
Footnotes
Published ahead of print 26 March 2014
REFERENCES
- 1.Ben-Israel H, Sharf R, Rechavi G, Kleinberger T. 2008. Adenovirus E4orf4 protein downregulates MYC expression through interaction with the PP2A-B55 subunit. J. Virol. 82:9381–9388. 10.1128/JVI.00791-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bondesson M, Ohman K, Mannervik M, Fan S, Akusjarvi G. 1996. Adenovirus E4 open reading 4 protein autoregulates E4 transcription by inhibiting E1A transactivation of the E4 promoter. J. Virol. 70:3844–3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Estmer Nilsson C, Petersen-Mahrt S, Durot C, Shtrichman R, Krainer AR, Kleinberger T, Akusjarvi G. 2001. The adenovirus E4-ORF4 splicing enhancer protein interacts with a subset of phosphorylated SR proteins. EMBO J. 20:864–871. 10.1093/emboj/20.4.864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanopka A, Muhlemann O, Petersen-Mahrt S, Estmer C, Ohrmalm C, Akusjarvi G. 1998. Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR proteins, Nature 393:185–187 [DOI] [PubMed] [Google Scholar]
- 5.Mannervik M, Fan S, Strom AC, Helin K, Akusjarvi G. 1999. Adenovirus E4 open reading frame 4-induced dephosphorylation inhibits E1A activation of the E2 promoter and E2F-1-mediated transactivation independently of the retinoblastoma tumor suppressor protein. Virology 256:313–321. 10.1006/viro.1999.9663 [DOI] [PubMed] [Google Scholar]
- 6.Muller U, Kleinberger T, Shenk T. 1992. Adenovirus E4orf4 protein reduces phosphorylation of c-fos and E1A proteins while simultaneously reducing the level of AP-1. J. Virol. 66:5867–5878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Shea C, Klupsch K, Choi S, Bagus B, Soria C, Shen J, McCormick F, Stokoe D. 2005. Adenoviral proteins mimic nutrient/growth signals to activate the mTOR pathway for viral replication. EMBO J. 24:1211–1221. 10.1038/sj.emboj.7600597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shtrichman R, Sharf R, Barr H, Dobner T, Kleinberger T. 1999. Induction of apoptosis by adenovirus E4orf4 protein is specific to transformed cells and requires an interaction with protein phosphatase 2A. Proc. Natl. Acad. Sci. U. S. A. 96:10080–10085. 10.1073/pnas.96.18.10080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Afifi R, Sharf R, Shtrichman R, Kleinberger T. 2001. Selection of apoptosis-deficient adenovirus E4orf4 mutants in Saccharomyces cerevisiae. J. Virol. 75:4444–4447. 10.1128/JVI.75.9.4444-4447.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kornitzer D, Sharf R, Kleinberger T. 2001. Adenovirus E4orf4 protein induces PP2A-dependent growth arrest in S. cerevisiae and interacts with the anaphase promoting complex/cyclosome. J. Cell Biol. 154:331–344. 10.1083/jcb.200104104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maoz T, Koren R, Ben-Ari I, Kleinberger T. 2005. YND1 interacts with CDC55 and is a novel mediator of E4orf4-induced toxicity. J. Biol. Chem. 280:41270–41277. 10.1074/jbc.M507281200 [DOI] [PubMed] [Google Scholar]
- 12.Roopchand DE, Lee JM, Shahinian S, Paquette D, Bussey H, Branton PE. 2001. Toxicity of human adenovirus E4orf4 protein in Saccharomyces cerevisiae results from interactions with the Cdc55 regulatory B subunit of PP2A. Oncogene 20:5279–5290. 10.1038/sj.onc.1204693 [DOI] [PubMed] [Google Scholar]
