Abstract
The reprogramming of parental methylomes is essential for embryonic development. In mammals, paternal 5-methylcytosines (5mCs) have been proposed to be actively converted to oxidized bases. These paternal oxidized bases and maternal 5mCs are believed to be passively diluted by cell divisions. By generating single-base resolution, allele-specific DNA methylomes from mouse gametes, early embryos and primordial germ cell (PGC), as well as single-base resolution maps of oxidized cytosine bases for early embryos, we report the existence of 5hmC and 5fC in both maternal and paternal genomes and find that 5mC or its oxidized derivatives, at the majority of demethylated CpGs, are converted to unmodified cytosines independent of passive dilution from gametes to 4-cell embryos. Therefore, we conclude that paternal methylome and at least a significant proportion of maternal methylome go through active demethylation during embryonic development. Additionally, all the known imprinting control regions (ICRs) were classified into germ-line or somatic ICRs.
Introduction
The epigenomes of sperm and oocyte are dramatically different. The paternal and maternal epigenomes reprogram to the same state during early embryogenesis. The reprogramming of parental epigenomes is essential for the compatibility of totipotency during embryonic development (Hackett and Surani, 2013). Recent studies show that paternal DNA methylome is stably inherited during early embryogenesis in zebrafish, while maternal methylome undergoes significant reprogramming to the sperm pattern (Jiang et al., 2013; Potok et al., 2013). In mammals, two waves of genome-wide DNA demethylation take place during primordial germ cell (PGC) development and early embryogenesis (Seisenberger et al., 2013; Wu and Zhang, 2014). However, our knowledge on genome-wide demethylation is still limited due to the lack of single-base resolution DNA methylomes for mouse oocyte and early embryos.
Currently, it is generally believed that paternal DNA is actively demethylated by oxidizing 5-methylcytosine (5mC) to 5-hydroxymethylcytsine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) by Tet3 (Gu et al., 2011; He et al., 2011; Inoue et al., 2011; Inoue and Zhang, 2011; Ito et al., 2011). Studies using cell immunostaining suggested that the oxidized derivatives of 5mC is further diluted passively by DNA replication over early cell divisions (Inoue et al., 2011; Inoue and Zhang, 2011; Wu and Zhang, 2014). Alternatively, the oxidized 5mC bases could be replaced to unmodified cytosines through the base excision repair pathway similar to what has been found in mouse embryonic stem cells (He et al., 2011; Zhang et al., 2012). Moreover, it is claimed that the oxidized 5mC bases only exist in paternal genome but not in maternal genome during early embryogenesis (Gu et al., 2011; Inoue et al., 2011; Inoue and Zhang, 2011; Iqbal et al., 2011; Xie et al., 2012). Previous study also proposed that 5mC in maternal DNA is protected from the oxidization by Stella in early embryos (Nakamura et al., 2012). Therefore, it has been generally believed that 5mC on maternal DNA is passively diluted through early cell divisions during mammalian early embryogenesis (Seisenberger et al., 2013; Wu and Zhang, 2014). Nevertheless, these conclusions lack the support from the sequencing data.
Genomic imprinting in mammals is important for embryonic development (Surani et al., 1990). Loss of imprinting is associated with many human diseases (Lalande, 1996). Imprinting control regions (ICRs) can be classified into germ-line ICRs (gICRs) and somatic ICRs (sICRs). The allele-specific methylation status of gICRs is set during gametes development and is maintained after fertilization throughout the development (Reik and Walter, 2001). The allele-specific methylation status of sICRs is achieved during the mammalian development after fertilization, often in a tissue-specific manner (Hayashizaki et al., 1994; Hiura et al., 2010; Kelsey et al., 1999; Peters et al., 1999; Plass et al., 1996; Xie et al., 2012). Until now, 55 ICRs have been identified in mouse genome (Xie et al., 2012). Due to the lack of oocyte DNA methylome at base-resolution, about half of ICRs could not be classified definitively as gICRs or sICRs. Additionally, very little is known about the DNA methylation status of the ICRs and the expression patterns of imprinted genes in gametes and early embryos.
To address these questions, we performed comprehensive analyses on allele-specific, single-base resolution maps of 5mC, 5hmC and 5fC as well as gene expression profiling in early embryos, to investigate the reprogramming and inheritance of parental methylomes in mammals. In contrast to previous reports, we find that 5hmC and 5fC present in both paternal and maternal genomes, and illustrate that at least a significant proportion of maternal methylome undergoes active demethylation during early embryonic development. This study refines the current knowledge on methylation reprogramming in mammals, and provides a powerful resource for early developmental studies.
Results
Base-resolution DNA methylome of gametes, early embryos and PGCs
We characterized paternal and maternal methylomes using the single nucleotide polymorphisms (SNPs) between DBA/2J (DBA) and C57BL/6J (C57) mice in the hybrid embryos. We performed genome re-sequencing of the DBA and C57 mice, and identified about 4.4 million SNPs. We collected mouse sperm (DBA), oocytes (C57), as well as early stage embryos (male DBA × female C57). The different stages of embryos include 2-cell and 4-cell cleavage stages, the early inner cell mass (ICM), E6.5 and E7.5 embryos. We also purified the primordial germ cells (PGCs) from E13.5 male and female embryos, respectively. As more than 80% of zygotes were contaminated with sperm in zona pellucida, we were unable to collect enough zygotes to map the methylome.
All samples were extensively washed and purified to remove any somatic or gametic contaminants (Smith et al., 2012). Base-resolution methylomes were generated using MethylC-Seq (Lister et al., 2013; Lister et al., 2009; Xi and Li, 2009; Xie et al., 2013; Ziller et al., 2013). For each stage, at least two independent biological replicates were sequenced. The average genomic depth of uniquely mapped reads (Hon et al., 2013) is 16-fold per strand. Around 90% of all CpGs in mouse genome were covered at least five times in each sample (Table S1). The distribution of CpG methylation level of each site across the genome at different stages is shown in Figure S1A. Only CpGs that were covered at least five times were considered for subsequent analyses.
The dynamics of functional genomic elements during early embryogenesis
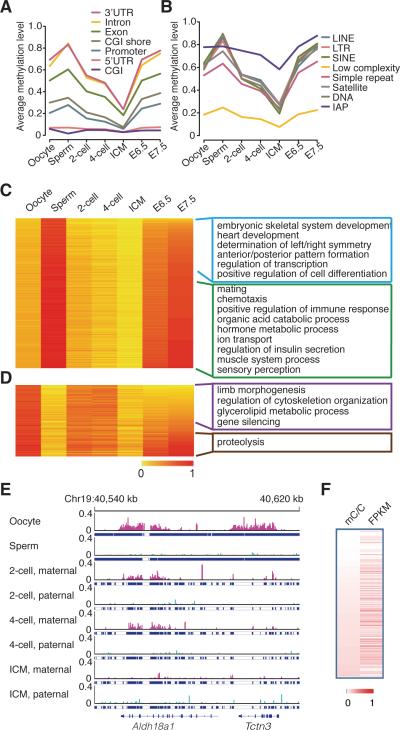
We plotted the average methylation levels of all different functional elements in the gametes and early embryos (Figure 1A, 1B, S1B), and revealed that most functional genomic elements undergo significant demethylation, except CpG Islands (CGIs) and 5’ untranslated regions (UTRs) whose methylation levels are already very low in gametes (Figure 1A). The average methylation levels of different functional elements gradually increase from ICM to E7.5 embryos, and the levels observed in E7.5 embryos are similar to levels seen in the sperm (Figure 1A, 1B, S1B). Only exception being CGI whose methylation level in E7.5 is similar to that of oocyte (Figure 1A). Interestingly, the average methylation level of CGIs in oocyte is higher than that of sperm, which is different from any other genomic elements (Figure 1A). We observed 297 oocyte-specific highly methylated CGIs, but only 20 sperm-specific highly methylated CGIs. Notably, 14 oocyte-specific highly methylated CGIs are located in the maternal ICRs (Table S2). Additionally, the methylation status of CGIs around transcriptional start sites (TSS) is stable, and the status of CGIs in genic region is more dynamic during early embryogenesis (Figure S1B right panel).
