Abstract
Staufen (STAU)1-mediated mRNA decay (SMD) degrades mammalian-cell mRNAs that bind the double-stranded (ds)RNA-binding protein STAU1 in their 3′-untranslated region. We report a new motif, which typifies STAU homologs from all vertebrate classes, that is responsible for human (h)STAU1 homodimerization. Our crystal structure and mutagenesis analyses reveal that this motif, now named the Staufen-swapping motif (SSM), and dsRNA-binding domain 5 (‘RBD’5) mediate protein dimerization: the two SSM α-helices of one molecule interact primarily through a hydrophobic patch with the two ‘RBD’5 α-helices of a second molecule. ‘RBD’5 adopts the canonical α-β-β-β-α fold of a functional RBD, but it lacks residues and features needed to bind duplex RNA. In cells, SSM-mediated hSTAU1 dimerization increases the efficiency of SMD by augmenting hSTAU1 binding to the ATP-dependent RNA helicase hUPF1. Dimerization regulates keratinocyte-mediated wound-healing and, undoubtedly, many other cellular processes.
INTRODUCTION
In mammals, STAU1 mediates embryonic stem-cell differentiation1, mRNA transport and localization2,3, mRNA translational activation4, human immunodeficiency virus type 1 assembly5,6 and SMD7–10. During SMD, STAU1 triggers the translation-dependent degradation of specific mRNAs that contain a STAU1-binding site (SBS) within their 3′-untranslated region (3′UTR) as a means to regulate gene expression during myogenesis7, keratinocyte motility10, adipogenesis11 and, most likely, other mammalian cellular pathways. In human cells, SBSs can be created in cis by intramolecular base-pairing within an mRNA 3′UTR9 or in trans by base-pairing between partially complementary Alu elements within an mRNA 3′UTR and a long noncoding RNA10. When translation terminates sufficiently upstream of an SBS so as not to disrupt the SBS, association of the UPF1 RNA helicase with SBS-bound STAU1 triggers mRNA decay (reviewed in ref. 12).
Generally, similarly numbered STAU RBDs from different species are more identical than are differently numbered RBDs within the same protein13, suggesting a common overall design of RBDs in STAU homologs. Human (h)STAU1 has 496- and 577-amino acid isoforms (NCBI Gene ID:6780; hSTAU155 and hSTAU163, respectively), each of which contains RBDs 2–5 (refs. 14,15), and an additional isoform with six amino acids inserted into hSTAU155 RBD3 that diminish dsRNA binding in the mouse ortholog16. Only RBD3 and RBD4 bind dsRNA in mammalian cells15,17(thus, we hereafter refer to RBD2 and RBD5 as, respectively, ‘RBD’2 and ‘RBD’5), and RBD3 binds dsRNA with higher affinity than does RBD4 (refs. 15,17). All three hSTAU1 isoforms also contain a tubulin-binding domain (TBD) situated between RBD4 and ‘RBD’5, which binds tubulin in in vitro studies of the mouse STAU1 (ref. 15).
The hSTAU1 paralog, hSTAU2, has 479-, 504-, 538- and 570-amino acid isoforms (NCBI Gene ID: 27067; hSTAU252, hSTAU256, hSTAU259 and hSTAU262, respectively), each of which contains RBDs 2, 3 and 4, and only the N- and C-terminal regions of what would be hSTAU1 ‘RBD’5 (ref. 18); additionally, hSTAU256 and hSTAU262 have a complete RBD1, whereas hSTAU252 and hSTAU259 contain a truncated RBD1 (refs. 3,18,19). Like hSTAU1, hSTAU2 mediates not only mRNA decay20 but also mRNA localization3. Each paralog and even some of their isoforms may function and localize differently within cells3,19,21.
The three-dimensional analyses of STAU proteins have been limited to two RBD structures. The first is the NMR structure of Drosophila melanogaster STAU RBD3 bound to a 12-bp stem-loop RNA, which revealed the interaction of the canonical α-β-β-β-α RBD fold with dsRNA22,23. The second is of mouse STAU2 RBD4 in the absence of dsRNA (PDB ID: 1UHZ; RIKEN Structural Genomics Initiative), which also showed the α-β-β-β-α fold. In general, evidence for structure- or sequence-specific recognition of cognate RNAs by RBDs remains elusive. RBD1 and RBD2 of mouse adenosine deaminase ADAR2 recognize distinct bases within a human pre-mRNA GluR-2 stem-loop because of subtle sequence and structural differences in their RNA-interacting regions24. However, what hSTAU1 recognizes when it binds dsRNA remains unknown.
Recently, Martel et al.25 demonstrated using cultured cells that multiple hSTAU155 molecules can bind to the SMD target encoding human ADP ribosylation factor (hARF)1 (ref. 9). Using yeast two-hybrid analyses, the authors identified a region in ‘RBD’2 and a region containing ‘RBD’5 that separately interact with full-length hSTAU155; and using cultured cells, ‘RBD’5 appeared to mediate the stronger interaction25. We recently discovered that some SBSs consist of intermolecular duplexes of partially complementary Alu elements that range from 86 to 298 nucleotides10 and might support the binding of more than one hSTAU1 molecule. Thus, we set out to investigate the details of hSTAU1–hSTAU1 interactions to understand the role of hSTAU1 dimerization in SMD.
We identified a region of hSTAU1 that includes a new motif, which we call the STAU-swapping motif (SSM). We found that the SSM (i) is conserved in all vertebrate STAU homologs examined, (ii) resides N-terminal to ‘RBD’5, to which it is connected by a flexible linker, and (iii) is responsible for forming hSTAU1 dimers in cells. Our crystal structure reveals that the two SSM α-helices interact with the two ‘RBD’5 α-helices. Mutagenesis data demonstrate that the interaction is ‘domain-swapped’ between two molecules so as to result in hSTAU1 dimerization. This capacity for dimerization is a previously unappreciated role for an RBD that no longer binds dsRNA. In cells, disrupting hSTAU1 dimerization by introducing deletion or point mutations into full-length hSTAU1 or by expressing exogenous ‘RBD’5 reduced the ability of hSTAU1 to co-immunoprecipitate with hUPF1 thereby reducing the efficiency of SMD. Remarkably, inhibiting SMD by disrupting hSTAU1 dimerization promoted keratinocyte-mediated wound-healing, suggesting that dimerization also inhibits the epithelial-to-mesenchymal transition during cancer metastasis.
RESULTS
Vertebrate STAU has a conserved motif N-terminal to ‘RBD’5
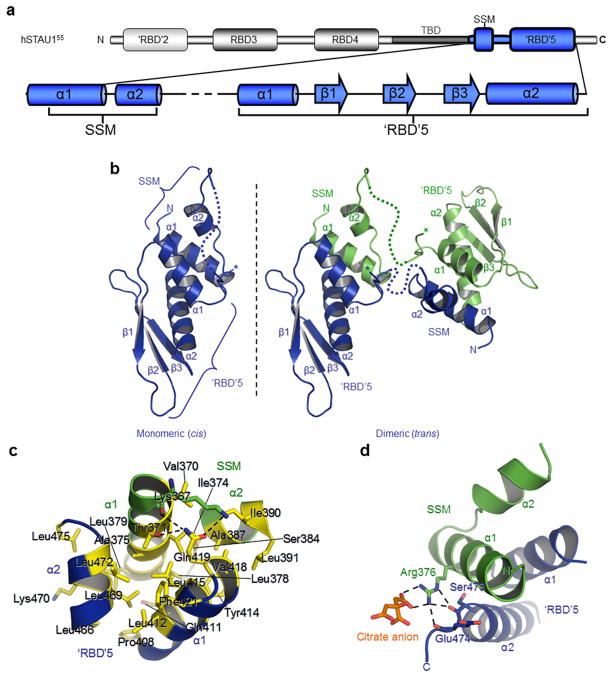
Using yeast two-hybrid analyses, Martel et al.25 demonstrated that full-length hSTAU155 interacts with amino acids 408–496 of another hSTAU155 molecule. These amino acids consist of the C-terminus of hSTAU155 and include ‘RBD’5 (Fig. 1a and Supplementary Fig. 1a), which has only 18% sequence identity to the prototypical hSTAU1 RBD3 and fails to bind dsRNA15,17. Using ClustalW26, multiple sequence alignments of full-length hSTAU1 with hSTAU2 and STAU orthologs from representatives of the five major vertebrate classes revealed a conserved sequence residing N-terminal to ‘RBD’5 that consists of hSTAU155 amino acids 371–390 (Supplementary Fig. 1a). We call this motif the Staufen-swapping motif (SSM; Fig. 1a and Supplementary Fig. 1a) for reasons explained below. Despite an identifiable ‘RBD’5, an SSM is absent from, e.g., D. melanogaster or Caenorabditis elegans STAU (Supplementary Fig. 1b). However, STAU in other invertebrates contain both SSM and ‘RBD’5 regions (Supplementary Fig. 1b). The SSM is proximal to the TBD, which spans amino acids 282–372 (ref. 15) (Fig. 1a), and it overlaps with amino acids 272–405, at least part of which recruits hUPF1 during SMD7.
Figure 1.
