Abstract
Potent DNA-damaging activities were seen in vitro from dietary chemicals found in coffee, tea, and liquid smoke. A survey of tea varieties confirmed genotoxic activity to be widespread. Constituent pyrogallol-like polyphenols (PLPs) such as epigallocatechin-3-gallate (EGCG), pyrogallol, and gallic acid were proposed as a major source of DNA-damaging activities, inducing DNA double-strand breaks in the p53R assay, a well characterized assay sensitive to DNA strand breaks, and comet assay. Paradoxically, their consumption does not lead to the kind of widespread cellular toxicity and acute disease that might be expected from genotoxic exposure. Existing physiological mechanisms could limit DNA damage from dietary injurants. Serum albumin and salivary α-amylase are known to bind EGCG. Salivary α-amylase, serum albumin, and myoglobin, but not salivary proline-rich proteins, reduced damage from tea, coffee, and PLPs, but did not inhibit damage from the chemotherapeutics etoposide and camptothecin. This represents a novel function for saliva in addition to its known functions including protection against tannins. Cell populations administered repeated pyrogallol exposures had abatement of measured DNA damage by two weeks, indicating an innate cellular adaptation. We suggest that layers of physiological protections may exist toward natural dietary products to which animals have had high-level exposure over evolution.
Keywords: DNA damage, genotoxin, clastogen, p53, liquid smoke, tea, coffee, pyrogallol, gallic acid, EGCG, saliva, α-amylase, serum albumin, myoglobin
1. Introduction
The consumption of hot teas and their constituents (Morton, 1987), high-tannin foods (Kirby, 1960; Kapadia et al., 1976; Morton, 1987, 1992), and processed meats (Santarelli et al., 2008; Chan et al., 2011) has been associated with an elevated risk of cancer. However, it is not clear how these substances promote carcinogenesis.
We recently evaluated potent strand-breaking genotoxic activities in coffee, tea, and liquid smoke flavoring (Hossain et al., 2013). Liquid smoke induced DNA double-strand breaks (Hossain et al., 2013) as detected in the p53R assay and neutral comet assay as well as DNA single-strand breaks in rat gastric mucosa in vivo (Ohshima et al., 1989). Flavonoids are well described to inhibit DNA topoisomerases (Bandele et al., 2008; Neukam et al., 2008; Lopez-Lazaro et al., 2011; Shiomi et al., 2013; Yoshida et al., 2013). Constituent pyrogallol-like polyphenols (PLPs) such as EGCG (found in green tea), gallic acid (green tea, black tea, and coffee), and pyrogallol (green tea, black tea, coffee, and liquid smoke) caused DNA strand breaks (Hossain et al., 2013). Because potential consequences of such potent DNA-damaging activity include cellular toxicity, mutagenesis, and carcinogenesis, it is relevant that damage occurred at concentrations consumed dietarily: etoposide near 5 μg/ml produced responses similar to a 1:1000 dilution of liquid smoke or a 1:20 dilution of coffee.
The divergence between natural intake patterns and a paucity of observed toxic effects in people suggests that physiological mechanisms might have evolved to handle dietary DNA-damaging agents. There is evidence for co-evolution of dietary habits and salivary proteins. For instance, between species the secretion level of salivary proline-rich proteins (PRPs) responds to the amount of tannin in the diet (carnivores < omnivores < herbivores) (McArthur et al., 1995). The binding of some injurious chemicals to proteins has been reported. PRPs have evolved so as to bind tannins in large amounts per unit of protein, serving to increase the amount of dietary protein and nitrogen available for nutrition (McArthur et al., 1995). Additionally, tissue proteins, dietary proteins, and albumin in blood might limit DNA damage. Serum albumin, for example, binds quercetin (Manach et al., 1995), EC (Papadopoulou and Frazier, 2004; Pal et al., 2012;), ECG (Pal et al., 2012), and EGCG (Nozaki et al., 2009).
In view of these observations, we posed some questions. How general, or widespread, is the potent dietary strand-breaking genotoxic activity we uncovered? How might strand-breaking genotoxic activity be handled physiologically? We hypothesized that physiologically relevant proteins might have a protective role against DNA-damaging agents from the diet. We tested candidate proteins for their ability to inhibit DNA-damage response in the p53R assay, a well characterized cellular biological assay sensitive to DNA strand breaks (Sohn et al., 2002; Cunningham et al., 2004; Gallmeier et al., 2005; Hossain et al., 2013;). This assay utilizes a human cell line in which luciferase expression is driven by a stably integrated p53 reporter construct.
2. Materials and Methods
2.1. Cell lines and cell culture
p53R cells were created and characterized in our laboratory (Sohn et al., 2002; Cunningham et al., 2004; Gallmeier et al., 2005; Hossain et al., 2013;). p53R and HeLa (ATCC) cells were grown in DMEM with 10% (v/v) FBS, 1% (v/v) penicillin/streptomycin, and 20 mM HEPES. MCF 10A cells were grown in DMEM/F-12 medium with 5% (v/v) horse serum, 1% (v/v) penicillin/streptomycin, 20 ng/ml epidermal growth factor, 0.5 μg/ml hydrocortisone, 0.1 μg/ml cholera toxin, and 10 μg/ml insulin.