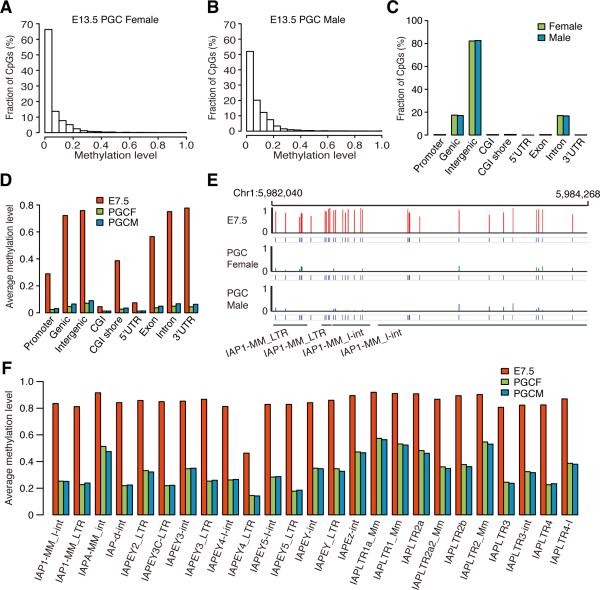
Figure 1. Dynamics of DNA methylomes for gametes and early embryos.
(A) The dynamics of methylation levels of different genomic elements (genic related classification) during early embryogenesis. The average methylation is the mean value of the methylation levels of all CpGs located in the specific element.
(B) The dynamics of methylation levels of different repeat elements. IAP is the sub-category of LTR.
(C) and (D) Heat map of the methylation reprogramming of oocyte-specific hypomethylated promoters and sperm-specific hypo-methylated promoters during early embryogenesis respectively. GO term enrichment in genes with E7.5 hypomethylated promoters or hyper methylated promoters. DNA methylation level is colored from orange to red to indicate low to high.
(E) Graphical representation of a genomic region showing the methylation level of non-CpGs for tracked maternal DNA (pink) and paternal DNA (light Blue) separately. Tracked non-CG cytosines are highlighted by short blue lines. “pa” means paternal DNA; “ma” means maternal DNA.
(F) Positive-correlations between gene expression and methylated non-CG cytosines in the genic regions. Pearson correlation coefficiency is 0.20 (p value < 10−5). Each row represents one gene from the total 22,742 expressed genes. mC/C means the density of methylated non-CG cytosines. FPKM represents gene expression level. Both DNA methylation density and gene expression are colored from white to red to indicate low to high.
See also Figure S1 and S2, Table S1, S2 and S3.
Previous studies have indicated that the methylation level of IAPs in oocyte is lower than that in sperm, and the methylation level of IAP in ICM is same as that in oocyte (Seisenberger et al., 2013). In contrast, our data reveals that the methylation levels of IAPs in sperm and oocyte are the same (0.78) (Figure 1B), and the methylation level of IAPs in ICM is 0.58. The methylation levels of different IAPs show a decreasing trend, particularly some sub-classes of IAPs such as IAP1-MM_I-int and IAPEY4_LTR (Figure S1C).
Dynamics of DNA methylation in promoters
Given that the DNA methylation at promoters is related to gene expression (Feng et al., 2010; Lister et al., 2009; Zemach et al., 2010), we characterized the dynamics of the gametic-specific promoters from gametes to early embryos, and performed the gene ontology (GO) analyses using the list of genes with differentially methylated promoter regions.
Our results show genes with sperm-specific hypomethylated promoters are enriched in spermatogenesis pathways; and oocyte-specific hypomethylated promoters are enriched in metabolism and hemostasis pathways (Table S3). Furthermore, genes with sperm-specific highly methylated hypo-methylated but hyper-methylated promoters in E7.5 embryos are enriched in the pathways that play important roles in fertilization and host defenses during early development, such as mating, ion transport, defense response and immune response pathways (Figure 1C green window). While genes with oocyte-specific methylated promoters but hyper-methylated promoters in E7.5 are enriched in proteolysis which is important for the sperm entrance into the oocyte (Figure 1D brown window). And genes with hypomethylated promoters in E7.5 embryos are enriched in processes regulating embryogenesis and organ development, such as anterior/posterior and left/right pattern formation, limb morphogenesis, skeletal system development, cardiac development, and cell differentiation (Figure 1C blue window, Figure 1D purple window, and Table S3).
It is known that growing oocyte preferentially metabolize pyruvate (one kind of organic carboxylic acid) over glucose, and embryos gradually switch to glycerolipid metabolic process during embryogenesis (Collado-Fernandez et al., 2012). Interestingly, GO enrichment results are consistent with this observation showing that genes with hypo-methylated promoters in oocyte but hyper-methylated promoters in E7.5 embryos are enriched with GO terms organic acid catabolic process (Figure 1C green window), and genes with hypo-methylated promoters in both sperm and E7.5 embryos are enriched in glycerolipid metabolic process (Figure 1D purple window). These data suggest that DNA methylation reprogramming associates with the embryonic development.
CpGs with stable methylation states during early embryogenesis
Single-base resolution methylomes of gametes and early embryos enable us to investigate how much of gametic epigenetic information can be stably maintained during early embryogenesis. We defined a CpG site as stably methylated CpG only if the standard deviation of the methylation level in sperm (or oocyte), ICM and E7.5 embryos is less than 0.05 (FDR < 10%). Our analyses revealed that 6.8% of CpGs in the mouse methylome are maintained stably among sperm, ICM and E7.5 embryos. Most of the stable CpGs are either unmethylated sites (methylation level ≤ 0.2), or highly methylated CpGs (methylation level ≥ 0.8) (Figure S2A). The majority of highly methylated CpGs are located in the introns and repeat regions (Figure S2B). In contrast, the majority of unmethylated CpGs are located in the promoters and CGIs (Figure S2B), suggesting that maintenance of these regions in unmethylated states is important for gene expression. The similar result was observed for the stably maintained methylated CpGs among oocyte, ICM and E7.5 embryos (Figure S2C and S2D).