Comparison of vertebrate STAU sequences, and X-ray crystal structure of hSTAU1 SSM-‘RBD’5. (a) Modular organization of human (h)STAU155. Relative positions are indicated for the true dsRNA-binding domains (RBDs) 3 and 4, degenerate ‘RBD’2, the region that binds tubulin (TBD) in vitro (amino acids 282–372), and the STAU-swapping motif (SSM)-‘RBD’5 region (amino acids 367–476). The secondary structure of hSTAU155 represented in blue derives from data reported here, and the dotted line denotes amino acids 397–402 that are missing from the crystal structure. (b) X-ray crystal structure represented as a single hSTAU155 SSM-‘RBD’5 molecule (left, blue) and as a domain-swapped dimer formed with another molecule in the crystal lattice (right, green). Asterisks denote an alternative conformation of the linker (Supplementary Fig. 2c). Connectivity between an SSM and ‘RBD’5 that could not be modeled due to multiple conformations and/or disorder in the crystal structure are indicated by dashed lines. ‘RBD’5 structural elements are underlined. (c) Close-up of the interaction between the SSM α-helices (green α1 and α2) and the ‘RBD’5 α-helices (blue α1 and α2). Important residues are shown as stick representations and labeled. Residues that contribute to the hydrophobic core are yellow. Hydrogen bonds between polar atoms are shown as dashed lines. (d) Polar interactions between SSM Arg376 and ‘RBD’5 main-chain oxygens, where SSM is green, ‘RBD’5 is blue, and key residues and the citrate ion ligand (orange) are shown as sticks and labeled. Hydrogen-bonds are shown as dotted lines.
Structure of hSTAU1 SSM-‘RBD’5
A search of the NCBI Conserved Domain Database27 did not identify hSTAU1 ‘RBD’5 as an RBD. To understand the atomic details of SSM-‘RBD’5, we purified hSTAU1 amino acids 367–476 from E. coli (Supplementary Fig. 2a), produced crystals that we verified were intact using SDS-polyacrylamide electrophoresis and also silver-staining (Supplementary Fig. 2a), and solved its X-ray crystal structure at 1.7 Å (Table 1). Our structure revealed that ‘RBD’5 adopts the α-β-β-β-α topology of a prototypical RBD and that the SSM forms two α-helices (hereafter called SSM α1 and α2) that are connected by a tight turn (Fig. 1b and Supplementary Fig. 2b). Electron density was clearly interpretable for the SSM and ‘RBD’5 but not for amino acids 397–402 that constitute the linker (393–406) between SSM and ‘RBD’5 (Fig. 1a,b and Supplementary Fig. 1a). Two conformations were observed at the C-terminal or ‘RBD’5 side of the linker, each hinged at L405 so that the position of P404 was variable (Supplementary Fig. 2c). The observed variability raises the possibility that SSM may interact with ‘RBD’5 as a monomer (cis), dimer (trans), or both in the crystal structure (Fig. 1b), but we cannot correlate either linker conformation with a monomeric or dimeric state. Each 649 Å2 interface is created when the ‘V’-shape formed by SSM α1 and α2 straddles ‘RBD’5 α1, while the ‘V’-shape created by ‘RBD’5 α1 and α2 straddles SSM α1 (Fig. 1b–d).
Table 1.
Crystallographic data collection and refinement statistics
| Ethyl-Hg SAD a | Native 1 a | Native 2 a | |
|---|---|---|---|
| Data collection | |||
| Space group | P41212 | P41212 | P41212 |
| Cell dimensions | |||
| a (=b), c (Å) | 47.4, 80.9 | 48.5, 82.1 | 45.7, 86.1 |
| α(=β,=γ)(°) | 90.0 | 90.0 | 90.0 |
| Resolution (Å) b | 25.81–2.92 (2.94–2.92) | 50–2.20 (2.24–2.20) | 50–1.70 (1.76–1.70) |
| Rsym or Rmerge b | 34.2 (69.0) | 6.9(42.7) | 8.5 (26.3) |
| I/σI b | 11.2 (3.4) | 34.4 (6.7) | 28.9 (10.6) |
| Completeness (%) b | 99.6 (96.7) | 99.5(100.0) | 99.8(99.7) |
| Redundancy b | 10.5 (3.5) | 19.3 (20.0) | 13.8 (13.3) |
| Refinement | |||
| Resolution (Å) | 31.35–1.70 | ||
| No. reflections | 10,601 | ||
| Rwork/Rfree | 16.3/20.2 | ||
| No. atoms | |||
| Protein | 864 | ||
| Ligand/ion | 14 | ||
| Water | 63 | ||
| B-factors | |||
| Protein | 27.5 | ||
| Ligand/ion | 46.0 | ||
| Water | 33.9 | ||
| r.m.s deviations | |||
| Bond lengths (Å) | 0.011 | ||
| Bond angles (°) | 1.49 | ||
Dataset was obtained from a single respective hSTAU1 SSM-‘RBD’5 crystal.
Highest resolution shell values are in parentheses.
SAD, single-wavelength anomalous dispersion.
The intramolecular interactions of an SSM and an ‘RBD’5 form a core composed of residues with hydrophobic side chains (Fig. 1c). The external solvent boundary of this core is defined by Thr371 of the longer of the two SSM α-helices, α1; Ser384 of SSM α2; Gln411, Tyr414, and Gln419 of ‘RBD’5 α1; and Lys470 of ‘RBD’5 α2 (Fig. 1c). Each of these residues amphipathically contributes hydrophobic portions of their side chains to the core, with their polar component pointed outward. Val370, Ile374, Ala375, Leu378 and Leu379 of SSM α1 also contribute to the hydrophobic core as do Ala387, Ile390 and Leu391 of SSM α2; ‘RBD’5 α1 constituents Pro408 (which starts α1), Leu412, Leu415 and Val418; and Phe421 of L1 (Fig. 1c). Additionally, ‘RBD’5 α2 contributes Leu466, Leu469, Leu472 and Leu475 (Fig. 1c).
Of the two polar interactions at the SSM–‘RBD’5 interface, one a basic charge is contributed by SSM Arg376: its two η-amine groups hydrogen-bond with two carboxyl groups of the citrate anion present in the crystal structure, while its η- and ε-amines interact with the main-chain oxygens of, respectively, Glu474 and Ser473 that are positioned near the C-terminus of ‘RBD’5 α2 (Fig. 1d). SSM Arg376 is conserved in those vertebrates analyzed except for D. rerio, where the residue is Asn, and Glu474 and Ser473 are invariant in vertebrates that contain the ‘RBD’5 α2 C-terminus (Supplementary Fig. 1a). In the other polar interaction, the side-chain hydroxyl group of SSM Thr371 and the main-chain oxygen of Lys367 hydrogen-bond with the amine group of ‘RBD’5 Gln419, while the ε-amine of Lys367 hydrogen-bonds with the hydroxyl group of Gln419 (Fig. 1c). SSM residues lacking strict conservation, i.e., Met373, Tyr380, Gly381, Thr383 and Pro385, are positioned on the solvent-exposed side, opposite to the interface that interacts with ‘RBD’5 (Supplementary Fig. 2d).
Comparison of ‘RBD’5 to an RBD that binds dsRNA
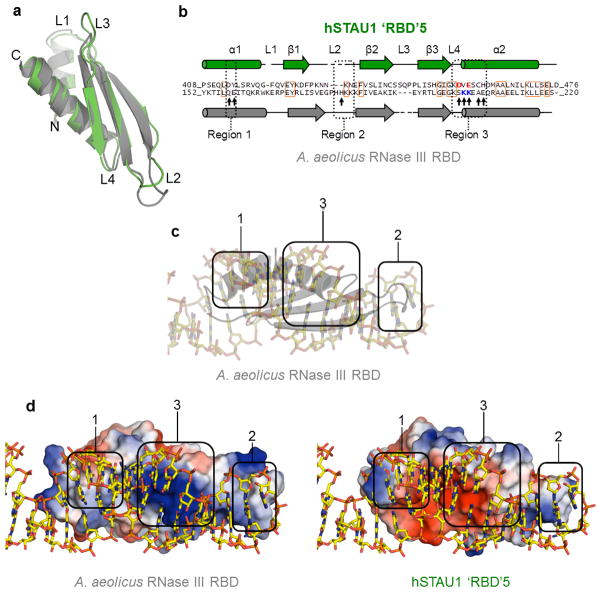
We were surprised that the three RBD structures identified by the Dali server28 to be structurally most similar to ‘RBD’5 do bind dsRNA (Supplementary Table 1). Of the three, Aquifex aeolicus RNase III RBD29 provides the most complete comparison. A structure-based sequence alignment of this RBD with hSTAU1 ‘RBD’5 revealed that while the two structures are nearly identical, hSTAU1 ‘RBD’5 has a slightly shorter loop (L)1, an altered L2, and a longer L3 (Fig. 2a,b). Furthermore, hSTAU1 ‘RBD’5 lacks key residues that typify the three RNA-binding regions (Regions 1, 2 and 3) of canonical RBDs23 and that are present in the A. aeolicus RNase III RBD (Fig. 2b). The most obvious differences reside in Region 2 (within L2) and Region 3. hSTAU1 ‘RBD’5 L2, which does not extend as far as A. aeolicus RNase III RBD L2 (Fig. 2a) and thus may be unable to reach the minor groove of dsRNA, lacks a His residue that in the A. aeolicus RNase III RBD29 and true RBDs23 interacts with the dsRNA minor groove (Fig. 2c). The importance of an L2 His residue derives from studies of D. melanogaster STAU RBD3 (Supplementary Fig. 3a), where RNA binding was lost when the sole L2 His was changed to Ala22. With regard to Region 3, the positively charged residues in the A. aeolicus RNase III RBD that interact with the negatively charged phosphate backbone spanning the dsRNA major groove are negatively charged in hSTAU1 ‘RBD’5 and might actually repel dsRNA (Figs. 2b–d). Consistent with this view, D. melanogaster STAU RBD3 (ref. 22) also maintains a basic charge in Region 3 (Supplementary Fig. 3a,b).