- 13.Pechkovsky A, Lahav M, Bitman E, Salzberg A, Kleinberger T. 2013. E4orf4 induces PP2A- and Src-dependent cell death in Drosophila melanogaster and at the same time inhibits classic apoptosis pathways. Proc. Natl. Acad. Sci. U. S. A. 110:E1724–1733. 10.1073/pnas.1220282110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pechkovsky A, Salzberg A, Kleinberger T. 2013. The adenovirus E4orf4 protein induces a unique mode of cell death while inhibiting classical apoptosis. Cell Cycle 12:2343–2344. 10.4161/cc.25707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brestovitsky A, Sharf R, Mittelman K, Kleinberger T. 2011. The adenovirus E4orf4 protein targets PP2A to the ACF chromatin-remodeling factor and induces cell death through regulation of SNF2h-containing complexes. Nucleic Acids Res. 39:6414–6427. 10.1093/nar/gkr231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kleinberger T, Shenk T. 1993. Adenovirus E4orf4 protein binds to protein phosphatase 2A, and the complex down regulates E1A-enhanced junB transcription. J. Virol. 67:7556–7560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lavoie JN, Champagne C, Gingras M-C, Robert A. 2000. Adenovirus E4 open reading frame 4-induced apoptosis involves dysregulation of Src family kinases. J. Cell Biol. 150:1037–1055. 10.1083/jcb.150.5.1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shtrichman R, Sharf R, Kleinberger T. 2000. Adenovirus E4orf4 protein interacts with both Bα and B′ subunits of protein phosphatase 2A, but E4orf4-induced apoptosis is mediated only by the interaction with Bα. Oncogene 19:3757–3765. 10.1038/sj.onc.1203705 [DOI] [PubMed] [Google Scholar]
- 19.Marcellus RC, Chan H, Paquette D, Thirlwell S, Boivin D, Branton PE. 2000. Induction of p53-independent apoptosis by the adenovirus E4orf4 protein requires binding to the Balpha subunit of protein phosphatase 2A. J. Virol. 74:7869–7877. 10.1128/JVI.74.17.7869-7877.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eichhorn PJ, Creyghton MP, Bernards R. 2009. Protein phosphatase 2A regulatory subunits and cancer. Biochim. Biophys. Acta 1795:1–15. 10.1016/j.bbcan.2008.05.005 [DOI] [PubMed] [Google Scholar]
- 21.Kleinberger T. 2004. Induction of transformed cell-specific apoptosis by the adenovirus E4orf4 protein. Prog. Mol. Subcell. Biol. 36:245–267. 10.1007/978-3-540-74264-7_12 [DOI] [PubMed] [Google Scholar]
- 22.Mui MZ, Kucharski M, Miron MJ, Hur WS, Berghuis AM, Blanchette P, Branton PE. 2013. Identification of the adenovirus E4orf4 protein binding site on the B55alpha and Cdc55 regulatory subunits of PP2A: implications for PP2A function, tumor cell killing and viral replication. PLoS Pathog. 9:e1003742. 10.1371/journal.ppat.1003742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li S, Brignole C, Marcellus R, Thirlwell S, Binda O, McQuoid MJ, Ashby D, Chan H, Zhang Z, Miron MJ, Pallas DC, Branton PE. 2009. The adenovirus E4orf4 protein induces G2/M arrest and cell death by blocking PP2A activity regulated by the B55 subunit. J. Virol. 83:8340–8352. 10.1128/JVI.00711-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Champagne C, Landry MC, Gingras MC, Lavoie JN. 2004. Activation of adenovirus type 2 early region 4 ORF4 cytoplasmic death function by direct binding to Src kinase domain. J. Biol. Chem. 279:25905–25915. 10.1074/jbc.M400933200 [DOI] [PubMed] [Google Scholar]
- 25.Landry MC, Sicotte A, Champagne C, Lavoie JN. 2009. Regulation of cell death by recycling endosomes and golgi membrane dynamics via a pathway involving Src-family kinases, Cdc42 and Rab11a. Mol. Biol. Cell 20:4091–4106. 10.1091/mbc.E09-01-0057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robert A, Smadja-Lamere N, Landry MC, Champagne C, Petrie R, Lamarche-Vane N, Hosoya H, Lavoie JN. 2006. Adenovirus E4orf4 hijacks rho GTPase-dependent actin dynamics to kill cells: a role for endosome-associated actin assembly. Mol. Biol. Cell 17:3329–3344. 10.1091/mbc.E05-12-1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong X, Guidotti G. 1999. A yeast Golgi E-type ATPase with an unusual membrane topology. J. Biol. Chem. 274:32704–32711. 10.1074/jbc.274.46.32704 [DOI] [PubMed] [Google Scholar]