Non-CG methylation in early embryos
Besides CpG methylation, non-CG cytosine methylation can also be observed in mammalian oocytes (Lister et al., 2013; Xie et al., 2012). But their functional role is unclear. Using whole genome bisulfite sequencing approach, we were able to quantify non-CG cytosine methylation in the gametes and early embryos. We did not observe non-CG cytosine methylation in the sperm DNA. The average level of non-CG cytosine methylation in maternal DNA is 0.027. The methylation level of non-CG cytosine in maternal DNA is gradually decreased during early embryogenesis (Figure S2E). In oocyte, the average non-CG methylation level of genic regions including exons and introns is 0.042, while the level of intergenic regions is 0.012, suggesting enrichment of non-CG methylated cytosines in the genic regions (Figure 1E). To investigate whether the non-CG cytosine methylation is associated with gene expression, we performed mRNA-seq on oocytes from C57 mice. Three independent libraries were generated. Our analyses show that non-CG cytosine methylation in genic regions is positively correlated with gene expression (Figure 1F). In contrast, negative correlation is observed between gene expression and CpG methylation in promoters (Figure S2F).
Allele-specific dynamics of CpG methylation during early embryogenesis
Using the SNPs between DBA and C57 mice, we were able to distinguish allele-specific CpG methylation patterns in the hybrid embryos. The number of tracked CpGs at each stage is listed in Table S4. Our data show that the average methylation levels at each specific stage are similar between genome-wide CpGs and SNP tracked CpGs (paternal reads and maternal reads merged together) (Figure S3A). In addition, the distribution of CpGs in different genomic elements is also similar between SNP tracked CpGs and genome-wide CpGs (Figure S3B, S3C). Therefore, the SNP tracked CpGs provide an unbiased representation of the genome-wide CpGs.
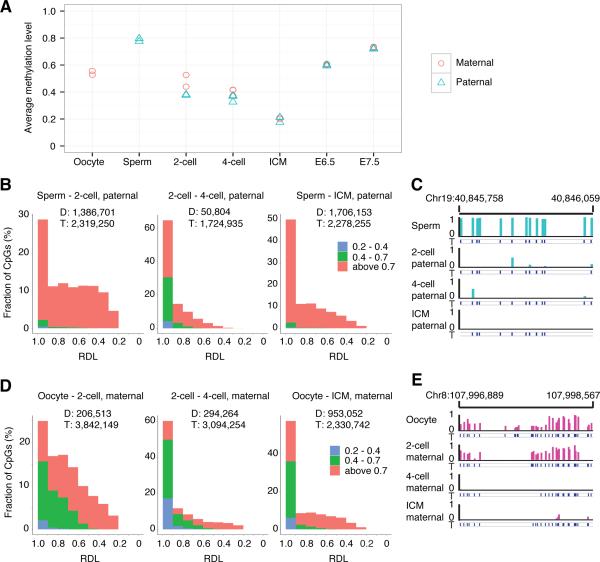
Next, we plotted the dynamics of the average methylation levels of SNP tracked paternal and maternal genomes, respectively (Figure 2A). Paternal DNA methylome is largely demethylated from sperm (0.80) to 2-cell embryos (0.37). In E3.5 ICM, paternal methylome reaches the lowest level (0.21). Surprisingly, limited change was observed in the methylation levels of maternal DNA between oocyte (0.54) and 2-cell embryos (0.49) (Figure 2A). Upon the specification to ICM, maternal methylome reaches the lowest level (0.20), similar to the level observed in paternal methylome (Figure 2A). Comparison of paternal and maternal methylomes in ICM shows similar patterns, except limited regions such as imprinting control regions. We also sequenced the methylome of the whole embryos of blastocysts (E3.5). The methylation level of blastocysts is 0.20 similar to that of ICM. With further embryonic development, the methylation levels of both paternal and maternal methylomes synchronously increase to around 0.73 in E7.5 embryos (Figure 2A).
Figure 2. Both paternal and maternal DNAs are actively demethylated.
(A) The dynamics of the average methylation level of paternal and maternal genome during early embryogenesis, respectively.
(B) Distribution of paternal demethylated CpGs according to the RDL between two compared stages. Y axis represents the fraction of demethylated CpGs. X axis represents the relative demethylation level. “D” means the number of the demethylated CpGs between two compared stages; “T” means the number of CpGs covered in both two compared stages with at least 5 reads. The CpGs were categorized into three classes according to the methylation level indicated by different colors. For example, in the left panel, red indicates the highly methylated CpGs in sperm (methylation level (ML) >0.7), green indicates intermediate methylated CpGs in sperm (0.7≥ML>0.4), and blue indicates low methylated CpGs in sperm (0.4≥ML>0.2). 63% demethylated CpGs with RDL higher than 0.6 from sperm to 2-cell embryos; 95% demethylated CpGs with RDL higher than 0.6 from 2-cell embryos to 4-cell embryos; 82% demethylated CpGs with RDL higher than 0.6 from sperm to ICM.
(C) Graphical representation of methylation pattern in paternal DNA at a locus in sperm, 2-cell embryos, 4-cell embryos, and ICM. The tracked CpGs are highlighted by short blue lines.
(D) Distribution of maternal demethylated CpGs according to the RDL between two compared stages. 74% demethylated CpGs with RDLs higher than 0.6 from oocyte to 2-cell embryos; 86% demethylated CpGs with RDL higher than 0.6 from 2-cell embryos to 4-cell embryos; 84% demethylated CpGs with RDL higher than 0.6 from oocyte to ICM. (E) Graphical representation of methylation pattern from maternal DNA at a locus for oocyte, 2-cell, 4-cell, and ICM.
See also Figure S3 and S4 and Table S4
Active DNA demethylation in both paternal and maternal CpG methylomes
To validate whether paternal methylome undergoes active demethylation, we compared the methylation level of each paternal tracked CpG site between sperm and 2-cell embryos. We called a CpG site as demethylated CpG if its methylation level decreases more than 0.2 between two compared stages (p value < 0.05 according to Fisher's exact test; FDR < 10%). Out of 2.32 million tracked CpGs covered in both sperm and 2-cell stage embryos, we observed 1.39 million CpGs to be demethylated (Figure 2B left panel). We calculated the relative demethylation level (RDL) for all paternal tracked CpGs. The RDL of sperm versus 2-cell embryos is defined as [(MLsperm – ML2-cell) / MLsperm], and ML means methylation level. Then, we plotted the distribution of the RDLs for 1.39 million demethylated CpGs (Figure 2B), and the rest of CpGs (Figure S4A). The results show there are 63% demethylated CpGs with RDL higher than 0.6 from sperm to 2-cell embryos in the paternal methylome (Figure 2B, left panel). In contrast, the distribution of RDL for non-demethylated sites is different, and Figure S4A shows that the RDLs of the majority sites are around 0. We also applied the RDL analyses to the demethylated CpGs from 2-cell embryos to 4-cell embryos, and observed 95% demethylated CpGs with RDL higher than 0.6 from 2-cell to 4-cell embryos (Figure 2B middle panel). These data indicate that paternal methylome is actively demethylated (Figure 2B). A representative region is shown in Figure 2C.
We performed the similar analysis for the maternal DNA. As the methylation change between oocytes to 2-cell embryos is very small, we focused on the RDL analysis between 2-cell and 4-cell embryos. Out of 3.09 million maternal tracked CpGs, 0.294 million CpGs are demethylated between 2-cell and 4-cell embryos. The results show there are 86% demethylated CpGs with RDL higher than 0.6 (Figure 2D middle panel), which is similar to paternal DNA demethylation. Therefore, if a CpG site in maternal DNA is demethylated, it usually undergoes active DNA demethylation (Figure 2D, 2E, S4B). Our results are in contrast to previous reports that maternal DNA demethylation is through passive dilution by inhibiting DNMT1 in early embryogenesis (Seisenberger et al., 2013; Wu and Zhang, 2014). We also compared how many demethylated sites overlapped between the transition of 2-cell to 4-cell stages and the transition of 4-cell to ICM stages in maternal genome, and found a small number of demethylated sites are overlapped (Figure S4C). This result indicates that demethylation of different CpG sites are initiated at different stages. For example, one locus is demethylated from 2-cell to 4-cell stages (Figure 2E), and another locus is demethylated from 4-cell stage to ICM stages (Figure S4D).