Figure 2.
Comparison of hSTAU155 ‘RBD’5 with an RBD that binds dsRNA. (a) Superimposition of hSTAU1 ‘RBD’5 (green) and A. aeolicus RNase III RBD29 (grey) using Dali28. (b) Structure-based sequence alignment of hSTAU155 ‘RBD’5 (top green sequence) and A. aeolicus RNase III RBD29 (bottom grey sequence). Conserved amino acids are in orange boxes. Key residues discussed in the text are indicated with an arrow. Structurally corresponding residues within Region 3 between the two proteins are blue if positively charged or red if negatively charged. (c) X-ray crystal structure of A. aeolicus RNase III RBD in complex with dsRNA within a hairpin29. Protein is grey and in cartoon-form, and dsRNA is in stick-representation. The three major dsRNA-interacting regions of typical RBDs23 are approximated using round-edged enclosures, numbered 1–3, to illustrate the important secondary structures. (d) Cartoon of the structure of A. aeolicus RNase III RBD bound to dsRNA (left) or STAU155 ‘RBD’5 superimposed on the structure of A. aeolicus RNase III RBD and shown in relation to the dsRNA in the structure of A. aeolicus RNase III RBD bound to dsRNA (right). Vacuum electrostatic potentials were generated using PyMOL to illustrate charge-variance, where on the surface representation of each protein blue is positive, red is negative and white is neutral. The three major dsRNA-interacting regions are encompassed as in c.
Human SSM-‘RBD’5 homodimerizes in solution and in cells
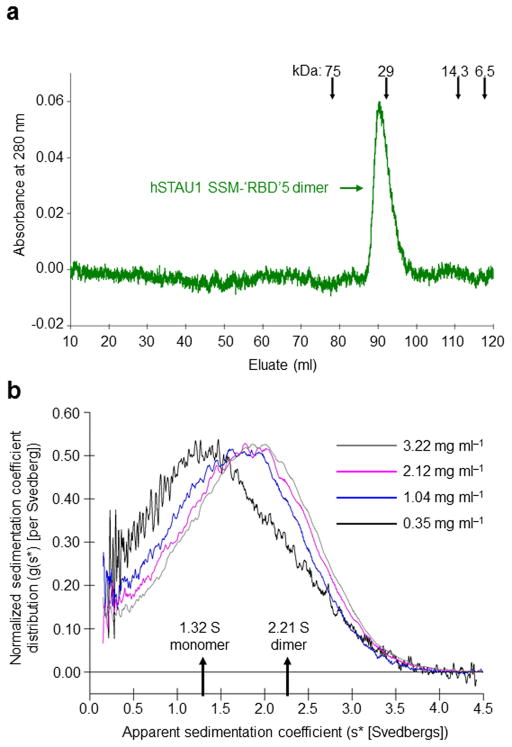
The crystal structure raised the possibility that the SSM could mediate hSTAU1 dimerization by trans interactions with ‘RBD’5. Thus, we tested whether the SSM-‘RBD’5 is sufficient to mediate dimerization of hSTAU1. After purifying GST-SSM-‘RBD’5 from E. coli and removing the GST tag, SSM-‘RBD’5 migrated during gel filtration at the size of a dimer (Fig. 3a). Sedimentation velocity determinations using analytical ultracentrifugation confirmed that the average weight-distribution of SSM-‘RBD’5 shifted to lower Svedberg values at lower concentrations (Fig. 3b). The best-fit model for SSM-‘RBD’5 [0.0090 mg ml−1 root mean standard deviation (rmsd) with 95% confidence limits] was one of rapid monomer (1.32 +0.02/−0.03 S)-dimer (2.21 ± 0.01 S) equilibrium where the dimer Kd was 79 ± 9 μM. That purified SSM-‘RBD’5 assumes a dimeric solution-state supports the existence of a trans, swapped interaction between the SSM of one hSTAU1 molecule and the ‘RBD’5 of another.
Figure 3.
hSTAU155 SSM-‘RBD’5 is a dimer in solution (a) Gel-filtration analysis of SSM-‘RBD’5, where size markers and the position of the SSM-‘RBD’5 dimer are indicated. (b) Analytical ultracentrifugation results support a model for the existence of a SSM-‘RBD’5 monomer–dimer equilibrium in solution. The normalized apparent sedimentation coefficient distribution (g(s*)) is plotted as a function of apparent Svedbergs (s*).
To determine if the SSM mediates dimerization of full-length hSTAU1 in vivo, human embryonic kidney (HEK)293T cells were transiently transfected with a mixture of two plasmids: (i) pEGFP-‘RBD’5, which produces monomeric enhanced green fluorescence protein (EGFP)-tagged ‘RBD’5, and either pmRFP-SSM-‘RBD’5 or pmRFP-‘RBD’5, which produces monomeric red fluorescence protein (mRFP)-tagged SSM-‘RBD’5 or mRFP-‘RBD’5, respectively; or (ii) pEGFP-SSM-‘RBD’5 and either pmRFP-SSM-‘RBD’5 or pmRFP-‘RBD’5 (Supplementary Fig. 4a). The results of IPs in the presence of RNase A using anti-GFP or, as a negative control, mouse (m) IgG revealed that dimerization cannot occur between two ‘RBD’5 molecules but can occur if one of two ‘RBD’5 molecules contributes an SSM (Supplementary Fig. 4a,b; see Supplementary Note 1 for extended details; see Supplementary Table 2 for IP and co-IP efficiencies).
To exclude the possibility that linker residues 393–406 contribute to the interaction between the SSM of one hSTAU1 molecule and ‘RBD’5 of another, we tested whether EGFP-SSM interacts with mRFP-‘RBD’5. HEK293T cells were transiently transfected with a mixture of two plasmids: one that produces EGFP-SSM, and the other that produces mRFP-SSM-‘RBD’5, pmRFP-‘RBD’5 or, as a negative control, pmRFP (Fig. 4a). Cell lysates were then generated and analyzed in the presence of RNase A before and after IP using anti-GFP or mIgG.
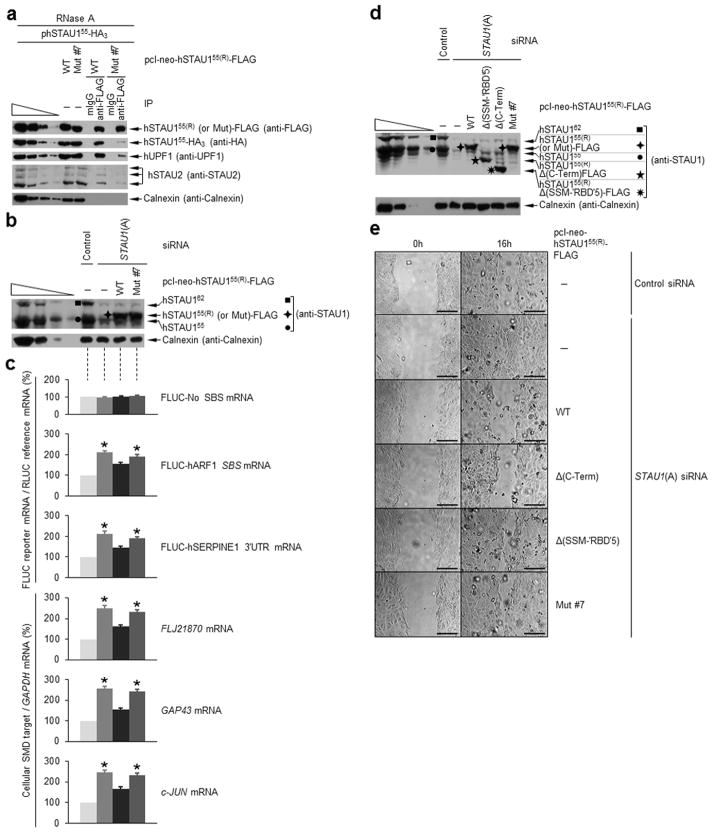
Figure 4.
