Abstract
Progressive HIV infection is characterized by dysregulation of the intestinal immune barrier, translocation of immunostimulatory microbial products, and chronic systemic inflammation that is thought to drive progression of disease to AIDS. Elements of this pathologic process persist despite viral suppression during highly active antiretroviral therapy (HAART) and drivers of these phenomena remain poorly understood. Disrupted intestinal immunity can precipitate dysbiosis that induces chronic inflammation in the mucosa and periphery of mice. However, putative microbial drivers of HIV-associated immunopathology versus recovery have not been identified in humans. Using high-resolution bacterial community profiling, we identified a dysbiotic mucosal-adherent community enriched in Proteobacteria and depleted of Bacteroidia members that was associated with markers of mucosal immune disruption, T cell activation, and chronic inflammation in HIV-infected subjects. Furthermore, this dysbiosis was evident among HIV-infected subjects undergoing HAART, and the extent of dysbiosis correlated with activity of the kynurenine pathway of tryptophan metabolism and plasma concentrations of the inflammatory cytokine interleukin-6 (IL-6), two established markers of disease progression. Gut-resident bacteria with capacity to metabolize tryptophan through the kynurenine pathway were found to be enriched in HIV-infected subjects, strongly correlated with kynurenine levels in HIV-infected subjects, and capable of kynurenine production in vitro. These observations demonstrate a link between mucosal-adherent colonic bacteria and immunopathogenesis during progressive HIV infection, which is apparent even in the setting of viral suppression during HAART. This link suggests that gut-resident microbial populations may influence intestinal homeostasis during HIV disease.
INTRODUCTION
Accumulating evidence from human and non-human primate studies supports the broad hypothesis that progression to AIDS during HIV infection is driven by chronically elevated T cell activation and systemic inflammation (1–4). While the etiology of such persistent immune activation is incompletely understood, the gastrointestinal mucosal immune disruption that follows progressive HIV and SIV infection is postulated to play a role. Specifically, compromised mucosal barrier function and increased translocation of immunostimulatory microbial products from the gut lumen into systemic circulation have been implicated in this process (5). Indeed, the presence of microbial products in the peripheral blood of HIV-infected subjects has been linked to immune activation and increased morbidity and mortality (6, 7). Among the reported defects in the mucosal barrier, a pronounced reduction in cell populations that are characterized by secretion of the enterocyte homeostasis-promoting factors, IL-17 and IL-22, has been associated with decreased gastrointestinal epithelial barrier integrity and accelerated disease (8, 9). Accompanying and apparently precipitating these changes are reported increases in activation of the kynurenine pathway of tryptophan metabolism through the interferon-inducible enzyme, indoleamine 2,3-dioxygenase 1 (IDO1) (9–12), which produces tryptophan catabolites that can inhibit the differentiation of IL-17 secreting CD4+ T cells (13, 14). IDO1 activity is induced during the course of pathologic HIV and SIV infection, fails to normalize during progressive disease, and is associated with impaired mucosal immunity and microbial translocation, the postulated drivers of chronic inflammation that precipitate AIDS (15). Although highly active anti-retroviral treatment (HAART) results in a partial diminution in inflammatory markers, including IDO1 activity, residual markers of T cell activation and inflammation (e.g., interleukin-6 [IL-6] and interferon-inducible protein 10 [IP-10]) persist and the extent of their elevation correlates with incomplete immune recovery and an increased risk for non-AIDS related morbidities (16–18). Nonetheless, the molecular and cellular mechanisms contributing to this inflammatory process during early infection and antiretroviral-treated infection remain poorly understood.
Gut-resident bacteria can modulate the mucosal immune system in ways that overlap with the salient features of HIV pathogenesis. For instance, the absence of a Clostridia class commensal bacterium, segmented filamentous bacteria (SFB), in the murine gut results in a decrease in IL-17-secreting CD4+ T cell subpopulations (19), while other members of the class Clostridia (Clostridium clusters IV and XIVa) have been found to induce mucosal expression of IDO1 (20). Indeed, Enterobacteriaceae family members belonging to the genera Klebsiella (21), Citrobacter (22), and Salmonella (23, 24) can actively induce an inflammatory microenvironment in the gastrointestinal tract that supports their proliferation and persistence. Additionally, disruptions in the mucosal innate immune system can result in the outgrowth of a “dysbiotic” pro-inflammatory community that can be sufficient to sustain pathologic, chronic inflammation in the mucosa and the periphery (21, 25–27). These findings suggest not only that gut bacterial species may modulate mucosal immunity in humans, but also that inflammatory imbalance can promote outgrowth of pathobionts that further exacerbate mucosal immune disequilibrium. Since HIV disease is characterized by pronounced disruptions to mucosal immunity as well as a sustained chronic inflammatory state, we sought to understand whether the gut microbiota is altered during HIV infection, and to assess relationships between these assemblages and characteristic immunologic states of HIV disease progression.
RESULTS
Gut bacterial microbiota composition differs between HIV- and VU subjects
To capture a wide range of stages of HIV disease progression and treatment, a cohort of 32 male subjects was assembled to include six viremic, untreated (VU) HIV-infected subject, 18 virally-suppressed subjects on HAART with varying degrees of CD4+ T cell recovery (HAART), one HIV-infected long-term non-progressor, and nine uninfected risk-matched controls (HIV−); see Table S1 for defining characteristics of the cohort. Rectosigmoid biopsy specimens and peripheral blood were collected for immunophenotyping of immune cell subpopulations and for profiling of microbial communities. Biopsy tissue was chosen over stool sampling based on evidence that immunomodulatory bacterial species adhere closely to the gastrointestinal mucosal epithelium in the mouse (19, 20), comprising a community that is unique from that within the gut lumen (28). Microbial profiling was performed using a standardized, high-density microarray containing oligonucleotide probes targeted against hypervariable regions of the bacterial 16S rRNA gene (G3 PhyloChip (29)). This approach is ideally suited for high-resolution microbiome comparative analyses and detection of lower-abundance species that may not be identified using sequencing approaches of relatively shallow read depth (30). This is particularly relevant given recent evidence demonstrating the presence of low-abundance “keystone” species that despite their low numbers are pivotal in modulating commensal community composition and host-microbe interactions in the oral cavity (31).
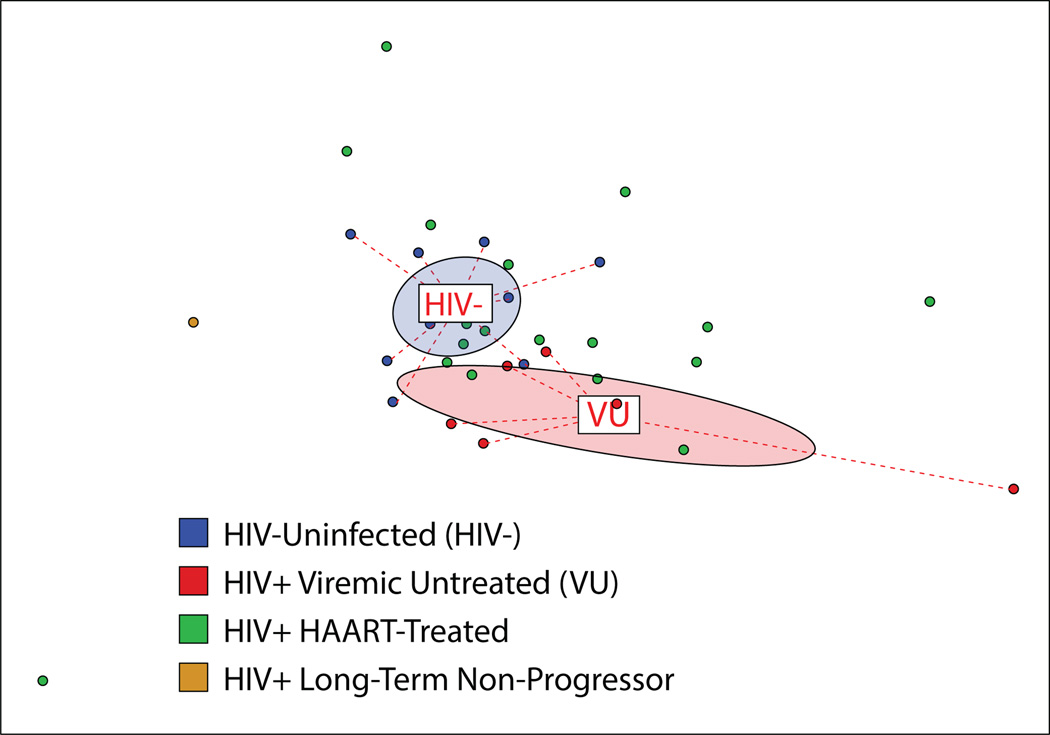
Analysis of rectosigmoid biopsies showed that total bacterial load was similar across all subjects, regardless of infection status (Fig. S1A), and that total bacterial load measures were concordant across different biopsies from a given individual as well as across different DNA extraction methods (Fig. S1B). Additionally, comparative analysis of gross microbial community metrics showed no significant differences between HIV-infected and uninfected subjects in community evenness or richness (Fig. S1C). By contrast, analysis of between-subject microbiota composition using a Canberra community dissimilarity matrix revealed distinct clustering of the subject groups representing extremes of health and disease: the HIV- and the VU group, respectively (Fig. 1). Permutational multivariate analysis of variance confirmed that the observed variability in community composition was significantly related to subjects belonging to the VU or HIV- group (Adonis; P = 0.002), indicating that untreated HIV infection is associated with a distinct mucosal microbiota composition. Interestingly, HAART-treated patients, though uniformly exhibiting viral suppression (i.e., plasma viral load < 40 copies/ml), exhibited highly dissimilar community compositions: the microbiota of some patients clustered closely with VU subjects while others were more similar to HIV- subjects.
Figure 1. Gut bacterial microbiota composition in HIV-infected viremic untreated (VU) subjects differs from that of HIV-uninfected risk-matched controls.
Community composition dissimilarity was analyzed using microbiota profiles generated using PhyloChip. Non-metric dimensional scaling (NMDS) was used to plot each sample community based on a Canberra community dissimilarity matrix. Each dot represents the microbiome of a single subject, and color indicates subject group as denoted in legend. Ellipses represent 95% confidence intervals for standard error of weighted NMDS score means of respective groups, and are colored based on subject group (red denotes HIV+VU subjects, while blue denotes HIV-). Community differences were verified using Adonis, P = 0.002.