2.2. Substances tested
2.2.1. Protein preparations
BSA, horse myoglobin, human salivary α-amylase, and protein A from Staphylococcus aureus were obtained from Sigma-Aldrich. An aliquot of human saliva was centrifuged at 2000 g for 15 min, and the supernatant tested at various concentrations in the p53R assay. To extract PRPs, equal volumes of saliva supernatant and 10% (w/v) TCA were mixed and centrifuged at 18000 × g for 10 min at 4°C to remove TCA-insoluble material (adapted from (Robbins et al., 1987)). The PRP-enriched supernatant was diluted 1:5 in DMEM containing 20 mM HEPES, without FBS or antibiotics. The remaining TCA was neutralized by adding 1 M NaOH drop-wise until the color of the phenol red-containing medium changed. Using a Slide-A-Lyzer 10 K MWCO dialysis cassette (Thermo Scientific), the PRP solution was dialyzed against a 300× excess volume of DPBS initially for 2 h at RT, for another 2 h at RT after changing the dialysate, and finally overnight at 4° C after changing the dialysate again. Graded concentrations of the PRP solution were tested in the p53R assay after TCA extraction, neutralization, and dialysis.
2.2.2. DNA-damaging agents
Tested injurants included a panel of 45 different green, white, black, and pu-erh teas (Lin et al., 2008; Zhao et al., 2011) (Supplementary Table), previously characterized for their phenolic content and provided by Drs. James Harnly and Pei Chen of the United States Department of Agriculture Agricultural Research Service. In addition, black tea (Twinings Irish Breakfast), coffee (Me Latte), liquid smoke flavoring (Wright's Hickory), food constituents (from Sigma-Aldrich): pyrogallol, gallic acid, and EGCG, and chemotherapeutic agents (from Sigma-Aldrich): etoposide and camptothecin were tested.
2.3. p53R assay
For each treatment, cells were plated in triplicate and incubated with each protein-chemical combination as described in figure legends. Matched combinations of medium and a given chemical were included on the same plate to facilitate direct comparison. The luciferase reporter assay was performed with the Promega Steady-Glo Luciferase Assay System according to the manufacturer's protocol. The PerkinElmer Microbeta Trilux plate reader was used to record luminescence signals. The average for each triplet was plotted for each treatment. A non-mammalian protein, protein A, was included as a negative control for inhibition of strand-breaking genotoxic activity. We extensively tested culture medium ± 10% FBS without any effect on assay results. The amount of BSA in 10% FBS might be insufficient to perturb the assay. To calculate % inhibition, the area under the curve was calculated for the protein-chemical combination as well as the corresponding control vehicle-chemical combination from untreated to the concentration at which the vehicle-chemical combination reached its peak activity, using trapezoidal integration in (Excel, Microsoft). The difference was expressed as a fraction of the area under the curve for the vehicle-chemical combination.
2.4. Protein assays
Cells were lysed by rocking in a detergent containing 50 mM Tris-Hcl, 150 mM NaCl, 1 mM EDTA, 1% (v/v) Triton X-100, and a protease inhibitor cocktail (Roche) for 1 h at 4°C. The cell lysate was clarified by centrifugation. We used the Lowry method (DC protein assay, Bio-Rad) to estimate protein concentrations. Whole cell lysates or salivary protein samples were denatured with a denaturant containing 2% (m/v) SDS, 10% (v/v) glycerol, 0.002% (m/v) bromophenol blue, 2 mM EDTA, 50 mM Tris pH 6.8, and 1% (v/v) β-mercaptoethanol, boiled, and resolved on a 4-12% Bis-Tris gel (NuPAGE, Invitrogen). The gel was stained (0.25% Coomassie brilliant blue in 50% methanol and 10% acetic acid) for 1h. To remove background, the gel was washed with 30% methanol, 10% acetic acid. Alternatively, for immunoblotting the protein samples were transferred onto a polyvinylidene fluoride (PVDF) membrane (Pierce). Blots were incubated with primary antibodies: γ-H2AX (Millipore) and GAPDH (Santa Cruz), followed by horseradish peroxidase (HRP)-conjugated anti-mouse IgG secondary antibodies (Santa Cruz). Membranes were developed with the Immobilon substrate (Millipore), and signals recorded on film.
2.5. Picogreen assay
After treatment with protein-chemical combination as described in figure legends, cells were washed and lysed in 0.03% SDS, and 0.5% picogreen (Molecular Probes) was added. Fluorescence was measured and the relative cell numbers calculated, defining the untreated sample as 1.
2.6. Neutral comet assay
After treatment with protein-chemical combination (exposures as given in figure legends), cells were detached using 0.05% trypsin, diluted to 100,000 cells/ml, mixed at 37°C with 0.75% low-melting agarose (1:10, v/v), and immediately layered onto pre-treated slides (Trevigen). Gels were incubated at 4°C in the dark for 30 min. Cells were lysed in pre-chilled lysis buffer containing 2.5 M NaCl, 100 mM EDTA, 10 mM Tris, 1% (m/v) sodium lauroyl sarcosinate, 1% (v/v) Triton X-100 overnight at 4°C and then washed with neutral TBE for 15 min. Electrophoresis was performed in neutral TBE at 23 V for 15 min at 4°C. Slides were washed in water for 5 min, submerged in 70% ethanol for 5 min, and air-dried overnight. Slides were then stained with SYBR Green (Molecular Probes) and imaged using a Zeiss Axiovert microscope. Images were acquired and analyzed with the MetaMorph software version 6.5. The CometScore software (TriTek) was used to morphometrically integrate the tail moment of at least 70 randomly selected comets from each gel.