‘RBD’5 and SSM interact in trans, and expression of either inhibits hSTAU1 dimerization and SMD. (a) Diagrams of the EGFP-tagged and mRFP-tagged proteins analyzed. Unshaded regions derive from hSTAU155. (b) Western blotting using the specified antibody of HEK293T-cell lysates before (−) or after IP using anti-GFP or, to control for nonspecific IP, mouse (m)IgG (Supplementary Table 2). Lysates derived from cells (1 × 107 cells per 150-mm dish) that had been transiently transfected with pEGFP-SSM (5 μg) and pmRFP (5 μg), pmRFP-‘RBD’5 (5 μg) or pmRFP-SSM-‘RBD’5 (5 μg) and subsequently treated with RNase. The four leftmost lanes analyze three-fold dilutions of lysate prior to IP. (c) As in b using lysates of HEK293T cells (1 × 107) transiently transfected with pcI-neo-hSTAU155(R)-FLAG (1 μg), phSTAU155-HA3 (10 μg) and pmRFP-‘RBD5’(5 μg) or pmRFP (5 μg). (d) Western blotting (upper) and histogram of RT-PCR (lower; Supplementary Fig. 4d) of lysates of HEK293T cells (1 × 107) transiently transfected with no plasmid (−) or 5 μg of pmRFP, pmRFP-‘RBD’5, pEGFP or pEGFP-SSM. After RT-PCR, the level of each cellular SMD target was normalized to the level of GAPDH mRNA, and the normalized level in the presence of no plasmid is defined as 100. All results are representative of three independently performed experiments. Error bars, s.e.m. *, n = 3, P < 0.05, determined using a one-tailed t-test.
Each mRFP-tagged protein or mRFP alone was expressed at a comparable level (Fig. 4b), and anti-GFP immunoprecipitated comparable amounts of EGFP-SSM (Fig. 4b). While EGFP-SSM did not co-immunoprecipitate with mRFP, it did co-immunoprecipate with mRFP-SSM-‘RBD’5 and mRFP-‘RBD’5 with comparable efficiencies, indicating that the linker does not significantly contribute to the interaction of the SSM and ‘RBD’5 when each derives from a distinct molecule (Fig. 4b). As expected, EGFP-SSM also co-immunoprecipitated with cellular hSTAU1 isoforms but not with cellular hUPF1 (Fig. 4b).
Disrupting hSTAU1 dimerization inhibits UPF1 binding and SMD
We next expressed mRFP-‘RBD’5 with the goal of inhibiting hSTAU1 dimerization. To this end, HEK293T cells were transiently transfected with siRNA-resistant (R) plasmids producing hSTAU155(R)-FLAG, hSTAU155-HA3 and either mRFP-‘RBD’5 or, as a negative control, mRFP. Cell lysates were then generated and analyzed in the presence of RNase A before and after IP using anti-FLAG or, as a negative control, mIgG.
Comparable amounts of hSTAU155(R)-FLAG were expressed and immunoprecipitated using anti-FLAG in the presence of a comparable amount of either mRFP or mRFP-‘RBD’5 (Fig. 4c). Furthermore, hSTAU155(R)-FLAG and hSTAU155-HA3 were not overexpressed relative to cellular hSTAU155 (Supplementary Fig. 4c). mRFP-‘RBD’5 expression reduced the amount of hSTAU155-HA3 that co-immunoprecipitated with hSTAU155(R)-FLAG to 35–40% of the amount that co-immunoprecipitated in the presence of mRFP alone (Fig. 4c). Our finding that mRFP-‘RBD’5 expression also reduced the amount of cellular hUPF1 that co-immunoprecipitated with hSTAU155(R)-FLAG to 35–40% of the amount that co-immunoprecipitated in the presence of mRFP alone (Fig. 4c), together with the finding that ‘RBD’5 does not bind hUPF1 (Fig. 4b)7, indicates that hUPF1 binds hSTAU1 dimers more efficiently than it binds hSTAU1 monomers.
We additionally examined the effect of mRFP-‘RBD’5 or EGFP-SSM, which we predicted would also inhibit hSTAU1-dimerization, on the efficiency of SMD by assaying the HEK293T-cell SMD targets FLJ21870, GAP43 and c-JUN mRNAs7,9. Each tagged protein was expressed in HEK293T cells comparably to its tag-only control (Fig. 4d). Transfections using plasmids expressing EGFP-SSM or mRFP-‘RBD’5 increased the abundance of each SMD target 2–2.5-fold relative to transfections using empty vector (pcI-neo) or plasmid expressing, respectively, EGFP or mRFP, none of which affected SMD target abundance (Fig. 4d and Supplementary Fig. 4d). Thus, hSTAU1 dimerization is critical for efficient SMD because dimerization augments hSTAU1 binding to hUPF1.
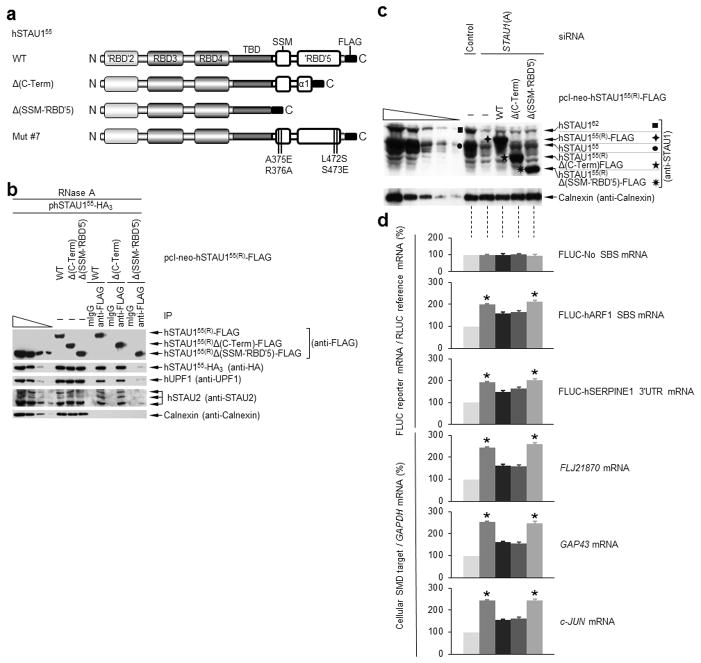
To define the minimal segment necessary for hSTAU1 dimerization in vivo, HEK293T cells were transiently transfected with pcI-neo-hSTAU155-HA3 and one of three siRNA-resistant plasmids that produce hSTAU155(R)-FLAG, hSTAU155(R)Δ(C-Term)-FLAG or hSTAU155(R)Δ(SSM-‘RBD’5)-FLAG, hereafter called WT, Δ(C-Term) or Δ(SSM-‘RBD’5), respectively (Fig. 5a). Cell lysates were generated and analyzed in the presence of RNase A before and after IP using (i) anti-FLAG or, as a negative control, mIgG or (ii) anti-HA or, as a negative control, rat (r)IgG.
Figure 5.
Sequences C-terminal to ‘RBD’5 α1 are not required for hSTAU155 dimerization, and dimerization promotes hUPF1 binding and SMD. (a) Diagrams of hSTAU155(R)-FLAG variants. (b) Western blotting of RNase A-treated HEK293T-cell lysates before (−) and after IP using anti-FLAG or mouse (m)IgG. Cells (1 × 107 cells per 150-mm dish) had been transiently transfected with 10 μg of phSTAU155-HA3 and 1 μg of pcI-neo-hSTAU155(R)-FLAG WT, Δ(C-Term) or Δ(SSM-‘RBD’5). (c) Western blotting (Supplementary Fig. 5a), where black symbols denote a hSTAU1 band, and (d) histogram of the RT-PCR analyses (Supplementary Fig. 5e) of HEK293T cells (1 × 107) that had been transiently transfected with the specified siRNA (100 nM) and, two days later, with the specified siRNA-resistant hSTAU155(R)-FLAG expression vector or pcI- neo (1 μg); a pFLUC-SMD reporter construct (3 μg), and the pRLUC reference plasmid (3μg). For the histograms, the level of each mRNA was normalized to the level of RLUC (for FLUC-SMD reporter mRNAs) or GAPDH mRNA (for cellular mRNAs), and normalized levels are presented as a percentage of the normalized level in the presence of Control siRNA, which is defined as 100. Results are representative of three independently performed experiments. Error bars, s.e.m. *, n = 3, P < 0.05, determined using a one-tailed t-test.
The three FLAG-tagged proteins were expressed at comparable levels prior to IP relative to each other (Fig. 5b) and relative to cellular hSTAU155 (Supplementary Fig. 5a) and were immunoprecipitated with comparable efficiencies using anti-FLAG (Fig. 5b). The level with which hSTAU155-HA3 or cellular hUPF1 co-immunoprecipitated with Δ(SSM-‘RBD’5) was only ~10% the level with which hSTAU155-HA3 or cellular hUPF1 co-immunoprecipitated with either WT or Δ(C-Term) (Fig. 5b). IPs of the same transfections using either anti-HA or, as negative control, rIgG revealed that the level with which Δ(SSM-‘RBD’5) co-immunoprecipitated with hSTAU155-HA was only ~10% the level with which WT or Δ(C-Term) co-immunoprecipitated with hSTAU155-HA3 (Supplementary Fig. 5b). Thus, domain-swapping between SSM and ‘RBD’5 is the major determinant of hSTAU1 dimerization and can be achieved even when one of the interacting proteins lacks residues C-terminal to ‘RBD’5 α1. Consistent with this conclusion, assays of the three detectable cellular hSTAU2 isoforms demonstrated that hSTAU2 co-immunoprecipitated with each hSTAU155(R)-FLAG variant, including Δ(C-Term), with the same relative efficiency as did hSTAU155-HA3 (Fig. 5b). Thus, hSTAU1 can homodimerize or heterodimerize with hSTAU2. Using anti-FLAG to immunoprecipitate a hSTAU155(R)-FLAG variant or anti-HA to immunoprecipitate hSTAU155-HA3, the co-IP of hUPF1 correlated with homodimerization ability (Fig. 5b and Supplementary Fig. 5b), in agreement with data obtained using mRFP-‘RBD’5 to disrupt dimerization (Fig. 4c). However, homodimerization did not augment the binding of hSTAU155 to an SBS because FLJ21870 mRNA and c-JUN mRNA each co-immunoprecipitate with WT, Δ(C-Term) or Δ(SSM-‘RBD’5) to the same extent (Supplementary Fig. 5c).