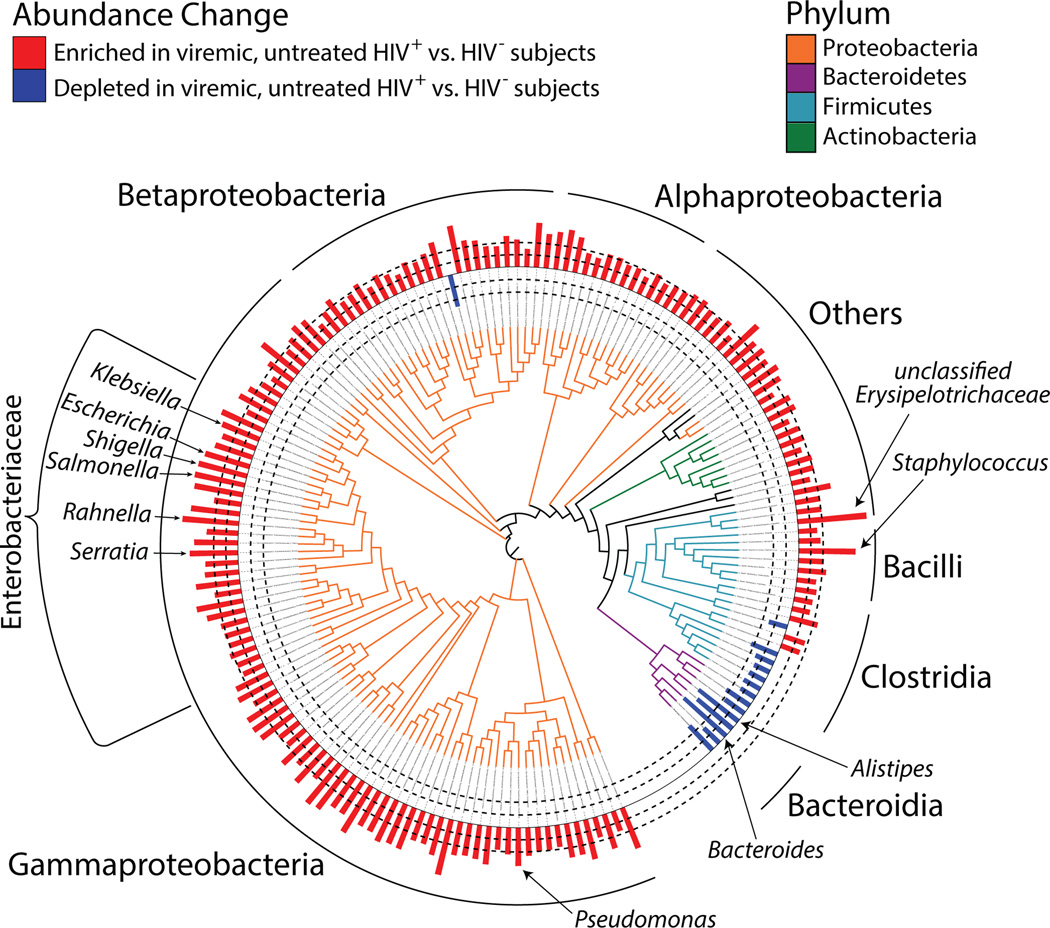
To identify differences in the adherent bacterial communities of HIV- and VU subjects, a permutational analysis algorithm with integrated false-discovery correction (SAM (32)) was used to detect specific bacterial taxa in differential fold abundance across these two subject groups. Of the 33,951 taxa (defined as a group of bacteria having at least 99% 16S rRNA sequence identity) that were detected in at least one of the samples studied, 625 exhibited significantly different relative abundance between VU and HIV- after stringent data filtering. This list comprised 579 taxa that were enriched and 45 taxa that were depleted in VU subjects compared with HIV- subject samples (Fig. 2 and Table S2 for list of taxa). Among the taxa enriched in VU subjects, the most enriched was Erysipelotrichaceae in the class Mollicutes, a class that has been associated with obesity and heightened cardiovascular morbidity (33–35). A majority of taxa enriched in VU subjects included members of the phylum Proteobacteria, with a notably high representation of the Enterobacteriaceae family, a finding verified by quantitative polymerase chain reaction (PCR) (Fig. S2) and supported by a prior report (36). Enriched genera from within this family included known pro-inflammatory pathobionts such as Salmonella, Escherichia, Serratia, Shigella, and Klebsiella species. Additionally, Staphylococcus, Pseudomonas, and Campylobacter spp., known opportunistic pathogens and sources of bacteremia in HIV-infected subjects (37–39), were highly enriched in the mucosae of VU subjects. Significant reductions in the relative abundance of specific members of Clostridia and Bacteroidia were observed in VU subjects, with the greatest degree of depletion amongst members of the Bacteroides and Alistipes genera, which have been previously found to be depleted in inflammatory bowel disease (40). We refer to this community of bacterial taxa (identified as differing significantly in abundance between VU subjects and HIV-uninfected subjects) as the “disease-associated microbial community” (DMC).
Figure 2. Phylogenetic distribution of the disease-associated microbial community (DMC), defined as significantly depleted or enriched in VU subjects compared with HIV- subjects.
Phylogenetic tree representing 16S rRNA gene sequence relatedness of taxa identified as significantly enriched or depleted in relative abundance between VU and HIV- subjects (see Materials & Methods for statistical analyses). The 625 taxa identified belonged to 172 unique genera, and representative taxa for each unique genus are depicted. Tree branches are colored-coded by phyla as indicated in legend. Red and blue bars depict bacterial genera in greater (red bars) and lesser (blue bars) abundance in VU subjects. Dashed lines indicate magnitude of relative abundance change (inner dashed circle denotes 500 MFI, outer denotes 2000 MFI difference between comparative groups; 1,000 MFI change represents a log2 difference in relative abundance).
Intestinal dysbiosis correlates with markers of disease progression among HIV-infected subjects
While levels of T cell activation (as measured by co-expression of CD38 and HLA-DR) and viral load are well-accepted markers of HIV disease progression (3), it is now also appreciated that other immune parameters are linked to morbidity and mortality, including plasma markers of inflammation, IDO1 activity, and markers of microbial translocation (7, 11, 17). Of note, specific murine gut commensal species have been shown to adhere to mucosal epithelia and induce altered inflammatory cytokine secretion patterns (19, 20), some in association with IDO1 upregulation (20). To determine whether associations exist between the mucosa-associated DMC and HIV disease progression, peripheral blood was drawn at the time of mucosal sampling and additional biopsies were obtained for immunophenotyping (see Table S3 for a complete list of immune parameters measured). Correlations between the relative abundance of taxa within the DMC and immunologic parameters among all HIV-infected subjects (HAART and VU) were examined (Fig. 3A). Strong associations were not found between taxon abundances and peripheral blood CD4+ T cell count or gut HIV RNA or DNA levels. However, taxa that were enriched in the DMC correlated with elevated levels of T cell activation, of catabolism of tryptophan through the kynurenine pathway (a marker of IDO1 activity measured by the ratio of plasma concentrations of the downstream product, kynurenine, to the parent compound, tryptophan [Kyn:Trp]), and of soluble plasma markers of inflammation (such as IP-10, soluble TNF receptor II, and IL-6), and diminished levels of mucosal T cells secreting IL-17 and IL-22. Though relative abundances of many taxa were found to correlate with these immune parameters (Fig. 3A), phylogenetically distinct members of the Enterobacteriaceae family were among the strongest correlates of elevated gut CD4+ and CD8+ T cell activation (unadjusted P = 0.00004, RS = 0.82; unadjusted P = 0.00006, RS = 0.80; Table S3; RS, Spearman rho correlation coefficient) while members of Staphylococcaceae were among the strongest correlates of higher Kyn:Trp and of elevated plasma IP-10 levels (unadjusted P = 0.00007, RS = 0.76; unadjusted P = 0.0014, RS = 0.64). Taxa within the families Micrococcaceae, Rhodobacteraceae, Halomonadaceae, and Pasteurellaceae also exhibited strong correlations with numerous markers of disease progression. Conversely, those taxa depleted in the DMC were found to correlate with lower levels of T cell activation and higher levels of mucosal T cells secreting IL-17, with the strongest correlates of lower T cell activation belonging to Bacteroidaceae (unadjusted P = 0.0017, RS = −0.68) and the strongest correlates of higher gut IL-17 production belonging to taxa within the Rikenellaceae family (unadjusted P = 0.0004, RS = 0.72; see Fig. S3 and Table S3 for full list of all correlation coefficients). Collectively, these findings indicate higher abundance of multiple, diverse microbial taxa within the DMC is linked to immune activation and to poor gut IL-17 responses, and that this state is concomitantly characterized by an apparent depletion of taxa linked to immune homeostasis and conditions which promote gastrointestinal integrity, i.e., lower levels of T cell activation and higher levels of IL-17 secretion.
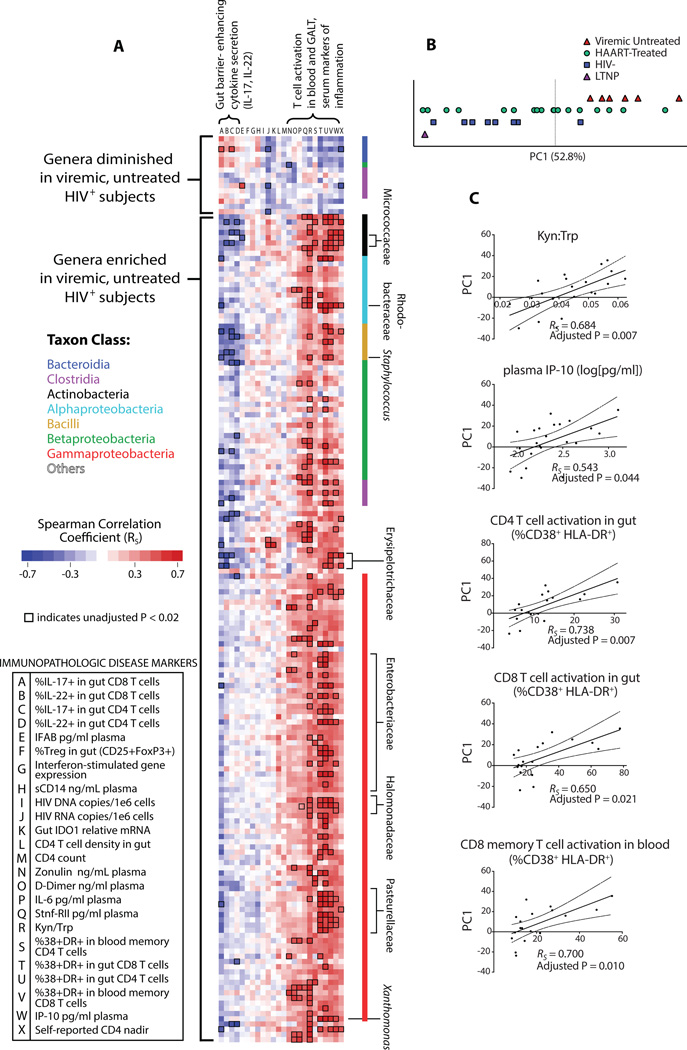
Figure 3. Bacterial community enriched in untreated infection associates with immunopathologic markers of HIV disease progression within HIV-infected subjects.