3.Results
In an earlier study, we uncovered potent genotoxic activities in the contemporary diet. DNA strand breaks were detected in vitro at doses achievable from routine dietary exposure (Hossain et al., 2013). A survey of distinct preparations of tea (Supplementary Table) now showed that the potent dietary DNA-damaging activity was widespread and rather uniform among a large and diverse panel of 45 different green, white, black, and pu-erh teas, excluding the possibility of this activity having affected only the few sources used in the initial report (Hossain et al., 2013).
To investigate the physiology plausibly protecting mucosal cells from PLP-induced DNA damage, we tested the ability of saliva to inhibit the strand-breaking genotoxic activity of DNA-injurants in the p53R assay. We tested human saliva at various concentrations. A 1:20 dilution of saliva was sufficient to inhibit DNA damage induced by dietary PLPs (structures shown in Fig. 1) such as pyrogallol, gallic acid, and EGCG (Supplementary Fig. 1). As part of quality control, we assayed the total protein concentration. We also resolved salivary proteins by SDS-PAGE, obtaining visual estimates by Coomassie staining (Supplementary Fig. 2). To exclude the possibility of saliva-induced cytotoxicity in the p53R assay, we assessed cell survival in the presence of saliva by picogreen assay. Even a 1:2 dilution of saliva was not toxic to p53R cells (Supplementary Fig. 3). When cells were treated with a DNA-injurant (pyrogallol), saliva also promoted cell survival (Supplementary Fig. 3).
Fig. 1.
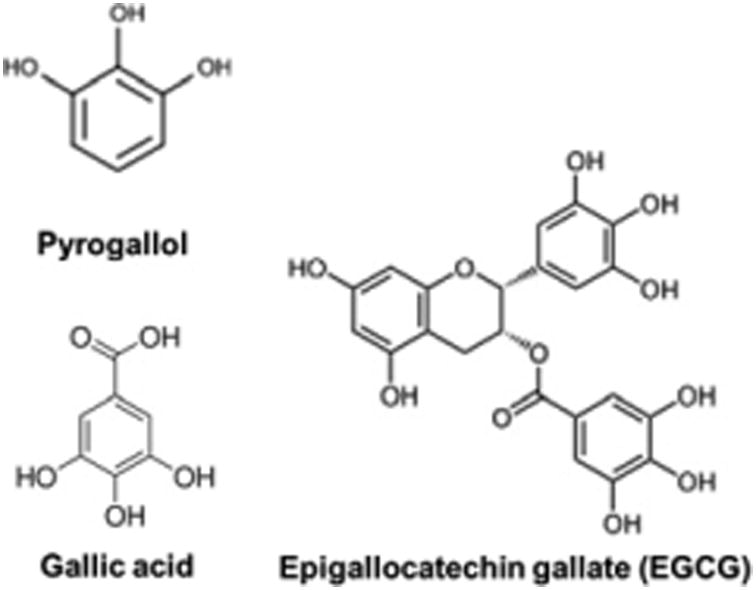
Structures of pyrogallol-like polyphenols (PLPs): pyrogallol, gallic acid, and EGCG.
In an effort to identify the major protein(s) responsible for the protective property in saliva, we prepared enriched salivary PRPs, tested at graded concentrations in the p53R assay. PRPs did not protect against the strand-breaking genotoxic activity of the constituent PLPs pyrogallol, gallic acid, and EGCG (Supplementary Fig. 1). For quality control, we assayed the protein content in PRP preparations by the DC assay and by visual estimation when resolved upon SDS-PAGE and Coomassie staining (Supplementary Fig. 2).
In contrast, human salivary α-amylase at 50 units/ml inhibited DNA damage from the constituent PLPs pyrogallol (Tables 1 and 2 and Fig. 2), gallic acid (Table 1 and Supplementary Fig. 1), and EGCG (Tables 1 and 2 and Fig. 2). α-amylase also inhibited DNA damage from coffee and tea (Table 1 and Supplementary Fig. 1), but did not inhibit DNA damage induced by liquid smoke flavoring even when a higher concentration (200 units/ml) of α-amylase was tested (Table 1 and Supplementary Fig. 1) or the chemotherapeutics etoposide (Table 1 and Supplementary Fig. 1) and camptothecin (Tables 1 and 2 and Fig. 2). α-amylase-mediated inhibition of EGCG-induced DNA damage was confirmed by the neutral comet assay in HeLa and p53R cells (Fig. 3). We used HeLa cells in addition to p53R cells to demonstrate that protein-mediated protection against DNA damage was not cell line-specific. HeLa cells were chosen for their familiarity.
Table 1.
Patterns of protein-mediated inhibition of clastogenic DNA damage.
| DNA-injurant | Whether DNA damage was inhibited in the p53R assaya | ||
|---|---|---|---|
|
| |||
| α-Amylase (50 units/ml) |
Albumin (2%) |
Myoglobin (0.01%) |
|
| Black tea | + | + | + |
| Coffee | + | + | + |
| Liquid smoke | - | + | - |
| Pyrogallol | + | + | + |
| Gallic acid | + | + | + |
| EGCG | + | + | + |
| Etoposide | - | - | - |
| Camptothecin | - | - | - |
+, inhibition detected. -, inhibition not detected.
Table 2.
