Abstract
A novel haplotype association method is presented, and its power is demonstrated. Relying on a statistical model for linkage disequilibrium (LD), the method first infers ancestral haplotypes and their loadings at each marker for each individual. The loadings are then used to quantify local haplotype sharing between individuals at each marker. A statistical model was developed to link the local haplotype sharing and phenotypes to test for association. We devised a novel method to fit the LD model, reducing the complexity from putatively quadratic to linear (in the number of ancestral haplotypes). Therefore, the LD model can be fitted to all study samples simultaneously, and, consequently, our method is applicable to big data sets. Compared to existing haplotype association methods, our method integrated out phase uncertainty, avoided arbitrariness in specifying haplotypes, and had the same number of tests as the single-SNP analysis. We applied our method to data from the Wellcome Trust Case Control Consortium and discovered eight novel associations between seven gene regions and five disease phenotypes. Among these, GRIK4, which encodes a protein that belongs to the glutamate-gated ionic channel family, is strongly associated with both coronary artery disease and rheumatoid arthritis. A software package implementing methods described in this article is freely available at http://www.haplotype.org.
Keywords: haplotype, association, local haplotype sharing, two-layer HMM, LD
DETECTING genetic variants in association with phenotypes is central to statistical genetics. Current genome-wide association studies (GWAS) test single genetic markers, usually single-nucleotide polymorphisms (SNP), one at a time, and this is effective in detecting common variants in association with phenotypes (e.g., Scott et al. 2007; Wellcome Trust Case Control Consortium 2007; Willer et al. 2008). For the majority of complex phenotypes, however, single-SNP common variants explained only <10% of phenotypic variations (Manolio et al. 2009). This “missing heritability” (Maher 2008) flummoxed the field and many directions have been suggested to search for it, including structure variations, gene–environment interactions, parental origin and phase-dependent interaction, and rare variants, among others (Eichler et al. 2010). On the other hand, a significant amount of phenotypic variation can be explained by common variants, as long as genome-wide SNPs are jointly analyzed (Yang et al. 2010; Zhou et al. 2013). Combined, they attest to the necessity for developing new association methods, in addition to assaying more genetic variants.
As a fundamental form of genetic variation and the unit of inheritance, a haplotype may affect phenotypes either directly through influencing promoter activity and protein structure (Drysdale et al. 2000; Joosten et al. 2001) or indirectly through tagging nearby untyped causal variants (Clark 2004; Servin and Stephens 2007). Thus, haplotype association is of great interest for unveiling the etiology of complex phenotypes. Haplotype association takes into account allelic heterogeneity—different mutations within a gene cause a similar phenotype, which is a blind spot for the single-SNP test. An association method that takes into account allelic heterogeneity is more powerful than the single-SNP analysis (Pritchard 2001), as demonstrated in both haplotype analysis (Zöllner and Pritchard 2005) and multi-SNP analysis (Guan and Stephens 2011). (In fact, multi-SNP analysis takes into account not only allelic heterogeneity, but also locus heterogeneity—mutations at different genes cause a similar phenotype.) Arguably, haplotype analysis is more powerful than multi-SNP analysis within a gene region because it accounts for not only allelic heterogeneity, but also possible statistical interactions among markers. Consider a two-marker haplotype and suppose that the C-T haplotype increases disease risk. A haplotype method may detect the association, while a multi-SNP method has to invoke an interaction term between the two markers.
A primitive haplotype association method first phases diploid genomes, specifies a window of arbitrary length to define haplotypes, and then tests each haplotype in turn for association. This approach suffers drawbacks in every aspect. First, it requires phasing a diploid genome and it is difficult to account for phase uncertainty in subsequent statistical testing. Second, using a fixed window to define haplotypes is both arbitrary and unsatisfactory because the size of the linkage disequilibrium (LD) block varies along the genome. Third, compared to testing for each haplotype in turn, grouping haplotypes is necessary because a large number of haplotypes increases the degree of freedom for a test statistic (Schaid 2004). Existing haplotype methods improve various aspects of the primitive haplotype method, particularly in grouping haplotypes before testing (Zöllner and Pritchard 2005; Browning and Browning 2007; Feng and Zhu 2010; Li et al. 2010a). In both Feng and Zhu (2010) and Li et al. (2010a), authors selected a subset of individuals as a training data set to identify and group haplotypes into disease causal and protective categories; Zöllner and Pritchard (2005) grouped haplotypes according to the posterior estimates of the coalescent trees; and Browning and Browning (2007) used their LD model to cluster local haplotypes and used cluster membership as a surrogate for haplotype grouping. These methods require phasing to obtain haplotypes and ignore the phasing uncertainty in the association testing, and Zöllner and Pritchard (2005), Feng and Zhu (2010), and Li et al. (2010a) require a window to define haplotypes.
Here we describe a novel method to detect associations between haplotypes and phenotypes. Our method relies on a hidden Markov model developed previously to model LD and haplotype variation (Guan 2014), from which we can infer ancestral haplotypes and their loadings at each marker for each individual. (Note that although the loadings are estimated at each marker, they are determined by local haplotypes around the core marker.) Then, local haplotype sharing (LHS)—the probability of two diploid individuals descending from the same ancestral haplotypes—can be quantified using the loadings. LHS reflects genetic similarity between individuals and it is a natural extension of identity by descent, a measure of genetic similarity for individuals in a pedigree, to unrelated samples. By testing whether the genetic similarity is associated with the phenotypes, we can identify associations—at each (core) marker—between local haplotypes and phenotypes.
In a case–control design, haplotypes conferring higher disease risk are expected to be more abundant in cases than in controls. Inevitably, the amount of local haplotype sharing is expected to be higher between two case individuals than that between a case individual and a control individual or that between two control individuals, which forms the basis for the association testing. The same argument applies to disease-protective haplotypes. And the rationale applies to quantitative phenotypes as well. Compared to existing haplotype association methods, our LHS method has the following novelties: (1) we worked directly with diploid genotypes and integrated out phase uncertainty that plagues other haplotype methods; (2) we avoided arbitrariness in specifying a window to define haplotypes (the extent of haplotypes is learned from the data through the LD model); (3) each SNP is a core SNP for its local haplotypes and is tested for association, so that our LHS method has the same number of tests as the single-SNP analysis; and (4) our LHS method is computationally efficient. We developed a linear algorithm that can fit the LD model to all study samples simultaneously.
Materials and Methods
The LD model and local haplotype sharing
The LD model was originally developed to infer local ancestries of admixed individuals (Guan 2014). It is an extension of the fastPHASE model (Scheet and Stephens 2006) to more than one source population. Briefly, it is a hidden Markov model that uses two layers of latent clusters to approximate coalescence with recombination [two-layer hidden Markov model (HMM)]. In each layer, clusters are labeled to represent ancestral alleles, and multiple clusters of the same label over adjacent loci represent an ancestral haplotype. Each cluster associates with an allele frequency parameter; the upper-layer clusters emit lower-layer cluster allele frequencies and the lower-layer clusters emit the observed genotypes. Recombination is approximated by cluster switching within each layer. Although only the lower-layer cluster loadings (defined later) were used in the association testing and the upper-layer clusters appeared irrelevant, they are indeed helpful in inferring both ancestral allele frequencies and loading matrix through enforcing structure on lower-layer clusters.
The structure of local haplotypes is a ubiquitous phenomenon in genetic data, and the two-layer model is effective in inferring such structure of haplotypes. The local ancestry of admixed individuals is a more apparent example of structure of haplotypes, where the upper-layer clusters represent the source population and lower-layer clusters represent ancestral haplotypes (Guan 2014). In fact, the two-layer model is also effective for haplotypes that are sampled from a single source population, where the upper-layer clusters represent subtle structure of more similar haplotypes that are represented by the lower-layer clusters. For example, the upper-layer clusters may represent two-digit human leukocyte antigen (HLA) allele classes (such as HLA-A02 and HLA-A03), which enforce structure on the lower-layer clusters that represent four-digit HLA alleles (such as HLA-A0202, HLA-A0203, HLA-A0301, and HLA-A0303). The ability to detect subtle structure of haplotypes, particularly that among haplotypes sampled from a single source population, makes the two-layer model useful for genetic association studies.
An HMM contains a set of parameters to model Markov transitions of latent states and a set of parameters for ancestry allele frequencies at each marker, collectively denoted by ξ. We assume individuals are unrelated, and, thus, conditional on ξ individuals are independent, so we may compute each individual in turn. Denote g(i) the collection of genotypes of individual i, which are assumed to be biallelic and are coded as 0, 1, or 2 counts of a reference allele. Let and be two sets of latent states at marker m, where X represents the upper-layer cluster and Y the lower-layer cluster (index i omitted). Assuming the numbers of upper- and lower-layer clusters are S and K, respectively, then and take values in 1 … S and and take values in 1 … K. The conditional likelihood for the ith individual is (recall that observed genotypes are assumed to be emitted from the lower-layer clusters Y and thus upper-layer cluster X plays no role in this likelihood), and the emission is modeled as
| (1) |
where θmk is the allele frequency associated with the lower-cluster k. The Markov transitions of the two sets of latent states are independent a priori and are modeled as
| (2) |
where I(a = b) is an indicator function and vectors ρ and r are cluster-switch probabilities for the upper and lower layers, respectively, and
| (3) |
where α(i) is an S vector to denote the admixture proportion, and βm is an S × K matrix shared by all individuals.