Taken together, both the paternal and maternal methylomes, at the majority of demethylated CpGs, go through active demethylation from gametes to 4-cell embryos.
5hmC exists in both paternal and maternal genomes
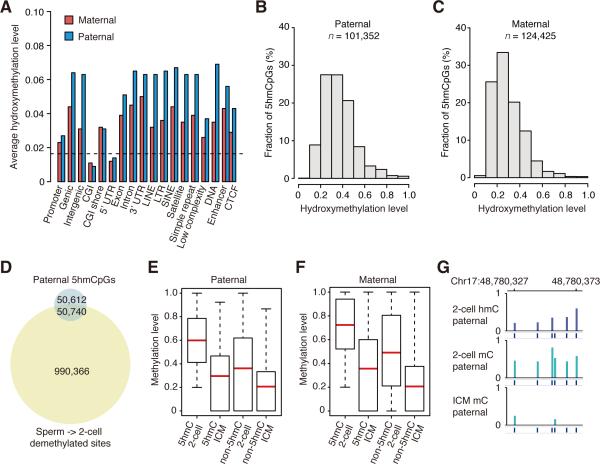
It has been proposed that paternal DNA demethylation is mediated by the passive dilution of oxidized 5mC bases (Inoue et al., 2011; Inoue and Zhang, 2011; Wu and Zhang, 2014). We performed single-base resolution 5hmC mapping in 2-cell embryos with TAB-seq (Yu et al., 2012). The sequencing depth is 18-fold per strand, covering 93% of CpGs in the mouse genome. The Tet non-conversion rate is 3.56% (Table S5). Because Tet oxidation can only take place at the methylated CpGs, the background hydroxymethylation level of 2-cell genome is 0.016. This is achieved by multiplying non-conversion rate with the methylation level at 2-cell stage (0.45). After the subtraction of the genomic background, the average hydroxymethylation level in 2-cell genome is 0.030.
In contrast to previous studies, the allelic specific analyses revealed that 5hmC exists not only in the paternal DNA, but also in the maternal DNA (Table S5). After the subtraction of the genomic background, the average hydroxymethylation levels of paternal and maternal tracked CpGs are 0.049 and 0.020 respectively. We determined the average hydroxymethylation levels of different genomic elements, and the results suggest that most of the genomic elements have higher than the background levels in both paternal DNA and maternal DNA, except CGIs and 5’UTRs (Figure 3A and S5A). This is consistent with the fact that the DNA demethylation occurs at most genomic elements, except CGIs and 5’UTRs. Additionally, hydroxymethylation levels of paternal DNA in different genomic elements are higher than those of maternal DNA (Figure 3A and S5A), consistent with the observation that average hydroxymethylation level of paternal DNA is higher than that of maternal DNA.
Figure 3. 5hmC in both maternal and paternal genomes.
(A) Average hydroxymethylation level of different genomic elements. The black dash line indicates the genomic background level (0.016). The average hydroxymethylation level is the mean value of the hydroxymethylation levels of all CpGs located in the specific element. Since there is no enhancer, CTCF data available for mouse 2-cell embryos, we used the data of mESC as the reference (Shen et al., 2012).
(B) Distribution of 5hmCpGs in paternal genome according to the hydroxymethylation level. Y axis represents the fraction of 5hmCpGs. X axis represents the hydroxymethylation level.
(C) Distribution of 5hmCpGs in maternal genome according to the hydroxymethylation level.
(D) Venn diagram shows that a small proportion of CpGs are overlapped between 5hmCpGs in 2-cell embryos and paternal demethylated CpGs from sperm to 2-cell embryos.
(E) Box plots of methylation levels of 5hmCpG sites or non-5hmCpG sites in paternal tracked DNA in 2-cell embryos and ICM. “5hmC” means 5hmCpGs. “non-5hmC” means non-5hmCpGs. Red line indicates the average methylation level, edges stand for the 25th/75th percentile, and whiskers stand for the 2.5th/97.5th percentile.
(F) Box plots of methylation levels of 5hmCpG sites or non-5hmCpG sites in maternal tracked DNA in 2-cell embryos and ICM.
(G) Graphical representation of 5hmC, 5mC pattern at a locus with paternal tracked CpGs in 2-cell embryos and ICM.
The hydroxymethylation modification is not universally distributed at each CpG site (Figure S5B and S5C). Using binomial distribution analysis (FDR < 5%), we show that only 101,352 CpGs in paternal DNA can be called as enriched 5-hydroxymethylated CpGs (5hmCpGs) (Figure 3B, Table S5). The average hydroxymethylation level of these enriched 5hmCpGs is 0.38 (Figure 3B). Similarly, there are 124,425 CpGs called as enriched 5hmCpGs in maternal genome, and the average hydroxymethylation level of these sites is 0.31 (Figure 3C, Table S5). We also plotted the distribution of the enriched 5hmCpG sites in different genomic elements (Figure S5D). In addition, 1,649 and 1,227 5hmCpG-enriched regions are identified in maternal and paternal genomes respectively (Figure S5E). Further analyses also show that 5hmCpG distribution is asymmetric between Watson and Crick strands (Figure S5F), consistent with the previous observations (Yu et al., 2012).
If demethylation of a CpG site is through the passive dilution of 5hmC, this demethylated CpG site should also be a 5hmCpG site. To identify how many CpGs may be demethylated by the passive dilution of 5hmC, we compared the demethylated CpGs with enriched 5hmCpGs. We found that 4.9% demethylated CpGs are also enriched 5hmCpGs (CpGs are covered by at least 5 reads in each dataset) (Figure 3D). Given that the sequencing depth can affect the calling of enriched 5hmCpGs and demethylated CpGs, the number of overlapped CpGs between these two sets is under-estimated with the cut-off of at least 5 reads covered for each CpG site, because some CpGs with sufficient coverage in mCpG dataset do not have sufficient coverage in 5hmCpG dataset. To reduce the effect of the sequencing depth, we raised the depth of cut-offs to at least 10, 15 and 20 reads covered for each CpG site, and found that 7.1%, 9.1% and 10.8% of paternal demethylated CpGs are enriched 5hmCpGs respectively. In anyway, the demethylation of the majority CpGs is not through the passive dilution of 5hmC. Further analyses show that 5hmCpG sites are usually located at the highly methylated CpG sites in both paternal and maternal DNAs (Figure 3E, 3F). Representative 5hmCpG enriched regions are shown in Figure 3G and S5G. In summary, our data show that 5hmC exists in both maternal and paternal genomes, and demethylation of the majority of CpGs is not mediated by the passive dilution of 5hmC.