Quantification of inhibition in the p53R assay.
| DNA-injurant | Inhibition (%)a: Mean ± SEM | ||
|---|---|---|---|
|
| |||
| α-amylase (50 units/ml) |
Albumin (2%) |
Myoglobin (0.01%) |
|
| Pyrogallol | 90 ± 2 | 88 ± 1 | 85 ± 5 |
| EGCG | 72 ± 7 | 83 ± 3 | 83 ± 5 |
| Camptothecin | -8 ± 4 | -2 ± 5 | -3 ± 4 |
Measured as described in the Materials and Methods section.
Fig. 2.
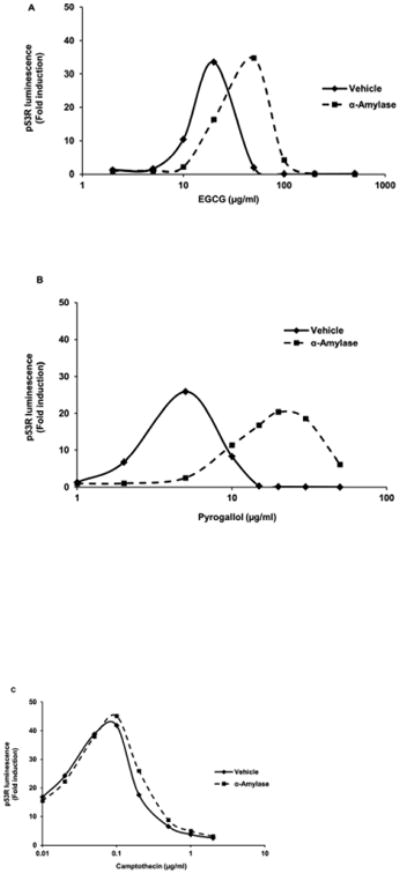
α-amylase inhibited strand-breaking genotoxic DNA damage induced by PLPs such as EGCG (A) and pyrogallol (B), but did not inhibit strand-breaking genotoxic DNA damage induced by the chemotherapeutic camptothecin (C) in the p53R assay. In each experiment, p53R cells were plated in triplicate and treated with various doses of each injurant for 18 h in the presence or absence of α-amylase at 50 units/ml. p53 reporter activity was measured in a luciferase assay. Luminescence was calculated relative to the untreated sample. The mean for each triplet was plotted for each treatment. Each experiment was performed three times with similar results. Data from representative experiments are shown.
Fig. 3.
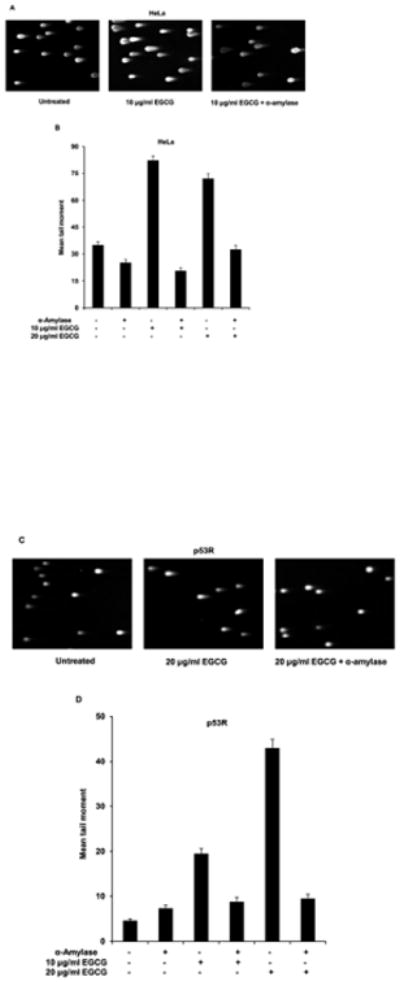
EGCG induced fewer DNA double-strand breaks, as detected in the neutral comet assay, in the presence of α-amylase in HeLa (A and B) and p53R (C and D) cells. To confirm our findings from the p53R assay, the neutral comet assay was performed on HeLa or p53R cells, treated with two different doses of EGCG (10 μg/ml and 20 μg/ml: doses at which the peak activity was seen in individual p53R assays) for 30 min in the presence or absence of α-amylase at 50 units/ml. At least 70 cells were analyzed per sample to calculate the mean tail moment of each cell population. Representative images of the cells are shown in (A) and (C). The mean tail moment for each cell population and the standard error of the mean reflecting between-cell variation in the same experiment are shown in (B) and (D).
To determine whether other physiologically prominent proteins have a similar protective property, we tested the dominant serum protein, albumin (as BSA), and a major muscle protein, myoglobin, in the p53R assay. Both inhibited DNA damage induced by the beverages (coffee: Table 1 and Supplementary Fig. 1; tea: Table 1 and Supplementary Fig. 1) and their constituent PLPs (EGCG: Tables 1 and 2 and Supplementary Fig. 1; pyrogallol: Tables 1 and 2 and Supplementary Fig. 1; gallic acid: Table 1 and Supplementary Fig. 1), but did not inhibit DNA damage from the chemotherapeutics (etoposide: Table 1 and Supplementary Fig. 1; camptothecin: Tables 1 and 2 and Supplementary Fig. 1). Serum albumin, but not myoglobin, inhibited the strand-breaking genotoxic activity of liquid smoke flavoring (Table 1 and Supplementary Fig. 1).
