Abstract
How common is balancing selection, and what fraction of phenotypic variance is attributable to balanced polymorphisms? Despite decades of research, answers to these questions remain elusive. Moreover, there is no clear theoretical prediction about the frequency with which balancing selection is expected to arise within a population. Here, we use an extension of Fisher’s geometric model of adaptation to predict the probability of balancing selection in a population with separate sexes, wherein polymorphism is potentially maintained by two forms of balancing selection: (1) heterozygote advantage, where heterozygous individuals at a locus have higher fitness than homozygous individuals, and (2) sexually antagonistic selection (a.k.a. intralocus sexual conflict), where the fitness of each sex is maximized by different genotypes at a locus. We show that balancing selection is common under biologically plausible conditions and that sex differences in selection or sex-by-genotype effects of mutations can each increase opportunities for balancing selection. Although heterozygote advantage and sexual antagonism represent alternative mechanisms for maintaining polymorphism, they mutually exist along a balancing selection continuum that depends on population and sex-specific parameters of selection and mutation. Sexual antagonism is the dominant mode of balancing selection across most of this continuum.
Keywords: intralocus sexual conflict, sexual antagonism, heterozygote advantage, pleiotropy, maintenance of polymorphism
ONE of the primary goals of population genetics is to evaluate how processes of mutation, selection, migration, recombination, and genetic drift account for the maintenance of genetic variation in natural populations. This “great obsession” with genetic variation (Gillespie 2004, p. 154) is reflected by a massive body of theoretical and empirical research that seeks to connect empirical patterns of population and quantitative genetic variability with the specific evolutionary processes that might account for them.
Balancing selection—selection that maintains genetic variation at a locus—could potentially account for an important fraction of observed quantitative genetic variability (Dobzhansky 1955; Lewontin 1974; Charlesworth and Hughes 1999). Although relatively few unambiguous cases of balancing selection have been documented to date (see Charlesworth 2006; Hedrick 2012; Johnston et al. 2013; Leffler et al. 2013), empirical methods for detecting their population genetic signatures are highly conservative and may fail to identify many (or most) instances of balancing selection within a genome (Charlesworth 2006; Charlesworth and Charlesworth 2010). Furthermore, balanced polymorphisms may plausibly account for several empirical observations that are not easily explained by alternative models of variation maintained by recurrent mutation. For example, the high genetic variance in life history and reproductive traits (Houle 1992; Pomiankowski and Moller 1995) and the presence of intermediate-frequency alleles with large phenotypic effects (Long et al. 2000) are each potentially attributable to a subset of loci that segregate for balanced alleles (Charlesworth and Hughes 1999; Turelli and Barton 2004).
Population genetic theory provides a complementary approach for assessing the plausibility of different scenarios of balancing selection, and indeed there are many formal models that delineate the parameter conditions required for balancing selection to operate (e.g., Wright 1969; Crow and Kimura 1970; Prout 2000). However, most theory cannot predict the probability of balancing selection because it does not incorporate details of the fitness effect distribution among random alleles and genotypes in a population. Such details ultimately determine the relative likelihood of different forms of selection at individual loci.
A recent study by Sellis et al. (2011) provides new insight into the probability of balancing selection by modeling the frequency of heterozygote advantage in Fisher’s geometric model (FGM) (Fisher 1930; Orr 2005a,b)—an influential theoretical framework for the fitness effect distribution of random mutations. Sellis et al. (2011) showed that, among loci contributing to adaptive evolutionary change, the probability of heterozygote advantage tends to increase with the “standardized size” (Orr 1998) of adaptive mutations. Under Fisher’s original scaling, mutation size is given by where r is the absolute phenotypic effect size of the mutation, n is the number of traits, and z represents the displacement of the population from a fitness optimum within phenotypic space (see Fisher 1930, pp. 38–41; Orr 1998). A mutation of size r is effectively small when the distance to the optimum is large (e.g., in the limit r/z → 0) and effectively large when the distance to the optimum is small (e.g., r → z) (for a clear discussion of x and its biological meaning, see Orr 1998, pp. 937–938). Adaptation—the evolutionary movement of a population toward its optimum—causes the distance to the optimum to shrink (Orr 1998) and thereby increases the scaled sizes of random mutations. Poorly adapted populations that are far from an optimum may initially experience little opportunity for heterozygote advantage, yet adaptive evolution will ultimately overturn such initially unfavorable conditions. During the course of an adaptive walk within phenotypic space, mutations eventually become sufficiently “large” for heterozygote advantage to be likely (Sellis et al. 2011; for related results, see Manna et al. 2011).
In species with separate sexes, balancing selection can potentially arise by way of heterozygote advantage or by sexual antagonism (a.k.a. “intralocus sexual conflict”), in which the best genotype for females differs from the best genotype for males (Rice 1992; Chippindale et al. 2001). Whether balanced, sexual antagonistic alleles are common within populations has important and wide-ranging biological implications for the genetic basis of quantitative traits (Rice 1984; Turelli and Barton 2004; Bonduriansky and Chenoweth 2009), genetic loads and extinction risk (Kokko and Brooks 2003; Whitlock and Agrawal 2009; Connallon et al. 2010; Holman and Kokko 2013), mating system evolution (Seger and Trivers 1986; Albert and Otto 2005; Blackburn et al. 2010; Roze and Otto 2012), and the evolution of genomes and genetic systems (Charlesworth and Charlesworth 1980; Day and Bonduriansky 2004; Ellegren and Parsch 2007; van Doorn and Kirkpatrick 2007, 2010; Mank 2009; Connallon and Clark 2010, 2011, 2013a; Parsch and Ellegren 2013; Wright and Mank 2013; Charlesworth et al. 2014; Kirkpatrick and Guerrero 2014). Although prior theory clearly defines the parameter criteria for balancing selection by sexual antagonism (e.g., Kidwell et al. 1977; Pamilo 1979; Patten and Haig 2009; Unckless and Herren 2009; Fry 2010; Patten et al. 2010, 2013; Arnqvist 2011; Connallon and Clark 2012; Mullon et al. 2012; Jordan and Charlesworth 2012), it remains unclear how often such conditions might be expected to arise in dioecious populations. A recent extension of Fisher’s geometric model provides a theoretical framework for predicting the sex-specific distribution of mutant fitness effects (Connallon and Clark 2014), yet this theory has not addressed opportunities for balancing selection.
Here, we present new theoretical predictions for the probability of balancing selection in a species with distinct sexes. Our models build upon a recent extension of Fisher’s geometric model (Connallon and Clark 2014) that incorporates two evolutionarily important features of dioecious species: (1) sexually dimorphic fitness landscapes, which generate sex-specific patterns of selection on phenotypes expressed by each sex, and (2) sex-by-genotype interactions (a form of gene-by-environment interaction during development), in which mutations have sexually dimorphic effects on female and male phenotypes. We first extend Fisher’s original scaling function by developing a mathematical expression for the standardized mutation size in a dioecious population. We show that this new scaling function can be used to calculate several interesting metrics in FGM, including (1) the probability that random mutations experience positive, purifying, or balancing selection and (2) the critical mutation size that maximizes the probability of balancing selection. We show that sex differences in selection on phenotypes and genotype-by-sex interactions each tend to inflate the standardized mutation size and that these effects increase opportunities for balancing selection in the FGM framework. Finally, we demonstrate that, for most of the parameter space of sex-specific selection and gene-by-sex interactions, sexual antagonism is a pervasive feature of balancing selection.
Model
FGM with two sexes
The basic model analyzed here is a diploid extension of the haploid, two-sex FGM model that was recently developed by Connallon and Clark (2014). Male and female phenotypes are each characterized by a vector of n trait values, with each vector representing a specific location in n-dimensional phenotypic space. For mathematical convenience, and following prior work in Fisher’s model (Fisher 1930; Orr 1998; Sellis et al. 2011; Connallon and Clark 2014), we assume that (1) sex-specific fitness is a function of the Euclidean distance to the relevant optimum and (2) each mutation has a random, unbiased orientation within phenotypic space. Relaxation of these assumptions—for example, by allowing for elliptical fitness surfaces and/or mutational biases—often decreases the “effective” dimensionality in Fisher’s geometric model (see Waxman and Welch 2005; Martin and Lenormand 2006; Martin 2014), such that n may be interpreted as the effective number (rather than the total number) of independent traits.
The initial population is assumed to be fixed for a wild-type genotype, for which males and females express phenotypic values of Am and Af, respectively, where Aj = {x1j, x2j, … , xnj} is a vector of n trait values for the jth sex; j = {f, m}. Optimal trait combinations for males and females are depicted by vectors Om and Of, which describe the locations of sex-specific optima for the n traits. The direction of selection acting in the two sexes can be described using a pair of vectors (one for each sex) that point from Aj to Oj (Figure 1). The lengths of the vectors are given by zm and zf and represent the displacement of each sex from its optimum (i.e., the Euclidean distance between Aj and Oj). The angle between vectors is θsel, which potentially takes any value between zero (where directions of sex-specific selection are the same in males and females) and π (where the sexes are selected in opposite directions). The degree of correlation between male and female orientations of directional selection is ρsel = cos(θsel) (Rodgers and Nicewander 1988; Connallon and Clark 2014). Sex-specific fitness of females and males, respectively, follows a Gaussian function
| (1a) |
and
| (1b) |
where ωf and ωm are positive constants that describe the rate of fitness decline away from each optimum.