Base resolution 5fC map of 2-cell embryos
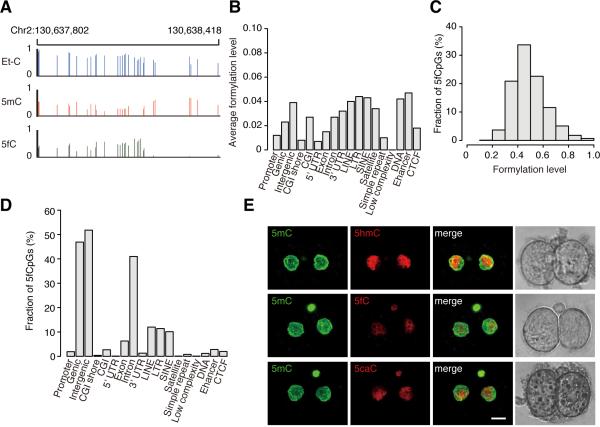
We generated a base-resolution 5fC map of 2-cell embryos by using the 5fC chemically assisted bisulfite sequencing (fCAB-seq) method (Song et al., 2013). The fCAB-seq method is based on the O-ethylhydroxylamine (EtONH2) protection of 5fC against bisulfite-mediated deamination, followed by bisulfite sequencing (BS-seq). The mapped CpGs from EtONH2-treated BS-seq data will be referred to Et-CpG in this paper. Given that Et-CpGs are the sum of 5mCpGs, 5hmCpGs and 5fCpGs, and the mapped CpGs from traditional BS-seq are the sum of 5hmCs and 5mCs, we generated the base-resolution 5fC map of 2-cell embryos by comparing EtONH2-treated BS-seq and traditional BS-seq datasets of the same DNA sample. The sequencing depth of EtONH2-treated BS-seq is 18-fold per strand. A representative locus with 5fC sites is shown in Figure 4A. Since the average Et-CpG level of genome-wide CpGs is 0.457 and average methylation level of the same sample is 0.432, the average formylation level for genome-wide CpGs is 0.025 (0.457-0.432). We further calculated the average formylation level (Figure 4B, S6A, S6B, and S6C) and plotted the distribution of CpGs at different genomic elements (Figure S6D and S6E), showing that the highly formylated elements are also the demethylated elements (Figure 1A, 1B, S1B).
Figure 4. 5fC and 5caC in 2-cell embryos.
(A) Graphical representation of the Et-CpG level, methylation level and formylation level at a locus in 2-cell embryos.
(B) The average formylation level of different genomic elements in genome-wide DNA. The average formylation level is the mean value of the formylation levels of all CpGs located in the specific element.
(C) Distribution of significant enriched 5fCpG sites according to the formylation level. Y axis represents the fraction of 5fCpGs. X axis represents the formylation level.
(D) Distribution of the 5fCpG sites in the different genomic elements in 2-cell embryos. Y axis represents the fraction of 5fCpGs map to each genomic element versus all 5fCpGs.
(E) Comparing the relative amount of 5hmC, 5fC and 5caC in 2-cell embryos with immunostaining. Antibody dilution is 1:2000 for Ab-5fC, 1:4000 for Ab-5hmC, and 1:5000 for Ab- 5caC. Scale bar is 10μm.
Our analyses show that under the current sequencing depth 0.95M CpGs are called as enriched 5-formylated CpGs (5fCpGs) in genome-wide according to Fisher's exact test (p value < 0.05, FDR < 5%), and the average formylation level of these enriched 5fCpGs is 0.50 (Figure 4C). We further determined the distribution of enriched 5fCpGs in different genomic elements (Figure 4D) and our results suggest that their distribution is similar to the distribution of 5hmC (Figure S5D).
5fC exists in both paternal and maternal genomes
We were able to determine the presence of 5fC in the parental genomes using our model system (Table S6). The average formylation level of 2-cell maternal genome is 2.8%, and the average formylation level of paternal genome is 2.0%. These data indicate that 5fC presents in both maternal and paternal DNA, consistent with the presence of 5hmC.
Previously, it was believed that 5fC is passively diluted during early embryogenesis. In contrast, our data show that 5%, 10% and 12% of paternal demethylated CpGs are also enriched 5fCpGs in 2-cell embryos with the cut-offs of 5, 10 and 15 reads covered for each CpG respectively. Therefore, the majority of CpGs are not demethylated through the passive dilution of 5fC, similar to the results of 5hmC.
5caC in 2-cell embryos
We explored whether the passive dilution of 5caC has any major contribution to the DNA demethylation in early embryogenesis. Until now, there is no reliable method to detect the 5caC at base-resolution level. Therefore, we performed the immunostaining of 5fC, 5caC and 5hmC to compare the relative amount of the 5caC, 5hmC and 5fC in 2-cell embryos. We titrated the antibody (Ab) concentrations for 5hmC, 5fC and 5CaC with the same amount of 5hmC, 5fC, and 5caC oligos by using dot blot assay. Our results show that 1:4000 Ab-5hmC, 1:2000 Ab-5fC, and 1:5000 Ab-5caC can generate similar signals in the presence of the same amount of 5hmC, 5fC, and 5caC oligos respectively (Figure S6F). Then 5fC, 5caC and 5hmC immunostaining images of 2-cell embryos were taken under the same conditions, including the exposure time and laser power. Image analysis results suggest that 5caC is less than 5hmC in 2-cell embryos (Figure 4E). Therefore, the contribution of passive dilution of 5caC to DNA demethylation would also be very limited.
In summary, 5hmC, 5fC, 5caC results together with RDL analyses show that 5mC or its oxidized derivatives, at the majority of the demethylated sites, are converted to unmodified cytosines independent of the passive dilution from gametes to 4-cell embryos. But, we cannot rule out the possibility that demethylation of limited loci can be partially mediated by the passive dilution.
DNA methylomes of E13.5 PGCs
It has been reported that the methylation level of E13.5 PGCs is the lowest point during PGCs development (Ficz et al., 2011; Hackett et al., 2013; Seisenberger et al., 2012). Therefore, we measured the methylomes of E13.5 PGCs from male and female progenies separately (Table S1). The average methylation level of female and male E13.5 PGCs is 0.063 and 0.077 respectively. Further analyses show that almost all of the CpGs are unmethylated in both male and female progenies (Figure 5A and 5B) and only about 0.3% of CpGs are highly methylated CpGs (methylation level ≥ 0.8), which is significantly different from the patterns observed in ICM (Figure S1A). Most of the highly methylated sites are located in the intergenic regions, especially in LTR (Figure 5C, S7A, and S7B). Our results show that almost all the different genomic elements appear to be unmethylated state (methylation level < 0.1) (Figure 5D and S7C). Interestingly, the methylation levels of all the different IAP subclasses are largely decreased (Figure 5E and 5F). The overall average methylation level of IAPs decreased from 0.88 in E7.5 embryos to 0.41 in E13.5 PGCs. In IAPs, only about 6% of CpGs are highly methylated CpGs. These observations are different from previous reports that IAPs are resistant to DNA demethylation (Morgan et al., 1999; Popp et al., 2010; Seisenberger et al., 2012; Seisenberger et al., 2013). In summary, our results indicate that DNA methylome is almost completely erased before germ cell specification.
Figure 5. DNA methylation pattern for E13.5 PGCs.
(A) Distribution of CpGs according to the methylation level in female E13.5 PGCs.
(B) Distribution of CpGs according to the methylation level in male E13.5 PGCs.
(C) The distribution of highly methylated CpGs (methylation level ≥ 0.8) in different genomic elements (genic related classification) for PGCs.