Occasionally, a higher level of activity was seen with the protective agent; this was observed at some very high concentrations of the injurant and presumably was due to increased cell viability in the presence of the protective agent (Supplementary Fig. 1). This accompanied a graphical shift in the peak activity towards requiring higher injurant exposure when in the presence of the protective agent.
Next, we wondered whether there were other physiological mechanisms of protection against dietary DNA damage. When p53R cells were chronically exposed (see Fig. 4, legend) to pyrogallol, the level of DNA damage decreased by day 15 as demonstrated by the neutral comet assay (Fig. 4A) and p53R assay (Fig. 4B), indicating activation of cell-intrinsic mechanisms of protection. The picogreen assay was additionally performed to exclude the possibility that p53R activity in chronically exposed cells was masked by reduced cellularity (Fig. 4C). Similarly, when non-tumorigenic MCF 10A cells were chronically exposed to pyrogallol, the extent of DNA damage, as assessed by H2AX phosphorylation, was drastically reduced by day 12, reflecting innate cellular adaptation (data not shown).
Fig. 4.
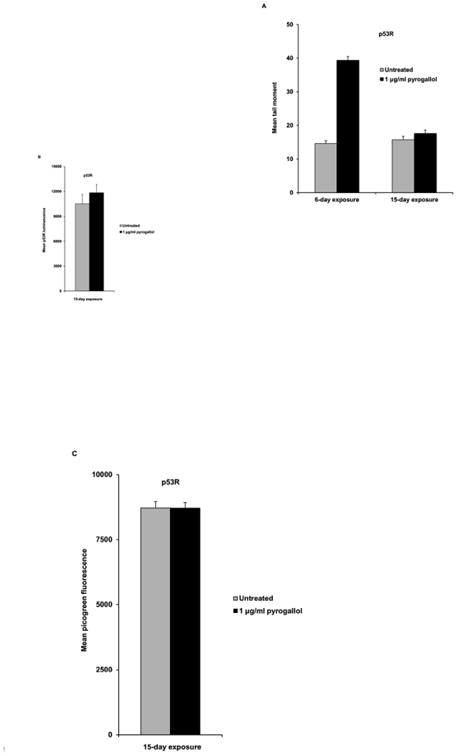
Chronic exposure to pyrogallol reduced the level of DNA damage in p53R cells. The neutral comet assay was performed on p53R cells, chronically exposed to pyrogallol at 1 μg/ml for 6 days or 15 days. Pyrogallol was added to the culture for the first time on day 0. The culture medium was replaced, and fresh pyrogallol was added on days 2 and 4 to cells exposed for 6 days. For cells treated for 15 days, the culture medium was replaced, and fresh pyrogallol was added on days 2, 4, 6, 8, and 13. At least 200 cells were analyzed per sample to calculate the mean tail moment of each cell population. The mean tail moment for each cell population the standard error of the mean reflecting between-cell variation in the same experiment are shown in (A). p53 reporter activity of cell populations chronically exposed to pyrogallol for 15 days was measured in a luciferase assay (B). The picogreen assay was performed at the same time to exclude the possibility that p53 reporter activity in chronically exposed cells was masked by elevated cytotoxicity (C).
4. Discussion
Our study extends prior evidence (Manach et al., 1995; Papadopoulou and Frazier, 2004; Nozaki et al., 2009; Hara et al., 2012; Pal et al., 2012;) that animals have evolved physiological protection against dietary polyphenols and, in particular, PLPs such as EGCG, gallic acid, and pyrogallol. We thus discovered a novel function for saliva in mitigating PLP-induced DNA damage; this is in addition to the known functions of enzymatic activity, moistening, diluting/washing, salivary PRPs binding tannins, and maintenance of tooth integrity through antibacterial activity and specialized interactions with bacterial metabolism in dental plaque (Humphrey and Williamson, 2001). We also implicated a high-quantity protein as responsible for this novel function. Saliva contains α-amylase at 400-900 units/ml (Granger et al., 2007; Hara et al., 2012). α-amylase from a purified source inhibited dietary DNA strand-breaking genotoxins at 50 units/ml in our studies, and a 1:20 dilution of saliva produced a similar level of inhibition. α-amylase could, therefore, largely explain our protective effect of human saliva against dietary DNA injurants. EGCG was already known to bind to α-amylase, but not to PRP (Hara et al., 2012). An evolutionary significance of this interaction was not, however, previously proposed.
Our findings suggest a division of labor among salivary proteins. PRPs have evolved, at least in part, to provide protection from tannins, as indicated by reported evidence for the co-evolution of dietary habits and salivary PRPs. For example, the level of salivary PRP secretion corresponds to the amount of tannin in the diet (carnivores < omnivores < herbivores) (McArthur et al., 1995). Moreover, in the same animal, levels of salivary PRPs are regulated to match to changing levels of tannins in the diet (McArthur et al., 1995). Tannic acid may not directly induce DNA double-strand breaks in the absence of degradative breakdown to its constituent PLP building blocks (Hossain et al., 2013). In contrast, α-amylase may have evolved to provide protection specifically against PLP-induced DNA damage, among other functions.
The protective role we uncovered was not limited to α-amylase. Serum albumin and myoglobin also inhibited dietary strand-breaking genotoxic activities. Consistent with a reported interaction between serum albumin and EC (Papadopoulou and Frazier, 2004); Pal et al., 2012, ECG (Pal et al., 2012), and EGCG (Nozaki et al., 2009), albumin might have an important role in providing protection against PLPs to the extent that they enter the blood. Our observations suggest a potential complementary role of dietary myoglobin, from meat consumption by carnivores and omnivores, in limiting DNA damage from dietary PLPs.