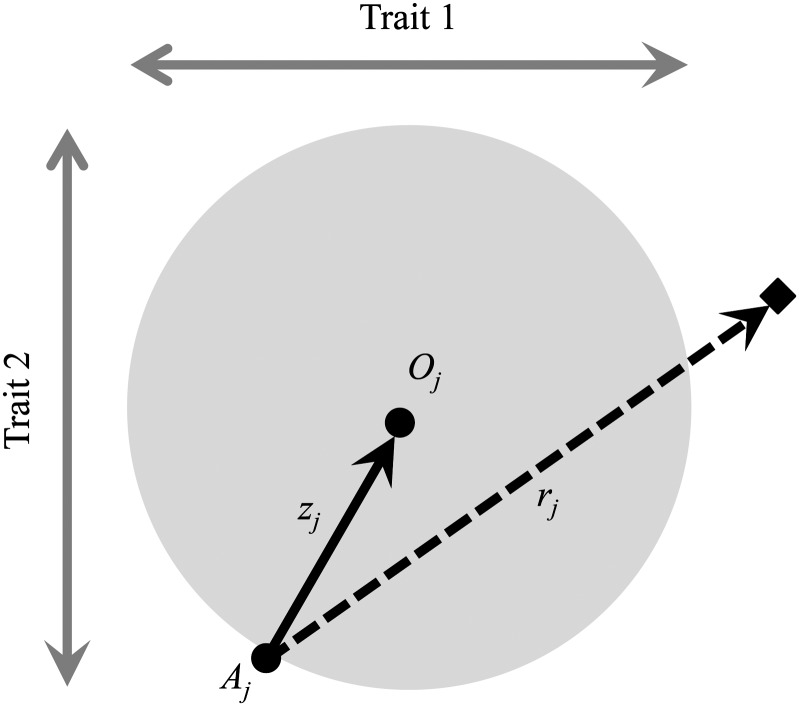
Figure 1.
Fisher’s geometric model with two sexes (an example with two traits). For the jth sex, individuals homozygous for a wild-type allele express phenotype Aj and are separated from their optimum (Oj) by distance zj. Mutations alter the phenotype of their carriers, with mutant homozygotes expressing a phenotype at a distance rj from the ancestral homozygote (an example mutant homozygote is represented by the solid diamond). Heterozygotes are similarly oriented away from Aj, but have a modified distance of hrj, where h is the dominance coefficient with respect to the mutant phenotypic effect. For the example shown, heterozygotes will express an intermediate phenotype between Aj and the solid diamond, and for most values of h (0 < h < 1), the heterozygote phenotype will be closer to the optimum than either homozygote genotype will be. In such cases, there will be heterozygote advantage for fitness in the jth sex. Females and males may be characterized by distinct fitness optima and unique positions in phenotypic space with respect to each genotype (see Connallon and Clark 2014).
The phenotypic effects of each mutation can be similarly summarized using sex-specific vectors (Figure 1). Each mutation has a magnitude, with rj representing the phenotypic (i.e., Euclidean) distance between Aj and the location of individuals that are homozygous for the mutation. The magnitude in a heterozygote is rjh, where h represents the dominance of the mutation with respect to the phenotype (i.e., 0 < h < 1, with h = 0, h = 1/2, and h = 1 corresponding to recessive, additive or codominant, and dominant effects, respectively). Mutational orientations are assumed to be random and unbiased in n-dimensional space. The degree of similarity between the sexes, for each mutation’s orientation in phenotypic space, depends on the between-sex correlation of phenotypic effects within individual trait axes. For a mutation with magnitude rm and rf, ρmut represents both the phenotypic correlation between the sexes for each trait and the mean degree of correlation between mutant vectors {ρmut = E[cos(θmut)], where θmut is the angle between the vectors of a mutation, and E[] denotes the expectation (see Connallon and Clark 2014)}.
Taking both selection and mutation into consideration, we can calculate distances between the optimum Oj and the phenotypic position of individuals that carry zero, one, or two copies of a given mutation. Letting Oj′ = Oj – Aj = {o1j, o2j, … , onj}, the distance to the optimum in heterozygous males and females will be
| (2a) |
and
| (2b) |
(Appendix A), where the values of mi and fi are obtained from n independent draws from a bivariate standard normal distribution with cov(mi, fi) = ρmut (Connallon and Clark 2014). These relationships naturally emerge from the mutation algorithm described in the Simulations section, with approximations on the right-hand side of Equations 2a and 2b accurate for values of n ≥ 10 (Waxman and Welch 2005; Connallon and Clark 2014). Distances to the optima for homozygous carriers of the mutation, zj(hom), can be obtained by setting h = 1 in Equation 2. Heterozygous and homozygous selection coefficients for mutations can be calculated directly from Equations 1 and 2 (Appendix A), with homozygous selection coefficients defined as sj2 = exp[ωj(zj2 – zj(hom)2)] – 1 and heterozygous selection coefficients defined as sj1 = exp[ωj(zj2 – zj(het)2)] – 1.
Criteria for positive selection and balancing selection
Balancing selection criteria follow the well-established theory of sex-specific selection at a diploid locus with two alleles (e.g., Kidwell et al. 1977). Let B1 represent an ancestral allele and B2 be the derived allele. The relative fitnesses for the three genotypes are provided in Table 1. We assume that generations are discrete and follow the life cycle of birth, selection prior to mating, and random mating among breeding adults. In cases where selection differs between males and females, the frequency of alleles inherited from mothers and from fathers will differ. Following convention, we define qm and qf as the frequency of B2 alleles among the male and female gametes that contribute to a given generation.
Table 1. Sex-specific fitnesses and genotype frequencies.
| Genotype | |||
|---|---|---|---|
| B1B1 | B1B2 | B2B2 | |
| Female fitness: | 1 | 1 + sf1 | 1 + sf2 |
| Male fitness: | 1 | 1 + sm1 | 1 + sm2 |
| Zygotic frequency: | (1 – qm)(1 – qf) | qm(1 – qf) + qf(1 – qm) | qmqf |
Following standard criteria for determining evolutionary stability (see Otto and Day 2007), a new (rare) mutation will experience selection to increase in frequency when the boundary equilibrium qm, qf = 0 is unstable. The criterion for selection to favor invasion of a rare mutation is
| (3) |
Mutations that meet this criterion can be further divided into mutually exclusive subsets: (1) positively selected mutations that are also favored to fix in the population and (2) mutations under balancing selection, which can invade from low frequency, but are not favored to fix. For the latter case, selection favors the maintenance of ancestral (wild-type) and mutant alleles. Balancing selection occurs when both boundary equilibria (qm, qf = 0; qm, qf = 1) are unstable, a condition often referred to as a “protected polymorphism” (Prout 1968). The condition for balancing selection is
| (4) |
We define positive selection when qm, qf = 0 is unstable (B2 invades when rare) and qm, qf = 1 is stable (B1 does not invade when rare).
Simulations
In our subsequent analytical results, we assume that (1) mutations have small fitness effects, as is generally expected (e.g., Orr 2005a,b, 2006; analytical results are appropriate for selection coefficients within the approximate range sf1, sf2, sm1, sm2 < 0.1), and (2) dimensionality is sufficiently high so that the approximations in Equation 2 remain valid [i.e., that n > 10 (Waxman and Welch 2005; Connallon and Clark 2014)]. Analytical results were verified using an exact simulation approach described in Connallon and Clark (2014), with adjustments for diploid inheritance. Without any loss of generality, we define the phenotypic positions of wild-type males and females, and their respective optima, so that Af = Am = {0, 0, …, 0}, Of = {zf, 0, …, 0}, and Om = {zmcos(θsel), zmsin(θsel), 0, …, 0}. Mutation magnitudes for each sex (rm, rf) were generated using a bivariate gamma distribution with equal male and female marginal distributions (values of rm and rf were obtained using the “mixture approach” algorithm, described in Michael and Schucany 2002). Within the jth sex, the phenotypic location of individuals homozygous for a random mutation is described by a vector, yj = {y1j, y2j, … , ynj}, with the elements of the vector representing a set of phenotypic changes in the n traits. For a mutation with magnitude rm in males, yim = rmmi/M, where and the mi are independent draws from a standard normal distribution. This sampling strategy generates random, uniformly distributed points on the surface of an n sphere with radius rm (Muller 1959; Marsaglia 1972; Connallon and Clark 2014). For a mutation with magnitude rf in females, yif = rffi/F, where and the fi are independent draws from a standard normal distribution with cov(mi, fi) = ρmut. For a mutation with homozygous effect yj = {y1j, y2j, … , ynj}, the corresponding heterozygous effect is modified by dominance, so that yjh = {y1jh, y2jh, … , ynjh} (following Sellis et al. 2011). Once the yim and yif are specified for an individual mutation, it is straightforward to calculate heterozygous and homozygous selection coefficients for each sex, by using Equations 1 and 2 (see Appendix A).
Results
Mutation size and the probability of balancing selection
The proportion of random mutations that experience positive selection, purifying selection, and balancing selection can be directly calculated using the selection criteria outlined above (Equations 3 and 4) and the distribution of fitness effects that emerges from the two-sex Fisher’s geometric model (Equations 1 and 2) (see Appendix B). Following Fisher’s original concept of the standardized mutation size (Fisher 1930; Orr 1998), let xA represent an adjusted standardized “size” that accounts for sex differences in directional selection and sex-by-genotype phenotypic effects of mutations. This adjusted mutation size is defined as
| (5) |
Mutation size is now a function of sex-specific selection and mutation parameters (ωj, rj, zj), dimensionality (n), and the between-sex correlation of fitness effects: ρmf = ρselρmut (see Model section). Note that, by removing all possible sex differences (letting ρmf = 1, ωm = ωf, r = rm = rf, z = zm = zf), Equation 5 collapses to Fisher’s (1930) original expression: x = r√(n)/(2z).