To fit the LD model, we need to specify three parameters: the number of upper-layer clusters S, the number of lower-layer clusters K, and the number of admixing generations γ. The parameter γ controls the ratio between ρ and r, the cluster-switching probabilities of upper- and lower-layer clusters, respectively, which are used to model Markov transitions in Equation 2. The name, admixing generation, becomes a misnomer in the current context, but we keep it for consistency with the original model setup, which was designed for local ancestry inference (Guan 2014). The correct interpretation of γ in the current context is through its reciprocal, 1/γ, which provides an a priori average length of shared haplotype segments (in centimorgans) among ancestral haplotypes. In all data analyses, we used γ = 50 and 100, which correspond to 2 cM and 1 cM of average length of shared haplotype segments, respectively. And, unless otherwise noted, we used S = 2, K = 10 for our association analyses throughout the article.
After fitting the model, we obtain ξ∗. The details of fitting the two-layer model using the classical EM algorithm can be found in Guan (2014); a fast linear algorithm to fit the two-layer model is documented in Results. For individual i at marker m, define haplotype loading as
| (4) |
where because of symmetry between two sets of latent states, the loading needs to be defined only on one set of latent states. The imputed allele dosage can be computed as
Local haplotype sharing between individuals and between markers:
At each marker, the LHS between two individuals was defined as the probability of the two individuals descending from the same ancestral haplotypes, which is simply an inner product of two loading vectors: A high LHS value between two individuals at a marker implies similar local haplotype background between the two individuals at that marker. The LHS estimates are correlated between nearby markers.
LHS can be used to quantify the LD between markers. Intuitively, if two markers are in strong LD, then, for an arbitrary individual, the loadings at the two markers are expected to be similar and if two markers are in weak LD, they are expected to be less similar. Thus, with slight abuse of notation, we define LHS between two markers, indexed by h and j, as When markers are independent, our model produces an mLHS estimate, as a measure of LD background noise, with mean 1/K. [To see this, we may assume each K vector is Dirichlet distributed with mean (1/K, … , 1/K).]
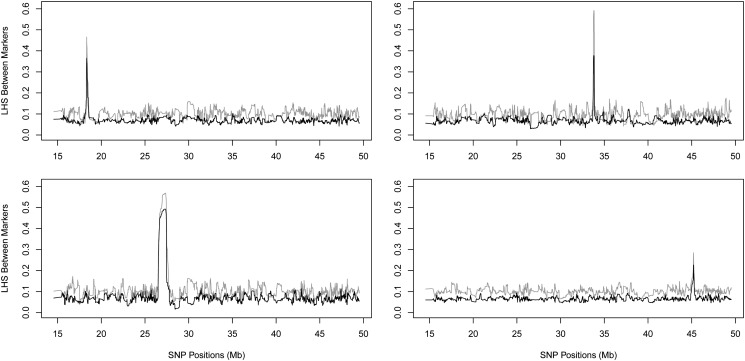
We randomly chose a marker and computed the mLHS between this marker and the rest. The LD block around a marker was defined as the largest region that has mLHS value larger than, say, 2.5 times the background mLHS value (i.e., 0.17 for K = 15 and 0.25 for K = 10). For 100 markers we sampled, the sizes of the LD block vary substantially in the HapMap3 CEU (Utah residents with Northern and Western European ancestry from the CEPH collection) samples (International HapMap Consortium 2010), ranging from 20 kb to 1.1 Mb with mean 170 kb. Figure 1 shows four examples of LD blocks with different strength and size. In all examples, the patterns of LD agreed with each other between two choices of parameters (K = 10, 15). The background noise level matched perfectly with the theoretical predictions: excluding the spike regions, the mean values are 0.101 for K = 10 and 0.067 for K = 15. For a significant association, we quantified an LD block around the core SNP, using mLHS, and located relevant genes. The mLHS was more informative to quantify a LD block than were the R2 values between SNPs.
Figure 1.
Examples of LD blocks quantified by mLHS, using chromosome 22 of CEU samples in HapMap3. In all four panels the shaded line corresponds to K = 10 and the solid line to K = 15. The four panels show LD blocks of different strengths and sizes. Note that the observed LD background noise matches the theoretical predictions. See Materials and Methods for details.
Testing for associations
Denote y = (y1, … , yn) a vector of phenotypic values. Let W be an n × q matrix representing q covariates such as age, sex, and principal components (PCs), including a column of 1 for grand mean, and a be a q vector. At an arbitrary marker m, use L (an n × K matrix) to denote the loading matrix and L⋅j to denote its jth column. We have
| (5) |
where Im denotes an identity matrix of dimension m, and MVN stands for multivariate normal. When multiple columns of L tag different markers that affect phenotypes, the model accounts for allelic heterogeneity.
Taking a Bayesian approach, we specify priors in model (5) as
| (6) |
where the Gamma density is in shape-rate parameterization, and βk are independent and identically distributed.
With the above prior specification, model (5) is equivalent to a random-effect model y = Wa + λb + e, where b ∼ MNV(0, LLt). To see this, without loss of generalization, assume each column of L is centered at 0 and denote β = (β1, … , βK); then for the first moment we have E(Lβ) = 0 = E(λb), and for the second moment we have and the right-hand side equals Var(λb) when λ = σ1/τ1/2. Thus, model (5) in fact captures the association between local haplotype sharing (LLt) and phenotypes.
Define X = (W, L⋅1, … , L⋅K) and following a standard normal-inverse-Gamma prior (cf. Servin and Stephens 2007), letting κ2 → 0, then κ1 → 0, and σ0 → ∞, we can compute the Bayes factor (BF) in a closed form
| (7) |
where Note here although improper priors (on nuisance parameters a and τ) are used, the Bayes factor is proper as priors associated with the nuisance parameters cancel out (cf. Servin and Stephens 2007). Specifying σ1 is required to compute a Bayes factor. We used σ1 = 0.2, 0.5 and averaged Bayes factors over two choices of priors. These priors put probability of 0.98 for effect sizes to be in the interval of [−1.0, 1.0], probability of 0.84 for effect sizes to be in [−0.5, 0.5], and probability of 0.50 for effect sizes to be in [−0.2, 0.2].
To extend the Bayesian linear regression model (5) to a case–control design is conceptually straightforward, but computationally difficult, particularly for the case of multiple covariates—the situation that we face. Because of the logistic (or probit) link function, the Bayes factor can no longer be evaluated in a closed form; instead, the integration over a prior distribution of β requires a numerical method such as the Laplace approximation (cf. Guan and Stephens 2008), which can be prohibitively slow for a genome-wide analysis. We therefore treated the binary phenotypes as quantitative ones and directly applied Equation 7 to compute Bayes factors. Treating binary phenotypes as quantitative ones has been used by others (Kang et al. 2008; Zhou and Stephens 2012) in genome-wide association studies.
Of course, for a case–control design we have a logistic regression model
| (8) |
This model can be fitted using standard iterative weighted least squares to obtain the likelihood l1 under the alternative. Setting β1 = … βK = 0 and refitting the model to obtain the likelihood l0 under the null, we obtained and thus a P-value.
Combining test statistics
To account for uncertainty of LD inferences, test statistics over multiple EM runs were combined. It is a nagging problem, however, to combine P-values: the minimum P-values (over multiple EM runs for each marker) cause inflated type I error; converting P-values to z scores (or chi-square values) and producing a P-value based on the mean z score (or chi-square value) is too conservative; and Fisher’s method has an independence assumption on P-values to be combined, which is not satisfied by our P-values. Combining test statistics is simple, however, for Bayesian analysis because Bayes factors over multiple EM runs can be directly averaged. In both power and real data analysis, we chose the Bayes factor as the test statistic and report minimum P-values for significant associations. The Bayes factor is the change of odds ratio in light of data (cf. Stephens and Balding 2009). We have ω1 = ωo ⋅ BF, where ω0 is the prior odds for association, and ω1 is the posterior odds, from which the posterior probability of association, denoted by π, can be calculated as π = ω1/(1 + ω1). For example, if we assume 10 loci of 1,000,000 are associated with a phenotype, then ω0 = 10−5; if a locus has a Bayes factor of 106, with ω0 = 10−5 we obtain ω1 = 10 and hence π = 0.91.