(A) Spearman correlations between individual taxa of the DMC and immunologic parameters were calculated among all HIV-infected subjects (treated and untreated), collated, and represented by color. Correlation coefficients for taxa with representative species belonging to the same genus and displaying similar FI trends were averaged for visualization purposes. Intersections of each genus within the DMC (rows) and immunologic parameters (columns) represent the strength of the positive (red) or negative (blue) correlation as determined by Spearman correlation coefficient (RS). Bacteria depleted in VU subjects correlate positively with protective immune markers (low T cell activation in the blood and gut, higher gut IL-17 secretion), while bacteria enriched in VU subjects correlate with markers of disease progression (converse of above, as well as lower T cell-mediated IL-22 secretion in the gut, higher Kyn:Trp, and higher concentrations of plasma markers of inflammation [IP-10, IL-6, and soluble TNF receptor II]). Families and genera of taxa that exhibited numerous strong correlations with immunologic parameters are indicated. Intersections enclosed by a black box represent associations with unadjusted P < 0.02. (B) First principal component (PC1) representing trends in DMC bacterial community composition accounts for 52.8% of bacterial community variance, and separates VU subjects from HIV-, while subjects undergoing HAART have PC1 values distributed amongst those for VU and HIV- subjects (PC1, Principal Component 1; LTNP, Long Term Non-Progressor). (C) First principal component correlates with markers of disease progression: plasma concentration of inflammatory marker IP-10, IDO activity (Kyn:Trp), gut T cell activation, and peripheral blood CD8 T cell activation (RS, Spearman rho correlation coefficient; dotted lines represent 95% confidence interval bands for linear regression coefficients).
Gut-resident microbial communities function as discrete ecosystems bound together by interdependent metabolic exchange and collective protective mechanisms. Accordingly, many taxa of the DMC exhibited similar patterns of correlation with immune parameters (Fig. 3A). Therefore, a holistic approach was taken to understand such trends in gross community abundance as they may relate to inflammatory states. Principal component analysis (PCA) was performed to consolidate patterns among co-varying microbial abundances into a handful of summary variables and to compare such variables to immune parameters. The first principal component (PC1) of this analysis comprised 52.8% of DMC variance (Fig. 3B and Fig. S4A), was able to differentiate HIV- from VU (Fig. 3C), and was chiefly characterized by opposing trends in the relative abundance of Proteobacteria (including Pasteurellaceae, Mycobacteriaceae, Pseudomonadaceae, and Enterobacteriaceae members) and Bacteroidaceae (Fig. S4B). We used PC1 as a measure of the degree to which a subject’s gut microbiota was similar to that of VU or HIV− (and therefore a surrogate marker for extent of dysbiosis), and related it to markers of immunopathology and disease progression among all HIV-infected subjects. PC1 was found to associate strongly with plasma IP-10 (adjusted P = 0.044), a marker of systemic inflammation shown to correlate with accelerated HIV disease progression (18, 41), with Kyn:Trp (adjusted P = 0.007), and with T cell activation in the gut and peripheral blood (respective significance values shown in figure) (Fig. 3D, Table S4).
Heterogeneity in microbiota of subjects undergoing HAART correlates with markers of disease progression
Even though HAART is effective in diminishing plasma viremia to nearly undetectable levels, subjects receiving long-term treatment can exhibit persistently elevated levels of chronic immune activation (16), of which the etiology is unknown. We hypothesized that persistence of even a subset of the DMC could continue to sustain pathologic chronic immune activation in this population despite suppression of viral replication. To address this hypothesis, the relative abundance of DMC members was averaged across each of the VU, HAART, and HIV- subject groups and compared between these groups. The DMC in HAART subjects was found to represent an intermediate state between that observed in VU subjects and uninfected controls (Fig. 4A). Additionally, longitudinal sampling of two HIV-infected subjects prior to and following nine months of effective HAART demonstrated a relative shift in the DMC to more closely resemble that of the HIV- group (Fig. S5). However, individual subjects within the HAART group exhibited a spectrum of relative abundance for members of the DMC, such that the microbial community composition of some subjects was more similar to that of HIV- subjects while the composition of others was more similar to that of VU subjects (Fig. 4B).
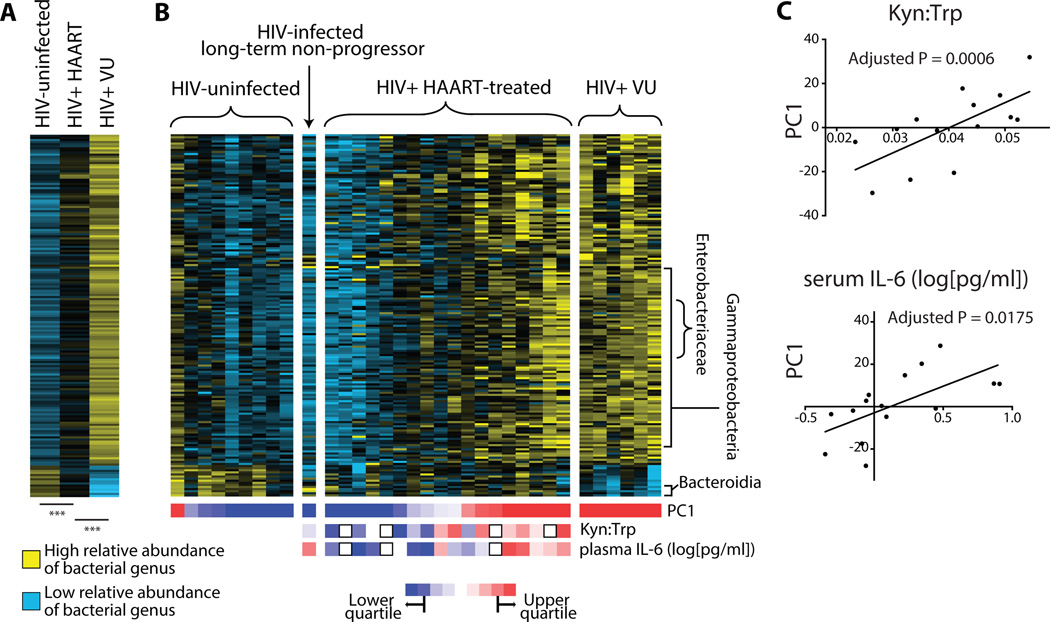
Figure 4. Relative abundance of DMC members is diminished in HAART subjects compared to VU and falls along a spectrum of VU-like or uninfected-like bacterial communities.
(A) Shown are relative abundances of genera within the DMC averaged within subject groups. Each row represents a unique genus. A higher relative abundance is denoted by yellow squares while blue denotes lower relative abundance. After assessment of data distribution, a two-tailed, unpaired T-test assuming unequal variance was used to test significance of differences (***P < 5×10−10). (B) Heatmap of DMC member relative abundances, ordered first by subject grouping, then by subject PC1 values (relative values denoted by color below each subject column). Rows represent genera of the DMC while columns represent individual subject samples. In the treated subject grouping, subjects were ranked for each variable (i.e. Kyn:Trp and IL-6 concentration), and these rankings are represented by colored squares below each subject column (red indicates high Kyn:Trp and high plasma IL-6 in the first and second rows, respectively). (C) Correlations between PC1 and Kyn:Trp or plasma IL-6 concentration. P values displayed represent the contribution of PC1 to the best-fit linear regression model that best predicts each immunologic variable, adjusted to account for multiple comparisons (see Materials & Methods for details).
We considered a number of possibilities to account for the presence of this spectrum in the DMC of subjects on HAART. Since prophylactic antibiotics are commonly used by subjects on HAART and may influence microbial community composition, relationships between PC1 and antibiotic administration history (for which data was available for 13 out of 18 subjects) were analyzed. No significant trends were observed, as was the case for comparisons to age and time on HAART (Fig. S6). Furthermore, gut inflammation itself has been found to stimulate the outgrowth of facultative anaerobes, including members of the Enterobacteriaceae family (24, 42, 43), by providing growth substrates in the form of anaerobic respiration terminal electron acceptors. Expression of genes involved in oxygen and nitrogen radical production (NADPH oxidase [CYBB subunit], arginase 1, nitric oxide synthase 2 [NOS2], type I interferons [IFN-α, IFN-β], type II interferon [IFN-γ], tumor necrosis factor-alpha [TNF-α]) was accordingly quantified in whole biopsies and compared to PC1, and no significant associations were found (Table S6). Rather, associations were found between PC1 and numerous markers of disease progression, including levels of IL-6 in plasma, Kyn:Trp, soluble TNF-α receptor II levels in plasma, blood coagulation marker D-Dimer levels in plasma, gut IDO1 transcript, blood memory CD8 T cell activation, plasma IP-10 levels, and gut CD4+ T cell activation (P < 0.05, |Rs| > 0.5; Table S5). After adjusting for multiple comparisons, significant relationships remained between PC1 and two disease markers: plasma Kyn:Trp and plasma IL-6 concentration (Fig. 4C). To understand whether these associations were influenced by potentially confounding variables and to incorporate a greater proportion of microbial community abundance trends, multiple linear regression analysis was performed. This approach allows for simultaneous consideration of multiple adjustment variables (antibiotics usage history, days on HAART, age, race) and the five most representative principal components of DMC variance (PC1, PC2, PC3, PC4, and PC5) toward understanding which variables are most predictive of the immunologic parameters of interest (Kyn:Trp and plasma IL-6). Out of all possible models involving these variables, the model with the best leave-one-out cross-validated (44) (LOO-CV) predictive accuracy was chosen and p-values were obtained by permutation. PC1 and ethnicity were significantly correlated with Kyn:Trp in the best scoring model (P = 0.0006 and P = 0.0036, respectively). In the case of plasma IL-6 levels, the variable lending greatest predictive power to the most predictive multivariate model was PC1 (P = 0.018), while four other variables contributed less significantly: PC4 (P = 0.032), ethnicity (P = 0.048), age (P = 0.027), and duration of HAART (P = 0.020). Comparisons of Kyn:Trp and plasma IL-6 to PC1 alone are shown in Fig. 4C.
Members of the DMC participate in immunoactive tryptophan catabolism
IDO1 is induced during inflammation and is thought to actively regulate immune activation by its numerous effects on T cells, including inhibition of IL-17 secretion and induction of Foxp3-expressing regulatory T cells (Treg) (45). These effects are mediated by its production of kynurenine pathway compounds from its enzymatic substrate tryptophan, a reaction for which IDO1 is thought to govern the rate-limiting step (46). Consistent with previous findings (47), we found that IDO1 is strongly associated with type II interferon expression in colonic mucosal tissue and the ratio of plasma concentrations of kynurenine to tryptophan (Kyn:Trp) was indeed associated with reduced IL-17 secretion in CD4+ T cells and higher abundance of Treg in the colon (Fig. S7A–B). However, though there was a significant positive correlation between IDO1 expression and plasma Kyn:Trp in VU subjects, there was no such association in the HAART HIV-infected subject group (Fig. S7C).