Since Δ(SSM-‘RBD’5) has residual dimerization activity (10% that of WT), and in view of reports that hSTAU1 ‘RBD’2 amino acids 37–79 interact with full-length hSTAU125, we assayed the ability of E. coli-produced hSTAU1-‘RBD’2-RBD3 (amino acids 43–173) to dimerize. Gel filtration demonstrated that hSTAU1-‘RBD’2-RBD3 indeed migrates at the position expected of an ‘RBD’2-RBD3–‘RBD’2-RBD3 dimer (Supplementary Fig. 5d). This low level of residual activity suggests that the contribution of ‘RBD’2 to hSTAU1 dimerization is relatively minor and as such was not pursued further.
Inhibiting hSTAU1 dimerization should inhibit SMD based on our finding that dimerization promotes the association of hSTAU1 with hUPF1. To test this hypothesis, HEK293T cells were transiently transfected with: (i) STAU1(A) siRNA8; (ii) plasmid expressing one of the three hSTAU155(R)-FLAG variants or, as a control, no protein; (iii) three plasmids that produce a firefly luciferase (FLUC) reporter mRNA, namely, FLUC-No SBS mRNA8, which lacks an SBS, FLUC-hARF1 SBS mRNA8, which contains the hARF1 SBS, and FLUC-hSERPINE1 3′UTR9, which contains the hSERPINE1 SBS; and (iv) a reference plasmid that produces renilla luciferase (RLUC) mRNA. In parallel, cells were transfected with (i) Control siRNA7, (ii) plasmid producing no hSTAU155(R)-FLAG protein, (iii) the three FLUC reporter plasmids, and (iv) the RLUC reference plasmid.
STAU1(A) siRNA reduced the abundance of cellular hSTAU1 to ~10% the level in Control siRNA-treated cells and that each hSTAU155(R)-FLAG variant was expressed at a comparable abundance that approximated the abundance of cellular hSTAU155 (Fig. 5c). After normalizing the level of each FLUC mRNA to the level of RLUC mRNA, the normalized level of FLUC-No SBS mRNA, which is not an SMD target, was found to be essentially identical in all transfections (Fig. 5d and Supplementary Fig. 5e), as expected. In contrast, the normalized level of FLUC-hARF1 SBS mRNA and FLUC-hSERPINE1 3′ UTR mRNA were increased ~2-fold in the presence of STAU1(A) siRNA alone, as were the normalized levels of mRNAs for FLJ21870, GAP43 and c-JUN mRNA, consistent with an inhibition of SMD (Fig. 5d). This inhibition was reversed by 50% when WT or Δ(C-Term) was expressed but not when Δ(SSM-‘RBD’5) was expressed (Fig. 5d). Thus, WT and Δ(C-Term) can functionally compensate for the siRNA-mediated downregulation of cellular hSTAU1 more efficiently than can Δ(SSM-‘RBD’5). These data indicate that hSTAU1 dimerization is important for SMD.
To define specific amino acids of hSTAU1 that contribute to domain-swapping, we used our X-ray crystal structure to design seven variants of hSTAU155(R)-FLAG that, relative to the deletion-bearing variants, would harbor more subtle changes (Fig. 5a and Supplementary Fig. 6a). Mutations were designed to target the SSM–‘RBD’5 interface and minimize any effects on the overlapping intramolecular hydrophobic interactions within ‘RBD’5 itself. When subjected to secondary structure predictions using PsiPred30,31, none of the mutations was predicted to disrupt the α-helical structure within which each resides.
Of the seven variants, only hSTAU155(R)-FLAG harboring A375E,R376A,L472S,S473E (called hereafter Mut #7) disrupted hSTAU155(R)-FLAG dimerization with hSTAU155-HA3 (Supplementary Fig. 6b). This variant contains a bulky substitution at residue 375, a change at residue 376 that disrupts one of the two polar interactions in the hSTAU1 SSM–‘RBD’5 interface, and L472S and S473E, both of which target residues within ‘RBD’5 α2 that interact with SSM α1 (Fig. 1c,d). Notably, T371R and Q419A, which disrupt the second polar interaction in the hSTAU1 SSM–‘RBD’5 interface, do not affect dimerization either individually or when combined in cis (Supplementary Fig. 6b).
Western blotting of lysates of HEK293T cells that transiently expressed comparable amounts of Mut #7 and hSTAU155-HA3 (Fig. 6a and Supplementary Fig. 6c) at a level that approximated the level of cellular hSTAU155 (Supplementary Fig. 6b) revealed that hSTAU155-HA3, cellular hUPF1 and isoforms of cellular hSTAU2 failed to co-immunoprecipitate efficiently with Mut #7 (Fig. 6a and Supplementary Fig. 6c). Also as expected, Mut #7 binding to FLJ21870 or c-JUN SMD targets was not compromised (Supplementary Fig. 6d). Consistent with the importance of hSTAU1 dimerization to SMD, Mut #7 was less able to reverse the STAU1(A) siRNA-mediated inhibition of SMD than was WT (Fig. 6b,c).
Figure 6.
hSTAU155 point mutations that disrupt dimerization inhibit hUPF1 binding and SMD, thereby precluding contributions of SMD toward inhibiting cell motility. (a–c) Essentially as in Fig. 5b–d, except using the specified siRNA-resistant hSTAU155(R)-FLAG expression plasmids. For (c), see Supplementary Fig. 6e for the RT-PCR analyses. Error bars, s.e.m. *, n = 3, P < 0.05, determined using a one-tailed t-test. (d,e) HaCaT keratinocytes (5 × 106 cells per 100-mm dish) were transiently transfected with the specified siRNA (100 nM) and, one day later, with the specified plasmid (3 μg). (d) Western blotting of lysates prior to scrape injury, and (e) phase-contrast microscopy of cells that were scrape-injured and analyzed immediately after scraping (0h) and at 16h. Scale bar represents 100 μm.
Disrupting STAU1 dimerization inhibits wound-healing
Downregulating the levels of SERPINE1 and RAB11FIP1 mRNAs, which are SMD targets, increases keratinocyte motility in a scrape-injury repair (i.e., wound-healing) assay10. To test the physiological importance of disrupting hSTAU1 dimerization, WT, Δ(C-Term), Δ(SSM-‘RBD’5) and Mut #7 were expressed individually at equal levels in human HaCaT keratinocytes that had been treated with STAU1(A) siRNA, which reduced cellular hSTAU1 abundance to 10% the level of Control siRNA-treated cells (Fig. 6d, where pcI-neo served as a control). After 16 hr, enhanced keratinocyte motility was evident in the presence of STAU1 siRNA alone, consistent with SERPINE1 and RAB11FIP1 proteins enhancing wound-healing10, and also when cellular hSTAU1 was replaced by Δ(SSM-‘RBD’5) or Mut #7, neither of which can dimerize to mediate SMD (Fig. 6e). From these findings together with data showing that replacing cellular hSTAU1 with either WT or Δ(C-Term), each of which supports hSTAU1 dimerization, had no effect on keratinocyte motility (Fig. 6e), we conclude that contributions of hSTAU1 dimerization to the efficiency of SMD are indeed important in promoting wound-healing.
DISCUSSION
hSTAU1 homodimerization is mediated by a new motif
Here we describe the hSTAU1 SSM, which is a two-helix motif (Fig. 1) that interacts with dsRNA-binding-deficient ‘RBD’5 of another hSTAU1 molecule (Figs. 1,3,4,5,6 and Supplementary Figs. 2 and 4–6). We propose that SSM is a modular adaptation in many and possibly all vertebrate STAU homologs that mediates STAU dimerization through its interaction with ‘RBD’5. Although the connectivity between SSM and ‘RBD’5 cannot be modeled, we suggest that the dynamic nature of the linker (Supplementary Fig. 2c) allows hSTAU1 SSM-‘RBD’5 to exist in both monomeric and dimeric states, and both states potentially exist in the crystal structure. We support our crystallographic model for dimerization by demonstrating that hSTAU1 SSM-‘RBD’5 dimers form in solution in vitro (Fig. 3) and in cells (Figs. 4–6 and Supplementary Figs. 4–6). If hSTAU1 multimerization were to occur in cells, it would likely involve not only SSM interacting with ‘RBD’5 in trans (Fig. 4) but also weaker contributions from ‘RBD’2 (ref. 25); Supplementary Fig. 5). Possibly, dimerization via intermolecular ‘RBD’2–‘RBD’2 interactions would promote trans over cis interactions between SSM and ‘RBD’5 interactions.