Simulating data to assess statistical power
A case–control design was assumed to assess the statistical power of our association method. Here we describe how we simulated data. We assumed two, four, and eight causal haplotypes that span 10 adjacent common SNPs (minor allele frequencies >5%), and the aggregated causal haplotype frequencies were chosen to be >0.05. Following Browning and Thompson (2012), we assumed the penetrance was 0.1 and the sporadic rate was 0.01 (an individual carrying one or multiple disease alleles has a probability of 0.1 to be a case and a wild-type individual has a probability of 0.01 to be a case). Our assumption put the prevalence of disease at ∼2–3%. As noted by Browning and Thompson (2012), the power is determined by the ratio between penetrance and the sporadic rate, and our choice of parameters was somewhat realistic and had a reasonable power. We also compared with a sporadic rate of 0.02.
We first simulated 10,000 haplotypes in a 200-kb region; the causal haplotypes were drawn from the middle 20-kb region. After causal alleles were determined, haplotypes were grouped as wild types (with count n0) and carriers (with count n1). We used sampling with replacement to obtain diplotypes of cases and controls separately. For cases, we first defined p = n1/(n1 + n0) and then computed two weights w1 = p(2 − p) × 0.1 and w2 = (1 − p)2 × 0.01; with probability w1/(w1 + w2) we sampled one haplotype from carriers and another from all haplotypes to form a case diplotype, and with probability w2/(w1 + w2) we sampled two haplotypes from wild types to form a case diplotype. Similarly for controls, we computed two weights w1 = p(2 − p) × (1 − 0.1) and w2 = (1 − p)2 × (1 − 0.01); with probability w1/(w1 + w2) we sampled one haplotype from carriers and another from all haplotypes to form a control diplotype, and with probability w2/(w1 + w2) we sampled two haplotypes from wild types to form a control diplotype.
Data quality control
We first excluded individuals and SNPs as suggested by the Wellcome Trust Case Control Consortium (2007). To select SNPs to perform principal component analysis (PCA), we started with the full set of autosomal SNPs, thinned SNPs so that the remaining SNPs were spaced at least 0.001 cM apart (HapMap estimates), and removed SNPs in the MHC and the lactase regions (2q21) and known inversions of 8p23 and 17q21.31. The number of SNPs used for the PCA was ∼208,000. Based on the PCA analysis, we further removed outlier individuals. The final numbers of samples for each phenotype can be found in Supporting Information, Table S1. Based on these individuals, we further excluded SNPs if the Hardy–Weinberg equilibrium exact test P-value was <1 × 10−6, or minor allele frequency was <1%, or the proportion of missing genotypes was >5%. The final number of SNPs used for association analysis was ∼395,000 (exact numbers are in Table S1).
Results
A linear algorithm to fit the two-layer LD model
We needed to fit the two-layer HMM (see Materials and Methods) for thousands of individuals over a few hundred thousand to several million SNPs and needed to do so multiple times to average over uncertainty of LD inferences. A diploid individual requires two sets of latent states (one for each haplotype), and, consequently, fitting a two-layer model is quadratic in number of haplotype clusters (Guan 2014). This poses a serious computational challenge for a genome-wide analysis. We now describe a linear algorithm to fit the two-layer HMM.
The model setup can be found in Materials and Methods, and more details can be found in Guan (2014). Following the notations in Materials and Methods, we have that are two sets of latent states, each for a haplotype; we are interested in computing their posterior distribution
| (9) |
where g is the genotypes of an arbitrary individual (superscript dropped) and ξ is a collection of parameters. This computation requires K2 operations because each joint state needs to be computed (recall K is the number of lower-layer clusters). To make the computation linear in K, we first marginalized (integrated) over under the prior distribution and then predicated allele frequencies at each locus, using the marginal posterior distribution of Conditional on the we computed the marginal posterior distribution of (and obtained updated as well). The first marginalization is exact and the second involves approximations. We first marginalize over to obtain
| (10) |
where M is the total number of markers, and is the expected allele frequency at maker m for the latent state We may compute forward and backward probabilities from the marginalization to get for all m and obtain the expected allele frequency at marker m for the latent state after marginalizing over Then conditional on we marginalize over to obtain
| (11) |
The joint posterior was approximated by two conditional marginals in the sense that for any linear function we have
| (12) |
Intuitively, at each EM iteration, the ancestral allele dosages and the Markov transition parameters provided an a priori average haplotype (in our model, this a priori average haplotype is different for each individual). Conditioning on the average haplotype, we computed the marginal forward and backward probabilities of one set of latent states. The forward and backward probabilities gave rise to a posterior mean haplotype, conditioning on which we computed the marginal forward and backward probabilities again for another set of latent states. Using these two sets of forward and backward probabilities, we approximated the joint probabilities of the two sets of latent states, from which we updated the ancestry allele frequencies and the Markov transition parameters. We call this algorithm stochastic linear iterative marginalization (SLIM).
Performance of the linear algorithm
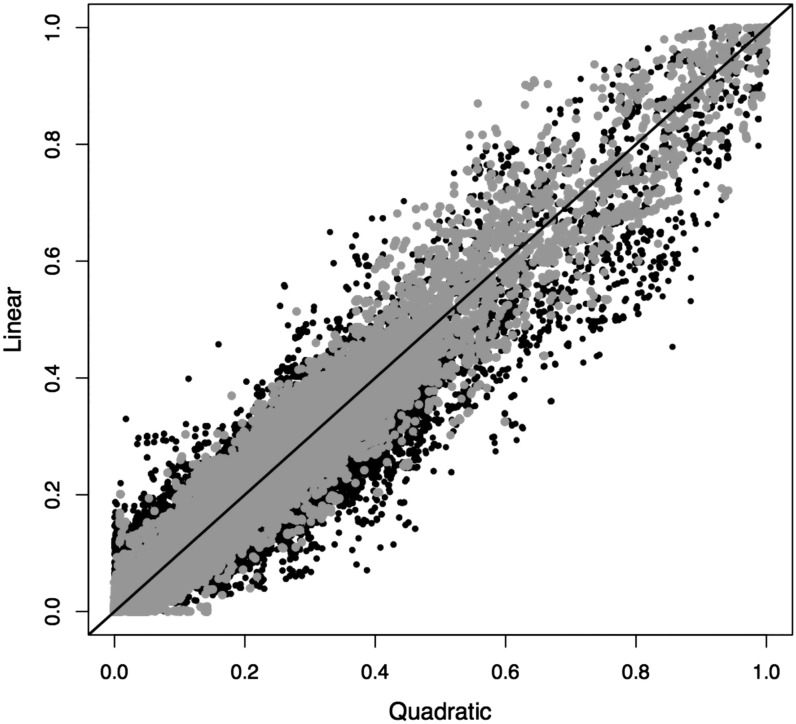
The linear algorithm SLIM plays a key role in our LHS method, allowing the genome-wide LD inference using all study samples simultaneously. The accuracy and consistency of L and θ estimates are critical to the power of our association method. We therefore assessed the performance of SLIM in inferring L (and θ) and compared it with the quadratic method. The first comparison is the LHS, which reflects the accuracy and consistency of L estimates. Between different EM runs, there were substantial variations in LHS inferences for both linear and quadratic methods. When averaged over 10 independent EM runs, however, the two methods showed high concordance: the Pearson correlation was 0.96 between two LHS inferences. As a comparison, between two trials of the quadratic method—each trial was obtained by averaging over 10 EM runs—the Pearson correlation was 0.98 (Figure 2).
Figure 2.
Consistency between the linear approximation and the quadratic method (solid circles). The results were obtained by averaging over 10 independent EM runs for each method. The straight line denotes x = y. As a comparison, shaded circles show consistency between two trials of quadratic methods, each trial averaging over 10 independent EM runs. The data set is chromosome 22 of CEU samples in HapMap3.
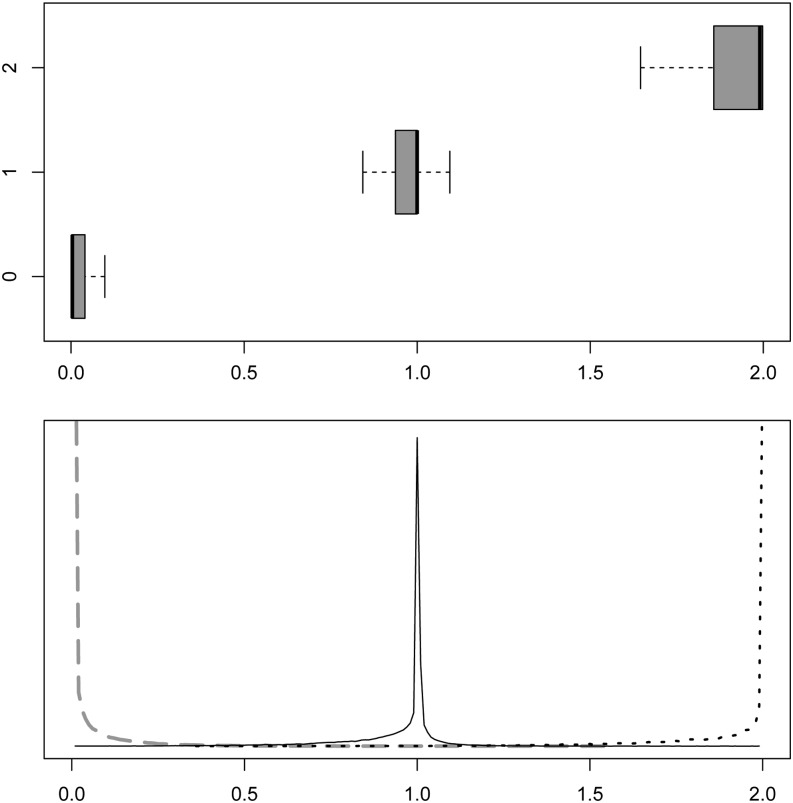
Next we computed the imputed allele dosages x = 2Lθ and compared them with the actual genotypes. The mean (median) Hamming distance between genotypes and imputed allele dosages was 0.058 (0.005) for the SLIM and 0.040 (0.002) for the quadratic method (Figure 3). Given that L was inferred consistently and x was accurate, we concluded that the θ estimates were sensible.