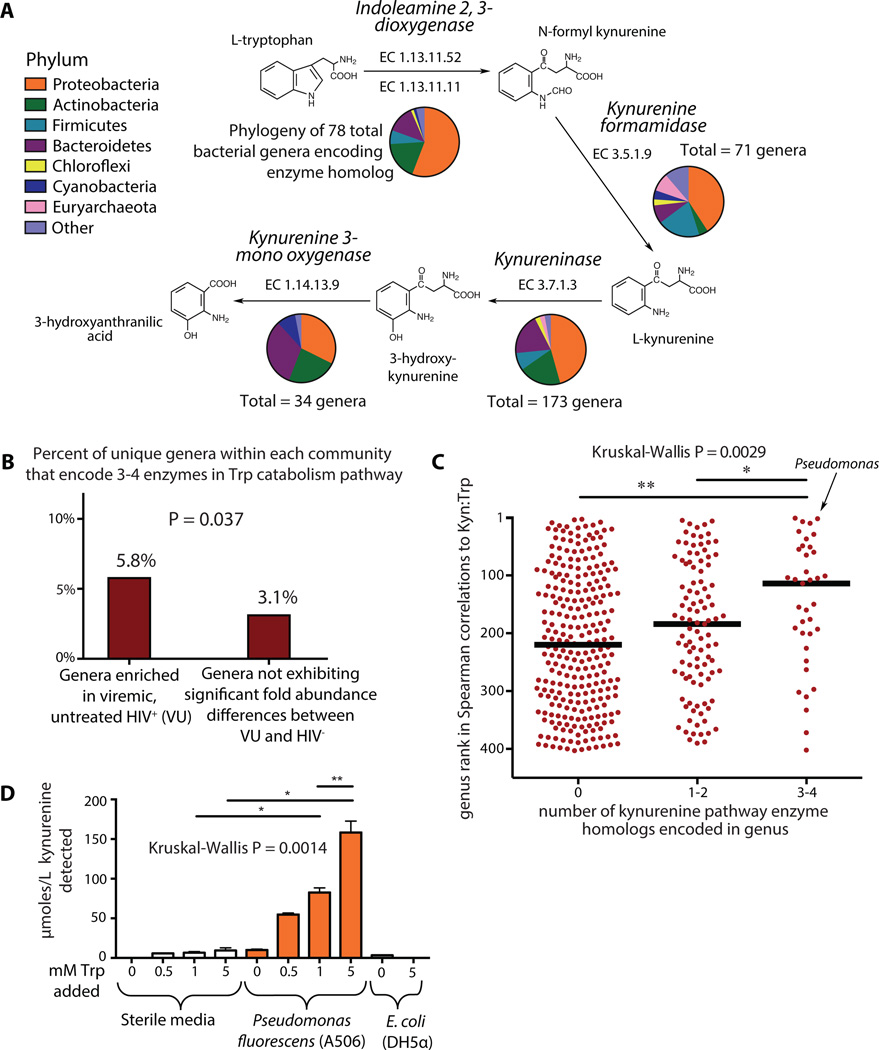
Given that a significant association was found between our marker of dysbiosis (PC1) and Kyn:Trp within the HAART subject group, we postulated that the DMC may directly contribute to immunomodulatory tryptophan catabolism through the kynurenine pathway. Comparing taxon abundances to plasma Kyn:Trp, we found that taxa within 140 genera positively correlated significantly with Kyn:Trp (Table S7). While bacteria are among the primary producers of the essential amino acid tryptophan, relatively few are known to encode the enzymatic machinery for tryptophan catabolism (48–51). Indeed, analysis of genomic and proteomic annotations from the UniProt Consortium (52) revealed several bacterial genera within Proteobacteria that encode enzymes homologous to those necessary for catabolism of tryptophan to the most potent immunomodulatory product of the kynurenine pathway (9, 13), 3-hydroxyanthranilic acid (3-HAA; Fig. 5A, Table S7). Comparing this list to that of genera enriched in the DMC, we found that genera that bore genetic homologs to tryptophan catabolism enzymes of the kynurenine pathway were preferentially enriched in the DMC (Fig. 5B), including Pseudomonas, Xanthomonas, Burkholderia, Stenotrophomonas, Shewanella, and Bacillus, as well as members of the families Rhodobacteraceae, Micrococcaceae, and Halomonadaceae (see Table S7 for full list). Furthermore, genera for which at least one member bore genetic homologs to 3–4 enzymes within the immunoactive kynurenine pathway correlated more strongly with Kyn:Trp ratios than genera that bore 1–2 tryptophan catabolism enzyme homologs, and even more so than genera that had no genetic evidence for kynurenine pathway metabolic machinery (Fig. 5C, Fig. S8).
Figure 5. Bacterial tryptophan catabolism machinery is genetically and functionally homologous to IDO1 enzymatic activity and is enriched in the DMC.
(A) Homologs to tryptophan catabolism pathway enzymes are genetically encoded in numerous bacterial genera as denoted by ‘Totals.’ Phylogenetic distribution of annotated enzymes in the kynurenine pathway was collated and represented using pie charts as defined in legend. (B) A Fisher’s exact test for enrichment found that a greater proportion of genera overrepresented in VU encoded 3–4 genes involved in the tryptophan catabolism pathway as compared to genera not differing abundance between VU and HIV- (P = 0.037). (C) All genera that were both detected in our cohort and contained members for which genome sequence data was available were ranked based on their strength of non-parametric Spearman correlation to Kyn:Trp ratios in HIV-infected subject plasma. Genera with 3 or 4 genetically encoded homologs to kynurenine pathway enzymes exhibited significantly stronger correlations to plasma Kyn:Trp ratios than those with only 1 or 2 homologs, and even more so than those with no annotated homologous enzymes. (D) Pseudomonas fluorescens strain A506 and a lab strain of E. coli (DH5α) were grown in King’s B Medium for 20 hours in the presence of varying concentrations of tryptophan. Supernatants from these cultures were assayed for the presence of kynurenine. Error bars indicate standard deviations calculated from three independent experiments. Non-parametric Mann-Whitney tests were used for group comparisons (*P < 0.05, **P < 0.005).
To confirm the capacity for production of kynurenine pathway catabolites among members of the DMC, a representative species of the strongest bacterial correlate to plasma Kyn:Trp ratios (genus Pseudomonas) was assayed for kynurenine production in pure culture. Pseudomonas fluorescens strain A506 was found to produce significant amounts of kynurenine from tryptophan (Fig. 5D) in vitro, further supporting the hypothesis that gut mucosal-adherent bacteria associated with HIV infection might directly contribute to immunoactive tryptophan catabolism during HIV disease.
DISCUSSION
Here, we report observations of differences in the composition of the colonic mucosal-adherent bacterial microbiota of untreated HIV-infected subjects as compared to uninfected risk-matched controls. This altered community prominently includes known pathobionts, such as Staphylococcus spp., Pseudomonas spp., and Enterobacteriaceae family members with pro-inflammatory potential. While these bacteria are documented sources of bacteremia in HIV-infected subjects with advanced disease (37, 39), it is possible that they assume an important but overlooked pathologic role outside the setting of overt bacteremia. Within all HIV-infected subjects that we studied (n=24), the composition of this community correlated with markers of disease progression and correlates of morbidity and mortality in HIV-infected populations (11, 16–18, 53), namely, higher levels of T cell activation in the blood and gut as well as higher levels of tryptophan metabolism and elevated plasma markers of inflammation. Strikingly, when considering aviremic subjects on effective HAART (n=18), correlations remained evident between dysbiosis and two immunologic correlates of disease, production of kynurenine and plasma levels of the inflammatory cytokine IL-6, suggesting a relationship between the composition of the gut microbial community and markers of disease outcomes even in treated subjects. Finally, numerous bacterial taxa of the DMC that were enriched in untreated HIV infection were found to encode the enzymatic machinery that performs the same catabolic function as human IDO1. Subsequent in vitro experiments confirmed the capacity of a model member of this community in the genus Pseudomonas to transform tryptophan to kynurenine.
While the mechanistic relationships defining the observed associations are not fully understood, a complex crosstalk between gut immune states and the microbiome in the context of HIV disease progression and treatment is tenable. Previous studies in the mouse support the idea that HIV-associated gut inflammation may contribute to the observed dysbiotic states (22–24, 42, 54). Gut inflammation may induce changes in nutrient microenvironments via production of oxygen and nitrogen radicals that allow outgrowth of facultative anaerobes such as members of the Enterobacteriaceae family (24, 42). While our current investigation did not find correlations between enzymes responsible for reactive oxygen and nitrogen species production and dysbiosis, we were limited by measurements of enzyme expression in whole tissue as opposed to direct measurement of enzymatic endproducts or measurement of enzyme expression in specific cell lineages (e.g. tissue macrophages). Perturbations to gut immune barrier function such as disruptions in microbial pattern recognition sensors have also been found to result in the outgrowth of dysbiotic communities in mice (26, 27). Numerous cells that participate in microbial recognition and response are dysregulated during progressive HIV/SIV disease, including dendritic cells (55), macrophages (56), Th17 cells (57), enterocytes (58), and Paneth cells (59). Such cells are critical regulators of gut microbial community composition and their disruption may allow for outgrowth of the dysbiotic community described herein.
While gut inflammation may well engender dysbiosis, murine studies also suggest that the dysbiotic communities that result from inflammation and immune barrier defects can reciprocally act to stimulate chronic inflammation in the gut and systemic circulation (21, 25–27). Although bacterial species that can directly alter or augment IDO1 activity, T cell differentiation, or gut inflammation in humans have not yet been identified, analogous immunomodulatory relationships are likely to exist between gut bacteria and human hosts. If so, the capacity for the mucosal immune system to regulate the abundance of such immunomodulatory species may be impaired during HIV disease. Indeed, we find that HIV-infected subjects are likely to harbor a community of enteropathogenic bacteria that can catabolize tryptophan into immunomodulatory kynurenine derivatives known to correlate with disease progression and thought to contribute to mucosal immune disruption (9, 15). Although our 16S rRNA-based profiling cannot directly confirm the functional capacity of this community, existing bacterial genome data suggest that members of the phylogenetic groups described herein indeed possess the functional capacity to produce kynurenine. We show here that a model member of this community, a strain from within the genus bearing strongest correlation to Kyn:Trp catabolism (Pseudomonas), is indeed able to catabolize tryptophan to kynurenine. Furthermore, members of this disease-associated community (including Pseudomonas, Xanthomonas, Bacillus, and Burkholderia spp.) have been shown to utilize kynurenine pathway catabolites as growth substrates (50, 60–64). In aggregate, these observations suggest that the outgrowth of this community may be driven by the persistent host IDO1 activity found during chronic HIV infection and may in turn exacerbate IDO1-mediated immune disruption by accelerating tryptophan catabolism along the kynurenine pathway. If so, the potential exists for a self-amplifying cycle of dysbiosis and inflammation. A re-envisioning of the pathogenic mechanisms associated with HIV disease progression may thus include dysregulation of the balance between homeostasis-promoting versus gut barrier-disrupting microbes, contributing to weakened mucosal immune function, microbial translocation, and subsequent chronic inflammation. This process may further select for functionally distinct, enteropathogen-enriched communities in the gut mucosa that aggravate these immunologic phenomena. Intriguingly, the one HIV-infected long-term non-progressor assayed in this study (classified as such by a stable peripheral blood CD4+ count despite over 21 years of untreated infection) revealed a gut community similar to that of uninfected subjects, presenting the interesting possibility that non-progressive disease may be linked to host-microbiome homeostasis.