(D) Average methylation levels of different genomic elements (genic related classification) in E7.5, male PGCs and female PGCs.
(E) Graphical representation of methylation pattern of four IAPs in E7.5 embryos, male E13.5 PGCs, and female E13.5 PGCs.
(F) Average methylation levels of all the different IAPs in E7.5 embryos, female E13.5 PGCs, and male E13.5 PGCs.
See also Figure S7.
Classification of ICRs into gICRs and sICRs
Genomic imprinting in mammals is crucial for embryonic development (Surani et al., 1990). Around 55 ICRs have been identified in mouse (Xie et al., 2012). However, due to the absence of high coverage and single-base resolution methylomes for oocyte and early embryos, 25 ICRs haven't been classified as gICRs or sICRs. Comparative analyses of the oocyte, sperm and early embryos methylomes confirmed that all the previous known gICRs are indeed differentially methylated regions (DMRs) between oocyte and sperm (DMR > 0.5), and all the known sICRs are not DMRs between oocyte and sperm (DMR < 0.3, Table S7). Moreover, for those 25 unclassified ICRs, our data show that 11 of them are maternal gICRs, 10 are maternal sICRs, and the remaining 4 are paternal sICRs (Table 1, Table S7). In addition, the high-resolution methylome data provided additional information to correctly annotate the gICRs that were poorly defined (Table S7). Classifying the ICRs into the gICRs and sICRs is essential to determine their roles in embryonic development or in specific tissues.
Table 1.
ICRs in mouse
| Maternal | Paternal | ||
|---|---|---|---|
| Known as glCR | Mcts2/H13, Nespas/Gnasxl, Gnasla, Peg10/Sgce, Mest (Pegl), Herc3/Nap1l5, Peg3/Usp29 , Inpp5f, Snurf/Snrpn, Kcnq1ot1, Plagl1, Grb10, Zrsr1/Commd1, Slc38a4, Airn/Igf2r, Impact, Peg13 | H19 ICR, Dlk1-Gtl2 IG, Gpr1/Zdbf2, Rasgrf1 | |
| glCRs | New classified glCR | H13 DMR2 (3' end), Casc1 intragenic, 6330408a02Rik 3' end, AK086712 promoter, Neurog3 upstream, Grb10 DMR2 (intragenic), FR149454 promoter, FR085584 promoter, Nhlrc1 downstream, Myo10 intragenic, Pvt1 promoter | |
| sICRs | Known as sICR | Ndn, Mkrn3, H19 promoter, Dlk1 | Nesp, Cdkn1c, Gtl2, Igf2r |
| New classified slCR | Snrpn U exon, U80893 5' upstream, mir344b, mir344, mir344-2, mir344g, Magel2-Mrkn3 intergenic, Peg12, Commd1 DMR2 (intragenic), Magel2 | Vwde promoter, Cdkn1c upstream, Gtl2-Mirg diffuse DMR, Eif2c2 diffuse DMR |
All the known ICRs are classified into gICR or sICR according to the methylation states in sperm and oocyte.
Xie et.al identified an imprinting control site mir344c (locating on Chromosome 7: 68982367) in 129X1/SvJ and Cast/EiJ crossed mice. We did not find this CpG site in either DBA or C57 mouse. The SNP in 129 and CAST strains results in a strain-specific imprinting control site (Xie et al., 2012). These data suggest that DNA sequence variations sometimes could have big impact on the epigenetic state, which could further impact development.
Dynamics of ICR
It is known that the methylation of gICRs is resistant to demethylation during early embryogenesis. Our data support this observation (Figure 6A, Table S7). However, three gICRs namely Gnas1a, H13 DMR2 (3’ end) and Gpr1/Zdbf2, undergo DNA demethylation upon ICM (Figure 6B, Table S7). In addition, maternal sICR Commd1 DMR2 (intragenic) is sperm-specific highly methylated region (methylation level in sperm is 0.95, and methylation level in oocyte is 0.23), which is fully demethylated by ICM (Table S7), and becomes a maternal sICR in neuron (Xie et al., 2012).
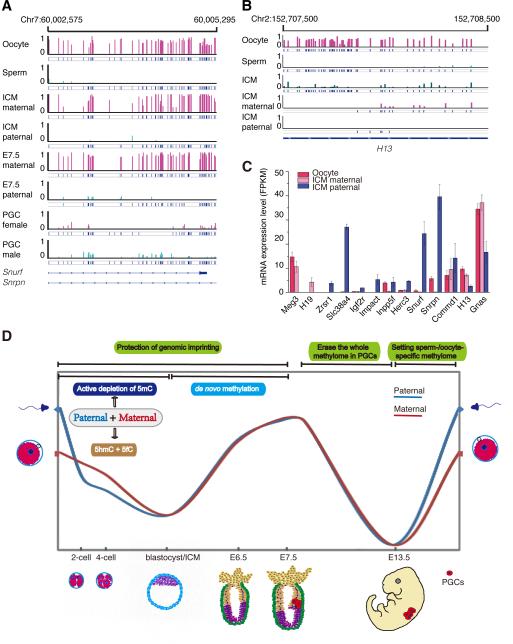
Figure 6. gICRs dynamics and imprinted gene expression in gametes and early embryos.
(A) Graphical representation of the methylation pattern of gICR Snurf/Snrpn in gametes, ICM, E7.5 embryos and PGCs. The graph shows that the ICR is maintained in early embryos, but erased in E13.5 PGCs.
(B) Graphical representation of the methylation pattern for gICR H13 DMR2 (3’ end) in gametes and ICM. The graph shows that this gICR is erased during early embryogenesis.
(C) Allele-specific gene expression of previously defined imprinted genes in oocyte and ICM. Data are represented as mean +/- SEM of three independent repeats.
(D) Two waves of genome-wide demethylation in mammals. 5hmC and 5fC present in both maternal and paternal genomes. Genome-wide demethylation is mainly through active demethylation in both paternal and maternal genomes, which is independent of the passive dilution of 5mC or its oxidized derivatives.
Expression of the imprinted genes in ICM
gICRs are known to be essential for early development. We investigated the expression pattern of imprinted genes during early embryogenesis. We performed mRNA-seq on ICM cells from hybrids of C57 strain and PWK/PhJ mice. Three independent libraries were generated. The allele-specific expression shows that two paternal imprinted genes H19 and Meg13 are expressed in the ICM cells, and many maternal germ-line imprinted genes are specifically expressed from paternal genome in ICM (Figure 6C). But three previously defined maternal germ-line imprinted genes, Commd1, H13 and Gnas, express from both maternal and paternal DNA. Our data show that two ICRs are related to gene H13, and one ICR H13 DMR2 (3’end) is demethylated in ICM (Table S7). Meanwhile, two ICRs are related to gene Commd1, and one ICR Commd1 DMR2 (intragenic) is demethylated in ICM (Table S7). For gene Gnas1a, its ICR is also demethylated in ICM (Table S7). Perhaps, it remains to be determined whether these three genes belong to germ-line imprinted genes.