The DNA-damaging property of liquid smoke was inhibited by serum albumin, but not by myoglobin or α-amylase. Liquid smoke is a complex mixture, which might potentially contain compounds saturating and masking any myoglobin- or α-amylase-mediated protective functions in the p53R assay. Higher concentrations of myoglobin were toxic to the cells, which precluded our testing of a full concentration range. Myoglobin contains iron, accounting for its toxicity (Papanikolaou and Pantopoulos, 2005). It is tempting to speculate that myoglobin or α-amylase fails to counteract the DNA-damaging activity of liquid smoke because the latter was not encountered during evolutionary history. The absence of protection against the chemotherapeutics etoposide and camptothecin – substances that were also not encountered over the course of evolution – can be interpreted similarly within this argument, suggesting a specificity of physiological protection against strand-breaking genotoxic dietary constituents encountered during evolution. Evolutionary pressure might have selected for elevated levels of inhibition by physiologically relevant proteins toward common dietary genotoxins (altered binding affinities, etc). We note that in most cases, the DNA-damaging activity was not completely eliminated; protective proteins mitigated its harmful effects.
In this study, chronic exposure to PLPs activated cell-intrinsic mechanisms of protection against DNA damage. There is evidence for a mammalian SOS response in which exposure to DNA-damaging agents leads to enhancement of DNA repair (Protic et al., 1988; Eller et al., 1997). A similar mechanism might reduce the level of DNA damage with chronic exposure to PLPs. Alternatively, cells might activate pathways that might reduce the uptake of PLPs, increase their export, or break them down more rapidly as part of an innate cellular adaptation (Lord et al., 2013).
Our study does not address the fate of DNA-injurant PLP chemicals after the binding protein dissociates or becomes degraded during digestive transit. Our in vitro models might also differ in responses from the cells most directly experiencing the natural in vivo exposures: native gastrointestinal mucosal epithelial cells. The p53R assay is performed in the absence of hepatic and other bodily metabolism. Despite limitations, our study may offer insights into the physiological protection against major dietary strand-breaking genotoxins.
Physiologically relevant proteins such as salivary α-amylase, serum albumin, and myoglobin are effective in vitro at counteracting the strand-breaking genotoxic activity of natural dietary constituents. Future studies should determine whether these findings could be replicated in vivo and should elucidate the biochemical mechanisms by which bodily proteins may bind DNA-injurants and limit the potential for dietary DNA damage. They should also address whether protein-PLP interactions have evolutionary or epidemiologic implications and whether protein binding reduces absorption and systemic exposure to DNA-damaging PLPs.
Supplementary Material
Supplementary Fig. 1. p53R assay plot for each protein-chemical combination tested. p53R cells were plated in triplicate and treated with each injurant over a wide dose range for 18 h in the presence or absence of a candidate protective agent. p53 reporter activity was measured in a luciferase assay. Luminescence was calculated relative to the untreated sample. The mean for each triplet was plotted for each treatment. To prepare black tea for use in the p53R assay, 10g (5 tea bags) of black tea was extracted in 100 ml of boiling water for 5 min. The tea solution was vacuum-dried until around 75% of original volume evaporated. The resulting concentrated stock tea solution was arbitrarily designated 40×. Freshly brewed coffee from a local vendor was vacuum-dried until around 90% of original volume evaporated. The resulting concentrated stock coffee solution was arbitrarily designated 10×. Manufacturer's stock of liquid smoke was considered 1×.
Supplementary Fig. 2. Coomassie staining of saliva and PRP preparations. The protein content in saliva and PRP preparations (before and after dialysis) was assessed by visual estimation when resolved upon SDS-PAGE and Coomassie staining.
Supplementary Fig. 3. Picogreen assay for cell survival in the presence of saliva. p53R cells were plated in triplicate and treated with various doses of pyrogallol for 18 h in the presence or absence of saliva at 1:2 dilution. Cell survival was assessed in a picogreen assay. The mean for each triplet was plotted for each treatment.
Highlights.
We used a cellular biological (p53R) assay to investigate physiological protection against dietary genotoxins.
Salivary α-amylase, serum albumin, and myoglobin inhibited the DNA-damaging activity in tea, coffee, and pyrogallol-like polyphenols (PLPs).
These proteins did not, however, inhibit the genotoxic activity in the chemotherapeutics etoposide and camptothecin.
Cell populations adapt to reduce DNA damage from repeated exposure to PLPs.
Physiological protection may preferentially exist for natural food products because of our dietary exposure to them during evolution.
We, thus, identified a novel function for saliva in mitigating PLP-induced DNA damage.
Acknowledgments
This study was supported by the National Institutes of Health grant CA62924 and the Everett and Marjorie Kovler Professorship in Pancreas Cancer Research. Tea varieties were kindly provided by Drs. James Harnly and Pei Chen of the United States Department of Agriculture Agricultural Research Service.