The individual probabilities of purifying, positive, and balancing selection are simple functions of the new mutation size scaling (Appendix B). The probability that selection favors invasion of a random mutation with size xA is
| (6) |
where Φ(xAh) is the cumulative standard normal distribution, and h is the dominance of the mutation relative to the wild-type allele (0 < h < 1). In the absence of sex differences, where xA = r√(n)/(2z), Equation 6 is identical to previous, high-dimension approximations (n > 10) for the probability of a beneficial mutation in Fisher’s geometric model (Fisher 1930; Kimura 1983; Orr 1998; Sellis et al. 2011).
Alleles selected to invade the population may go to fixation or evolve to an intermediate balanced polymorphic state. These outcomes—positive and balancing selection, respectively—are subsets of the probability space described by Equation 6 and are mutually exclusive. The probability that a random mutation evolves under balancing selection is
| (7) |
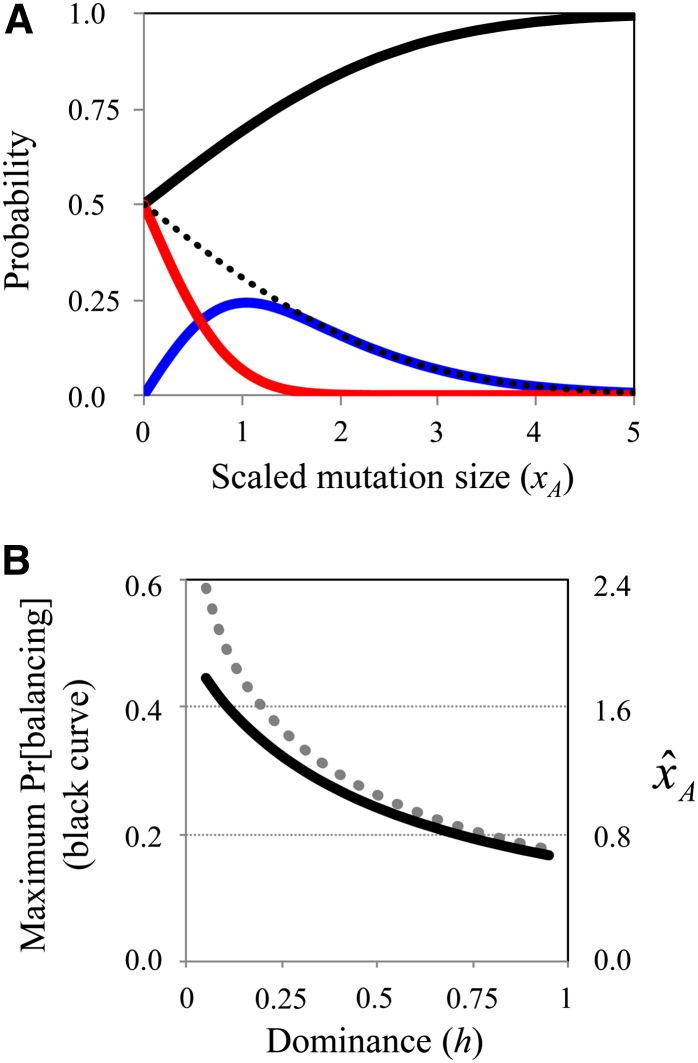
where Φ again refers to the cumulative standard normal. The probability of positive selection is therefore Pr(positive) = Pr(sf1 + sm1 > 0) – Pr(balancing) = 1 – Φ[xA(1 – h2)/(1 – h)]. Relative probabilities of the three modes of selection—purifying, positive, and balancing—are shown in Figure 2A. Positive selection is more common than balancing selection when scaled mutation size is small (∼xA < 1/2). For larger values of xA, a majority of adaptive mutations evolve under balancing selection, rather than positive selection, with the probability of balancing selection ultimately converging, with increasing mutation size, to Pr(balancing) = 1 – Φ(xAh). In other words, as mutation size increases, an ever-increasing share of adaptive mutations (mutations favored to invade when rare) will experience balancing selection.
Figure 2.
Balancing selection in Fisher’s geometric model. (A) Probabilities of purifying selection (black), positive selection (red), or balancing selection (blue), as a function of the scaled mutation of size, xA. The dotted line represents the probability that selection will favor evolutionary invasion of a random mutation of size xA (this is the sum of individual probabilities of positive and balancing selection). Results are for the case of codominant expression in heterozygotes (h = 1/2), and curves were generated using Equations 6 and 7. Simulations (not shown) confirm that these results are extremely accurate at moderate to high dimensionality (i.e., n > 10). (B) The maximum probability of balancing selection (black curve) and the critical value of xA that maximizes the probability of balancing selection (dotted gray line). Results are based on Equations 7 and 8.
As originally noted by Fisher (1930), mutations with small phenotypic effects (xA → 0) are likely to improve fitness, whereas large mutations have small probabilities of doing so. A scaled mutation size near zero will therefore maximize the probability of positive selection and minimize the probability of purifying selection. In contrast, the probability of balancing selection is maximized for mutations of intermediate size. The value of xA that maximizes the probability of balancing selection is
| (8) |
(Appendix C). In the absence of dominance between mutant and wild-type alleles (h = 1/2), this reduces to with partial recessivity (h < 1/2) increasing this critical mutation effect size and partial dominance decreasing it (Figure 2B, gray dotted line). The maximum probability of balancing selection can also be calculated by evaluating Equation 7, using the critical mutation size of Equation 8. When phenotypic effects are codominant (h = 1/2), a maximum of ∼24% of mutations experience balancing selection; the probability of balancing selection further increases with partial recessivity and decreases with partial dominance (Figure 2B).
Mean mutation size in species with separate sexes
Mutation size (xA) clearly influences the probability that random mutations experience purifying, positive, or balancing selection. Small-sized mutations evolve primarily under positive or purifying selection (in the limit, xA → 0, there is an ∼50% probability of each type of selection). For larger xA values, most mutations experience purifying selection, but those that do not generally evolve under balancing selection (see above; Figure 2). How will sex-specific selection and sex-by-genotype effects influence the average magnitude of xA? As demonstrated below, both factors inflate the scaled mutation size and should thereby expand opportunities for balancing selection relative to positive selection.
An intuitive way to view the scaled mutation size is by rewriting it as a function of the strength of directional selection in each sex. The strength of directional selection in the jth sex can be quantified as βj = |dln(wj)/dzj| = 2ωjzj, which is conceptually similar to the selection gradient of quantitative genetics (Lande 1980; Falconer and Mackay 1996) and is roughly proportional to the expected rate of adaptation in Fisher’s geometric model (Orr 2000; Connallon and Clark 2014). Mean mutation size, as a function of the strength and orientation of sex-specific selection, is
| (9) |
which takes into account the probability density of male and female mutational magnitudes among random mutations. To illustrate how sex differences in selection or mutational effects will affect E(xA), it is useful to consider two extremes of the possible values of βm relative to βf: (1) highly asymmetric directional selection (i.e., βm/βf → 0 or βm/βf → ∞), which may be plausible immediately after a change in the environment, and (2) equally strong directional selection in each sex (βm/βf → 1), which is the long-run expectation during adaptive evolution (Lande 1980; Connallon and Clark 2014).
When directional selection is highly asymmetric, the scaled mutation size reduces to xA ≈ (ωmrm2 + ωfrf2)(n)0.5/(βjrj), where j refers to the sex experiencing directional selection (there is no directional selection in the other sex if it is at its optimum, which we assume here). We arbitrarily present results for the case where males alone experience directional selection (βm/βf → ∞; similar results are obtained for the opposite case of βm/βf → 0). The average scaled size of a mutation can be approximated using a Taylor series expansion of xA, to second order with respect to the mean mutation size of each sex. We assume that mutation size follows a bivariate gamma distribution of mutational magnitudes, with equal male and female marginal distributions [E(r) = E(rm) = E(rf); var(r) = var(rm) = var(rf)] and a between-sex correlation of mutational magnitudes of corr(rm, rf) = cov(rm, rf)/var(r). The mean size of a random mutation is then
| (10) |
(Appendix D), where k is the shape parameter of the gamma distribution (k > 0), and is the male-specific scaled mutation size. From Equation 10 we can visualize three factors that inflate the mean scaled mutation size: E(xA) increases as the relative strength of stabilizing selection in females increases (ωf/ωm increases), and as the distribution of mutation magnitudes becomes increasingly leptokurtic (where k decreases, which skews the distribution toward small values), and the correlation between sexes becomes small to negative [corr(rm, rf) decreases]. The scaled mutation size is always elevated as a consequence of selection in females, and the magnitude of this elevation is substantial when the ratio ωf/ωm is moderate to large [e.g., for ωf/ωm > 1/2 and corr(rm, rf) > 0, E(xA) is inflated by at least 50% relative to the case of no selection in females].