Notably, a potentially detrimental host-microbiome interaction in the setting of HIV disease is not always reversed by effective antiretroviral treatment. Indeed, HAART subjects exhibited higher relative abundances of the identified disease-associated microbial community than did uninfected controls, and those with the greatest degrees of dysbiosis were characterized by elevated markers of chronic inflammation and disease progression, i.e., high levels of kynurenine production and of the plasma inflammatory marker IL-6 (17). Previous findings support the possibility that the pathobiont community of Enterobacteriaceae enriched in untreated infection can indeed potentiate immunologic disruptions similar to those found in HIV disease (21, 23–25). Bacteroides genus members, which can protect mice from colitis (65) and that are notably depleted in sufferers of inflammatory bowel disease (40) and asthma (66), were similarly found in relative depletion among our cohort of HIV-infected untreated subjects.
Finally, we present data suggesting that a diverse community of gut-resident bacteria may augment IDO1-mediated mucosal disruption by direct catabolism of tryptophan through the kynurenine pathway. Targeting these microbial communities may present new avenues for therapeutic interventions aimed at preventing or reversing HIV-associated immunopathology. Indeed, recent reports of trials using agents that modulate the composition of the microbial community in HIV-infected populations or SIV-infected macaques (e.g., wtih antibiotics (67), probiotics (68–70), and prebiotics (71)) have shown improvements in clinical outcomes as measured by CD4+ T cell recovery, reduced mortality, and decreased markers of microbial translocation. However, microbial population dynamics during HIV disease progression remain poorly understood and efforts at developing therapeutics targeting the microbiome will certainly be augmented by a greater understanding of the host-microbiome relationship in HIV-infected subjects. Further efforts to determine the mechanistic processes underlying the complex relationships between the gut microbiome and its host will be critical toward developing the potential of gut microbial community modulation as a therapeutic strategy, and may lead to new approaches for the management of HIV disease as well as of other chronic inflammatory conditions.
MATERIALS & METHODS
Study design
The primary aims of the present study were to understand whether differences in mucosal-adherent bacterial communities exist between HIV-infected untreated, HIV-infected on HAART, or HIV-uninfected subjects, and whether such differences are associated with immunological perturbations characteristic of HIV disease. To accomplish these aims, microarray-based microbial community profiling was employed and assessment of immunologic status of subjects was performed using clinical data, flow cytometry, ELISA, liquid chromatography-tandem mass spectrometry, and quantitative polymerase chain reaction. The investigators were not blinded to serostatus nor to viral load measurements, medication history, or peripheral blood CD4+ T cell counts. The control subjects were seronegative risk-matched men who had a history of having sex with men (MSM) and/or with histories of exposure to HIV from interactions with seropositive partners. The initial cohort of HIV-infected subjects on HAART comprised HIV-infected patients who had maintained undetectable viral loads on stable antiretroviral regimens for at least one year, with equal numbers of individuals with peripheral blood CD4+ T cell counts > 500 cells/mm3 and with CD4+ T cell counts < 350 cells/mm3, though CD4+ T cell counts at times of sample collection spanned the range between these benchmark CD4+ T cell counts. All subjects were included that met the criterion of having had no recorded antibiotics usage within two months of mucosal sampling, regardless of peripheral blood CD4+ T cell count, and the final list of subjects included nearly equivalent numbers of subjects with < 350 CD4+ T cells/mm3 and > 500 CD4+ T cells/mm3. Triplicate 16S amplicon hybridizations using separate G3 PhyloChip microarrays (Second Genome, Inc.) were performed using mucosal biopsy samples from two different study subjects. All triplicates were found to cluster closest together in a global hierarchical clustering analysis based on a Canberra community dissimilarity matrix.
Subject sample collection
All participants gave written informed consent for research sigmoidoscopy and biopsies, a protocol approved by the Committee on Human Research, UCSF. Study participants underwent a blood draw and received a Fleet enema prior to the procedure, and rectosigmoid biopsies (each ~3 mm in diameter) were obtained between 10 and 20 cm from the anus using jumbo forceps. Four biopsies were formalin-fixed and paraffin-embedded for immunohistochemistry; 12 biopsies were placed immediately in 15 ml of RPMI 1640 with 15% fetal calf serum, and processed for flow cytometric analysis within 4 hours of collection; two biopsies were placed in RNAlater (Life Technologies) and frozen at −80°C for quantitation of inflammatory gene expression; finally, another two allocations of two biopsies each were snap-frozen in liquid N2 and stored at −80°C for HIV viral nucleic acid quantitation and bacterial community profiling by G3 PhyloChip microarray.
Nucleic acid extraction and quantitative polymerase chain reaction (qPCR)
For quantitation of inflammatory and interferon-stimulated gene expression, RNA was extracted from snap-frozen biopsies frozen in RNAlater (Life Technologies) after homogenization in Trizol (Life Technologies) and purification using the RNeasy mini-kit (Qiagen). RNA was also prepared from whole blood, via collection using the PaxGene Blood RNA Kit (Qiagen). cDNA was prepared from RNA using Omniscript RT Kit (Qiagen) using random hexamer primers. qPCR was carried out using TaqMan Gene Expression MasterMix (Applied Biosystems) in a StepOne Plus Real Time PCR system (Applied Biosystems). IDO1 transcript was measured using custom primers and probe (F: 5’- GCCAAATCCACAGGAAAATCT, R: 5’- GCTGTGACTTGTGGTCTGTGA, P: 5’- AAACATCTGCCTGATCTCATAGAGT). Interferon stimulated genes (ISG) were measured using commercially available primer-probe sets from Applied Biosystems: GBP1 Hs00977005_m1, IFI27 HSs00271467_m1, MX1 Hs00895601_m1, OAS1 Hs00973637_m1. ISG expression was consolidated using the geometric mean of relative expression values for all four aforementioned genes. Quantitation of expression of inflammatory genes (NADPH oxidase [CYBB subunit], arginase 1, nitric oxide synthase 2, type I interferons [IFN-α, IFN-β], type II interferon [IFN-γ], interferon-stimulated gene expression, tumor necrosis factor-alpha) was performed using the Fluidigm platform, with gene-specific pre-amplification using proprietary computationally designed primers (DELTAgene from Fluidigm). Relative expression was calculated using the ΔΔCT method, using HPRT as housekeeping gene.
Total bacterial burden quantification was carried out using DNA extracted from separate snap-frozen rectosigmoid biopsies that were collected at the same time as those above. Biopsies were resuspended in RNAlater ICE (Life Technologies), drained, and transferred to Lysis Matrix B (MP Biomedicals) tubes containing 600 ul RLT buffer and subjected to 30 seconds of bead beating at 5.5 m/s. Samples were briefly spun down and DNA was extracted using the AllPrep Kit (Qiagen). TaqMan qPCR was performed, as above, using the following primers: P891F, 5’-TGGAGCATGTGGTTTAATTCGA; P1033R, 5’-TGCGGGACTTAACCCAACA, and UniProbe 5’-CACGAGCTGACGACARCCATGCA. A standard curve of known 16S rRNA copy numbers was included with each run and was used to calculate total bacterial burden normalized to total 16S rRNA genes. Enterobacteriaceae were quantitated using Brilliant II SYBR Green (Agilent Technologies) and the following primers: F: 5’- ATGGCTGTCGTCAGCTCGT; R: 5’- CCTACTTCTTTTGCAACCCACTC (72). Bacteroides fragilis was also quantified using Brilliant II SYBR Green, and the following primers: F:5’- GAAAGCATTAAGTATTCCACCTG; R: 5’- CGGTGATTGCTCACTGACA (73).
T cell phenotyping analysis by flow cytometry
Mononuclear cells were prepared from peripheral blood by density gradient centrifugation and isolation using Ficoll-Paque (STEMCELL Technologies). Rectosigmoid biopsy mononuclear cells were isolated from biopsy specimens using a protocol optimized for lymphocyte viability and yield (74). Biopsy pieces were digested in three rounds of 0.5 mg/mL collagenase type II (Sigma-Aldrich), after which tissue was disrupted with a syringe bearing a 16-gauge blunt-end needle and passaged through a 70mm cell strainer.
Cells were then stained directly or after phorbol 12-myristate 13-acetate (PMA)/ionomycin stimulation in the presence of brefeldin and monensin for surface marker expression, and then permeabilized and stained intracellularly for FoxP3 or cytokine production using the FoxP3 Fixation/Permeabilization Staining Set (eBiosciences) or Cytofix/Cytoperm (BD Biosciences), respectively. Acquisition was performed on a Becton Dickinson LSRII (BD Biosciences) and analyzed using FlowJo software, version 9 (Treestar). Naive T cells were identified as CD27+ cells lacking CD45RO expression, and were excluded to yield total memory T cells. Proportions of activated CD4+ and CD8+ T cell were measured by the fraction of these cell populations staining positive for HLA-DR and CD38. Proportions of regulatory T cells (Treg) from total CD4+ cells were determined by co-staining of CD25 and FoxP3.
Immunohistochemistry and viral load quantification in rectosigmoid tissues
Four rectosigmoid biopsies were collected into 4% paraformaldehyde, embedded in paraffin, cut into 5 µm sections and mounted on glass slides. Slides were deparaffinized in Clear-Rite 3 and rehydrated in an ethanol series, and antigenic epitopes were then retrieved by placing slides in a pressure cooker for 20 minutes at 121°C in pH6 citrate buffer. Slides were blocked and stained with polyclonal rabbit anti-CD3 (DAKO), and stain developed with DAB chromogen (anti-rabbit Polink-2 Plus HRP Detection Kit, GBI Labs). Whole slide composite images were acquired on a ScanScope (Aperio) and analyzed with the accompanying software to quantify CD3 stained cells per area of tissue. To determine the number of CD4+ cells per area, the CD3+ cells/mm2 value was multiplied by the fraction of CD3+ cells that stained with CD4 as determined by flow cytometry. Two rectosigmoid biopsies were snap-frozen in liquid N2 for quantification of HIV DNA and RNA as previously described (75).
Plasma inflammatory marker measurements
Plasma was collected from peripheral blood by centrifugation and stored at −80°C. The following soluble plasma proteins were measured by ELISA using commercially available kits and according to the manufacturer’s instructions: IP-10, sTNF-RII, IL-6, FABP2, sCD14 (all from R&D Systems), zonulin (Immundiagnostik), D-dimer (Diagnostica Stago). Concentrations of kynurenine, tryptophan, and 3-hydroxyanthranilic acid were quantitated in plasma and in bacterial growth cultures by liquid chromatography-tandem mass spectrometry, as previously described (9).