Discussion
Active DNA demethylation in both paternal and maternal genome
Recently, genome-scale methylomes were achieved in mouse gametes and early embryos by using reduced representation bisulfite sequencing, which covered about 5% of the genome (Smith et al., 2012). The results from Smith et.al support the genome-wide DNA demethylation in mammals. These studies presented limited information to distinguish the dynamic difference between paternal and maternal methylome reprogramming. Currently, it is believed that that paternal DNA is actively demethylated and the maternal DNA is passively demethylated (Seisenberger et al., 2013; Wu and Zhang, 2014). In contrast, by generating single-base resolution, allele-specific whole-genome methylomes, we demonstrate that paternal methylome and at least a significant proportion of maternal methylome goes through active demethylation during embryonic development (Figure 2). There are still a number of demethylated CpGs whose relative demethylation levels are around 0.5 between two compared stages. Our data cannot exclude the possibility of passive dilution for these CpGs. Another possibility is that the erasure of methylation is cell-specific at each embryonic stage and there are populations of cells at different stages of this process.
It has been hypothesized that oxidized 5mC bases only exist in paternal DNA but not in maternal DNA (Inoue and Zhang, 2011; Iqbal et al., 2011; Nakamura et al., 2012), and that active DNA demethylation in early embryos is mediated by the passive dilution of oxidized 5mC bases. In contrast, our data clearly demonstrate that 5hmC and 5fC exist in both paternal and maternal genomes. 5mC or its oxidized derivatives, at the majority of demethylated CpGs, are converted to unmodified cytosines independent of passive dilution from gametes to 4-cell embryos (Figure 3 and 4). Recent studies have suggested that 5hmC or 5caC can be directly converted back to cytosine (Liutkeviciute et al., 2009; Zhang et al., 2012), suggesting that an enzyme may exist in mammals which can directly convert the oxidized 5mC bases back to cytosines during mammalian early embryogenesis.
Evolutionary comparisons of methylation reprogramming and the establishment of germ-line imprinting in mammals
Genome-wide demethylation is essential for mammalian early embryogenesis. In zebrafish and Xenopus, no genome-wide demethylation occurs in early embryogenesis (Jiang et al., 2013; Potok et al., 2013; Veenstra and Wolffe, 2001). These data suggest that the genome-wide demethylation is acquired only for some special functions unique to mammals, but not to all the vertebrates. Additionally, it has also been proposed that genome-wide demethylation may be only required for the regulation on a few important genes (Hackett and Surani, 2013).
A limited number of imprinted genes, such as Igf2r, play a very important role in development by preventing the overgrowth of embryos in the placenta (Iwasa, 1998; Ludwig et al., 1996; Moore et al., 1995). gICRs are stably maintained during mammalian embryonic development. To erase the parental allele-specific methylation state before resetting the sperm- or oocyte-specific methylome, genome-wide complete demethylation takes place during PGC development (Figure 6A). During early embryogenesis, genome-wide DNA methylation reprogramming happens in order to fulfill two goals. Firstly, the epigenetic states of sperm and oocyte are dramatically different, which need to reset to the same state during early embryogenesis. The majority of the DMRs between maternal and paternal genomes can be erased by global DNA demethylation. Secondly, the ICRs need to be maintained to regulate the embryonic size and growth during development. During early embryogenesis, gICRs which bind with Stella are resistant to the demethylation machinery (Nakamura et al., 2007). In addition, Tet3 has the sequencing selectivity that cannot target Stella binding regions (Nakamura et al., 2012), suggesting that Tet3 cannot oxidize the imprinted regions. Our data show that the demethylation of different sites is initiated at different stages, which also suggests the existence of the sequencing selectivity for DNA demethylation. Taken together, using the demethylation strategy with the sequencing selectivity, the parental allele-specific methylated regions (genomic imprinting) can be protected from reprogramming and be maintained during early embryogenesis. Therefore, two waves of genome-wide demethylation are necessary for imprinting in mammals (Figure 4D).
In contrast to other vertebrates, such as zebrafish and Xenopus, whose embryonic development occurs in vitro, there is no need for imprinting to avoid the embryos overgrowth, and does not require genome-wide demethylation. During zebrafish embryogenesis, paternal methylome is stably maintained in paternal genome; and maternal genome acts as one unit to reprogram to a methylome identical to the sperm methylome (Jiang et al., 2013). In another word, during zebrafish embryogenesis, no sequences at parental allele-specific methylated regions are able to be selected to avoid the reprogramming. This strategy cannot generate the parental allele-specific DMRs. Therefore, mammals cannot employ the strategy of zebrafish to establish the genomic imprinting.
Therefore, evolutionary comparisons of methylation reprogramming between mammals and other vertebrates indicate that the two waves of genome-wide demethylation are required for setting up genomic imprinting in mammals, which in turn facilitate the embryonic development in the placenta. It suggests that genome-wide demethylation is a major innovation in the evolutionary transition to placental viviparity.
In summary, this study refines the knowledge of the inheritance and reprogramming of parental methylomes in mammals, and also provides a powerful resource for the future developmental studies.
EXPERIMENTAL PROCEDURES
Sample Collection
The cross of two mouse strains was performed using DBA/2J as the paternal strain and C57BL/6J as the maternal strain. Gametes and embryos were collected. Samples were serially washed with KSOM (Millipore) to deplete any somatic contaminants and collected at minimal volume before snap freezing. Female and male E13.5 PGC samples (B6; 129S4-Pou5f1tm2Jae/J) were collected separately, digested with collagenase and purified using a FACSAria cell sorter (see Extended Experimental Procedures).
MethylC-Seq Library Preparation
Sonicated DNA was subjected to end-repair, A-tailing, and ligation using the NEBNext Kit. The size of the adaptor-ligated DNA was selected by gel electrophoresis. Bisulfite conversion was performed using the EZ DNA methylation-Gold Kit (Zymo Research) according to the manufacturer's instructions (see Extended Experimental Procedures).
TAB-Seq Library Preparation
Purified and sonicated genomic DNA with spike-in controls was glucoslyated and oxidized using 5hmC TAB-Seq Kit (WiseGene) according to the manufacturer's instructions. Prepared DNA was subjected to library construction following the standard protocol of methylC-Seq library preparation.
fCAB-Seq Library Preparation
Hydroxylamine protection of 5fC was performed following the standard protocol (Song et al., 2013). Purified hydroxylamine-protected DNA was treated with bisulfite conversion and subsequent methylC-Seq library preparation procedures (see Extended Experimental Procedures).
Supplementary Material
Highlight.
At least a significant proportion of maternal methylome is actively demethylated
5hmC and 5fC exist in both maternal and paternal genome
DNA demethylation is independent of the passive dilution of oxidized 5mC bases
Classification of all the known ICRs into germ-line ICRs or somatic ICRs
ACKNOWLEDGMENTS
This work was supported by grants from the 973 Program of China to X.H. (2010CB945101), to J.L. (2011CB510101); from National Natural Science Foundation of China to J.L. (91219104 and 81171902), to W.C. (91231112 and 31171244), to J.Z. (31200958). C.H. is supported by National Institutes of Health (HG006827). We thank the sequencing facility in BIG, CAS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
X.H. and J.L. conceived the study and L.W., X.H. and J.L. facilitated its designs. L.W., J.Z. and X.G. collected samples. L.W. performed methylation and hydroxymethylation profiling. L.W. and X.L. performed formylation profiling. L.Y. performed the mRNA library construction. L.W., J.D., W.Z., and J.Z. performed the bioinformatics analyses. G.L., W.L., Q.Z., N.A., F.T., and C.H. provided critical technical assistance and expertise. L.W., J.Z., J.D., X.G., W.C., X.H. and J.L. interpreted the data. L.W., N.A., X.H. and J.L. wrote the paper with the assistance of other authors.