For the scenario of equally strong directional selection in each sex (β = βm = βf), the average scaled mutation size becomes
| (11) |
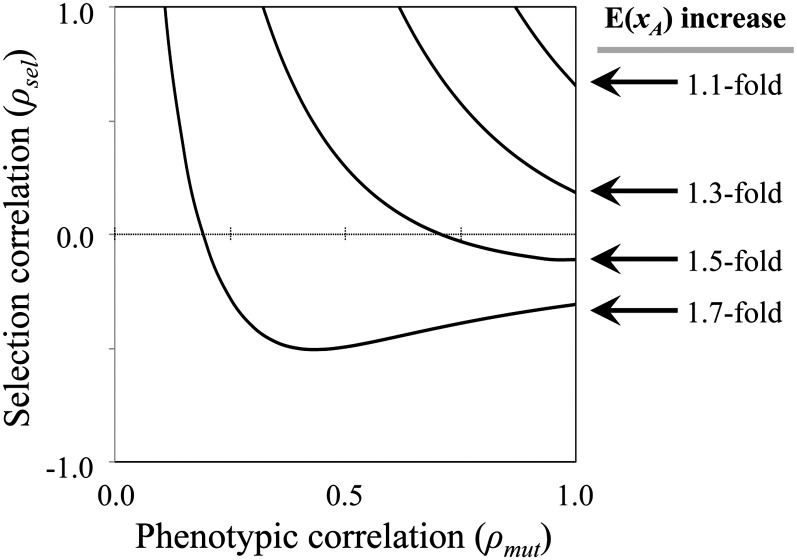
(Appendix D), where ω = (ωm + ωf)/2. E(xA) now depends on the interaction between three forms of correlation between the sexes: (1) the correlation of mutant magnitudes, corr(rm, rf); (2) the correlation of male and female selection orientations, cos(θsel); and (3) the mean correlation of mutational orientations, E[cos(θmut)] {recall that ρmf = cos(θsel)E[cos(θmut)]}. E(xA) is inflated by sex differences in selection or mutational effects when the bracketed term in Equation 11 is >1. When mutant phenotypic effects are strongly correlated between the sexes [corr(rm, rf), cos(θmut) → 1], the bracketed term reduces to 2[2 + 2cos(θsel)]–1/2, which increases as the orientations of selection become increasingly divergent between the sexes (θsel > 0). When phenotypic correlations between the sexes are weak [corr(rm, rf), cos(θmut) → 0], E(xA) is elevated by a factor of (1 + 4k)/(2k√2) or at least ∼1.4-fold. Figure 3 shows the magnitude of increase for E(xA), across a range of conditions for sex-specific selection and mutational effects. E(xA) is inflated across the entire range of parameter space and particularly so when the sexes are selected in different directions (cos(θsel) << 1) or the phenotypic effects of mutations are strongly decoupled between the sexes (0 < ρmut = corr(rm, rf) = E[cos(θmut)] << 1).
Figure 3.
Sex differences in the direction of selection, and sex-by-genotype effects of mutations, elevate mean mutation size, E(xA). Solid curves show the elevation of E(xA) relative to an idealized population in which the selection and mutational effects are identical between the sexes. The fold change to E(xA) is based on numerical evaluation of the bracketed term in Equation 11, with ρmut = corr(rm, rf).
Mechanisms of balancing selection
There are two general mechanisms that may account for individual episodes of balancing selection in the two-sex geometric model considered here. Variation may be maintained by heterozygote advantage in both sexes (sm2 < sm1 > 0 and sf2 < sf1 > 0) or by sexually antagonistic (SA) selection, which in its broadest sense occurs when male and female fitnesses are maximized by a different genotype at a locus. Sexual antagonism models can be further divided into two subforms: (1) “directional” SA selection, where the fitness of males and females is maximized by different homozygous genotypes at the locus, leading to positive selection in one sex and purifying selection in the other (sm2 < sm1 < 0 < sf1 < sf2 or sf2 < sf1 < 0 < sm1 < sm2), and (2) “mixed” SA selection, where there is positive or purifying selection in one sex and heterozygote advantage in the other (sm2 < sm1 > 0 or sf2 < sf1 > 0).
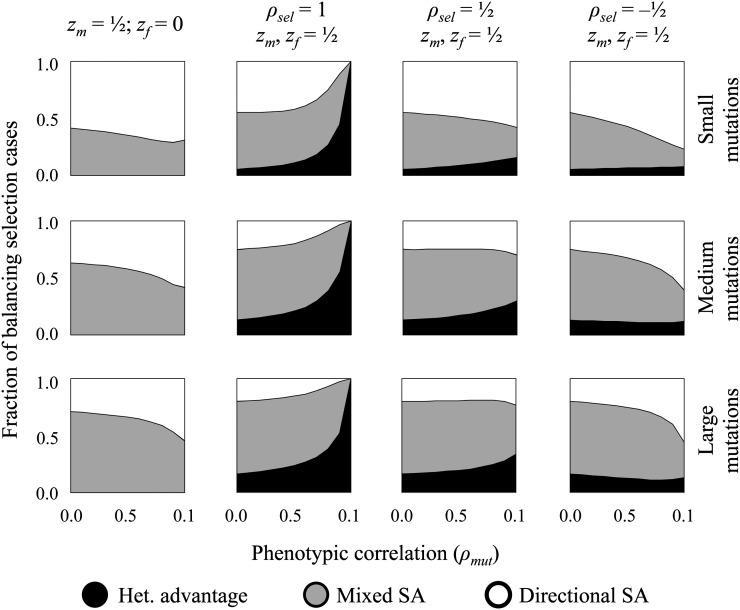
To estimate the relative frequency of each balancing selection mechanism, we simulated random mutations in Fisher’s geometric model and classified the proportion of balancing selection cases that were attributable to heterozygote advantage, directional SA selection, or mixed SA selection. We first considered a scenario where females were perfectly adapted and males were displaced from their fitness optimum (zf = 0 and zm > 0; equivalent results apply when zf > 0 and zm = 0). Under this strongly asymmetric scenario, all mutations are deleterious to females and heterozygote advantage in both sexes is not possible. Directional SA selection becomes the dominant mechanism of balancing selection when scaled mutation sizes are small or mutations have strongly correlated phenotypic effects between the sexes (e.g., Figure 4, top left). As phenotypic correlations decrease and the size of mutations increases, the mixed SA selection scenario predominates (Figure 4; Supporting Information, Figure S1, Figure S2, and Figure S3).
Figure 4.
Balancing selection mechanisms and their relative probabilities. SA selection mechanisms (open and shaded areas) account for the vast majority of balancing selection cases. Heterozygote advantage becomes the dominant form of balancing selection in the restrictive case where directional selection and mutant phenotypic effects are strongly concordant between the sexes (e.g., zm → zf and ρsel, ρmut → 1). For each parameter set (zm, zf, ρsel, ρmut), 500,000 balanced polymorphisms were randomly generated using the simulation approach described in the text. These results were used to calculate the proportion of balancing selection arising from heterozygote advantage, mixed SA selection, and directional SA selection. For each parameter combination, mutation magnitudes were generated using a bivariate exponential distribution (gamma with shape parameter k = 1), with equal marginal distributions and correlation of corr(rm, rf) = ρmut. Small mutations use E[r] = 0.05, medium mutations use E[r] = 0.2, and large mutations use E[r] = 0.4 {these absolute mutation sizes correspond to male-specific scaled sizes of E[xm] = E(r)(n0.5)/(2zm) = 0.25, E[xm] = 1, and E[xm] = 2, respectively}. Representative results are shown for n = 25 and ωm = ωf = 1/2, with additional results presented in Figure S1, Figure S2, and Figure S3).
We also considered scenarios where both sexes were under similarly strong directional selection (βf = βm). In this case, heterozygote advantage remains rare unless the orientation of directional selection and the phenotypic effects of mutations are both strongly correlated between the sexes (e.g., ρsel, ρmut → 1; Figure 4, Figure S1, Figure S2, and Figure S3). Under the remaining conditions, sexual antagonism is the predominant driver of balancing selection, with the mixed SA selection model representing the most common mechanism across most of the parameter space. Directional SA selection dominates when males and females are selected in different directions, mutational effects are strongly correlated between the sexes, and the scaled mutation size is small (Figure 4, Figure S1, Figure S2, and Figure S3).
The efficacy of balancing selection
The above results (e.g., Equation 7) describe the probability of balancing selection in Fisher’s geometric model, but they do not account for evolutionary dynamics under balancing selection, in which genetic drift is expected to play an important role (Robertson 1962; Ewens and Thomson 1970; Nei and Roychoudhury 1973). To identify criteria for effective balancing selection—the degree to which allele frequencies in a finite population converge to the deterministic equilibrium, —we analyzed the stationary distribution for random mutations within the FGM framework. For a diploid locus in a Wright–Fisher population of effective size Ne, the stationary distribution of a B2 allele is proportional to
| (12) |
(Ewens 2004; Rice 2004; Charlesworth and Charlesworth 2010), where q is the population frequency of B2, and ∆q is the expected frequency change of B2 over a single generation. Assuming selection coefficients are small (|sf1|, |sf2|, |sm1|, |sf2| << 1), the expected frequency change is
| (13) |
where represents the polymorphic equilibrium under balancing selection (see Appendix E).
The relative importance of selection vs. drift depends on the density of the stationary distribution near the equilibrium, relative to the density near q = 0 and q = 1, with effectively strong selection leading to a high relative density near (Ewens 2004, pp. 26 and 27). The ratio of the density near the equilibrium, relative to the density near the boundaries, can be described by the ratio
| (14) |
(Appendix F), where α = [2(sm1 + sf1) – (sf2 + sm2)] represents the net strength of selection at the locus, and 2ε is an arbitrarily small allele frequency interval. Equation 14 summarizes how the efficacy of balancing selection increases with the net strength of selection (α), as equilibrium values become increasingly intermediate ( → 1/2) and with increasing Ne. While there is no specific cutoff to delineate weak from strong balancing selection, values of R > 10 [assuming a sufficiently intermediate equilibrium, i.e., (1 – ) > 0.001] are generally sufficient for maintaining polymorphism, and we use this cutoff as a guideline for interpreting subsequent results. [Note that this guideline shares properties with other diffusion-based metrics, such as mean heterozygosity under balancing selection and allele fixation rates or times at polymorphic loci (cf. Robertson 1962; Ewens and Thomson 1970; Nei and Roychoudhury 1973; Hedrick 1974; Ewens 2004; Connallon and Clark 2012; Mullon et al. 2012).]