Figure 3.
Distribution of the imputed allele dosages (same data set as in Figure 2). Results were obtained by averaging over 10 independent EM runs. Top, box plots of imputed allele dosages for three genotype classes (outliers removed); bottom, corresponding density plots.
Power and comparison with other approaches
We studied the statistical power of our hapotype method with the presence of allelic heterogeneity. Arguably, this is when a haplotype method has an advantage. We used the software ms (Hudson 2002) to simulate haplotypes under the neutral model. Following the procedure described in Materials and Methods, we simulated samples for cases and controls in a 200-kb region. We removed SNPs whose minor allele frequencies (MAFs) are <0.05 and SNPs in the causal center (20 kb in the middle, see Materials and Methods), but allowed Whait (Li et al. 2010a) to use all SNPs, which effectively assumed that it has a perfect imputation to recover all variants, both common and rare. This process was repeated 100 times to create 100 regions for each simulation condition. The factors that determine simulation conditions include sample size, sporadic (or background) rate, and number of causal haplotypes. The last factor decides the degree of allelic heterogeneity.
For the benefit of comparing our LHS method with other frequentist approaches, we computed P-values using the logistic regression (8), in addition to BFs, based on a single inference of the loadings (or a single EM run). Using a single EM run avoided the headache of combining P-values, but at the cost of a reduced power. We used the minimum P-value in each region as the region P-value and the maximum BF in each region as the region BF. We used 106 as the BF significant threshold and 10−8 as the P-value significant threshold, the latter of which is comparable to the genome-wide threshold in a typical GWAS study. The power of the BF is defined as the proportion of regions (of 100 total) whose region BF is >106, and the power of the P-value is defined as the proportion of regions whose region P-value is <10−8. Table 1 shows that, although the power of the BF is slightly better than the power of the P-value in 3 of 12 simulation conditions examined, the two sets of power are comparable and agree with each other in most (9 of 12) simulation conditions.
Table 1. The power comparison between BF cutoff of 106 and P-value cutoff of 10−8 under different simulation conditions.
| Conditions | Power | |||
|---|---|---|---|---|
| Case/control | Sporadic ratea | Causal haplotypes | BF > 106 | P-value < 10−8 |
| 1000/1000 | 0.01 | 2 | 1.00 | 1.00 |
| 4 | 0.95 | 0.95 | ||
| 8 | 0.72 | 0.71 | ||
| 0.02 | 2 | 0.98 | 0.98 | |
| 4 | 0.93 | 0.93 | ||
| 8 | 0.59 | 0.53 | ||
| 2000/2000 | 0.01 | 2 | 1.00 | 1.00 |
| 4 | 0.98 | 0.98 | ||
| 8 | 0.84 | 0.83 | ||
| 0.02 | 2 | 0.99 | 0.99 | |
| 4 | 0.97 | 0.97 | ||
| 8 | 0.71 | 0.71 | ||
The result was obtained using a single EM run so that the P-value is valid. Under all simulation conditions considered, two sets of powers are close to each other, and the BF cutoff of 106 has slightly better power than the P-value cutoff of 10−8.
The penetrance is assumed to be fixed at 0.10, so that a higher sporadic rate results in a lower power.
Next, we compared our haplotype method (averaged over five EM runs) with a Bayesian single-SNP analysis (Servin and Stephens 2007) implemented in BIMBAM (Guan and Stephens 2008). In this comparison, both the haplotype method and the single-SNP method produced BFs, and we used the maximum BF in each region as the region BF. The power for both methods is defined as the proportion of regions whose region BF is >106. Table 2 reflects what we expected: a larger sample size produces more power; when the sporadic rate is higher, the power is lower; and when more allelic heterogeneity exists (e.g., more causal haplotypes), our LHS method has a greater margin over the single-SNP test.
Table 2. The power comparison between our LHS method and the single-SNP analysis for different simulation conditions.
| Conditions | Power | |||
|---|---|---|---|---|
| Case/control | Sporadic ratea | Causal haplotypes | LHS | Single SNP |
| 1000/1000 | 0.01 | 2 | 1.00 | 0.99 |
| 4 | 0.95 | 0.94 | ||
| 8 | 0.76 | 0.58 | ||
| 0.02 | 2 | 0.98 | 0.96 | |
| 4 | 0.95 | 0.88 | ||
| 8 | 0.59 | 0.32 | ||
| 2000/2000 | 0.01 | 2 | 1.00 | 1.00 |
| 4 | 0.99 | 0.99 | ||
| 8 | 0.84 | 0.75 | ||
| 0.02 | 2 | 0.99 | 0.97 | |
| 4 | 0.97 | 0.93 | ||
| 8 | 0.73 | 0.67 | ||
The BF for the LHS method is averaged over five EM runs. The single-SNP analysis was performed using BIMBAM and the BF was computed, instead of P-values. The BF threshold for both methods is 106.
The penetrance is assumed to be fixed at 0.10, so that a higher sporadic rate results in a lower power
Finally, we compared our LHS method with three other haplotype methods: the one described by Browning and Browning (2007) (henceforth Beagle), the one by Feng and Zhu (2010) (henceforth FZ), and the Whait method by Li et al. (2010a). All three methods require phased haplotypes as input, and we supplied them with the true haplotypes. For our own method, we used diplotypes, ignoring the phase information. For the Beagle method, we used the default setting in the software. Following the description of the FZ method, we used 30 SNPs to define haplotypes, used 30% of individuals to screen significant haplotypes and combined them, and used the remaining 70% of individuals to perform the test—both causal and protective haplotypes—and chose the minimum P-value as the test statistics. For Whait, we used all SNPs with true phasing (in other words, we assumed that the imputation and phasing are perfect for Whait) and tried different window sizes to define haplotypes and picked the best window size to report power. Beagle, FZ, and Whait are frequentist methods that produce only P-values. Therefore, we computed P-values for our LHS method based on a single EM run. For all methods, the region P-value is defined as the minimum P-value in each region, and the power is the proportion of regions whose P-values are <10−8. Table 3 shows that our LHS method outperforms (or performs as well as) Beagle, FZ, and Whait under 11 of 12 simulation conditions. In particular, when more allelic heterogeneity exists (e.g., more causal haplotypes), the advantage of our LHS method is more apparent.
Table 3. The power comparison (at P-value threshold of 10−8) between our LHS method (with a single EM run) and three other haplotype methods—Beagle in Browning and Browning (2007), FZ in Feng and Zhu (2010), and Whait in Li et al. (2010a)—under different simulation conditions.
| Conditions | Power | |||||
|---|---|---|---|---|---|---|
| Case/control | Sporadic ratea | Causal haplotypes | LHS | Beagle | FZ | Whait |
| 1000/1000 | 0.01 | 2 | 1.00 | 0.99 | 0.99 | 0.97 |
| 4 | 0.95 | 0.93 | 0.92 | 0.95 | ||
| 8 | 0.71 | 0.60 | 0.64 | 0.64 | ||
| 0.02 | 2 | 0.98 | 0.95 | 0.95 | 0.93 | |
| 4 | 0.93 | 0.74 | 0.89 | 0.87 | ||
| 8 | 0.53 | 0.33 | 0.32 | 0.41 | ||
| 2000/2000 | 0.01 | 2 | 1.00 | 0.99 | 1.00 | 1.00 |
| 4 | 0.98 | 0.98 | 0.98 | 0.99 | ||
| 8 | 0.83 | 0.71 | 0.81 | 0.73 | ||
| 0.02 | 2 | 0.98 | 0.96 | 0.98 | 0.98 | |
| 4 | 0.97 | 0.92 | 0.97 | 0.95 | ||
| 8 | 0.71 | 0.56 | 0.68 | 0.63 | ||
Note that Beagle, FZ, and Whait require phased haplotypes for association and we supplied them with the true haplotypes; our LHS method used diplotypes.