Data indicate that the minimal region of ‘RBD’5 from one molecule that is needed to interact with the SSM from another is ‘RBD’5 α1. First, sequences that reside C-terminal to ‘RBD’5 α1 are not required for hSTAU1–hSTAU1 dimerization (Fig. 5). Second, the smallest hSTAU2 isoform co-immunoprecipitates with hSTAU155 even though its ‘RBD’5 consists of only α1 and L1 (Figs. 1 and 5). Thus, all STAU1 isoforms can dimerize if not multimerize with themselves and/or with all STAU2 isoforms. We suggest that ‘RBD’5 α2 may stabilize dimer formation given that the SSM–‘RBD’5 interaction can be disrupted by simultaneously mutating both SSM and ‘RBD’5 α2 (Fig. 6). Additionally, mutations at the SSM–‘RBD’5 α1 interface fail to effectively disrupt dimerization, possibly due to the compensating presence of ‘RBD’5 α2 (Supplementary Fig. 6).
hSTAU1 homodimerization contributes to SMD
Compared to hSTAU1 monomers, hSTAU1 dimers bind hUPF1 more efficiently (and mediate SMD more effectively without promoting dsRNA binding (Figs. 4–6 and Supplementary Figs. 4–6). Thus, cells may regulate SMD by controlling hSTAU1 abundance32 and therefore dimer formation (Fig. 7). There is clear evidence that multiple hSTAU155 molecules can bind a single dsRNA. For example, multiple hSTAU155 molecules bind the hARF1 SMD target in cells25 and mRNA containing as many as 250 CUG repeats that typify patients with myotonic dystrophy in vitro33. Also, our finding that hSTAU155 stabilizes the relatively large (86–298 imperfectly base-paired) regions that constitute intermolecular SBSs formed between mRNAs and long noncoding RNA via Alu- element base-pairing10 suggest that multiple hSTAU1 molecules bind in tandem to the same dsRNA to efficiently recruit the ATP-dependent helicase hUPF1. Proteins known to dimerize and become activated on double-stranded nucleic acid are exemplified by transcriptional activators (for review, see ref. 34), the adenosine deaminases ADAR1 and ADAR2 (refs. 35,36), and the protein kinase PKR (for review see ref. 37).
Figure 7.
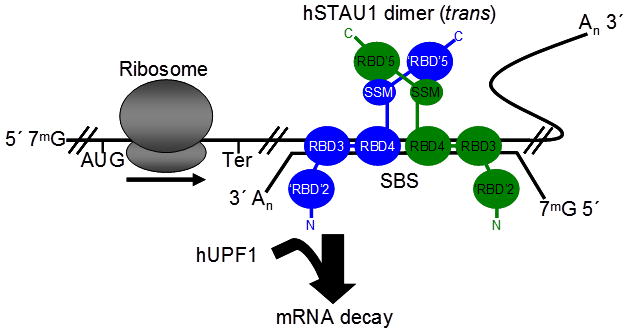
Model for how hSTAU1 dimers recruit hUPF1 to the 3′UTR of an mRNA that is targeted for SMD and promote its decay. hSTAU1 can bind to a STAU1-binding site (SBS), which is shown here as an intermolecular SBS formed by base-pairing of an mRNA 3′UTR (upper) with a long non-coding RNA (lower) but can also form through intramolecular base-pairing32, as a dimer if not multimer, depending on the length of the SBS. Dimerization is mediated by the domain-swapped interaction between the SSM of one hSTAU1 molecule (blue) and the ‘RBD’5 of another hSTAU1 molecule (green), and vice versa. Note that ‘RBD’2 may also facilitate hSTAU1–hSTAU1 dimerization25, which could change the orientation of the hSTAU1 dimer from what is illustrated. hSTAU1 dimerization promotes hUPF1 binding. Binding activates UPF1 helicase activity and initiates mRNA decay20. 7mG, 7-methyl guanosine cap; AUG, translation initiation codon; Ter, normal termination codon; An, poly(A) tail. For emphasis, molecules are not shown to scale.
hSTAU1 ‘RBD’5 has functionally diverged from a true RBD
Assuming hSTAU1 ‘RBD’5 evolved from a functional RBD, it not only lost the ability to bind dsRNA but gained the ability to interact with SSM. While RBD Regions 2 and 3 of true dsRBDs interact, respectively, with the minor groove and bridge the proximal major groove of dsRNA in true RBDs23, these Regions of ‘RBD’5 are mutated so as to be incapable of these functions (Fig. 2). Furthermore, in contrast to Region 1 of true RBDs, which determines RNA recognition specificity by binding the minor groove and possibly distinguishing features such as loops at the apex of dsRNA22,24, Region 1 of ‘RBD’5 specifies SSM recognition (Fig. 1). Notably, ‘RBD’5 Region 1 interacts with SSM using a face that is orthogonal to the face that would interact with dsRNA in a true RBD.
The RBD fold as a template for functional diversity
As reported here, the combination of a modified RBD, i.e., hSTAU1 ‘RBD’5, within the context of an adapter region, i.e., hSTAU1 SSM, can promote greater functionality within the larger, often modular and flexible framework of RBD-containing proteins. In support of this view, modifications that consist of an L1 Cys and an L3 His within the RBD of the Schizosaccharomyces pombe Dicer DCR1 protein work together with a 33-amino acid region that resides C-terminal to the RBD to form a zinc-coordination motif that is required for nuclear retention and possibly dsDNA binding38.
‘RBD’s that fail to bind dsRNA may also acquire new functions independently of adjacent regions. For example, ‘RBD’5 of D. melanogaster STAU has adapted to bind the Miranda protein required for proper localization of prospero mRNA39,40. Also, human TAR RNA-binding protein 2 contains three RBDs, the C-terminal of which binds Dicer instead of dsRNA41,42. Additionally, ‘RBD’3 of Xenopus laevis RNA-binding protein A, like its human homolog p53-associated cellular protein, appear to homodimerize independent of an accessory region43. It will be interesting to determine if hSTAU1 ‘RBD’2-mediated dimerization25 involves an adapter motif or occurs solely through the RBD-fold.
Online Methods
Sequence alignments
Sequences were obtained from NCBI. Multiple protein sequence alignments were performed using Clustal W26 (v.1.4) within BioEdit44, which was used to generate figures. To generate Figure 1b, STAU protein sequences from the following vertebrate classes were used for the alignment: fish (zebrafish, Danio rerio, NP_991124.1), amphibians (African clawed frog, Xenopus laevis, NP_001085239.1 for STAU-1, NP_001086918.1 for STAU-2), reptiles (Carolina anole; Anolis carolinensis, XP_003220668.1), birds (zebra finch, Taeniopygia guttata; XP_002188609.1) and mammals, i.e., human Homo sapiens (NP_004593.2 for STAU155,NP_001157856.1 for STAU252, STAU259; NP_001157853.1) and mouse Mus musculus (STAU1-2;NP_001103375.1, STAU2-2; NP_001104742.1).
Plasmid constructions
See Supplementary Note 2.
Protein expression in E. coli and protein purification
E. coli BL21(DE3) transformed with pGEX-6p-1-hSTAU1-SSM-‘RBD’5 was propagated in multiple l-liter cultures of Luria Broth supplemented with ampicillin (100 mg l−1) to an O.D.600 of ~0.5, at which time 300 μl of 1M isopropyl β-D-1- thiogalactopyranoside was added to each liter and the temperature was reduced from 37°C to 30°C. The following morning, cells were collected at ~7,000 × g and 4°C and either used directly or flash-frozen in liquid N2 for storage at −80°C.
Cell pellets were resuspended in ~40 ml of Buffer A (1M NaCl, 25 mM Tris-HCl pH 8) to which was added 55 μl of 0.93 M dithiothreitol (DTT), 500 μl of 100 mM PMSF, 50 μl of 0.5 M EDTA pH 8, 500 μl of 80 mg ml−1 lysozyme, and a protease inhibitor tablet (Roche). Cells were lysed using sonication, and lysates were cleared by centrifugation at 17,000 × g for 30 minutes at 4°C. The soluble portion was removed and loaded on a GSTrap™ HP column (GE Healthcare), washed with 1M NaCl, 25 mM Hepes pH 8 (which was sometimes replaced with Buffer A), washed with gel-filtration (GF) buffer (100 mM NaCl, 10 mM Tris-HCl pH 8, 1.3 mM DTT; this step was sometimes omitted), and then eluted with 0.3 g of glutathione (reduced, free acid) dissolved in 100 ml of GF buffer. A ~1 mg aliquot of PreScission™ Protease (GE Healthcare) was added to ~50 ml of eluted sample and left at 4 °C overnight. The following day, the sample was applied to a HiTrap™ Q HP column (GE Healthcare) to remove GST. The flow-through was concentrated to ~1 ml using a Corning® Spin-X® UF 20 5K column (MW cut-off at 5 kDa), and loaded using an ÄKTAFPLC™ system (GE Healthcare) onto a 120-ml HiLoad™ Superdex™ 75 16/60 prep-grade gel-filtration column (GE Healthcare) that was pre-equilibrated with GF buffer. hSTAU1-SSM-‘RBD’5 peak fractions were concentrated as above and used immediately or stored for short periods at 4°C.