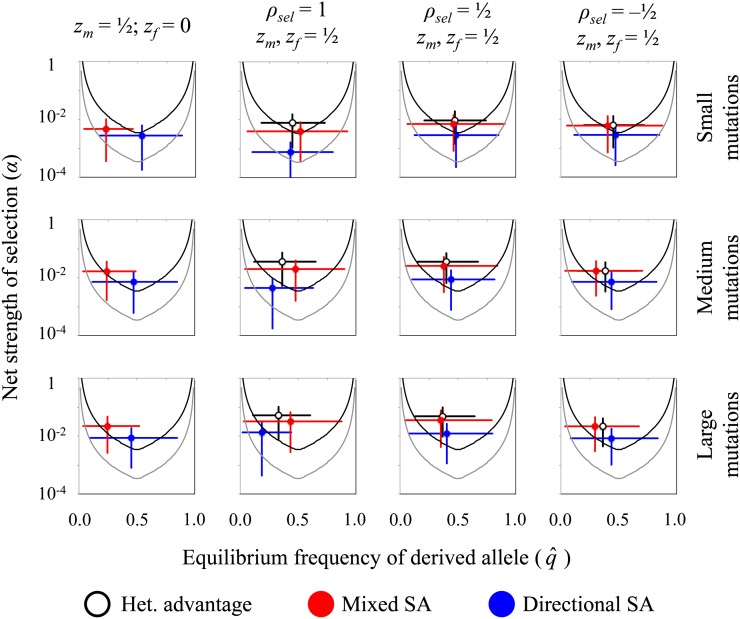
To evaluate opportunities for effective balancing selection, and to contrast the efficacy under distinct mechanisms of sex-specific selection, we simulated random mutations and characterized the distribution of and α for mutations meeting balancing selection criteria (Equation 4). We again considered cases of symmetric vs. asymmetric directional selection (βf/βm = 0 and βf/βm = 1) across a broad range of mutation sizes, selection and mutation correlations between the sexes (ρsel, ρmut), and dominance (h). Specific values of and α vary widely among mutations subject to balancing selection, with sexually antagonistic mutations consistently more susceptible to drift than mutations under heterozygote advantage (Figure 5 and Figure S4). Mutations subject to directional SA selection (positive selection in one sex, in purifying selection in the other) are associated with substantially lower values of net selection (α), on average, and are the most likely to evolve primarily by drift (Figure 5 and Figure S4; note that the α-values are plotted on a logarithmic scale in Figure 5, Figure S4, Figure S5, Figure S6, Figure S7, and Figure S8). Mutations causing heterozygote advantage in one sex (mixed SA selection) or both sexes (heterozygote advantage) have similar equilibria and net selection strengths, although mixed SA mutations are somewhat more susceptible to drift. Overall, these qualitative differences among balancing selection mechanisms are robust to variability in male and female fitness landscapes (zm, zf, ρsel), the strength of genetic correlations between the sexes (ρmut), dimensionality (n), and dominance (h) (see Figure S4, Figure S5, Figure S6, Figure S7, and Figure S8).
Figure 5.
The efficacy of selection differs between mechanisms of balancing selection. Results show the distributions (means and 75% confidence intervals) of α and for mutations under each of the three forms of balancing selection. The areas above the solid curves represent the parameter space for effectively strong selection, based on Equation 14 with criteria R > 10 (see the text surrounding Equation 14). Gray curves (lower) represent the parameter space for a population with effective size Ne = 100,000; black curves (upper) are for populations with Ne = 10,000. For each balancing selection mechanism and for each parameter set (zm, zf, ρsel), 10,000 balanced polymorphisms were randomly generated using the simulation approach described in the main text. These results were used to calculate means and confidence intervals. For each parameter combination, mutation magnitudes were generated using a bivariate exponential distribution (gamma with shape parameter k = 1), with equal marginal distributions and correlation of ρmut = corr(rm, rf). Three different mean mutation sizes were used: small mutations use E[r] = 0.05, medium mutations use E[r] = 0.2, and large mutations use E[r] = 0.4 {these absolute mutation sizes correspond to male-specific scaled sizes of E[xm] = E(r)(n0.5)/(2zm) = 0.25, E[xm] = 1, and E[xm] = 2, respectively}. Representative results are shown for ρmut = 0.9, n = 25, and ωm = ωf = 1/2, with additional results presented in Figure S4, Figure S5, Figure S6, Figure S7, and Figure S8.
Discussion
Our analysis yields four theoretical insights. First, there are ample opportunities for balancing selection in dioecious populations. Criteria for balancing selection at a locus are easily met for mutations of intermediate size (on the relevant scale of Fisher’s geometric model: xA, for a dioecious population), with a maximum probability typically on the order of 25% (conditional on dominance; Figure 2). For all but small-effect mutations, balancing selection is more common than positive selection and may therefore be an important feature of adaptive evolutionary change, as also recently demonstrated for the monoecious case by Sellis et al. (2011). Second, mean mutation size, E(xA), is elevated by divergent directional selection on male and female phenotypes and by sexually dimorphic phenotypic effects of random mutations (sex-by-genotype effects). Consequently, the relative probability of balancing vs. positive selection should increase in species with separate sexes. Third, under most of the parameter space of sex-specific selection and sex-by-genotype effects, balancing selection primarily involves sexually antagonistic alleles. Finally, in a finite population, the effective strength of sexually antagonistic balancing selection is expected to generally be weaker than that of heterozygote advantage, particularly under directional SA selection (the mixed SA selection model shows a smaller departure from the het. advantage model). Therefore, SA alleles under balancing selection may often exhibit population frequencies that are far from equilibrium (see Livingstone 1992; Connallon and Clark 2013b), evolutionary dynamic properties that mimic those of neutral or weakly deleterious alleles (Connallon and Clark 2012), and relatively weak molecular population genetic signatures of partial selective sweeps (Connallon and Clark 2013b; see Sellis et al. 2011 for an additional discussion of balancing selection signals in FGM).
Fitness trade-offs and opportunities for balancing selection
Several studies have reported sexually antagonistic genetic variation for fitness, or fitness components, in animal and plant populations (reviewed in Bonduriansky and Chenoweth 2009; Kirkpatrick 2009; van Doorn 2009; Pennell and Morrow 2013), yet the genetic basis of this quantitative genetic variation is not well known. Much of this phenotypic variation could be attributable to many loci that segregate for low-frequency sexually antagonistic alleles that are maintained at a balance between recurrent mutation, net purifying selection, and genetic drift. Alternatively, the variation could be primarily attributable to a small set of loci that segregate for intermediate-frequency sexually antagonistic alleles under balancing selection.
Population genetics models are often used to evaluate the plausibility of sexual antagonism as a common mechanism for balancing selection (Kidwell et al. 1977; Pamilo 1979; Curtsinger 1980; Prout 2000; Patten and Haig 2009; Unckless and Herren 2009; Fry 2010; Arnqvist 2011; Jordan and Charlesworth 2012; Patten et al. 2013), and most of this theory has focused on “SA directional” selection models (but see Kidwell et al. 1977; Gavrilets and Rice 2006; Mokkonen et al. 2011), where only a small fraction of the conceivable parameter space for sex-specific fitness and dominance will generate balancing selection at a diploid locus (Prout 2000; Patten and Haig 2009). A broader class of fitness trade-off scenarios for balancing selection—e.g., between environments that vary over time or space (Levene 1953; Haldane and Jayakar 1963) and antagonistic pleiotropy between traits (Rose 1982; Curtsinger et al. 1994)—suffers from similarly severe parameter restrictions (Prout 2000). On the other hand, fitness landscape topography biases the range of fitness effect values that beneficial alleles are likely to take (Orr 2005a,b, 2010; Manna et al. 2011; Sellis et al. 2011), with mutations that are subject to fitness trade-offs (including sexually antagonistic alleles) preferentially occupying portions of the conceivable parameter space that are the most amenable to balancing selection (Gillespie 1978; Fry 2010).
Our analysis of the dioecious Fisher’s geometric model, coupled with the recent monoecious analysis by Sellis et al. (2011), suggests that balancing selection may arise naturally by way of the structure of multidimensional fitness landscapes. Because high-dimensional systems like FGM ensure that improvements along some trait axes are coupled with maladaptation along others (Orr 1998), adaptive mutations and alleles subject to balancing selection are nearly always subject to fitness trade-offs. Sex differences in selection (ρsel < 1) and sex-by-genotype effects on trait variation (ρmut < 1) increase the effective dimensionality of Fisher’s geometric model and thereby expand opportunities for fitness trade-offs between individuals of a population. In models that ignore the effects of sex, balancing selection by heterozygote advantage requires that the population be relatively close to a local optimum in the fitness landscape (Sellis et al. 2011). Such restrictions are alleviated in the two-sex FGM, whose net effect is to elevate the standardized mutation size and consequently the importance of balancing relative to positive selection during the process of adaptation.
