Abstract
The forkhead box transcription factor FOXM1 is an essential effector of G2/M-phase transition, mitosis and the DNA damage response. As such, it is frequently deregulated during tumorigenesis. Here we report that FOXM1 is dynamically modified by SUMO1 but not by SUMO2/3 at multiple sites. We show that FOXM1 SUMOylation is enhanced in MCF-7 breast cancer cells in response to treatment with epirubicin and mitotic inhibitors. Mutation of five consensus conjugation motifs yielded a SUMOylation-deficient mutant FOXM1. Conversely, fusion of the E2 ligase Ubc9 to FOXM1 generated an auto-SUMOylating mutant (FOXM1-Ubc9). Analysis of wild-type FOXM1 and mutants revealed that SUMOylation inhibits FOXM1 activity, promotes translocation to the cytoplasm and enhances APC/Cdh1-mediated ubiquitination and degradation. Further, expression of the SUMOylation-deficient mutant enhanced cell proliferation compared with wild-type FOXM1, whereas the FOXM1-Ubc9 fusion protein resulted in persistent cyclin B1 expression and slowed the time from mitotic entry to exit. In summary, our findings suggest that SUMOylation attenuates FOXM1 activity and causes mitotic delay in cytotoxic drug response.
Keywords: FOXM1, SUMO, chemotherapy, drug resistance, breast cancer, cell cycle
Introduction
Anthracyclines and taxanes are some of the most effective and commonly used classes of anticancer drugs. However, resistance to these agents is also common and significantly restricts their therapeutic efficacy. The forkhead transcription factor FOXM1 has a crucial role in mediating the DNA damage response and resistance to genotoxic agents, including epirubicin, through transcriptional control of a family of DNA double-strand break-sensing and homologous recombination repair genes, including CHEK1 (CHK1), EXO1, RAD51 and BRIP1.1, 2, 3 Similarly, FOXM1 has also been implicated in mediating the actions of taxanes, but the underlying mechanisms are less clearly defined.4 Nevertheless, the efficacy of these genotoxic and cytotoxic agents depends on their ability to modulate the DNA damage response and/or the G2/M cell cycle checkpoint.5, 6
Transition through the cell cycle is tightly co-ordinated at multiple levels, including at gene expression and in response to post-translational modifications of key regulators. Deregulation of the cell cycle underpins multiple diseases, including cancer, where the loss of checkpoint control can lead to uncontrolled proliferation, resistance to apoptosis and genomic instability. In Saccharomyces cerevisiae and mammalian cells, coordination of the G2/M checkpoint is mediated through the forkhead transcription family members Fkh2p7 and FOXM1,8 respectively. Loss of FOXM1 is associated with defects in chromosome segregation and cytokinesis,8 and is homozygous lethal in mouse models.9 We have shown previously that genotoxic agents such as epirubicin induce FOXM1 transcription.10 FOXM1 is further regulated at the post-translational level in response to epirubicin,10, 11 although this level of regulation remains poorly understood.
SUMOylation is a post-translational modification critical for DNA damage response,12, 13 mitosis and cell cycle progression.14 It is also required for the regulation of mitotic spindle asymmetry in S. cerevisiae.15 SUMOylation has an established role in the mitosis of mammalian cells, in part by regulating the association of CENP-E to the kinetochores16 and by mobilising topoisomerase II from the mitotic chromatin.17 Deconjugation of SUMO is also essential for cell cycle progression and mitosis in both yeast and mammals, and SUMO-specific proteases (SENPs) display cell cycle-phase-specific expression patterns.18, 19
On the basis of these observations, we speculated that SUMOylation could have a key role in regulating FOXM1 activity in the response to anticancer drugs. We show that treatment of MCF-7 breast cancer cells with epirubicin, paclitaxel or spindle poison nocodazole enhances SUMOylation of FOXM1. We further report that this dynamic modification governs the transcriptional activity, cellular localization and turnover of FOXM1.