- 28.Zimmermann H. 2001. Ectonucleotidases: some recent developments and a note on nomenclature. Drug Dev. Res. 52:44–56. 10.1002/ddr.1097 [DOI] [Google Scholar]
- 29.Mittelman K, Ziv K, Maoz T, Kleinberger T. 2010. The cytosolic tail of the Golgi apyrase Ynd1 mediates E4orf4-induced toxicity in Saccharomyces cerevisiae. PLoS One 5:e15539. 10.1371/journal.pone.0015539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biederbick A, Kosan C, Kunz J, Elsasser HP, Rose S. 2000. First apyrase splice variants have different enzymatic properties. J. Biol. Chem. 275:19018–19024. 10.1074/jbc.M001245200 [DOI] [PubMed] [Google Scholar]
- 31.Biederbick A, Rose S, Elsasser HP. 1999. A human intracellular apyrase-like protein, LALP70, localizes to lysosomal/autophagic vacuoles. J. Cell Sci. 112:2473–2484 [DOI] [PubMed] [Google Scholar]
- 32.Wang TF, Guidotti G. 1998. Golgi localization and functional expression of human uridine diphosphatase. J. Biol. Chem. 273:11392–11399. 10.1074/jbc.273.18.11392 [DOI] [PubMed] [Google Scholar]
- 33.Biederbick A, Rosser R, Storre J, Elsasser HP. 2004. The VSFASSQQ motif confers calcium sensitivity to the intracellular apyrase LALP70. BMC Biochem. 5:8. 10.1186/1471-2091-5-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shtrichman R, Kleinberger T. 1998. Adenovirus type 5 E4 open reading frame 4 protein induces apoptosis in transformed cells. J. Virol. 72:2975–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hinds P, Finlay CA, Quartin RS, Baker SJ, Fearon ER, Vogelstein B, Levine AJ. 1990. Mutant p53 DNA clones from human colon carcinomas cooperate with ras in transforming primary rat cells: a comparison of the “hot spot” mutant phenotypes. Cell Growth Differ. 1:571–580 [PubMed] [Google Scholar]
- 36.Brestovitsky A, Sharf R, Kleinberger T. 2012. Preparation of cell-lines for conditional knockdown of gene expression and measurement of the knockdown effects on E4orf4-induced cell death. J. Vis. Exp. 2012:4442. 10.3791/4442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lavoie JN, Nguyen M, Marcellus RC, Branton PE, Shore GC. 1998. E4orf4, a novel adenovirus death factor that induces p53-independent apoptosis by a pathway that is not inhibited by zVAD-fmk. J. Cell Biol. 140:637–645. 10.1083/jcb.140.3.637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li S, Szymborski A, Miron MJ, Marcellus R, Binda O, Lavoie JN, Branton PE. 2009. The adenovirus E4orf4 protein induces growth arrest and mitotic catastrophe in H1299 human lung carcinoma cells. Oncogene 28:390–400. 10.1038/onc.2008.393 [DOI] [PubMed] [Google Scholar]
- 39.Livne A, Shtrichman R, Kleinberger T. 2001. Caspase activation by adenovirus E4orf4 protein is cell line-specific and is mediated by the death receptor pathway. J. Virol. 75:789–798. 10.1128/JVI.75.2.789-798.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robert A, Miron MJ, Champagne C, Gingras MC, Branton PE, Lavoie JN. 2002. Distinct cell death pathways triggered by the adenovirus early region 4 ORF 4 protein. J. Cell Biol. 158:519–528. 10.1083/jcb.200201106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller JP, Lo RS, Ben-Hur A, Desmarais C, Stagljar I, Noble WS, Fields S. 2005. Large-scale identification of yeast integral membrane protein interactions. Proc. Natl. Acad. Sci. U. S. A. 102:12123–12128. 10.1073/pnas.0505482102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marcellus RC, Lavoie JN, Boivin D, Shore GC, Ketner G, Branton PE. 1998. The early region 4 orf4 protein of human adenovirus type 5 induces p53-independent cell death by apoptosis. J. Virol. 72:7144–7153 [DOI] [PMC free article] [PubMed] [Google Scholar]