Whereas fitness trade-offs are expected to manifest across the full parameter space for balancing selection in Fisher’s geometric model, the nature of the underlying trade-offs varies as a function of specific properties of mutation and the phenotypic orientation of sex-specific selection. The classical heterozygote balance model (Fisher 1922) dominates when mutant phenotypic effects are strongly correlated between the sexes and directional selection is similar in strength and orientation between males and females (i.e., ρmut, ρsel >> 0). Sexual antagonism, in the absence of heterozygote advantage in either sex (SA directional), dominates as patterns of directional selection diverge between the sexes (ρsel decreases), but mutational effects remain tightly coupled (ρmut >> 0). A mixed scenario of sexual antagonism—with heterozygote advantage in one sex and positive/purifying selection in the other (SA mixed)—represents the primary mechanism for maintaining balanced polymorphisms when mutant phenotypic effects are poorly correlated between the sexes.
We can tentatively gauge opportunities for each mode of balancing selection by considering relevant empirical estimates of selection and mutational effects on phenotypes expressed by both sexes. Current estimates of sex-specific selection reveal widespread asymmetries in the strength or direction of selection on individual phenotypic traits (Cox and Calsbeek 2009; Stulp et al. 2012), and recent estimates of multivariate selection from humans and two insect species provide concrete examples of sex differential selection within multivariate trait space [ρsel ≈ –0.61 for three traits of the Indian meal moth (Lewis et al. 2011), ρsel ≈ –0.73 for seven traits in Drosophila serrata (Gosden et al. 2012), and ρsel ≈ 0.22 for seven traits in a human population (Stearns et al. 2012)]. Although this set of studies is small, selection patterns are consistently divergent between the sexes, which implies that mutations with sexually antagonistic fitness effects should be common (Connallon and Clark 2014) and that sexual antagonism should play an important role in generating balancing selection (i.e., SA directional or SA mixed mechanisms, as shown here). Estimates of quantitative genetic variability among male and female traits mostly show strong and positive phenotypic correlations between the sexes [e.g., detectable, but modest gene-by-sex effects (Poissant et al. 2010; Houle and Fierst 2012; Griffin et al. 2013)], which tentatively suggests an important potential role for SA directional balancing selection. There is, however, a great need for studies that measure the joint effects of random mutations on male and female traits (Houle and Fierst 2012). These should prove valuable for further evaluating predictions of balancing selection that emerge from Fisher’s geometric model, as well as the severity of genetic constraints to sex-specific adaptation and the evolution of sexual dimorphism.
Conclusions
Balancing selection may be common within diploids (Sellis et al. 2011) and typically involves alleles with sexually antagonistic fitness effects. The genetic architecture of sex-specific fitness is likely to be complex, with processes of recurrent mutation, balancing selection, and genetic drift each potentially contributing to the maintenance of fitness variation (Bonduriansky and Chenoweth 2009; Connallon and Clark 2012). Our results show that the parameter criteria for sexually antagonistic balancing selection are not difficult to meet, and consequently intermediate-frequency sexually antagonistic alleles may contribute substantially to the high levels of quantitative genetic variance that are commonly observed in life-history traits and fitness components (Houle 1992; Charlesworth and Charlesworth 1999; Charlesworth and Hughes 1999). However, the common assumption that balanced polymorphisms are maintained at or near equilibrium may be unrealistic, particularly with regard to sexually antagonistic alleles, which are particularly susceptible to genetic drift (see Hesketh et al. 2013).
Finally, both theory and data suggest that the severity of sexual antagonism should generally increase during the course of adaptation, with sexually antagonistic mutations being rare in poorly adapted populations and common in well-adapted ones (e.g., Long et al. 2012; Berger et al. 2014; Connallon and Clark 2014), but this is not always so (see Delcourt et al. 2009; Punzalan et al. 2014). The relative probability of balancing vs. positive selection similarly increases during the course of adaptation in Fisher’s geometric model [e.g., because the average mutation size increases as the distance to the optimum shrinks (Sellis et al. 2011); see above]. Taken together, we expect that both sexual antagonism and balancing selection will be most rare following an abrupt change in the environment, in which both sexes are displaced from their fitness optima. As the population adapts, sexual antagonism and balancing selection should become increasingly important in shaping patterns of population genetic variation, leading to a reconfiguration of the sex-specific distribution of fitness effects among random mutations (Connallon and Clark 2014) and of the genetic basis of phenotypic (including fitness) variance between poorly and well-adapted populations.
Supplementary Material
Acknowledgments
We thank two anonymous reviewers for valuable comments and suggestions on an earlier version of the manuscript and members of the Clark laboratory for discussion. This work was funded by National Institutes of Health grant GM64590 (to A. G. Clark and A. B. Carvalho).
Appendix A: Movements in Phenotypic Space and Selection Coefficients
Let Aj = {x1j, x2j, … , xnj} represent the phenotypic value expressed by wild-type individuals of the jth sex (j = {f, m}); Oj = {O1j, O2j, … , Onj} is the location of its optimum, and Oj′ = Oj – Aj = {o1j, o2j, … , onj} is a rescaled optimum that becomes useful elow. Consider a mutation with a homozygous magnitude of rm and rf and heterozygous magnitude of rmh and rfh. The phenotypic value expressed by a heterozygote will be Aj + hyj, and a homozygote will be Aj + yj, where yj = {y1j, y2j, … , ynj} is a mutation vector that describes the set of phenotypic changes in each of the n traits (this is described in the Simulations section of the main text). The Euclidean distance between a wild-type individual of sex j and the respective phenotypic optimum is
The distance between a heterozygote and its optimum is
The exact result in Equation 2 is obtained by substituting yim = rmmi/M, where M2 = ∑mi2, and yif = rffi/F, where F2 = ∑fi2 (as described in the Simulations section). With sufficiently high dimensionality (n > 10 works well) mi/M and fi/F converge to mi/√(n) and fi/√(n) (Connallon and Clark 2014), which accounts for the final approximations in Equation 2. The same approach can be taken to obtain values of zj(hom).
For a random mutation, the fitness of the three genotypes will be wj(wild type) = exp[–ωjzj2], wj(het) = exp[–ωjzj(het)2], and wj(hom) = exp[–ωjzj(hom)2]. Selection coefficients are scaled relative to the wild-type fitness. The heterozygote and homozygote selection coefficients for a random mutation are (respectively) sj1 = wj(het)/wj(wild type) – 1 = exp[ωj(zj2 – zj(het)2)] – 1, and sj2 = wj(hom)/wj(wild type) – 1 = exp[ωj(zj2 – zj(hom)2)] – 1.
Appendix B: Modes of Selection and Their Probabilities
Define t = ln(1 + s) as the natural logarithm of fitness for a genotype with relative fitness 1 + s. Under weak selection (s << 1), t is well approximated as t = ln(1 + s) ∼ s. The criterion for balancing selection at a diploid locus can be approximated as
| (B1) |
where tmhet = ln(1 + sm1), tfhet = ln(1 + sf1), tmhom = ln(1 + sm2), and tfhom = ln(1 + sf2). The criterion for invasion of a new mutation is modified to
| (B2) |
Let the mutation effect size in heterozygotes be rmh and rfh, where h is the dominance coefficient (assumed to fall in the range 0 < h < 1), and the mutation size in homozygotes be rm and rf. From Equation B1, along with Equation 2 and its surrounding text from the main text, the criteria for balancing selection can be restated as
| (B3) |
Terms mi and fi are standard normal variables with cov(mi, fi) = ρmut. The function in the middle is therefore normally distributed with respective mean of zero and variance of
where ρmf = ρmutcos(θsel). After dividing the terms of the inequality (B3) by √(var), the function in the middle is now distributed as a standard normal [N(0, 1)], and we can now solve for the probability of balancing selection,
where
and
which accounts for Equation 7 in the main text. The same basic approach can be taken to calculate the probability that selection will favor invasion of a rare mutation. In this case, Pr(sm1 + sf1 > 0) = 1 – Φ(b), as in Equation 6.
Appendix C: The Maximum Probability of Balancing Selection
To find the specific value of xA that maximizes the probability of balancing selection, we restate Equation 7 as
which has a first derivative, with respect to xA, of
The maximum probability of balancing selection is found by setting this function to zero and solving for xA, which yields
Appendix D: Mean Standardized Mutation Size
For βm > 0 and βf = 0 (zf = 0), we can approximate E[xA] by performing a second-order Taylor series expansion in the vicinity of rm, rf = E(r) and take the expectation, which leads to
Equation 10 in the main text can be found by substituting βm = 2ωmzm, E(r) = kθ, var(r) = kθ2, and corr(rm, rf) = cov(rm, rf)/var(r) and rearranging. Using the same approach for β = βm = βf, we get
Substituting E(r) = kθ, var(r) = kθ2, and corr(rm, rf) = cov(rm, rf)/var(r) and simplifying yields Equation 11 in the main text.
Appendix E: Allele Frequency Dynamics and Equilibrium of B2 Under Balancing Selection
Among zygotes, let qm be the frequency of B2 transmitted from males and qf be the frequency of B2 transmitted from females. The genotypic frequencies among zygotes are qmqf = [B2B2]; qm(1 – qf) + qf(1 – qm) = [B1B2]; (1 – qm)(1 – qf) = [B1B1]. Let q = (qm + qf)/2, and 2ε = qm – qf. Assuming weak selection (Nagylaki 1979; Charlesworth and Charlesworth 2010), we approximate the allele frequency dynamics to first order in ε. Following selection, the expected allele frequency change in males and females will be
The net change after a generation is (Δqm + Δqf)/2,
where is the balanced polymorphic equilibrium when it exists.
Appendix F: Criteria for Effective Balancing Selection
Following Ewens (2004, pp. 26 and 27), we wish to know the relative proportion of the density, f(q), that is found near the deterministic equilibrium value (), compared to the allele frequency boundaries q = 0 and q = 1. Using a second-order Taylor series expansion, we can approximate the area under the stationary distribution near arbitrary frequency point a:
By substituting a = 0.001, a = 0.999, and a = (as appropriate), we obtain Equation 14 of the main text.