Abbreviations
- BSA
bovine serum albumin
- DMEM
Dulbecco's modified Eagle's medium
- DMSO
dimethyl sulfoxide
- DPBS
Dulbecco's phosphate-buffered saline
- EC
epicatechin
- ECG
epicatechin gallate
- EGCG
epigallocatechin-3-gallate
- FBS
fetal bovine serum
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- PAGE
polyacrylamide gel electrophoresis
- PLP
pyrogallol-like polyphenol
- PRP
proline-rich protein
- SDS
sodium dodecyl sulfate
- SEM
standard error of the mean
- TBE
Tris/Borate/EDTA
- TCA
trichloroacetic acid
Footnotes
Conflict of Interest: The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bandele OJ, Clawson SJ, Osheroff N. Dietary polyphenols as topoisomerase II poisons: B ring and C ring substituents determine the mechanism of enzyme-mediated DNA cleavage enhancement. Chem Res Toxicol. 2008;21:1253–1260. doi: 10.1021/tx8000785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DS, Lau R, Aune D, Vieira R, Greenwood DC, Kampman E, Norat T. Red and processed meat and colorectal cancer incidence: meta-analysis of prospective studies. PloS One. 2011;6:e20456. doi: 10.1371/journal.pone.0020456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham SC, Ryu B, Sohn TA, Kern SE. Nonspecific enhancement of gene expression by compounds identified in high-throughput cell-based screening. Biotechniques. 2004;37:120–122. doi: 10.2144/04371DD01. [DOI] [PubMed] [Google Scholar]
- Eller MS, Maeda T, Magnoni C, Atwal D, Gilchrest BA. Enhancement of DNA repair in human skin cells by thymidine dinucleotides: evidence for a p53-mediated mammalian SOS response. Proc Natl Acad Sci U S A. 1997;94:12627–12632. doi: 10.1073/pnas.94.23.12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallmeier E, Winter JM, Cunningham SC, Kahn SR, Kern SE. Novel genotoxicity assays identify norethindrone to activate p53 and phosphorylate H2AX. Carcinogenesis. 2005;26:1811–1820. doi: 10.1093/carcin/bgi132. [DOI] [PubMed] [Google Scholar]
- Granger DA, Kivlighan KT, el-Sheikh M, Gordis EB, Stroud LR. Salivary alpha-amylase in biobehavioral research: recent developments and applications. Ann N Y Acad Sci. 2007;1098:122–144. doi: 10.1196/annals.1384.008. [DOI] [PubMed] [Google Scholar]
- Hara K, Ohara M, Hayashi I, Hino T, Nishimura R, Iwasaki Y, Ogawa T, Ohyama Y, Sugiyama M, Amano H. The green tea polyphenol (-)-epigallocatechin gallate precipitates salivary proteins including alpha-amylase: biochemical implications for oral health. Eur J Oral Sci. 2012;120:132–139. doi: 10.1111/j.1600-0722.2012.00947.x. [DOI] [PubMed] [Google Scholar]
- Hossain MZ, Gilbert SF, Patel K, Ghosh S, Bhunia AK, Kern SE. Biological clues to potent DNA-damaging activities in food and flavoring. Food Chem Toxicol. 2013;55:557–567. doi: 10.1016/j.fct.2013.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey SP, Williamson RT. A review of saliva: normal composition, flow, and function. J Prosthet Dent. 2001;85:162–169. doi: 10.1067/mpr.2001.113778. [DOI] [PubMed] [Google Scholar]
- Kapadia GJ, Paul BD, Chung EB, Ghosh B, Pradhan SN. Carcinogenicity of Camellia sinensis (tea) and some tannin-containing folk medicinal herbs administered subcutaneously in rats. J Natl Cancer Inst. 1976;57:207–209. doi: 10.1093/jnci/57.1.207. [DOI] [PubMed] [Google Scholar]
- Kirby KS. Induction of tumours by tannin extracts. Br J Cancer. 1960;14:147–150. doi: 10.1038/bjc.1960.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin LZ, Chen P, Harnly JM. New phenolic components and chromatographic profiles of green and fermented teas. J Agric Food Chem. 2008;56:8130–8140. doi: 10.1021/jf800986s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Lazaro M, Calderon-Montano JM, Burgos-Moron E, Austin CA. Green tea constituents (-)-epigallocatechin-3-gallate (EGCG) and gallic acid induce topoisomerase I- and topoisomerase II-DNA complexes in cells mediated by pyrogallol-induced hydrogen peroxide. Mutagenesis. 2011;26:489–498. doi: 10.1093/mutage/ger006. [DOI] [PubMed] [Google Scholar]
- Lord C, Ashworth A. Mechanisms of resistance to therapies targeting BRCA-mutant cancers. Nature Medicine. 2013;19:1381–1388. doi: 10.1038/nm.3369. [DOI] [PubMed] [Google Scholar]
- Manach C, Morand C, Texier O, Favier ML, Agullo G, Demigne C, Regerat F, Remesy C. Quercetin metabolites in plasma of rats fed diets containing rutin or quercetin. J Nutr. 1995;125:1911–1922. doi: 10.1093/jn/125.7.1911. [DOI] [PubMed] [Google Scholar]
- McArthur C, Sanson GD, Beal AM. Salivary Proline-Rich Proteins in Mammals -Roles in Oral Homeostasis and Counteracting Dietary Tannin. Journal of Chemical Ecology. 1995;21:663–691. doi: 10.1007/BF02033455. [DOI] [PubMed] [Google Scholar]
- Morton JF. Tannin and oesophageal cancer. Lancet. 1987;2:327–328. doi: 10.1016/s0140-6736(87)90908-1. [DOI] [PubMed] [Google Scholar]