The penetrance is assumed to be fixed at 0.10, so that a higher sporadic rate results in a lower power.
Analysis of Wellcome Trust Case Control Consortium data sets
We applied our LHS method to data from the Wellcome Trust Case Control Consortium (2007). We analyzed all seven phenotypes, but bipolar disorder and hypertension yielded no significant haplotype association that survived pruning (see below) and thus were excluded from the discussion. The remaining five phenotypes are coronary artery disease (CAD), Crohn’s disease (CD), rheumatoid arthritis (RA), type 1 diabetes (T1D), and type 2 diabetes (T2D). Each disease phenotype had ∼1800 cases, 2800 controls, and 395,000 autosomal SNPs after routine data quality control (QC) (described in Materials and Methods). We performed the principal component analysis after the QC and used the top 10 eigenvectors to control for population stratification in our regression models. (The histograms of the top six principal components, their pairwise plots, and the histogram for all eigenvalues for the RA phenotype are shown in Figure S1. Plots for other phenotypes were omitted because they were similar to that of RA.)
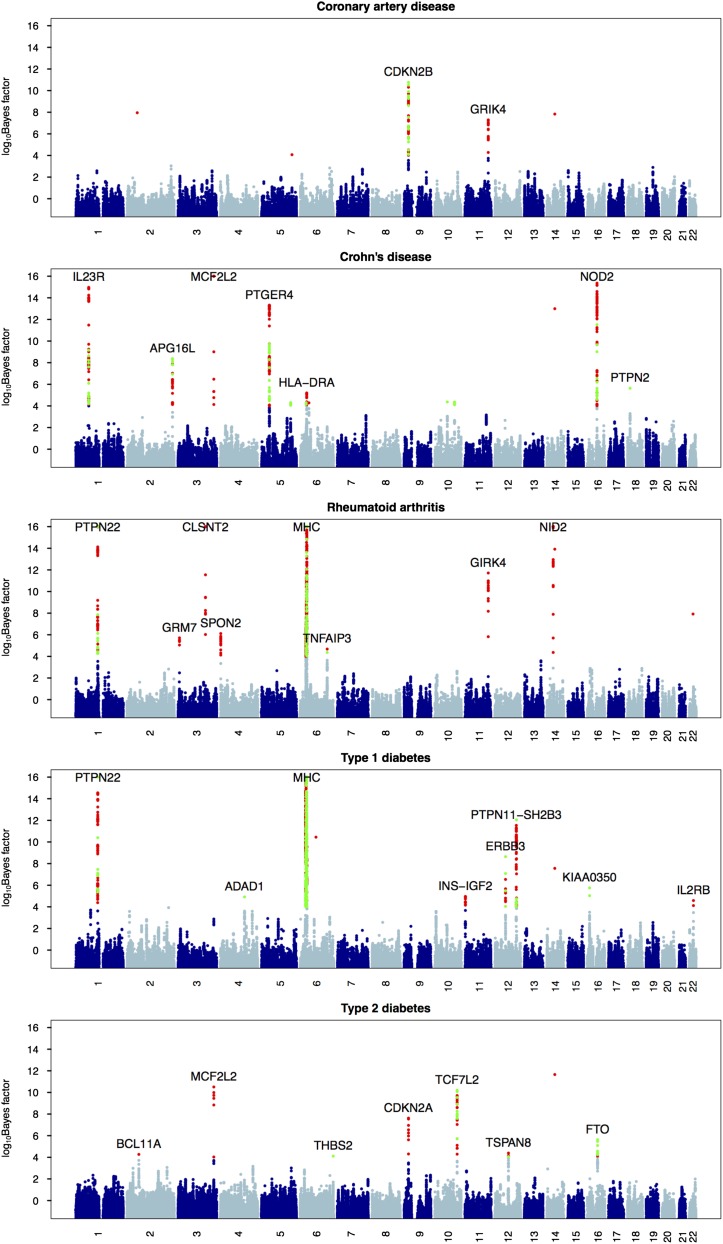
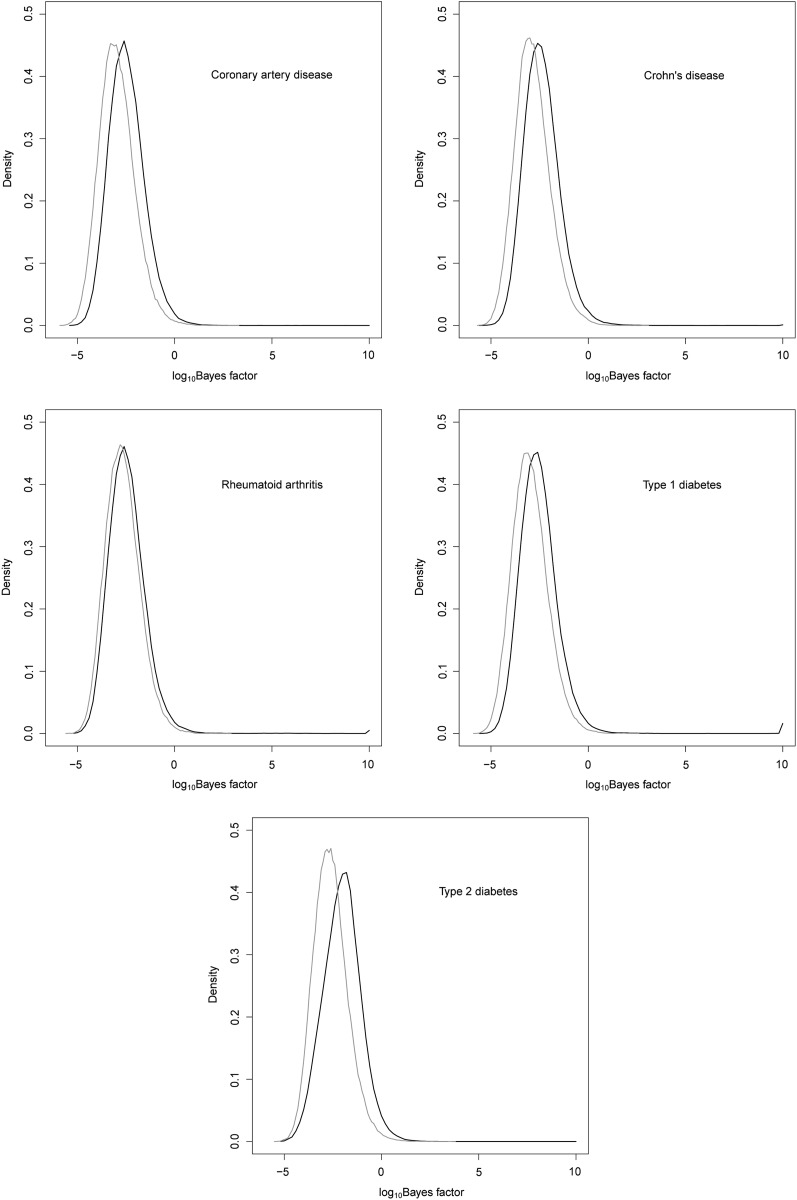
On a small cluster of 15 computing nodes with a total of 100 cores, one Wellcome Trust Case Control Consortium (WTCCC) data set can be analyzed overnight. This includes 10 independent replicates of LD model fitting and association testing (The LD model fitting used 30 EM steps and the number of upper-layer clusters S = 2 and the number of lower-layer clusters K = 10). Figure 4 shows association signals of our haplotype method: the core SNPs whose log10 BF > 4 are colored in red, and signals from the single-SNP analysis whose log10 BF > 4 are superimposed on them and colored in green. Many strong association signals could be detected by both the single-SNP analysis and the haplotype analysis; some strong associations were detected only by our haplotype method, and a few modest associations were detected by the single-SNP method but not by our haplotype method. This largely agrees with our intuition and the power analysis: for regions that have allelic heterogeneity, our haplotype method has more power; while for regions that have no allelic heterogeneity, the single-SNP method performs better due to a smaller degree of freedom in its test statistic. For each disease phenotype, we permuted case–control labels once and computed Bayes factors, treating these as Bayes factors under the null. Figure 5 compares distribution of Bayes factors under the alternative and under the null for five disease phenotypes. The maximum log10 BF under the null for all five phenotypes combined is 3.6, which corroborates our empirical threshold of 4 for log10 Bayes factors.
Figure 4.
Manhattan plots for five disease phenotypes in the WTCCC. The log10 Bayes factors are plotted at each marker (values were truncated at 16). The green circles are single SNPs whose log10 Bayes factors are >4; the red circles are core SNPs whose haplotype association log10 Bayes factors are >4. The relevant genes are marked for significant associations.
Figure 5.
Distribution of Bayes factors under the null (shaded line) and the alternative (solid line) for five disease phenotypes in the WTCCC. Bayes factors under the alternative were truncated at 10. Each null distribution was obtained by permuting the phenotype once.