Footnotes
Communicating editor: W. Stephan
Literature Cited
- Albert A. Y. K., Otto S. P., 2005. Sexual selection can resolve sex-linked sexual antagonism. Science 310: 119–121. [DOI] [PubMed] [Google Scholar]
- Arnqvist G., 2011. Assortative mating by fitness and sexually antagonistic genetic variation. Evolution 65: 2111–2116. [DOI] [PubMed] [Google Scholar]
- Berger D., Grieshop K., Lind M. I., Goenaga J., Maklakov A. A., et al. , 2014. Intralocus sexual conflict and environmental stress. Evolution DOI: 10.1111/evo.12439. [DOI] [PubMed] [Google Scholar]
- Blackburn G. S., Albert A. Y. K., Otto S. P., 2010. The evolution of sex ratio adjustment in the presence of sexually antagonistic selection. Am. Nat. 176: 264–275. [DOI] [PubMed] [Google Scholar]
- Bonduriansky R., Chenoweth S. F., 2009. Intralocus sexual conflict. Trends Ecol. Evol. 24: 280–288. [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Charlesworth D., 1999. The genetic basis of inbreeding depression. Genet. Res. 74: 329–340. [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Charlesworth D., 2010. Elements of Evolutionary Genetics. Roberts & Co., Greenwood Village, Colorado. [Google Scholar]
- Charlesworth B., Hughes K. A., 1999. The quantitative genetics of life history traits, pp. 369–392 in Evolutionary Genetics From Molecules to Morphology, edited by Singh R. S., Krimbas C. B. Cambridge University Press, Cambridge, UK. [Google Scholar]
- Charlesworth B., Jordan C. Y., Charlesworth D., 2014. The evolutionary dynamics of sexually antagonistic mutations in pseudoautosomal regions of sex chromosomes. Evolution 68: 1339–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D., 2006. Balancing selection and its effects on sequences in nearby genome regions. PLoS Genet. 2: e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D., Charlesworth B., 1980. Sex differences in fitness and selection for centric fusions between sex-chromosomes and autosomes. Genet. Res. 35: 205–214. [DOI] [PubMed] [Google Scholar]
- Chippindale A. K., Gibson J. R., Rice W. R., 2001. Negative genetic correlation for adult fitness between sexes reveals ontogenetic conflict in Drosophila. Proc. Natl. Acad. Sci. USA 98: 1671–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T., Clark A. G., 2010. Sex linkage, sex-specific selection, and the role of recombination in the evolution of sexually dimorphic gene expression. Evolution 64: 3417–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T., Clark A. G., 2011. The resolution of sexual antagonism by gene duplication. Genetics 187: 919–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T., Clark A. G., 2012. A general population genetic framework for antagonistic selection that accounts for demography and recurrent mutation. Genetics 190: 1477–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T., Clark A. G., 2013. a Sex-differential selection and the evolution of X inactivation strategies. PLoS Genet. 9: e1003440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T., Clark A. G., 2013b. Antagonistic versus nonantagonistic models of balancing selection: characterizing the relative timescales and hitchhiking effects of partial selective sweeps. Evolution 67: 908–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T., Clark A. G., 2014. The evolutionary inevitability of sexual antagonism. Proc. Biol. Sci. 281: 20132123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T., Cox R. M., Calsbeek R., 2010. Fitness consequences of sex-specific selection. Evolution 64: 1671–1682. [DOI] [PubMed] [Google Scholar]
- Cox R. M., Calsbeek R., 2009. Sexually antagonistic selection, sexual dimorphism, and the resolution of intralocus sexual conflict. Am. Nat. 173: 176–187. [DOI] [PubMed] [Google Scholar]
- Crow, J. F., and M. Kimura, 1970 An Introduction to Population Genetics Theory. Burgess Publishing Co., Minneapolis. [Google Scholar]
- Curtsinger J. W., 1980. On the opportunity for polymorphism with sex-linkage or haplodiploidy. Genetics 96: 995–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtsinger J. W., Service P. M., Prout T., 1994. Antagonistic pleiotropy, reversal of dominance, and genetic polymorphism. Am. Nat. 144: 210–228. [Google Scholar]
- Day T., Bonduriansky R., 2004. Intralocus sexual conflict can drive the evolution of genomic imprinting. Genetics 167: 1537–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcourt M., Blows M. W., Rundle H. D., 2009. Sexually antagonistic genetic variance for fitness in an ancestral and novel environment. Proc. Biol. Sci. 276: 2009–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T., 1955. A review of some fundamental concepts and problems of population genetics. Cold Spring Harb. Symp. Quant. Biol. 20: 1–15. [DOI] [PubMed] [Google Scholar]
- Ellegren H., Parsch J., 2007. The evolution of sex-biased genes and sex-biased gene expression. Nat. Rev. Genet. 8: 689–698. [DOI] [PubMed] [Google Scholar]
- Ewens W. J., 2004. Mathematical Population Genetics. I. Theoretical Introduction, Ed. 2 Springer-Verlag, New York. [Google Scholar]
- Ewens W. J., Thomson G., 1970. Heterozygote selective advantage. Ann. Hum. Genet. 33: 365–376. [Google Scholar]
- Falconer D. S., Mackay T. F. C., 1996. Introduction to Quantitative Genetics, Ed. 4 Longmans Green, Harlow, Essex, UK. [Google Scholar]
- Fisher R. A., 1922. On the dominance ratio. Proc. R. Soc. Edinb. 42: 321–341. [Google Scholar]
- Fisher R. A., 1930. The Genetical Theory of Natural Selection. Clarendon Press, Oxford. [Google Scholar]
- Fry J. D., 2010. The genomic location of sexually antagonistic genetic variation: some cautionary comments. Evolution 64: 1510–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilets S., Rice W. R., 2006. Genetic models of homosexuality: generating testable predictions. Proc. Biol. Sci. 273: 3031–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie J. H., 1978. A general model to account for enzyme variation in natural populations. V. The SAS-CFF model. Theor. Popul. Biol. 14: 1–45. [DOI] [PubMed] [Google Scholar]
- Gillespie, J. H., 2004 Population Genetics: A Concise Guide. Johns Hopkins University Press, Baltimore. [Google Scholar]
- Gosden T. P., Shastri K. L., Innocenti P., Chenoweth S. F., 2012. The B-matrix harbours significant and sex-specific constraints on the evolution of multicharacter sexual dimorphism. Evolution 66: 2106–2116. [DOI] [PubMed] [Google Scholar]
- Griffin R. M., Dean R., Grace J. L., Ryden P., Friberg U., 2013. The shared genome is a pervasive constraint on the evolution of sex-biased gene expression. Mol. Biol. Evol. 30: 2168–2176. [DOI] [PubMed] [Google Scholar]
- Haldane J. B. S., Jayakar S. D., 1963. Polymorphism due to selection of varying direction. J. Genet. 48: 237–242. [Google Scholar]
- Hedrick P. W., 1974. Genetic variation in a heterogeneous environment. I. Temporal heterogeneity and the absolute dominance model. Genetics 78: 757–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick P. W., 2012. What is the evidence for heterozygote advantage selection? Trends Ecol. Evol. 27: 698–704. [DOI] [PubMed] [Google Scholar]
- Hesketh J., Fowler K., Reuter M., 2013. Genetic drift in antagonistic genes leads to divergence in sex-specific fitness between experimental populations of Drosophila melanogaster. Evolution 67: 1503–1510. [DOI] [PubMed] [Google Scholar]
- Holman L., Kokko H., 2013. The consequences of polyandry for population viability, extinction risk and conservation. Philos. Trans. R. Soc. Lond. B 368: 20120053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houle D., 1992. Comparing evolvability and variability of quantitative traits. Genetics 130: 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houle D., Fierst J., 2012. Properties of spontaneous mutational variance and covariance for wing size and shape in Drosophila melanogaster. Evolution 67: 1116–1130. [DOI] [PubMed] [Google Scholar]
- Johnston S. E., Gratten J., Berenos C., Pilkington J. G., Clutton-Brock T. H., et al. , 2013. Life history trade-offs at a single locus maintain sexually selected genetic variation. Nature 502: 93–95. [DOI] [PubMed] [Google Scholar]
- Jordan C. Y., Charlesworth D., 2012. The potential for sexually antagonistic polymorphism in different genomic regions. Evolution 66: 505–516. [DOI] [PubMed] [Google Scholar]
- Kidwell J. F., Clegg M. T., Stewart F. M., Prout T., 1977. Regions of stable equilibria for models of differential selection in the two sexes. Genetics 85: 171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1983. The Neutral Theory of Molecular Evolution. Cambridge University Press, Cambridge, UK. [Google Scholar]
- Kirkpatrick M., 2009. Patterns of quantitative genetic variation in multiple dimensions. Genetica 136: 271–284. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M., Guerrero R. F., 2014. Signatures of sex-antagonistic selection on recombining sex chromosomes. Genetics 197: 531–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokko H., Brooks R., 2003. Sexy to die for? Sexual selection and the risk of extinction. Ann. Zool. Fenn. 40: 207–219. [Google Scholar]
- Lande R., 1980. Sexual dimorphism, sexual selection, and adaptation in polygenic characters. Evolution 34: 292–305. [DOI] [PubMed] [Google Scholar]