Procedures for expressing pGEX-6p-1-hSTAU1-‘RBD’2-RBD3 were identical to those used when expressing hSTAU-SSM-‘RBD’5. However, Buffer A contained 5% glycerol, and the GSTrap™ column elution employed a solution prepared by dissolving 0.3 g glutathione (reduced, free acid), a protease inhibitor tablet (Roche) and 405 μl of 0.93 M DTT in 100 ml of GF buffer. After PreScission™ Protease treatment overnight, the solution was loaded onto a HiTrap™ SP FF column (GE Healthcare) and eluted using a linear NaCl gradient made by mixing GF buffer and glycerol-containing Buffer A and a BioLogic DuoFlow™ FPLC system. Peak fractions were collected, concentrated as above, and loaded onto a HiTrap™ Q HP column to remove contaminating RNAs. The flow-through was concentrated and fractionated using the BioLogic DuoFlow™ FPLC system and a 120 ml HiLoad™ Superdex™ 200 16/60 prep-grade column (GE Healthcare) that was equilibrated with GF buffer containing 2.97 mM DTT.
Analytical ultracentrifugation
hSTAU1-SSM-‘RBD’5 was purified as above, except the final GF buffer contained 2.97 mM DTT, and submitted to the University of Connecticut Analytical Ultracentrifugation Facility for sedimentation velocity analysis. A Beckman-Coulter XL-I analytical ultracentrifuge with double-sector synthetic boundary cells having sapphire windows was used to take interference scans. Measuring refractive index rather than absorbance was especially useful considering the low extinction coefficient at A280 that typifies SSM- ‘RBD’5, which lacks tryptophan residues. Interference scans were collected at 55,000 rpm and 20 °C every minute for 7 hours. Data were analyzed using: 1) DcDt+, version 2.0.9 (refs. 45,46), to determine the sedimentation coefficient distribution that was independent of a model; 2) Sedfit, version 10.09beta47, to produce a model-based continuous sedimentation coefficient distribution using the Lamm equation or c(s) to identify the number of species (e.g., monomers vs. dimers) in solution; and 3) Sedanal, version 5.60 (ref. 48) to combine datasets from the three highest of four concentrations tested, perform a global analysis, and determine the protein association model using the Lamm equation.
Size determination using gel-filtration chromatography
Size standards were prepared by dissolving dried proteins in 2 ml of GF buffer containing 2.97 mM DTT. Proteins consisted of 3.8 mg of conalbumin (75 kDa), 2.3 mg of carbonic anhydrase (29 kDa) and 6.7 mg of aprotinin (6.5 kDa), each from the Low Molecular Weight Gel Filtration Calibration Kit (GE Healthcare; #28-4038-41), and 6 mg of lysozyme (14.3 kDa) (Sigma; #L6876-10G). The dissolved solution (1 ml, determined using a 1-ml loop) was loaded onto a 120 ml HiLoad™ Superdex™ 200 16/60 prep-grade column (GE Healthcare) and separated at a 1 ml min−1 flow rate using the BioLogic DuoFlow™ FPLC system. For size estimations, gel-filtrations of SSM-‘RBD’5 and ‘RBD’2-RBD3 were performed as described for the size standards. SSM-‘RBD’5 was loaded at a concentration of 7 mg ml−1, and ‘RBD’2-RBD3 was loaded at 6 mg ml−1.
Protein crystallization and structure determinations
Native crystals were produced from gel-filtration-purified hSTAU1 SSM-‘RBD’5 using either the sitting-drop method (Native 1 crystal) or the hanging-drop method (Native 2 crystal) (Table 1). The Native 1 crystal was collected at the Cornell High Energy Synchrotron Source (CHESS) beamline F1 under a cryostream at a wavelength of 0.9177 Å (Table 1). The Native 2 crystal was collected remotely at the Stanford Synchrotron Radiation Lightsource (SSRL) beamline 9-2 under a cryostream at a wavelength of 0.9793 Å (Table 1). An initial model was built using low-resolution SAD phases (0.432 figure of merit) from data collected in-house on an ethyl mercuric phosphate-soaked crystal (Ethyl-Hg SAD) under a cryostream at a wavelength of 1.5418 Å (Table 1). Model coordinates were used for molecular-replacement and refined against the 2.2 Å Native 1 dataset (Table 1), and the resulting coordinates were subsequently refined against the 1.7 Å Native 2 dataset. For the final structure, MolProbity49 reported a clashscore of 19.14 and that 97% of the residues were in the favored region of the Ramachandran plot with no outliers. Structure figures were generated using PyMOL (Schrödinger, LLC). See Supplementary Note 3 for crystallization and structure determination details.
HEK239T-cell transfections, and protein and RNA purification
Human HEK293T cells were grown in Dulbecco’s-modified eagle medium (Gibco-BRL) containing 10% fetal-bovine serum (Gibco-BRL). Cells were transiently transfected with plasmids using Lipofectamine 2000 (Invitrogen) or with siRNA using Oligofectamine (Invitrogen) as specified. siRNAs consisted of STAU1 siRNA(A)8 and Negative Control #1 siRNA (Ambion). Protein was isolated using Passive Lysis Buffer (Promega), and RNA was purified using TRIzol Reagent (Invitrogen).
Western blotting, RT-PCR and immunoprecipitations
Protein was electrophoresed in SDS-polyacrylamide, transferred to Hybond ECL nitrocellulose (Amersham), and probed with antibodies that recognize FLAG (Sigma, cat# F315, 1:5000), HA (Roche, cat# 11867423001, 1:1000), calnexin (StressGen, cat# SPA-860, 1:1000), UPF1 (ref. 7; 1:1000), STAU1 (a gift from the Ortín lab; 1:2400), RFP (Abcam, cat# ab65865, 1:1000), GFP (Abcam, cat# ab1218, 1:1000) or STAU2 (Sigma, cat# HPA019155, 1:500). Immunoreactivity was assessed using SuperSignal West Pico or Femto (Pierce Biotechnology). After autoradiography, films were quantitated using ImageQuant (Molecular Dynamics).
Reverse transcription (RT) and PCR amplification were performed as previously described7. RT-PCR products were electrophoresed in 5% polyacrylamide and quantitated by PhosphorImaging (Molecular Dynamics). The five leftmost lanes of each figure represent 2-fold serial dilutions of RNA. A standard curve was derived from these five lanes and used to calculate the relative abundance of each mRNA from different transfections. P values were determined using a one-tailed t-test.
Immunoprecipitations were performed7 using anti-GFP (Abcam), anti-HA (Roche) or anti-FLAG (Sigma). To determine IP and co-IP efficiencies, ImageQuant values that were obtained by western blotting samples before or after IP were superimposed on the values obtained for the 3-fold serial dilutions of protein prior to IP that are provided in the four leftmost lanes of each western blot. For each protein, the value after IP was normalized to the value before IP, and values were then compared. See Supplementary Table 2, which lists IP and co-IP efficiencies for each experiment.
Wound-healing assays
Methods were as described10. Cells were imaged with a Nikon Eclipse TE2000-U inverted fluorescence microscope.
Supplementary Material
Acknowledgments
We thank H. Kuzmiak for generating pSTAU155(R)-HA3; L. DesGroseillers (Université de Montréal, Montréal, Québec, Canada) for pSTAU155-HA3; K. Nehrke for microscope use; G. Pavlencheva and C. Hull for technical assistance; R. Singer (Albert Einstein College of Medicine, Bronx, NY, USA) for pmRFP; S. de Lucas and J. Ortín (Centro Nacional de Biotecnología, Madrid, Spain) for STAU1 antibody; J. Lary (UConn Analytical Ultracentrifugation Facility), J. Jenkins, J. Wedekind and M. Popp for helpful conversations. This work was made possible by NIH R01 GM074593 to L.E.M. M.L.G. was supported by a Ruth L. Kirschstein NRSA NIHF32 GM090479 Fellowship and NIH NCI T32 CA09363. C.G. was supported by a Messersmith Graduate Student Fellowship. The University of Rochester Medical Center Structural Biology & Biophysics Facility is supported by NIH NCRR grants 1S10 RR026501 and 1S10 RR027241, NIH NIAID P30 AI078498, and the School of Medicine and Dentistry. CHESS is supported by the NSF and NIH/NIGMS via NSF award DMR-0225180. MacCHESS is supported by NIH/NCRR RR-01646. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, the NIH National Center for Research Resources Biomedical Technology Program (P41RR001209), and the NIGMS.
Footnotes
Accession Code
The hSTAU1 SSM-‘RBD’5 coordinates and structure factors have been deposited in the Protein Data Bank with accession code 4DKK.
Author Contributions
M.L.G and L.E.M conceived the project and wrote the manuscript with input from C.L.K. M.L.G, C.G., and L.E.M designed the experiments. M.L.G carried out the structural work with input from C.L.K. and designed and constructed the plasmids needed for this study. C.G. undertook experiments using cultured cells. All authors contributed to data interpretation.