Pruning false positives:
Our LHS method is sensitive to possible batch effect and genotyping errors. We therefore checked cluster plots for all core SNPs that showed significant associations. Because LHS is correlated between nearby markers, we expected to see signal buildup near genuine associations. Figure 4 contains nine orphan signals that have no signal buildup. We examined cluster plots for these SNPs and, not surprisingly, discovered data quality problems with them all. Specifically, SNP rs7154773 on chromosome 14 appeared as an orphan signal in all five phenotypes. The cluster plot for this SNP revealed that it has a fourth cluster in both control samples and all five case samples (Figure S2), which might be caused by a third allele or probes in repeat regions. The same artifact was reported previously by Liu et al. (2011). The other four orphan signals—rs10167057 and rs7731936 for CAD, rs5755495 for RA, and rs2655693 for T1D—were also found problematic in their SNP cluster plots (Figure S3).
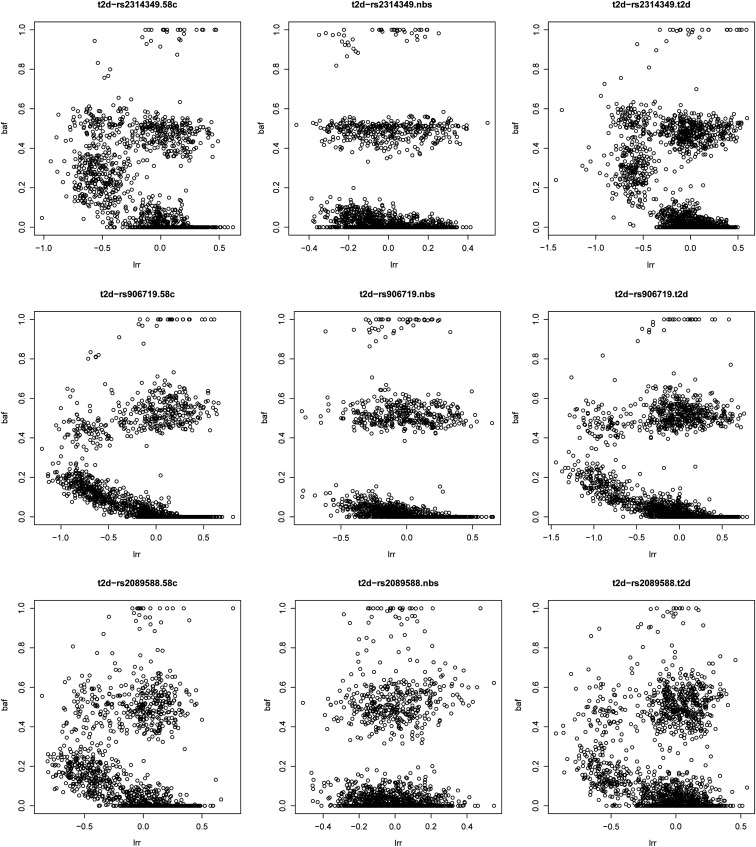
Next, for each novel association not discovered by the single-SNP analysis, we examined cluster plots for all core SNPs that showed strong haplotype associations (log10 BF > 4). The SNPs whose cluster plots indicated possible excessive genotyping errors or batch effects were removed, the remaining SNPs were refitted to the LD model, and Bayes factors were recomputed. This practice removed four more associations: MCF2L2 for CD, CLSNT2 and NID2 for RA, and MCF2L2 for T2D. In Figure 6, we plotted an example of such SNPs in gene MCF2L2 on chromosome 3, which showed strong associations with T2D (and CD). When these SNPs were removed, however, the association signals disappeared. (The cluster plots for SNPs in other gene regions can be found in Figure S4, Figure S5, and Figure S6.) Note that all these SNPs passed non-LD-based routine QC described in Materials and Methods.
Figure 6.
Cluster plots for three SNPs in gene MCF2L2 (on chromosome 3), which shows a strong association with type 2 diabetes. After these three SNPs were removed, the signal disappeared. In each panel, the x-axis is the logR-Ratio (LRR), and the y-axis is the B-allele frequency (BAF). The LRR is a normalized measure of the total signal intensity for two alleles of the SNP. The BAF is a normalized measure of the allelic intensity ratio of two alleles.
Annals of associations:
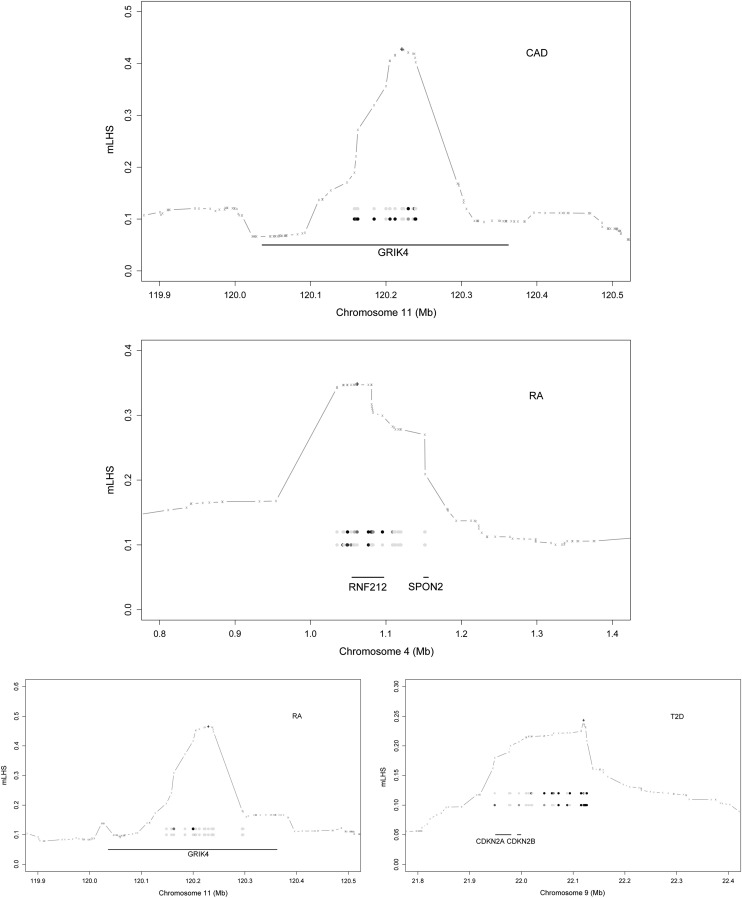
The remaining eight novel associations (not discovered by the single-SNP analysis) are summarized in Table 4. These were separated into strong and modest associations, at the Bayes factor cutoff of 106. Four of them were reported previously, but are novel to the WTCCC study. The association between CDKN2A/CDKN2B and T2D was reported by three GWAS in Asian populations (Takeuchi et al. 2009; Li et al. 2013; Tabassum et al. 2013) and a meta-analysis with European samples (Voight et al. 2010); the two gene regions—INS-IGF2 and IL2RB—are both well known to be associated with T1D (Hakonarson et al. 2007; Plagnol et al. 2011); and the association between HLA-DRA and CD was discovered by directly typing and testing HLA alleles (Stokkers et al. 1999). The remaining four associations appear to be novel in the GWAS context: these are GRIK4 for CAD and SPON2, GRM7, and GRIK4 for RA. Of course, these novel associations require confirmation from other data sets and functional studies. Here we briefly discuss their biological plausibility.
Table 4. Significant associations.
| Disease | SNP ID | Chr | Region (Mb) | Gene | BF2 | P2 | BF1 | P1 |
|---|---|---|---|---|---|---|---|---|
| CAD | rs7104543 | 11 | 120.16–120.24 | GRIK4 | 7.29 | 10.74 | 0.71 | 2.36 |
| RA | rs1010342 | 4 | 1.04–1.08 | SPON2 | 6.11 | 11.98 | 0.60 | 2.33 |
| RA | rs11218032 | 11 | 120.16–120.24 | GRIK4 | 11.60 | 14.50 | −0.90 | 0.52 |
| T2D | rs2383208 | 9 | 22.12–22.13 | CDKN2A, CDKN2B | 7.62 | 8.57 | 1.96 | 3.69 |
| CD | rs9268858 | 6 | 32.54–32.56 | HLA-DRA | 5.20 | 8.27 | 1.66 | 3.35 |
| RA | rs1605705 | 3 | 7.24–7.28 | GRM7 | 5.76 | 8.43 | 1.04 | 2.82 |
| T1D | rs6578246 | 11 | 2.18–2.26 | INS-IGF2 | 4.96 | 8.52 | 2.34 | 4.09 |
| T1D | rs3218256 | 22 | 35.87–35.87 | IL2RB | 4.60 | 6.47 | 2.44 | 4.26 |
P2 = −log10 P-value, which is the minimum P-value over 10 independent EM runs, and in each EM run a P-value was computed using logistic regression Equation 8. BF2 = log10 Bayes factor, which is averaged over 10 independent EM runs, and in each EM run a Bayes factor was computed using our haplotype method. P1 and BF1 are P-values and Bayes factors for the single-SNP test, respectively. The coordinates are from NCBI Build 35.