16S rRNA amplicon preparation, Phylochip hybridization, scanning, and data normalization
Nucleic acid extraction was performed on snap-frozen rectosigmoid biopsies by re-suspending in RNAlater ICE (Life Technologies), according to the manufacturer’s protocol and as previously described (76). G3 PhyloChip microarrays were obtained from Second Genome, Inc. PCR amplification of the bacterial 16S rRNA gene, Phylochip hybridization, Phylochip scanning, and fluorescence intensity normalization and initial probeset filtering of Phylochip data were performed as previously described (29), with the exception of performing 25 cycles of 16S PCR amplification rather than 30 and the usage of customized quartile ‘r score’ cutoff values. For each 16S rRNA gene probeset, an r score was independently calculated that incorporated mismatch probe fluorescence intensity as well as distribution of total chip fluorescence to account for per-chip background fluorescence. Quartile r score cutoffs were chosen based on r scores retrieved for control probes spiked-in at known concentrations, as recorded from all Phylochip microarrays of the current study, and were as follows: rQ1 > .30, rQ2 > .65, rQ3 > .88.
Phylochip analysis
Following probe intensity normalization and to reduce the dataset for analyses, probesets belonging to phylogenetically related (based on species-level identity) 16S rRNA sequences and bearing similar probeset response patterns across all samples were represented by a single taxon, while others were eliminated from the dataset. This process was performed by calculating the variance in fluorescence intensity for each probeset and its two most phylogenetically related neighbors, and was repeated for each taxon across multiple different samples; the sum of all variances was used to define groups of taxa whose probe sets responded similarly. The resulting measures were ranked to select for removal the top ~15% of probesets exhibiting similar response trends across all samples, allowing reduction of redundant probesets. Next, the ‘vegan’ package within the R programming environment was used to perform non-metric multidimensional scaling (NMDS) based on community dissimilarity using the Canberra dissimilarity metric. The ‘Adonis’ package was used to estimate strength of relationship between community dissimilarity and classification of subjects into either VU or HIV-uninfected. To identify bacterial taxa in differential abundance among viremic untreated and HIV- subjects, the software package Statistical Analysis of Microarrays (SAM (32)), which incorporates the Storey (77) method of false discovery rate estimation in the form of adjusted q-values, was used. Taxa were filtered down to a final list of 547 taxa by the following criteria: q-value < 0.05, raw MFI fold change > 2, and Mann-Whitney U test P value < 0.03. Genera to which belonged taxa within the final list were visualized using the Interactive Tree of Life (78, 79) (iTOL, http://itol.embl.de/).
Comparisons of disease-associated microbial genera to immunologic parameters
The R software package ‘multtest’ was used to calculate Spearman rho correlation coefficient (RS) values for comparisons of all 562 bacterial taxa that comprise the disease-associated microbial community (DMC) to each of the 24 immunologic parameters measured. Measures of correlations between immunologic variables and taxa were aggregated and condensed to genus-level classifications for visualization purposes in Fig. 3A, using the following methodology: for bacterial genera that were represented by more than one taxon in the DMC, RS values representing correlations between taxa belonging to the same genus and an immunologic parameter were averaged to yield a RSave that represents the correlation between relative abundance of that genus and each immune parameter. This was performed on the remaining immunologic parameters and all genera represented by more than one taxon, and was visualized using Cluster3 and Java TreeView (Version 1.1.6r2). To represent unadjusted significance values for each correlation shown in Fig. 3A, boxes were made to surround each correlation for which unadjusted P < 0.02. For genera that were represented by more than one taxon in the DMC, the median unadjusted P value for comparisons among immunologic parameters and taxa within that genus was used. Choice of mean for computing aggregated RS and median for aggregated unadjusted P values for each genus was informed by assessment of global RS and P value distributions (normal and non-normal, respectively, Fig. S9).
Principal component analysis
To quantify trends in co-variance among taxa that have similar patterns in relative abundance across all subject samples, a principal component analysis (PCA) was performed within the R programming environment (package ‘prcomp’). This allowed the condensation of trends in co-varying microbial abundances into discrete variables, thereby yielding single measures that represented broad trends in abundance of multiple taxa. To reduce the number of relationships tested, Spearman rank correlations with immunologic parameters were calculated only for the principal component that represented the greatest portion of co-variance (PC1), and the resultant P values were adjusted for multiple comparisons by the Benjamini-Hochberg technique (80).
Multiple linear regression model selection analysis
Among treated subjects, associations were confirmed using linear regression, allowing for the inclusion of a variety of adjustment variables comprising age, race, antibiotic use, and duration of HAART, as well as the first five principal components representing the greatest proportion of microbial abundance co-variance (PC1, PC2, PC3, PC4, PC5). All possible models were fit and compared according to their predictive accuracy. Predictive accuracy was defined as the mean squared error (MSE) calculated from leave-one-out cross-validation (LOO-CV). Under this approach, the model is fit repeatedly, leaving out one observation each time in order to assess a given model’s ability to predict new observations. For each outcome, the model with the lowest LOO-CV MSE was chosen. Given the small number of observations available, p-values were then obtained by permutation of residuals (81).
Analysis of kynurenine pathway enzyme annotations and associations with plasma Kyn:Trp ratios
The UniProt Consortium database (52) (www.uniprot.org) was queried for all annotated bacterial homologs of enzymes within the pathway to catabolize tryptophan to 3-hydroxyanthranilic acid (Enzyme Commission numbers: EC 1.13.11.11, EC 3.5.1.9, EC 3.7.1.3, EC 1.14.13.9). Phylum classifications for each annotation were combined, counted, and represented as pie charts with colors representing phyla as denoted in legend of Fig. 5A. ‘Totals’ represent numbers of unique genera within the domain Bacteria that bear annotated homologs in the highly curated and comprehensive UniProt database for each respective enzyme at the time of publication.
Spearman correlation P values were calculated for each taxon in relation to subject plasma Kyn:Trp ratios. For Fig. 5C, each genus was ranked based on P values of the taxon with the strongest correlation (as defined by the taxon with the lowest P value within a given genus). Annotations for tryptophan catabolism enzymes for all sequenced species and strains within each genus were considered when counting numbers of tryptophan catabolism enzymes for each genus.
Pseudomonas fluorescens kynurenine production in bacterial cultures
Pseudomonas fluorescens strain A506 (ATCC 31948) was grown in King’s B Medium (82) within U-bottomed 96 well polystyrene culture plates at an initial inoculum of 2.85 × 107 cfu per well. King’s B Medium was prepared using Bacto Proteose Peptone No. 3 which, according to manufacturer-provided information, would contain 0.3 mM in the King’s B Medium preparation. Thus, bacterial cultures contained the free tryptophan already present plus 0, 0.5, 1.0, and 5.0 mM tryptophan added exogenously. Cultures were grown at room temperature and, at 20 hours post inoculation, were spun at 12,000 rpm for 10 minutes in a tabletop microcentrifuge. Supernatants were filtered through a 0.22 µm filter and kanamycin was added to a final concentration of 50 µg/ml. Liquid chromatography-tandem mass spectrometry was used to assay concentrations of kynurenine and tryptophan in growth culture supernatants as described previously (9). Three temporally independent biological replicates were assessed for each condition.
Supplementary Material
Acknowledgements
We would like to acknowledge S. Yukl for HIV viral load measurements, M. Krone for cohort characteristics data, M. Segal for statistical analysis consultation, and J. Robinson, B. Clagett, S. Sirdeshmukh and D. Dorazio for assistance in plasma inflammatory marker analyses. We would like to additionally thank P. Baum, W. Yonemoto, J. Mar, E. Krow-Lucal, M. Lowe, A. Chitre, C. Baker, B. Kanwar, T. Burt, N. Nagalingam, K. Fujimura, M. Rauch, and S. Galang for helpful discussion.
Funding: This study was funded by NIH NRSA 5T32AI007334-23, NIH U19 AI96109, NSF 1144247, NIH UCSF-GIVI Center for AIDS Research P30-AI027763, and by the Harvey V. Berneking Living Trust. JMM is a recipient of the NIH Director’s Pioneer Award Program, part of the NIH Roadmap for Medical Research, through grant number DPI OD00329. Additional support was provided by the American Foundation for AIDS Research (107854-48-RGRL to JMM and SGD), the Hurlbut-Johnson Fund administered by the AIDS Research Institute at UCSF (to RMD), R37 AI040312 (to JMM), K23 CA157929 (to MS), F32 AI091534 (to RMD), the California HIV/AIDS Research Program (ID09-SF-067 to PWH), and the UCSF Clinical and Translational Science Institute (RR024131-01).
Footnotes
SUPPLEMENTARY MATERIALS
Figure S1: Gross bacterial community metrics do not differ significantly across subject groups or intra-subject samples.
Figure S2: Comparisons of quantitative PCR (qPCR) measurements and PhyloChip relative abundance measurements [mean fluorescence intensity (MFI)] for selected taxa of interest.
Figure S3: Taxon and immunologic parameter identities for each variable in correlative analyses relating DMC taxa and immunopathologic markers of HIV disease among all HIV-infected subjects.
Figure S4: Principal component analysis characteristics.
Figure S5: Highly active antiretroviral therapy causes a shift in DMC composition toward the healthy, uninfected state in two subjects.
Figure S6: Comparison of adjustment variables (antibiotics usage, days on HAART, age) to PC1.
Figure S7: Relationships between IDO1 expression, IDO1 activity, T cell differentiation, and type II interferon.
Figure S8: Bacterial taxa that predict plasma Kyn:Trp ratios in a multivariate machine learning analysis preferentially encode genetic homologs of tryptophan catabolism enzymes involved in the kynurenine pathway.
Figure S9: Data distribution of RS and P values for all taxa as compared to all immunologic variables in Spearman correlation tests.
Table S1. Patient cohort data.
Table S2: DMC taxon phylogenetic classifications and statistical analyses.
Table S3: Spearman correlations between individual taxa of the DMC and markers of disease progression, as calculated among all HIV-infected subjects.
Table S4: Spearman correlations between PC1 and immunologic variables among all HIV-infected subjects.
Table S5: Spearman correlations between PC1 and immunologic variables among HIV+ subjects on HAART.
Table S6: Comparisons of PC1 marker of dysbiosis to gut inflammatory gene expression in whole rectosigmoid biopsy specimens.
Table S7: Spearman correlation to Kyn:Trp genus ranks, and analysis of kynurenine pathway metabolic enzymes (those involved in metabolism of tryptophan to 3-hydroxyanthranilic acid) in bacteria.
Author Contributions: IVC, RMD, MJB, SVL, and JMM designed experiments. IVC and SI performed PhyloChip experiments. IVC, MCM, RDH, and SI performed PhyloChip analysis. RMD and RA performed flow cytometry and analysis. MS, PWH, and SGD recruited subjects and obtained human samples. YH and ML provided plasma inflammatory marker measurements. IVC, RMD, JMM, and SVL wrote the manuscript.
Competing interests: SVL is a member of the Scientific Advisory Board of Second Genome, Inc. The remaining authors declare they have no competing interests.
Data and materials availability: PhyloChip data used in this study are available at the Gene Expression Omnibus web site (https://www.ncbi.nlm.nih.gov/geo/) under accession number: GSE43746.