- Leffler E. M., Gao Z., Pfeifer S., Segurel L., Auton A., et al. , 2013. Multiple instances of ancient balancing selection shared between humans and chimpanzees. Science 339: 1578–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levene H., 1953. Genetic equilibrium when more than one ecological niche is available. Am. Nat. 87: 331–333. [Google Scholar]
- Lewis Z., Wedell N., Hunt J., 2011. Evidence for strong intralocus sexual conflict in the Indian meal moth, Plodia interpunctella. Evolution 65: 2085–2097. [DOI] [PubMed] [Google Scholar]
- Lewontin, R. C., 1974 The Genetic Basis of Evolutionary Change. Columbia University Press, New York. [Google Scholar]
- Livingstone F. B., 1992. Polymorphism and differential selection for the sexes. Hum. Biol. 64: 649–657. [PubMed] [Google Scholar]
- Long A. D., Lyman R. F., Morgan A. H., Langley C. H., Mackay T. F. C., 2000. Both naturally occurring insertions of transposable elements and intermediate frequency polymorphisms at the achaete-scute complex are associated with variation in bristle number in Drosophila melanogaster. Genetics 154: 1255–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long T. A. F., Agrawal A. F., Rowe L., 2012. The effect of sexual selection on offspring fitness depends on the nature of genetic variation. Curr. Biol. 22: 204–208. [DOI] [PubMed] [Google Scholar]
- Mank J. E., 2009. Sex chromosomes and the evolution of sexual dimorphism: lessons from the genome. Am. Nat. 173: 141–150. [DOI] [PubMed] [Google Scholar]
- Manna F., Martin G., Lenormand T., 2011. Fitness landscapes: an alternative theory for the dominance of mutation. Genetics 189: 923–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsaglia G., 1972. Choosing a point from the surface of a sphere. Ann. Math. Stat. 43: 645–646. [Google Scholar]
- Martin G., 2014. Fisher’s geometrical model emerges as a property of complex integrated phenotypic networks. Genetics 197: 237–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G., Lenormand T., 2006. A general multivariate extension of Fisher’s geometrical model and the distribution of mutation fitness effects across species. Evolution 6: 893–907. [PubMed] [Google Scholar]
- Michael J. R., Schucany W. R., 2002. The mixture approach for simulating bivariate distributions with specified correlations. Am. Stat. 56: 48–54. [Google Scholar]
- Mokkonen M., Kokko H., Koskela E., Lehtonen J., Mappes T., et al. , 2011. Negative frequency-dependent selection of sexually antagonistic alleles in Myodes glareolus. Science 334: 972–974. [DOI] [PubMed] [Google Scholar]
- Muller M. E., 1959. A note on a method for generating points uniformly on N-dimensional spheres. Comm. Assoc. Comp. Mach. 2: 19–20. [Google Scholar]
- Mullon C., Pomiankowski A., Reuter M., 2012. The effects of selection and genetic drift on the genomic distribution of sexually antagonistic alleles. Evolution 66: 3743–3753. [DOI] [PubMed] [Google Scholar]
- Nagylaki T., 1979. Selection in dioecious populations. Ann. Hum. Genet. 14: 143–150. [DOI] [PubMed] [Google Scholar]
- Nei M., Roychoudhury A. K., 1973. Probability of fixation and mean fixation time of an overdominant mutation. Genetics 74: 371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., 1998. The population genetics of adaptation: the distribution of factors fixed during adaptive evolution. Evolution 52: 935–949. [DOI] [PubMed] [Google Scholar]
- Orr H. A., 2000. Adaptation and the cost of complexity. Evolution 54: 13–20. [DOI] [PubMed] [Google Scholar]
- Orr H. A., 2005a The genetic theory of adaptation: a brief history. Nat. Rev. Genet. 6: 119–127. [DOI] [PubMed] [Google Scholar]
- Orr H. A., 2005b Theories of adaptation: what they do and don’t say. Genetica 123: 3–13. [DOI] [PubMed] [Google Scholar]
- Orr H. A., 2006. The distribution of fitness effects among beneficial mutations in Fisher’s geometric model of adaptation. J. Theor. Biol. 238: 279–285. [DOI] [PubMed] [Google Scholar]
- Orr H. A., 2010. The population genetics of beneficial mutations. Philos. Trans. R. Soc. B 365: 1195–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto, S. P., and T. Day, 2007 A Biologist’s Guide to Mathematical Modeling in Ecology and Evolution. Princeton University Press, Princeton, NJ. [Google Scholar]
- Pamilo P., 1979. Genic variation at sex-linked loci: quantification of regular selection models. Hereditas 91: 129–133. [DOI] [PubMed] [Google Scholar]
- Parsch J., Ellegren H., 2013. The evolutionary causes and consequences of sex-biased gene expression. Nat. Rev. Genet. 14: 83–87. [DOI] [PubMed] [Google Scholar]
- Patten M. M., Haig D., 2009. Maintenance or loss of genetic variation under sexual and parental antagonism at a sex-linked locus. Evolution 63: 2888–2895. [DOI] [PubMed] [Google Scholar]
- Patten M. M., Haig D., Ubeda F., 2010. Fitness variation due to sexual antagonism and linkage disequilibrium. Evolution 64: 3638–3642. [DOI] [PubMed] [Google Scholar]
- Patten M. M., Ubeda F., Haig D., 2013. Sexual and parental antagonism shape genomic architecture. Proc. Biol. Sci. 280: 2013117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell T. M., Morrow E. H., 2013. Two sexes, one genome: the evolutionary dynamics of intralocus sexual conflict. Ecol. Evol. 3: 1819–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poissant J., Wilson A. J., Coltman D. W., 2010. Sex-specific genetic variance and the evolution of sexual dimorphism: a systematic review of cross-sex genetic correlations. Evolution 64: 97–107. [DOI] [PubMed] [Google Scholar]
- Pomiankowski A., Moller A. P., 1995. A resolution of the lek paradox. Proc. Biol. Sci. 260: 21–29. [Google Scholar]
- Prout T., 1968. Sufficient conditions for multiple niche polymorphism. Am. Nat. 102: 493–496. [Google Scholar]
- Prout T., 2000. How well does opposing selection maintain variation? pp. 157–181 in Evolution Genetics: from Molecules to Morphology, edited by Singh R. S., Krimbas C. B. Cambridge University Press, Cambridge, UK. [Google Scholar]
- Punzalan D., Delcourt M., Rundle H. D., 2014. Comparing the intersex genetic correlation for fitness across novel environments in the fruit fly, Drosophila serrata. Heredity 112: 143–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, S. H., 2004 Evolutionary Theory: Mathematical and Conceptual Foundations. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Rice W. R., 1984. Sex chromosomes and the evolution of sexual dimorphism. Evolution 38: 735–742. [DOI] [PubMed] [Google Scholar]
- Rice W. R., 1992. Sexually antagonistic genes: experimental evidence. Science 256: 1436–1439. [DOI] [PubMed] [Google Scholar]
- Robertson A., 1962. Selection for heterozygotes in small populations. Genetics 47: 1291–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers J. L., Nicewander W. A., 1988. Thirteen ways to look at the correlation coefficient. Am. Stat. 42: 59–66. [Google Scholar]
- Rose M. R., 1982. Antagonistic pleiotropy, dominance, and genetic variation. Heredity 48: 63–78. [Google Scholar]
- Roze D., Otto S. P., 2012. Differential selection between the sexes and selection for sex. Evolution 66: 558–574. [DOI] [PubMed] [Google Scholar]
- Seger J., Trivers R., 1986. Asymmetry in the evolution of female mating preferences. Nature 319: 771–773. [Google Scholar]
- Sellis D., Callahan B. J., Petrov D. A., Messer P. W., 2011. Heterozygote advantage as a natural consequence of adaptation in diploids. Proc. Natl. Acad. Sci. USA 108: 20666–20671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns S. C., Govindaraju D. R., Ewbank D., Byars S. G., 2012. Constraints on the coevolution of contemporary human males and females. Proc. Biol. Sci. 279: 4836–4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stulp G., Kuijper B., Buunk A. P., Pollet T. V., Verhulst S., 2012. Intralocus sexual conflict over human height. Biol. Lett. 23: 976–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M., Barton N. H., 2004. Polygenic variation maintained by balancing selection: pleiotropy, sex-dependent allelic effects and G×E interactions. Genetics 166: 1053–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unckless R. L., Herren J. K., 2009. Population genetics of sexually antagonistic mitochondrial mutants under inbreeding. J. Theor. Biol. 260: 132–136. [DOI] [PubMed] [Google Scholar]
- van Doorn G. S., 2009. Intralocus sexual conflict. Ann. N. Y. Acad. Sci. 1168: 52–71. [DOI] [PubMed] [Google Scholar]
- van Doorn G. S., Kirkpatrick M., 2007. Turnover of sex chromosomes induced by sexual conflict. Nature 449: 909–912. [DOI] [PubMed] [Google Scholar]
- van Doorn G. S., Kirkpatrick M., 2010. Transitions between male and female heterogamety caused by sex-antagonistic selection. Genetics 186: 629–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman D., Welch J. J., 2005. Fisher’s microscope and Haldane’s ellipse. Am. Nat. 166: 447–457. [DOI] [PubMed] [Google Scholar]
- Whitlock M. C., Agrawal A. F., 2009. Purging the genome with sexual selection: reducing mutation load through selection on males. Evolution 63: 569–582. [DOI] [PubMed] [Google Scholar]
- Wright A. E., Mank J. E., 2013. The scope and strength of sex-specific selection in genome evolution. J. Evol. Biol. 26: 1841–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S., 1969 Evolution and the Genetics of Populations, Vol. 2. University of Chicago Press, Chicago. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.