- Morton JF. Widespread tannin intake via stimulants and masticatories, especially guarana, kola nut, betel vine, and accessories. Basic Life Sci. 1992;59:739–765. doi: 10.1007/978-1-4615-3476-1_45. [DOI] [PubMed] [Google Scholar]
- Neukam K, Pastor N, Cortes F. Tea flavanols inhibit cell growth and DNA topoisomerase II activity and induce endoreduplication in cultured Chinese hamster cells. Mutat Res. 2008;654:8–12. doi: 10.1016/j.mrgentox.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Nozaki A, Hori M, Kimura T, Ito H, Hatano T. Interaction of polyphenols with proteins: binding of (-)-epigallocatechin gallate to serum albumin, estimated by induced circular dichroism. Chem Pharm Bull (Tokyo) 2009;57:224–228. doi: 10.1248/cpb.57.224. [DOI] [PubMed] [Google Scholar]
- Ohshima H, Furihata C, Matsushima T, Bartsch H. Evidence of potential tumour-initiating and tumour-promoting activities of hickory smoke condensate when given alone or with nitrite to rats. Food Chem Toxicol. 1989;27:511–516. doi: 10.1016/0278-6915(89)90046-x. [DOI] [PubMed] [Google Scholar]
- Pal S, Saha C, Hossain M, Dey SK, Kumar GS. Influence of galloyl moiety in interaction of epicatechin with bovine serum albumin: a spectroscopic and thermodynamic characterization. PLoS One. 2012;7:e43321. doi: 10.1371/journal.pone.0043321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulou A, Frazier RA. Characterization of protein-polyphenol interactions. Trends in Food Science & Technology. 2004;15:186–190. [Google Scholar]
- Papanikolaou G, Pantopoulos K. Iron metabolism and toxicity. Toxicol Appl Pharmacol. 2005;202:199–211. doi: 10.1016/j.taap.2004.06.021. [DOI] [PubMed] [Google Scholar]
- Protic M, Roilides E, Levine AS, Dixon K. Enhancement of DNA repair capacity of mammalian cells by carcinogen treatment. Somatic Cell Mol Genet. 1988;14:351–357. doi: 10.1007/BF01534643. [DOI] [PubMed] [Google Scholar]
- Robbins CT, Mole S, Hagerman AE, Hanley TA. Role of Tannins in Defending Plants against Ruminants - Reduction in Dry-Matter Digestion. Ecology. 1987;68:1606–1615. doi: 10.2307/1939852. [DOI] [PubMed] [Google Scholar]
- Santarelli RL, Pierre F, Corpet DE. Processed meat and colorectal cancer: a review of epidemiologic and experimental evidence. Nutr Cancer. 2008;60:131–144. doi: 10.1080/01635580701684872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi K, Kuriyama I, Yoshida H, Mizushina Y. Inhibitory effects of myricetin on mammalian DNA polymerase, topoisomerase and human cancer cell proliferation. Food Chem. 2013;139:910–918. doi: 10.1016/j.foodchem.2013.01.009. [DOI] [PubMed] [Google Scholar]
- Sohn TA, Bansal R, Su GH, Murphy KM, Kern SE. High-throughput measurement of the Tp53 response to anticancer drugs and random compounds using a stably integrated Tp53-responsive luciferase reporter. Carcinogenesis. 2002;23:949–957. doi: 10.1093/carcin/23.6.949. [DOI] [PubMed] [Google Scholar]
- Yoshida N, Kuriyama I, Yoshida H, Mizushina Y. Inhibitory effects of catechin derivatives on mammalian DNA polymerase and topoisomerase activities and mouse one-cell zygote development. J Biosci Bioeng. 2013;115:303–309. doi: 10.1016/j.jbiosc.2012.10.001. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Chen P, Lin LZ, Harnly JM, Yu LL, Li ZW. Tentative identification, quantitation, and principal component analysis of green pu-erh, green, and white teas using UPLC/DAD/MS. Food Chemistry. 2011;126:1269–1277. doi: 10.1016/j.foodchem.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. p53R assay plot for each protein-chemical combination tested. p53R cells were plated in triplicate and treated with each injurant over a wide dose range for 18 h in the presence or absence of a candidate protective agent. p53 reporter activity was measured in a luciferase assay. Luminescence was calculated relative to the untreated sample. The mean for each triplet was plotted for each treatment. To prepare black tea for use in the p53R assay, 10g (5 tea bags) of black tea was extracted in 100 ml of boiling water for 5 min. The tea solution was vacuum-dried until around 75% of original volume evaporated. The resulting concentrated stock tea solution was arbitrarily designated 40×. Freshly brewed coffee from a local vendor was vacuum-dried until around 90% of original volume evaporated. The resulting concentrated stock coffee solution was arbitrarily designated 10×. Manufacturer's stock of liquid smoke was considered 1×.
Supplementary Fig. 2. Coomassie staining of saliva and PRP preparations. The protein content in saliva and PRP preparations (before and after dialysis) was assessed by visual estimation when resolved upon SDS-PAGE and Coomassie staining.
Supplementary Fig. 3. Picogreen assay for cell survival in the presence of saliva. p53R cells were plated in triplicate and treated with various doses of pyrogallol for 18 h in the presence or absence of saliva at 1:2 dilution. Cell survival was assessed in a picogreen assay. The mean for each triplet was plotted for each treatment.