REFERENCES
- 1.Giorgi JV, Hultin LE, McKeating JA, Johnson TD, Owens B, Jacobson LP, Shih R, Lewis J, Wiley DJ, Phair JP, Wolinsky SM, Detels R. Shorter Survival in Advanced Human Immunodeficiency Virus Type 1 Infection Is More Closely Associated with T Lymphocyte Activation than with Plasma Virus Burden or Virus Chemokine Coreceptor Usage. Journal of Infectious Diseases. 1999;179:859–870. doi: 10.1086/314660. [DOI] [PubMed] [Google Scholar]
- 2.Sousa AE, Carneiro J, Meier-Schellersheim M, Grossman Z, Victorino RMM. CD4 T cell depletion is linked directly to immune activation in the pathogenesis of HIV-1 and HIV-2 but only indirectly to the viral load. The Journal of Immunology. 2002;169:3400–3406. doi: 10.4049/jimmunol.169.6.3400. [DOI] [PubMed] [Google Scholar]
- 3.Deeks SG, Kitchen CM, Liu L, Guo H, Gascon R, Narváez AB, Hunt P, Martin JN, Kahn JO, Levy J, McGrath MS, Hecht FM. Immune activation set point during early HIV infection predicts subsequent CD4+ T-cell changes independent of viral load. Blood. 2004;104:942–947. doi: 10.1182/blood-2003-09-3333. [DOI] [PubMed] [Google Scholar]
- 4.Estes JD, Gordon SN, Zeng M, Chahroudi AM, Dunham RM, Staprans SI, Reilly CS, Silvestri G, Haase AT. Early resolution of acute immune activation and induction of PD-1 in SIV-infected sooty mangabeys distinguishes nonpathogenic from pathogenic infection in rhesus macaques. The Journal of Immunology. 2008;180:6798–6807. doi: 10.4049/jimmunol.180.10.6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Estes JD, Harris LD, Klatt NR, Tabb B, Pittaluga S, Paiardini M, Barclay GR, Smedley J, Pung R, Oliveira KM, Hirsch VM, Silvestri G, Douek DC, Miller CJ, Haase AT, Lifson J, Brenchley JM. Damaged intestinal epithelial integrity linked to microbial translocation in pathogenic simian immunodeficiency virus infections. PLoS Pathog. 2010;6:e1001052. doi: 10.1371/journal.ppat.1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, Kazzaz Z, Bornstein E, Lambotte O, Altmann D, Blazar BR, Rodriguez B, Teixeira-Johnson L, Landay A, Martin JN, Hecht FM, Picker LJ, Lederman MM, Deeks SG, Douek DC. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 7.Sandler NG, Wand H, Roque A, Law M, Nason MC, Nixon DE, Pedersen C, Ruxrungtham K, Lewin SR, Emery S, Neaton JD, Brenchley JM, Deeks SG, Sereti I, Douek DC. INSIGHT SMART Study Group, Plasma levels of soluble CD14 independently predict mortality in HIV infection. J Infect Dis. 2011;203:780–790. doi: 10.1093/infdis/jiq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raffatellu M, Santos RL, Verhoeven DE, George MD, Wilson RP, Winter SE, Godinez I, Sankaran S, Paixao TA, Gordon MA, Kolls JK, Dandekar S, Bäumler AJ. Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat Med. 2008;14:421–428. doi: 10.1038/nm1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Favre D, Mold J, Hunt PW, Kanwar B, Loke P, Seu L, Barbour JD, Lowe MM, Jayawardene A, Aweeka F, Huang Y, Douek DC, Brenchley JM, Martin JN, Hecht FM, Deeks SG, McCune JM. Tryptophan catabolism by indoleamine 2,3-dioxygenase 1 alters the balance of TH17 to regulatory T cells in HIV disease. Sci Transl Med. 2010;2:32ra36. doi: 10.1126/scitranslmed.3000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuchs D, Forsman A, Hagberg L, Larsson M, Norkrans G, Reibnegger G, Werner ER, Wachter H. Immune activation and decreased tryptophan in patients with HIV-1 infection. Journal of interferon research. 1990;10:599–603. doi: 10.1089/jir.1990.10.599. [DOI] [PubMed] [Google Scholar]
- 11.Huengsberg M, Winer JB, Gompels M, Round R, Ross J, Shahmanesh M. Serum kynurenine-to-tryptophan ratio increases with progressive disease in HIV-infected patients. Clinical chemistry. 1998;44:858–862. [PubMed] [Google Scholar]
- 12.Hunt PW, Rodriguez B, Shive C. Gut epithelial barrier dysfunction, inflammation, and coagulation predict higher mortality during treated HIV/AIDS; 19th Conference on Retroviruses and Opportunistic Infections; 2012. [Google Scholar]
- 13.Desvignes L, Ernst JD. Interferon-gamma-responsive nonhematopoietic cells regulate the immune response to Mycobacterium tuberculosis. Immunity. 2009;31:974–985. doi: 10.1016/j.immuni.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Romani L, Fallarino F, De Luca A, Montagnoli C, D'Angelo C, Zelante T, Vacca C, Bistoni F, Fioretti MC, Grohmann U, Segal BH, Puccetti P. Defective tryptophan catabolism underlies inflammation in mouse chronic granulomatous disease. Nature. 2008;451:211–215. doi: 10.1038/nature06471. [DOI] [PubMed] [Google Scholar]
- 15.Sandler NG, Douek DC. Microbial translocation in HIV infection: causes, consequences and treatment opportunities. Nat Rev Microbiol. 2012;10:655–666. doi: 10.1038/nrmicro2848. [DOI] [PubMed] [Google Scholar]
- 16.Hunt PW, Martin JN, Sinclair E, Bredt B, Hagos E, Lampiris H, Deeks SG. T cell activation is associated with lower CD4+ T cell gains in human immunodeficiency virus-infected patients with sustained viral suppression during antiretroviral therapy. Journal of Infectious Diseases. 2003;187:1534–1543. doi: 10.1086/374786. [DOI] [PubMed] [Google Scholar]
- 17.Kalayjian RC, Machekano RN, Rizk N, Robbins GK, Gandhi RT, Rodriguez BA, Pollard RB, Lederman MM, Landay A. Pretreatment levels of soluble cellular receptors and interleukin-6 are associated with HIV disease progression in subjects treated with highly active antiretroviral therapy. J Infect Dis. 2010;201:1796–1805. doi: 10.1086/652750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sauce D, Larsen M, Fastenackels S, Pauchard M, Ait-Mohand H, Schneider L, Guihot A, Boufassa F, Zaunders J, Iguertsira M, Bailey M, Gorochov G, Duvivier C, Carcelain G, Kelleher AD, Simon A, Meyer L, Costagliola D, Deeks SG, Lambotte O, Autran B, Hunt PW, Katlama C, Appay V. HIV disease progression despite suppression of viral replication is associated with exhaustion of lymphopoiesis. Blood. 2011;117:5142–5151. doi: 10.1182/blood-2011-01-331306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, Gordon JI, Onderdonk AB, Glimcher LH. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 23.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, Dougan G, von Mering C, Hardt WD. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Bäumler AJ. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, Glickman JN, Glimcher LH. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elinav E, Strowig T, Henao-Mejia J, Flavell RA. Regulation of the antimicrobial response by NLR proteins. Immunity. 2011;34:665–679. doi: 10.1016/j.immuni.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 27.Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, Thaiss CA, Kau AL, Eisenbarth SC, Jurczak MJ, Camporez JP, Shulman GI, Gordon JI, Hoffman HM, Flavell RA. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nava GM, Friedrichsen HJ, Stappenbeck TS. Spatial organization of intestinal microbiota in the mouse ascending colon. ISME J. 2011;5:627–638. doi: 10.1038/ismej.2010.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hazen TC, Dubinsky EA, DeSantis TZ, Andersen GL, Piceno YM, Singh N, Jansson JK, Probst A, Borglin SE, Fortney JL, Stringfellow WT, Bill M, Conrad ME, Tom LM, Chavarria KL, Alusi TR, Lamendella R, Joyner DC, Spier C, Baelum J, Auer M, Zemla ML, Chakraborty R, Sonnenthal EL, D'haeseleer P, Holman HY, Osman S, Lu Z, Van Nostrand JD, Deng Y, Zhou J, Mason OU. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science. 2010;330:204–208. doi: 10.1126/science.1195979. [DOI] [PubMed] [Google Scholar]
- 30.DeSantis TZ, Brodie EL, Moberg JP, Zubieta IX, Piceno YM, Andersen GL. High-density universal 16S rRNA microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microb Ecol. 2007;53:371–383. doi: 10.1007/s00248-006-9134-9. [DOI] [PubMed] [Google Scholar]
- 31.Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, McIntosh ML, Alsam A, Kirkwood KL, Lambris JD, Darveau RP, Curtis MA. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011;10:497–506. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y, Parameswaran P, Crowell MD, Wing R, Rittmann BE. Human gut microbiota in obesity and after gastric bypass. Proceedings of the National Academy of Sciences. 2009;106:2365–2370. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, Didonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WH, Bushman FD, Lusis AJ, Hazen SL. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013 doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ellis CL, Ma ZM, Mann SK, Li CS, Wu J, Knight TH, Yotter T, Hayes TL, Maniar AH, Troia-Cancio PV, Overman HA, Torok NJ, Albanese A, Rutledge JC, Miller CJ, Pollard RB, Asmuth DM. Molecular characterization of stool microbiota in HIV-infected subjects by panbacterial and order-level 16S ribosomal DNA (rDNA) quantification and correlations with immune activation. J Acquir Immune Defic Syndr. 2011;57:363–370. doi: 10.1097/QAI.0b013e31821a603c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tumbarello M, Tacconelli E, Caponera S, Cauda R, Ortona L. The impact of bacteraemia on HIV infection. Nine years experience in a large Italian university hospital. Journal of Infection. 1995;31:123–131. doi: 10.1016/s0163-4453(95)92110-9. [DOI] [PubMed] [Google Scholar]
- 38.Bouza E, Rodriguez-Créixems M. Bacteremic infections in the HIV-infected patient and recurrent bacteremia. Clinical microbiology and infection. 1999;5:2s33–2s39. [Google Scholar]
- 39.Manfredi R, Calza L, Chiodo F. Enteric and disseminated Campylobacter species infection during HIV disease: a persisting but significantly modified association in the HAART era. The American journal of gastroenterology. 2002;97:510–511. doi: 10.1111/j.1572-0241.2002.05522.x. [DOI] [PubMed] [Google Scholar]
- 40.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proceedings of the National Academy of Sciences. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kamat A, Misra V, Cassol E, Ancuta P, Yan Z, Li C, Morgello S, Gabuzda D. A plasma biomarker signature of immune activation in HIV patients on antiretroviral therapy. PLoS One. 2012;7:e30881. doi: 10.1371/journal.pone.0030881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, Laughlin RC, Gomez G, Wu J, Lawhon SD, Popova IE, Parikh SJ, Adams LG, Tsolis RM, Stewart VJ, Bäumler AJ. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013;339:708–711. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rivera-Chávez F, Monack DM, Winter SE, Lopez CA, Xavier MN, Winter MG, Nuccio S-P, Russell JM, Laughlin RC, Lawhon SD, Sterzenbach T, Bevins CL, Tsolis RM, Harshey R, Adams LG, Bäumler AJ. Salmonella Uses Energy Taxis to Benefit from Intestinal Inflammation. PLoS Pathogens. 2013;9:e1003267. doi: 10.1371/journal.ppat.1003267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Picard RR, Cook RD. Cross-validation of regression models. Journal of the American Statistical Association. 1984;79:575–583. [Google Scholar]