The gene GRIK4 encodes a protein that belongs to the glutamate-gated ionic channel family. Glutamate functions as the major excitatory neurotransmitter in the central nervous system through activation of ligand-gated ion channels and G protein-coupled membrane receptors, and multiple neurogenetrative diseases, such as bipolar disorder (Pickard et al. 2008), schizophrenia (Pickard et al. 2006), and depression (Paddock et al. 2007), are associated with GRIK4. Its association with bipolar disorder, however, is not shown in the WTCCC data set.
In our haplotype analysis, GRIK4 was associated with both CAD and RA, both of which are common chronic inflammatory diseases, and RA patients have an increased prevalence of CAD (Seferovic et al. 2006; Abou-Raya et al. 2007). A study suggests links between glutamate receptor and autoimmune interactions (Gahring et al. 1997). In addition, there is experimental evidence that connects GRIK4 with CAD: in rat cardiac tissue, the ionotropic glutamate receptors (iGluRs) were detected, and immunohistochemistry localized the iGluRs to cardiac nerve terminals, ganglia, conducting fibers, and some myocardiocytes (Gill et al. 1998). Meanwhile, glutamate connects GRIK4 and RA. Glutamate is relevant to RA in two ways: inflammation of the joint is accompanied by elevated levels of glutamate within the synovial fluid (Flood et al. 2004), and glutamate modulates bone cell phenotype (Chenu 2002). The gene GRM7 and RA also find their connections through glutamate, as GRM7 encodes the receptor for glutamate.
The association between RA and the gene SPON2 is also biologically plausible. SPON2 encodes extracellular matrix proteins that bind directly to bacteria and their components and functions as an opsonin for macrophage phagocytosis of bacteria. This gene is essential in the initiation of the innate immune response and represents a unique pattern-recognition molecule in the extracellular matrix for microbial pathogens. The connection between microbial infection and RA has been long established (Wilder and Corfford 1991).
Figure 7 provides details of four strongly associated regions. The result shown was obtained from a single EM run (of 10 total) that produced the most significant association. For each region, we quantified the LD block, using mLHS, around the core SNP that showed the strongest association; the plot illustrates that the genes of interest are indeed within the LD block. At the same core SNP, we obtained a loading matrix of K columns (recall K is the number of lower-layer clusters and we used K = 10 for the data analyses). We fitted a logistic regression between each column of the loading matrix and the case–controls status to obtain a P-value. Because each column of the loading matrix corresponds to an ancestral haplotype (informally, the columns are dosages of the corresponding ancestral haplotype that each individual carries), these P-values are evidence for whether the corresponding ancestral haplotypes affect disease risk. We declared an ancestral haplotype as significant if the P-value of its loading is <0.001. The two most significant haplotypes within the LD block are shown in the plot. The single-SNP analysis suggested that all four regions have negligible single-SNP associations. Put together, these four regions are examples of allelic heterogeneity and that our haplotype method, by aggregating association signals from multiple ancestral haplotypes, is able to take advantage of the allelic heterogeneity to detect associations, which is evidently more powerful than single-SNP analysis for such regions.
Figure 7.
Details of strong associations. The LD block (based on mLHS) around the most significant signal (marked by “+”) is denoted with a shaded line. The top two ancestral haplotypes whose loadings are strongly associated with phenotypes are expressed as circles, with darker circles representing larger ancestral allele frequencies. The genes within the LD block are shown as solid segments.
Discussion
We have developed a haplotype association method and demonstrated its power through both simulations and real data analysis. Our LHS method redefines the haplotype association. Compared to existing haplotype methods, our method integrates out phase uncertainty, avoids arbitrariness in specifying a window to define haplotypes, and aggregates haplotypes—through ancestral haplotypes—before testing. Each SNP serves as a core SNP for its local haplotypes. The extent of local haplotypes is not prespecified; rather, it is learned from the data and varies along the genome according to sample-specific LD patterns. In regions that have no allelic heterogeneity, our method loses power, compared to the single-SNP test, because its test statistic has a higher degree of freedom. This is more than compensated for, however, by the power gain at regions that have allelic heterogeneity. Thus, our method complements and improves upon the single-SNP analysis.
We compared our LHS method with three other haplotype methods (Browning and Browning 2007; Feng and Zhu 2010; Li et al. 2010a). We regret that the method described in Browning and Thompson (2012) was not included in the comparison; the significance level we examined requires 108 permutation tests for their method, which is not feasible with our current computational resources. Although the other three methods have advantages by assuming known haplotype phase (Browning and Browning 2007; Feng and Zhu 2010; Li et al. 2010a) and perfect imputation (Li et al. 2010a), our LHS method outperforms them all in all but one simulation scenario, and the margin increases with the allelic heterogeneity. The most singular advantage of our method is to aggregate haplotypes according to ancestral haplotypes and then to test aggregated haplotypes jointly, via the loadings, in association with a phenotype. And this haplotype aggregating and testing approach appears to be more effective than other competing methods that we compared with.
One might be tempted to combine the single-SNP test and the haplotype test, in hopes of creating a more powerful method. Indeed, we tried this. Adding an extra term in model (5), we obtained a new model where g is a vector of genotypes. This model appeared to be very powerful, producing a plethora of associations with very large effect sizes. But, the majority of these associations are genotyping artifacts. Let βj = 2γ2θj, where θj are ancestral allele frequencies, and the model reduces to y = Wa + gγ1 + xγ2 + e, where x = 2Lθ is a vector of imputed allele dosages. This model tends to capture SNPs that have different genotyping error rates between case and control, which seem to be more enriched than one would expect in the WTCCC data sets.
A new association method may have ample ways to produce false positives. We have seen two: genotyping artifacts and an extra term in the regression model. Allele flipping is a third. Our method is sensitive to allele flipping between case and control, even for those SNPs whose MAFs = 0.5. Suppose we have two adjacent markers, the first a C/G SNP and the second an A/T SNP, and both have MAFs of 0.5. Suppose the G-T haplotype dominates in both cases and controls when there is no allele flip, and suppose in the cases the two SNPs have their alleles flipped; the dominant haplotype in the cases then becomes C-A, and the dominant haplotype in the controls remains G-T. A well-behaved haplotype method can pick up this difference easily and report a strong association. On the other hand, prevailing QC procedures often flip alleles of A/T or C/G SNPs to match their allele frequencies between cases and controls. This practice is problematic for the haplotype method because some haplotype associations might be lost due to allele flipping—just imagine the opposite of the previous example. Therefore, it is prudent to ensure the consistency of allele codings between cases and controls, and the research community should provide strand information for every data set (as well as low-level data to produce cluster plots). The LD-based data QC (Scheet and Stephens 2008) is important for haplotype association methods and methods that detect epistatic interactions.
We developed a novel algorithm to fit the LD model that is linear in number of clusters; this is crucial for applying our method to big data, such as GWAS and resequencing studies. Our linear algorithm is different from the linear algorithm used in the phasing method SHAPEIT (Delaneau et al. 2012). SHAPEIT groups adjacent heterozygous markers into small windows, iterates all haplotypes within each window, and samples haplotypes across windows. Our linear algorithm conditioned on an average haplotype and achieved linear complexity without sampling. It can be adapted to fit other models for phasing and imputation, such as MaCH (Li et al. 2010b) and fastPHASE (Scheet and Stephens 2006). We also anticipate the fast algorithm will contribute to the LD-based data QC. Moreover, its theoretical property is of interest in its own right and we will investigate this further elsewhere.
Our LHS method can be extended to analyze rare variants. Aggregating rare variants to account for allelic heterogeneity to increase power is standard in analyzing rare variants (Li and Leal 2008). Our method suggests aggregating rare variants based on their local haplotype backgrounds. Accommodating rare variants in our LD model requires a large number of clusters to capture more subtle differences between more recent ancestral haplotypes. Our linear algorithm makes the computation feasible. Moreover, current methods aggregate rare variants based on gene annotation and are not applicable to intergenic regions. Our LD model can quantify an LD block, through the mLHS, around each marker and aggregate rare variants within the LD block.
Supplementary Material
Acknowledgments
We thank Yufeng Shen and Kai Wang for discussion of association results; Nancy Cox for discussion of the allele-flipping problem in GWAS; and Mark Meyer, at Department of Pediatrics, Baylor College of Medicine, for valuable editorial comments. We thank two anonymous referees for their comments that improved the clarity of the presentation. This study makes use of data generated by the Wellcome Trust Case Control Consortium (WTCCC). A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Funding for the WTCCC project was provided by the Wellcome Trust under awards 076113 and 085475. This study was supported in part by research grants from the U.S. Department of Agriculture/Agricultural Research Service (6250-51000-052) and the National Institutes of Health (HG005859).