References
- 1.Gautrey H, McConnell J, Lako M, Hall J, Hesketh J. Staufen1 is expressed in preimplantation mouse embryos and is required for embryonic stem cell differentiation. Biochim Biophys Acta. 2008;1783:1935–42. doi: 10.1016/j.bbamcr.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 2.Martel C, Macchi P, Furic L, Kiebler MA, Desgroseillers L. Staufen1 is imported into the nucleolus via a bipartite nuclear localization signal and several modulatory determinants. Biochem J. 2006;393:245–54. doi: 10.1042/BJ20050694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miki T, Takano K, Yoneda Y. The role of mammalian Staufen on mRNA traffic: a view from its nucleocytoplasmic shuttling function. Cell Struct Funct. 2005;30:51–6. doi: 10.1247/csf.30.51. [DOI] [PubMed] [Google Scholar]
- 4.Dugre-Brisson S, et al. Interaction of Staufen1 with the 5′ end of mRNA facilitates translation of these RNAs. Nucleic Acids Res. 2005;33:4797–812. doi: 10.1093/nar/gki794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chatel-Chaix L, Abrahamyan L, Frechina C, Mouland AJ, DesGroseillers L. The host protein Staufen1 participates in human immunodeficiency virus type 1 assembly in live cells by influencing pr55Gag multimerization. J Virol. 2007;81:6216–30. doi: 10.1128/JVI.00284-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatel-Chaix L, Boulay K, Mouland AJ, Desgroseillers L. The host protein Staufen1 interacts with the Pr55Gag zinc fingers and regulates HIV-1 assembly via its N-terminus. Retrovirology. 2008;5:41. doi: 10.1186/1742-4690-5-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gong C, Kim YK, Woeller CF, Tang Y, Maquat LE. SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes Dev. 2009;23:54–66. doi: 10.1101/gad.1717309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 9.Kim YK, et al. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J. 2007;26:2670–81. doi: 10.1038/sj.emboj.7601712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–8. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho H, et al. Staufen1-mediated mRNA decay functions in adipogenesis. Mol Cell. 2012;46:495–506. doi: 10.1016/j.molcel.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 12.Maquat LE, Gong C. Gene expression networks: competing mRNA decay pathways in mammalian cells. Biochem Soc Trans. 2009;37:1287–92. doi: 10.1042/BST0371287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Micklem DR, Adams J, Grunert S, St Johnston D. Distinct roles of two conserved Staufen domains in oskar mRNA localization and translation. EMBO J. 2000;19:1366–77. doi: 10.1093/emboj/19.6.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.St Johnston D, Brown NH, Gall JG, Jantsch M. A conserved double-stranded RNA-binding domain. Proc Natl Acad Sci U S A. 1992;89:10979–83. doi: 10.1073/pnas.89.22.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wickham L, Duchaîne T, Luo M, Nabi IR, DesGroseillers L. Mammalian staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol Cell Biol. 1999;19:2220–30. doi: 10.1128/mcb.19.3.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duchaîne T, et al. A novel murine Staufen isoform modulates the RNA content of Staufen complexes. Mol Cell Biol. 2000;20:5592–601. doi: 10.1128/mcb.20.15.5592-5601.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo M, Duchaîne TF, DesGroseillers L. Molecular mapping of the determinants involved in human Staufen-ribosome association. Biochem J. 2002;365:817–24. doi: 10.1042/bj20020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allison R, et al. Two distinct Staufen isoforms in Xenopus are vegetally localized during oogenesis. RNA. 2004;10:1751–63. doi: 10.1261/rna.7450204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duchaîne TF, et al. Staufen2 isoforms localize to the somatodendritic domain of neurons and interact with different organelles. J Cell Sci. 2002;115:3285–95. doi: 10.1242/jcs.115.16.3285. [DOI] [PubMed] [Google Scholar]
- 20.Park E, Gleghorn ML, Maquat LE. Staufen2, like Staufen1, functions in SMD by binding to itself and its paralog and promoting UPF1 helicase but not ATPase activity. Proc Natl Acad Sci U S A. 2013;110(2):405–412. doi: 10.1073/pnas.1213508110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furic L, Maher-Laporte M, DesGroseillers L. A genome-wide approach identifies distinct but overlapping subsets of cellular mRNAs associated with Staufen1-and Staufen2-containing ribonucleoprotein complexes. RNA. 2008;14:324–35. doi: 10.1261/rna.720308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramos A, et al. RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J. 2000;19:997–1009. doi: 10.1093/emboj/19.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian B, Bevilacqua PC, Diegelman-Parente A, Mathews MB. The double-stranded-RNA-binding motif: interference and much more. Nat Rev Mol Cell Biol. 2004;5:1013–23. doi: 10.1038/nrm1528. [DOI] [PubMed] [Google Scholar]
- 24.Stefl R, et al. The solution structure of the ADAR2 dsRBM-RNA complex reveals a sequence-specific readout of the minor groove. Cell. 2010;143:225–37. doi: 10.1016/j.cell.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martel C, et al. Multimerization of Staufen1 in live cells. RNA. 2010;16:585–97. doi: 10.1261/rna.1664210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchler-Bauer A, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–9. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–9. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gan J, et al. A stepwise model for double-stranded RNA processing by ribonuclease III. Mol Microbiol. 2008;67:143–54. doi: 10.1111/j.1365-2958.2007.06032.x. [DOI] [PubMed] [Google Scholar]
- 30.Buchan DW, et al. Protein annotation and modelling servers at University College London. Nucleic Acids Res. 2010;38:W563–8. doi: 10.1093/nar/gkq427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 32.Gong C, Maquat LE. “Alu”strious long ncRNAs and their role in shortening mRNA half-lives. Cell Cycle. 2011;10:1882–3. doi: 10.4161/cc.10.12.15589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ravel-Chapuis A, et al. The RNA-binding protein Staufen1 is increased in DM1 skeletal muscle and promotes alternative pre-mRNA splicing. J Cell Biol. 2012;196:699–712. doi: 10.1083/jcb.201108113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amoutzias GD, Robertson DL, Van de Peer Y, Oliver SG. Choose your partners: dimerization in eukaryotic transcription factors. Trends Biochem Sci. 2008;33:220–9. doi: 10.1016/j.tibs.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 35.Cho DS, et al. Requirement of dimerization for RNA editing activity of adenosine deaminases acting on RNA. J Biol Chem. 2003;278:17093–102. doi: 10.1074/jbc.M213127200. [DOI] [PubMed] [Google Scholar]
- 36.Valente L, Nishikura K. RNA binding-independent dimerization of adenosine deaminases acting on RNA and dominant negative effects of nonfunctional subunits on dimer functions. J Biol Chem. 2007;282:16054–61. doi: 10.1074/jbc.M611392200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cole JL. Activation of PKR: an open and shut case? Trends Biochem Sci. 2007;32:57–62. doi: 10.1016/j.tibs.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barraud P, et al. An extended dsRBD with a novel zinc-binding motif mediates nuclear retention of fission yeast Dicer. EMBO J. 2011;30:4223–35. doi: 10.1038/emboj.2011.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fuerstenberg S, Peng CY, Alvarez-Ortiz P, Hor T, Doe CQ. Identification of Miranda protein domains regulating asymmetric cortical localization, cargo binding, and cortical release. Mol Cell Neurosci. 1998;12:325–39. doi: 10.1006/mcne.1998.0724. [DOI] [PubMed] [Google Scholar]
- 40.Schuldt AJ, et al. Miranda mediates asymmetric protein and RNA localization in the developing nervous system. Genes Dev. 1998;12:1847–57. doi: 10.1101/gad.12.12.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haase AD, et al. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–7. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parker GS, Maity TS, Bass BL. dsRNA binding properties of RDE-4 and TRBP reflect their distinct roles in RNAi. J Mol Biol. 2008;384:967–79. doi: 10.1016/j.jmb.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hitti EG, Sallacz NB, Schoft VK, Jantsch MF. Oligomerization activity of a double-stranded RNA-binding domain. FEBS Lett. 2004;574:25–30. doi: 10.1016/j.febslet.2004.07.080. [DOI] [PubMed] [Google Scholar]
- 44.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 45.Philo JS. A method for directly fitting the time derivative of sedimentation velocity data and an alternative algorithm for calculating sedimentation coefficient distribution functions. Anal Biochem. 2000;279:151–63. doi: 10.1006/abio.2000.4480. [DOI] [PubMed] [Google Scholar]
- 46.Philo JS. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal Biochem. 2006;354:238–46. doi: 10.1016/j.ab.2006.04.053. [DOI] [PubMed] [Google Scholar]
- 47.Schuck P. Size-distribution analysis of macromolecules by sedimentation svelocity ultracentrifugation and Lamm equation modeling. Biophys J. 2000;78:1606–19. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stafford WF, Sherwood PJ. Analysis of heterologous interacting systems by sedimentation velocity: curve fitting algorithms for estimation of sedimentation coefficients, equilibrium and kinetic constants. Biophys Chem. 2004;108:231–43. doi: 10.1016/j.bpc.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 49.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.