- 45.Mezrich JD, Fechner JH, Zhang X, Johnson BP, Burlingham WJ, Bradfield CA. An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J Immunol. 2010;185:3190–3198. doi: 10.4049/jimmunol.0903670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujigaki S, Saito K, Sekikawa K, Tone S, Takikawa O, Fujii H, Wada H, Noma A, Seishima M. Lipopolysaccharide induction of indoleamine 2, 3-dioxygenase is mediated dominantly by an IFN-γ-independent mechanism. European journal of immunology. 2001;31:2313–2318. doi: 10.1002/1521-4141(200108)31:8<2313::aid-immu2313>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 47.Taylor MW, Feng GS. Relationship between interferon-gamma, indoleamine 2, 3-dioxygenase, and tryptophan catabolism. The FASEB Journal. 1991;5:2516–2522. [PubMed] [Google Scholar]
- 48.Lima WC, Varani AM, Menck CF. NAD biosynthesis evolution in bacteria: lateral gene transfer of kynurenine pathway in Xanthomonadales and Flavobacteriales. Mol Biol Evol. 2009;26:399–406. doi: 10.1093/molbev/msn261. [DOI] [PubMed] [Google Scholar]
- 49.Matthijs S, Baysse C, Koedam N, Tehrani KA, Verheyden L, Budzikiewicz H, Schäfer M, Hoorelbeke B, Meyer JM, De Greve H, Cornelis P. The Pseudomonas siderophore quinolobactin is synthesized from xanthurenic acid, an intermediate of the kynurenine pathway. Mol Microbiol. 2004;52:371–384. doi: 10.1111/j.1365-2958.2004.03999.x. [DOI] [PubMed] [Google Scholar]
- 50.Farrow JM, Pesci EC. Two distinct pathways supply anthranilate as a precursor of the Pseudomonas quinolone signal. J Bacteriol. 2007;189:3425–3433. doi: 10.1128/JB.00209-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kurnasov O, Goral V, Colabroy K, Gerdes S, Anantha S, Osterman A, Begley TP. NAD Biosynthesis: Identification of the Tryptophan to Quinolinate Pathway in Bacteria. Chemistry & Biology. 2003;10:1195–1204. doi: 10.1016/j.chembiol.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 52.UniProt Consortium, Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Giorgi JV, Hultin LE, McKeating JA, Johnson TD, Owens B, Jacobson LP, Shih R, Lewis J, Wiley DJ, Phair JP, Wolinsky SM, Detels R. Shorter Survival in Advanced Human Immunodeficiency Virus Type 1 Infection Is More Closely Associated with T Lymphocyte Activation than with Plasma Virus Burden or Virus Chemokine Coreceptor Usage. Journal of Infectious Diseases. 1999;179:859–870. doi: 10.1086/314660. [DOI] [PubMed] [Google Scholar]
- 54.Nagalingam NA, Kao JY, Young VB. Microbial ecology of the murine gut associated with the development of dextran sodium sulfate-induced colitis. Inflamm Bowel Dis. 2011;17:917–926. doi: 10.1002/ibd.21462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manches O, Munn D, Fallahi A, Lifson J, Chaperot L, Plumas J, Bhardwaj N. HIV-activated human plasmacytoid DCs induce Tregs through an indoleamine 2,3-dioxygenase-dependent mechanism. J Clin Invest. 2008;118:3431–3439. doi: 10.1172/JCI34823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Müller F, Rollag H, Frøland SS. Reduced oxidative burst responses in monocytes and monocyte-derived macrophages from HIV-infected subjects. Clinical & Experimental Immunology. 1990;82:10–15. doi: 10.1111/j.1365-2249.1990.tb05396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brenchley JM, Paiardini M, Knox KS, Asher AI, Cervasi B, Asher TE, Scheinberg P, Price DA, Hage CA, Kholi LM, Khoruts A, Frank I, Else J, Schacker T, Silvestri G, Douek DC. Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood. 2008;112:2826–2835. doi: 10.1182/blood-2008-05-159301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Somsouk M, Estes J, Hoh R, Gilman L, Martin J, Deeks S, Hunt P. Colonic Epithelial Cell Proliferation Is Elevated in Untreated HIV Infection and May Not Normalize During Suppressive ART; 19th Conference on Retroviruses and Opportunistic Infections, Seattle, Washington, Mar 5–8, 2012; 2012. Sess. 49. [Google Scholar]
- 59.Kelly P, Feakins R, Domizio P, Murphy J, Bevins C, Wilson J, McPhail G, Poulsom R, Dhaliwal W. Paneth cell granule depletion in the human small intestine under infective and nutritio nal stress. Clinical and Experimental Immunology. 2004;135:303–309. doi: 10.1111/j.1365-2249.2004.02374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Palleroni NJ, Stanier RY. Regulatory mechanisms governing synthesis of the enzymes for tryptophan oxidation by Pseudomonas fluorescens. Journal of general microbiology. 1964;35:319–334. doi: 10.1099/00221287-35-2-319. [DOI] [PubMed] [Google Scholar]
- 61.Rosenfeld H, Feigelson P. Synergistic and product induction of the enzymes of tryptophan metabolism in Pseudomonas acidovorans. Journal of bacteriology. 1969;97:697–704. doi: 10.1128/jb.97.2.697-704.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bouknight RR, Sadoff HL. Tryptophan catabolism in Bacillus megaterium. Journal of bacteriology. 1975;121:70–76. doi: 10.1128/jb.121.1.70-76.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chang K, Mohseni P, Zylstra J. Characterization and Regulation of the Genes for a Novel Anthranilate 1,2-Dioxygenase from Burkholderia cepacia DBO1. Journal of Bacteriology. 2003;185:5871–5881. doi: 10.1128/JB.185.19.5871-5881.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Davis D, Henderson LM, Powell D. The niacin-tryptophan relationship in the metabolism of Xanthomonas pruni. Journal of Biological Chemistry. 1951;189:543–549. [PubMed] [Google Scholar]
- 65.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 66.Björkstén B, Naaber P, Sepp E, Mikelsaar M. The intestinal microflora in allergic Estonian and Swedish 2-year-old children. Clinical and Experimental Allergy. 1999;29:342–346. doi: 10.1046/j.1365-2222.1999.00560.x. [DOI] [PubMed] [Google Scholar]
- 67.Walker AS, Ford D, Gilks CF, Munderi P, Ssali F, Reid A, Katabira E, Grosskurth H, Mugyenyi P, Hakim J. Daily co-trimoxazole prophylaxis in severely immunosuppressed HIV-infected adults in Africa started on combination antiretroviral therapy: an observational analysis of the DART cohort. Lancet. 2010;375:1278. doi: 10.1016/S0140-6736(10)60057-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anukam KC, Osazuwa EO, Osadolor HB, Bruce AW, Reid G. Yogurt containing probiotic Lactobacillus rhamnosus GR-1 and L. reuteri RC-14 helps resolve moderate diarrhea and increases CD4 count in HIV/AIDS patients. Journal of clinical gastroenterology. 2008;42:239–243. doi: 10.1097/MCG.0b013e31802c7465. [DOI] [PubMed] [Google Scholar]
- 69.Irvine SL, Hummelen R, Hekmat S, WN Looman C, Habbema JDF, Reid G. Probiotic yogurt consumption is associated with an increase of CD4 count among people living with HIV/AIDS. Journal of clinical gastroenterology. 2010;44:e201. doi: 10.1097/MCG.0b013e3181d8fba8. [DOI] [PubMed] [Google Scholar]
- 70.Klatt NR, Canary LA, Sun X, Vinton CL, Funderburg NT, Morcock DR, Quiñones M, Deming CB, Perkins M, Hazuda DJ. Probiotic/prebiotic supplementation of antiretrovirals improves gastrointestinal immunity in SIV-infected macaques. The Journal of Clinical Investigation. 2013:0. doi: 10.1172/JCI66227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gori A, Rizzardini G, Van't Land B, Amor KB, van Schaik J, Torti C, Quirino T, Tincati C, Bandera A, Knol J, Benlhassan-Chahour K, Trabattoni D, Bray D, Vriesema A, Welling G, Garssen J, Clerici M. Specific prebiotics modulate gut microbiota and immune activation in HAART-naive HIV-infected adults: results of the "COPA" pilot randomized trial. Mucosal Immunol. 2011;4:554–563. doi: 10.1038/mi.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Castillo M, Martín-Orúe SM, Manzanilla EG, Badiola I, Martín M, Gasa J. Quantification of total bacteria, enterobacteria and lactobacilli populations in pig digesta by real-time PCR. Vet Microbiol. 2006;114:165–170. doi: 10.1016/j.vetmic.2005.11.055. [DOI] [PubMed] [Google Scholar]
- 73.Malinen E, Kassinen A, Rinttilä T, Palva A. Compariso n of real-time PCR with SYBR Green I or 5 -nuclease assays and dot-blot hybridization with rDNA-targeted oligonucleotide probes in quantification of selected faecal bacteria. Microbiology. 2003;149:269–277. doi: 10.1099/mic.0.25975-0. [DOI] [PubMed] [Google Scholar]
- 74.Shacklett BL, Yang O, Hausner MA, Elliott J, Hultin L, Price C, Fuerst M, Matud J, Hultin P, Cox C. Optimization of methods to assess human mucosal T-cell responses to HIV infection. Journal of immunological methods. 2003;279:17–31. doi: 10.1016/s0022-1759(03)00255-2. [DOI] [PubMed] [Google Scholar]
- 75.Yukl SA, Gianella S, Sinclair E, Epling L, Li Q, Duan L, Choi AL, Girling V, Ho T, Li P, Fujimoto K, Lampiris H, Hare CB, Pandori M, Haase AT, Günthard HF, Fischer M, Shergill AK, McQuaid K, Havlir DV, Wong JK. Differences in HIV burden and immune activation within the gut of HIV-positive patients receiving suppressive antiretroviral therapy. J Infect Dis. 2010;202:1553–1561. doi: 10.1086/656722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Abreu NA, Nagalingam NA, Song Y, Roediger FC, Pletcher SD, Goldberg AN, Lynch SV. Sinus Microbiome Diversity Depletion and Corynebacterium tuberculostearicum Enrichment Mediates Rhinosinusitis. Sci Transl Med. 2012;4:151ra124. doi: 10.1126/scitranslmed.3003783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Storey JD. A direct approach to false discovery rates. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2002;64:479–498. [Google Scholar]
- 78.Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 79.Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995:289–300. [Google Scholar]
- 81.Freedman D, Lane D. A nonstochastic interpretation of reported significance levels. Journal of Business & Economic Statistics. 1983;1:292–298. [Google Scholar]
- 82.King EO, Ward MK, Raney DE. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.