Accession number
The GEO accession number for the sequencing data reported in this paper is GSE56697.
References
- Collado-Fernandez E, Picton HM, Dumollard R. Metabolism throughout follicle and oocyte development in mammals. The International journal of developmental biology. 2012;56:799–808. doi: 10.1387/ijdb.120140ec. [DOI] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. Conservation and divergence of methylation patterning in plants and animals. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, Surani MA. Germline DNA Demethylation Dynamics and Imprint Erasure Through 5-Hydroxymethylcytosine. Science. 2013;339:448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett Jamie A., Surani MA. Beyond DNA: Programming and Inheritance of Parental Methylomes. Cell. 2013;153:737–739. doi: 10.1016/j.cell.2013.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashizaki Y, Shibata H, Hirotsune S, Sugino H, Okazaki Y, Sasaki N, Hirose K, Imoto H, Okuizumi H, Muramatsu M, et al. Identification of an imprinted U2af binding protein related sequence on mouse chromosome 11 using the RLGS method. Nat Genet. 1994;6:33–40. doi: 10.1038/ng0194-33. [DOI] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiura H, Sugawara A, Ogawa H, John RM, Miyauchi N, Miyanari Y, Horiike T, Li Y, Yaegashi N, Sasaki H, et al. A tripartite paternally methylated region within the Gpr1-Zdbf2 imprinted domain on mouse chromosome 1 identified by meDIP-on-chip. Nucleic Acids Res. 2010;38:4929–4945. doi: 10.1093/nar/gkq200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon GC, Rajagopal N, Shen Y, McCleary DF, Yue F, Dang MD, Ren B. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nature genetics. 2013;45:1198–1206. doi: 10.1038/ng.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Shen L, Dai Q, He C, Zhang Y. Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res. 2011;21:1670–1676. doi: 10.1038/cr.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci U S A. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasa Y. The conflict theory of genomic imprinting: how much can be explained? Curr Top Dev Biol. 1998;40:255–293. doi: 10.1016/s0070-2153(08)60369-5. [DOI] [PubMed] [Google Scholar]
- Jiang L, Zhang J, Wang J-J, Wang L, Zhang L, Li G, Yang X, Ma X, Sun X, Cai J, et al. Sperm, but Not Oocyte, DNA Methylome Is Inherited by Zebrafish Early Embryos. Cell. 2013;153:773–784. doi: 10.1016/j.cell.2013.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsey G, Bodle D, Miller HJ, Beechey CV, Coombes C, Peters J, Williamson CM. Identification of imprinted loci by methylation-sensitive representational difference analysis: application to mouse distal chromosome 2. Genomics. 1999;62:129–138. doi: 10.1006/geno.1999.6022. [DOI] [PubMed] [Google Scholar]
- Lalande M. Parental imprinting and human disease. Annu Rev Genet. 1996;30:173–195. doi: 10.1146/annurev.genet.30.1.173. [DOI] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liutkeviciute Z, Lukinavicius G, Masevicius V, Daujotyte D, Klimasauskas S. Cytosine-5-methyltransferases add aldehydes to DNA. Nat Chem Biol. 2009;5:400–402. doi: 10.1038/nchembio.172. [DOI] [PubMed] [Google Scholar]
- Ludwig T, Eggenschwiler J, Fisher P, D'Ercole AJ, Davenport ML, Efstratiadis A. Mouse mutants lacking the type 2 IGF receptor (IGF2R) are rescued from perinatal lethality in Igf2 and Igf1r null backgrounds. Dev Biol. 1996;177:517–535. doi: 10.1006/dbio.1996.0182. [DOI] [PubMed] [Google Scholar]
- Moore T, Hurst LD, Reik W. Genetic conflict and evolution of mammalian X-chromosome inactivation. Dev Genet. 1995;17:206–211. doi: 10.1002/dvg.1020170305. [DOI] [PubMed] [Google Scholar]
- Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, et al. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nature cell biology. 2007;9:64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Liu YJ, Nakashima H, Umehara H, Inoue K, Matoba S, Tachibana M, Ogura A, Shinkai Y, Nakano T. PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. Nature. 2012;486:415–419. doi: 10.1038/nature11093. [DOI] [PubMed] [Google Scholar]
- Peters J, Wroe SF, Wells CA, Miller HJ, Bodle D, Beechey CV, Williamson CM, Kelsey G. A cluster of oppositely imprinted transcripts at the Gnas locus in the distal imprinting region of mouse chromosome 2. Proc Natl Acad Sci U S A. 1999;96:3830–3835. doi: 10.1073/pnas.96.7.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plass C, Shibata H, Kalcheva I, Mullins L, Kotelevtseva N, Mullins J, Kato R, Sasaki H, Hirotsune S, Okazaki Y, et al. Identification of Grf1 on mouse chromosome 9 as an imprinted gene by RLGS-M. Nat Genet. 1996;14:106–109. doi: 10.1038/ng0996-106. [DOI] [PubMed] [Google Scholar]
- Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potok Magdalena E., Nix David A., Parnell Timothy J., Cairns Bradley R. Reprogramming the Maternal Zebrafish Genome after Fertilization to Match the Paternal Methylation Pattern. Cell. 2013;153:759–772. doi: 10.1016/j.cell.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W, Reik W. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell. 2012;48:849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seisenberger S, Peat JR, Reik W. Conceptual links between DNA methylation reprogramming in the early embryo and primordial germ cells. Current Opinion in Cell Biology. 2013;25:281–288. doi: 10.1016/j.ceb.2013.02.013. [DOI] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012 doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C-X, Szulwach Keith E., Dai Q, Fu Y, Mao S-Q, Lin L, Street C, Li Y, Poidevin M, Wu H, et al. Genome-wide Profiling of 5-Formylcytosine Reveals Its Roles in Epigenetic Priming. Cell. 2013;153:678–691. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani MA, Kothary R, Allen ND, Singh PB, Fundele R, Ferguson-Smith AC, Barton SC. Genome imprinting and development in the mouse. Dev Suppl. 1990:89–98. [PubMed] [Google Scholar]
- Veenstra GJ, Wolffe AP. Constitutive genomic methylation during embryonic development of Xenopus. Biochim Biophys Acta. 2001;1521:39–44. doi: 10.1016/s0167-4781(01)00280-9. [DOI] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y, Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics. 2009;10:232. doi: 10.1186/1471-2105-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Barr Cathy L., Kim A, Yue F, Lee Ah Y., Eubanks J, Dempster Emma L., Ren B. Base-Resolution Analyses of Sequence and Parent-of-Origin Dependent DNA Methylation in the Mouse Genome. Cell. 2012;148:816–831. doi: 10.1016/j.cell.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Schultz Matthew D., Lister R, Hou Z, Rajagopal N, Ray P, Whitaker John W., Tian S, Hawkins RD, Leung D, et al. Epigenomic Analysis of Multilineage Differentiation of Human Embryonic Stem Cells. Cell. 2013;153:1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- Zhang L, Lu X, Lu J, Liang H, Dai Q, Xu GL, Luo C, Jiang H, He C. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat Chem Biol. 2012;8:328–330. doi: 10.1038/nchembio.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Muller F, Donaghey J, Tsai LTY, Kohlbacher O, De Jager PL, Rosen ED, Bennett DA, Bernstein BE, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.