Footnotes
Communicating editor: C. Kendziorski
Literature Cited
- Abou-Raya S., Abou-Raya A., Naim A., Abuelkheir H., 2007. Chronic inflammatory autoimmune disorders and atherosclerosis. Ann. N. Y. Acad. Sci. 1107: 56–67. [DOI] [PubMed] [Google Scholar]
- Browning B. L., Browning S. R., 2007. Efficient multilocus association testing for whole genome association studies using localized haplotype clustering. Genet. Epidemiol. 31(5): 365–375. [DOI] [PubMed] [Google Scholar]
- Browning S. R., Thompson E. A., 2012. Detecting rare variant associations by identity-by-descent mapping in case-control studies. Genetics 190: 1521–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenu C., 2002. Glutamatergic regulation of bone resorption. J. Musculoskelet. Neuronal Interact. 2(5): 423–431. [PubMed] [Google Scholar]
- Clark A. G., 2004. The role of haplotypes in candidate gene studies. Genet. Epidemiol. 27: 321–333. [DOI] [PubMed] [Google Scholar]
- Delaneau O., Marchini J., Zagury J., 2012. A linear complexity phasing method for thousands of genomes. Nat. Methods 9(1): 179–181. [DOI] [PubMed] [Google Scholar]
- Drysdale C. M., McGraw D. W., Stack C. B., Stephens J. C., Judson R. S., et al. , 2000. Complex promoter and coding region b2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc. Natl. Acad. Sci. USA 97: 10483–10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler E. E., Flint J., Gibson G., Kong A., Leal S. M., et al. , 2010. Missing heritability and strategies for finding the underlying causes of complex disease. Nat. Rev. Genet. 11(6): 446–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng T., Zhu X., 2010. Genome-wide searching of rare genetic variants in WTCCC data. Hum. Genet. 128(3): 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flood S., Duance V., Mason D., 2004. The role of glutamate signalling in rheumatoid arthritis. Int. J. Exp. Pathol. 85(1): A19–A20. [Google Scholar]
- Gahring L., Rogers S., Twyman R., 1997. Autoantibodies to glutamate receptor subunit glur2 in nonfamilial olivopontocerebellar degeneration. Neurology 48(2): 494–500. [DOI] [PubMed] [Google Scholar]
- Gill S. S., Pulido O. M., Mueller R. W., McGuire P. F., 1998. Molecular and immunochemical characterization of the ionotropic glutamate receptors in the rat heart. Brain Res. Bull. 46(5): 429–434. [DOI] [PubMed] [Google Scholar]
- Guan Y., 2014. Detecting structure of haplotypes and local ancestry. Genetics 196: 625–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Stephens M., 2008. Practical issues in imputation-based association mapping. PLoS Genet. 4(12): e1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Stephens M., 2011. Bayesian variable selection regression for genome-wide association studies, and other large-scale problems. Ann. Appl. Stat. 5(3): 1780–1815. [Google Scholar]
- Hakonarson H., Grant S. F. A., Bradfield J. P., Marchand L., Kim C. E., et al. , 2007. A genome-wide association study identifies kiaa0350 as a type 1 diabetes gene. Nature 448(7153): 591–594. [DOI] [PubMed] [Google Scholar]
- Hudson R. R., 2002. Generating samples under a Wright-Fisher neutral model. Bioinformatics 18: 337–338. [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium , 2010. Integrating common and rare genetic variation in diverse human populations. Nature 467(7311): 52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten P. H., Toepoel M., Mariman E. C., Van Zoelen E. J., 2001. Promoter haplotype combinations of the platelet-derived growth factor alpha-receptor gene predispose to human neural tube defects. Nat. Genet. 27: 215–217. [DOI] [PubMed] [Google Scholar]
- Kang H. M., Zaitlen N. A., Wade C. M., Kirby A., Heckerman D., et al. , 2008. Efficient control of population structure in model organism association mapping. Genetics 178: 1709–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Leal S. M., 2008. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am. J. Hum. Genet. 83(3): 311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Gan W., Lu L., Dong X., Han X., et al. , 2013. A genome-wide association study identifies grk5 and rasgrp1 as type 2 diabetes loci in Chinese Hans. Diabetes 62(1): 291–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Byrnes A. E., Li M., 2010a To identify associations with rare variants, just whait: weighted haplotype and imputation-based tests. Am. J. Hum. Genet. 87(5): 728–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Willer C. J., Ding J., Scheet P., Abecasis G. R., 2010b Mach: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 34(8): 816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Xu H., Chen S., Chen X., Zhang Z., et al. , 2011. Genome-wide interaction-based association analysis identified multiple new susceptibility loci for common diseases. PLoS Genet. 7(3): e1001338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher B., 2008. Personal genomes: the case of the missing heritability. Nature 456: 18–21. [DOI] [PubMed] [Google Scholar]
- Manolio T. A., Collins F. S., Cox N. J., Goldstein D. B., Hindorff L. A., et al. , 2009. Finding the missing heritability of complex diseases. Nature 461: 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddock S., Laje G., Charney D., Rush A. J., Wilson A. F., et al. , 2007. Association of grik4 with outcome of antidepressant treatment in the star*d cohort. Am. J. Psychiatry 164: 1181–1188. [DOI] [PubMed] [Google Scholar]
- Pickard B. S., Malloy M. P., Christoforou A., Thomson P. A., Evans K. L., et al. , 2006. Cytogenetic and genetic evidence supports a role for the kainate-type glutamate receptor gene, grik4, in schizophrenia and bipolar disorder. Mol. Psychiatry 11(9): 847–857. [DOI] [PubMed] [Google Scholar]
- Pickard B. S., Knight H. M., Hamilton R. S., Soares D. C., Walker R., et al. , 2008. A common variant in the 3′ UTR of the grik4 glutamate receptor gene affects transcript abundance and protects against bipolar disorder. Proc. Natl. Acad. Sci. USA 105(39): 14940–14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagnol V., Howson J. M. M., Smyth D. J., Walker N., Hafler J. P., et al. , 2011. Genome-wide association analysis of autoantibody positivity in type 1 diabetes cases. PLoS Genet. 7(8): e1002216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., 2001. Are rare variants responsible for susceptibility to complex diseases? Am. J. Hum. Genet. 69(1): 124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid D. J., 2004. Evaluating associations of haplotypes with traits. Genet. Epidemiol. 27: 348–364. [DOI] [PubMed] [Google Scholar]
- Scheet P., Stephens M., 2006. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 78: 629–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheet P., Stephens M., 2008. Linkage disequilibrium-based quality control for large-scale genetic studies. PLoS Genet. 4(8): e1000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott L., Mohlke K., Bonnycastle L., Willer C., Li Y., et al. , 2007. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316: 1341–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seferovic P. M., Ristic A. D., Maksimovic R., Simeunovic D. S., Ristic G. G., et al. , 2006. Cardiac arrhythmias and conduction disturbances in autoimmune rheumatic diseases. Rheumatology 45(Suppl. 4): iv39–iv42. [DOI] [PubMed] [Google Scholar]
- Servin B., Stephens M., 2007. Efficient multipoint analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 3(7): e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M., Balding D. J., 2009. Bayesian statistical methods for genetic association studies. Nat. Rev. Genet. 10: 681–690. [DOI] [PubMed] [Google Scholar]
- Stokkers P. C. F., Reitsma P. H., Tytgat G. N. J., van Deventer S. J. H., 1999. Hla-dr and -dq phenotypes in inflammatory bowel disease: a meta-analysis. Gut 45: 395–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabassum R., Chauhan G., Dwivedi O. P., Mahajan A., Jaiswal A., et al. , 2013. Genome-wide association study for type 2 diabetes in Indians identifies a new susceptibility locus at 2q21. Diabetes 62(3): 977–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi F., Serizawa M., Yamamoto K., Fujisawa T., Nakashima E., et al. , 2009. Confirmation of multiple risk loci and genetic impacts by a genome-wide association study of type 2 diabetes in the Japanese population. Diabetes 58(7): 1690–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight B. F., Scott L. J., Steinthorsdottir V., Morris A. P., Dina C., et al. , 2010. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat. Genet. 42(7): 579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium , 2007. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447: 661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder R. L., Corfford L. J., 1991. Do infectious agents cause rheumatoid arthritis? Clin. Orthop. Relat. Res. 265: 36–41. [PubMed] [Google Scholar]
- Willer C., Sanna S., Jackson A., Scuteri A., Bonnycastle L., et al. , 2008. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 40: 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Benyamin B., McEvoy B. P., Gordon S., Henders A. K., et al. , 2010. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42(7): 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Stephens M., 2012. Genome-wide efficient mixed model analysis for association studies. Nat. Genet. 44(4): 821–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Carbonetto P., Stephens M., 2013. Polygenic modeling with Bayesian sparse linear mixed models. PLoS Genet. 9(2): e1003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zöllner S., Pritchard J. K., 2005. Coalescent-based association mapping and fine mapping of complex trait loci. Genetics 169: 1071–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.