Results
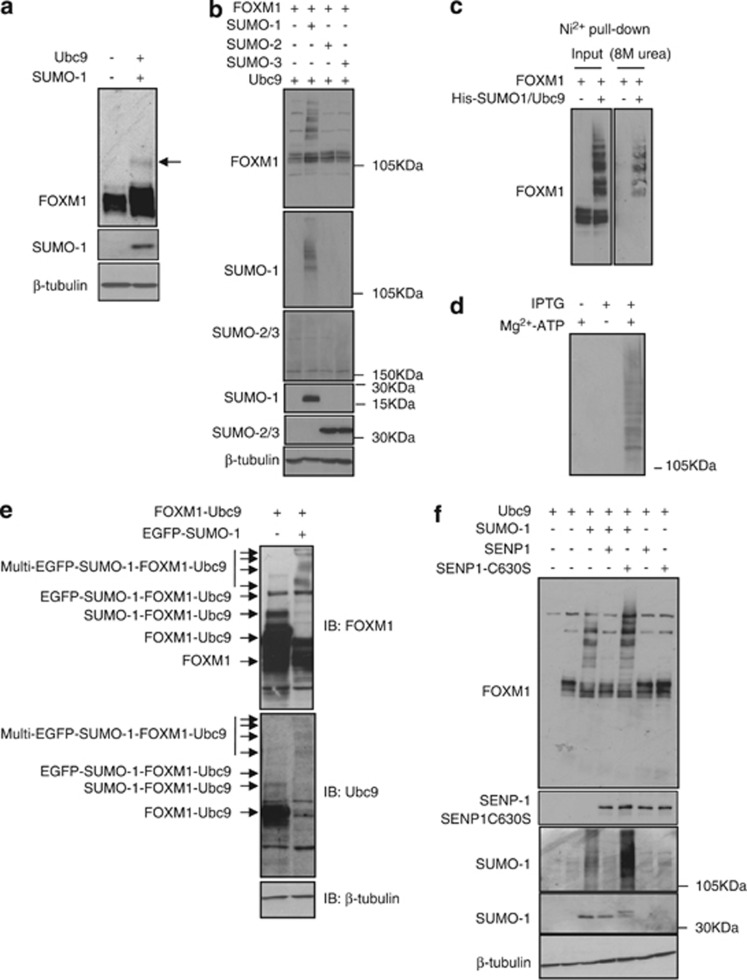
FOXM1 can be modified by SUMO1
SUMOylation has a critical role in the DNA damage response and G2/M cell cycle checkpoint. We postulated that FOXM1 is modified by SUMO in response to cytotoxic drug treatment. To test this idea, we first investigated whether FOXM1 is subjected to SUMOylated in vivo and in vitro. Expression of the SUMO E2 ligase Ubc9 in MCF-7 breast carcinoma cells resulted in the appearance of higher molecular weight FOXM1 species on western blot analysis, suggestive of SUMO conjugation (Figure 1a). In addition, coexpression of Ubc9 and SUMO1, but not of SUMO2 or -3, resulted in the detection of multiple SUMOylated FOXM1 species, indicating that free SUMO1 is a limiting factor in the level of modification of this transcription factor (Figure 1b). However, this result does not preclude the involvement of SUMO2 or -3 in FOXM1 SUMOylated. To confirm covalent linkage of FOXM1 to SUMO1, we coexpressed FOXM1 in the presence or absence of a His-tagged SUMO1, and purified the His-tagged proteins under denaturing conditions (Figure 1c). Although SUMO1 does not commonly form poly-SUMO chains, multiple SUMO-conjugated forms of FOXM1 were detected, suggesting the presence of several acceptor sites in FOXM1 (Figure 1c). An in vitro SUMOylation assay confirmed multi-SUMOylation of recombinant isopropyl β-D-thiogalactopyranoside (IPTG)-inducible FOXM1 in an ATP-dependent manner (Figure 1d). To increase specifically the proportion of modified FOXM1, we made use of the Ubc9 fusion-directed SUMOylation system.20 To this end, we fused FOXM1 to mouse Ubc9 via a five amino-acid linker sequence. This FOXM1-Ubc9 fusion protein was subjected to auto-SUMOylation, which was enhanced upon co-transfection of enhanced green fluorescent protein (eGFP)-tagged SUMO1 (Figure 1e). SUMOylation of FOXM1 was reversed upon coexpression of the SUMO deconjugase SENP-1 (Figure 1f). Moreover, coexpression of a dominant-negative SENP-1(C630S) mutant significantly enhanced FOXM1 SUMOylation, suggesting that modified FOXM1 is a substrate for endogenous SENP-1 (Figure 1f). These data support the conclusion that FOXM1 is dynamically modified by SUMO1 in vivo.
Figure 1.
FOXM1 is SUMOylated by SUMO1. (a) Protein lysates were prepared from MCF-7 cells with or without transfection of the SUMO E2-ligase Ubc-9. FOXM1 was detected by western blot analysis and a higher molecular weight form was observed consistent with SUMOylation of FOXM1. (b) MCF-7 cells were transfected with FOXM1, Ubc9 or SUMO1, -2 or -3, and FOXM1 was detected by western blot analysis. Multiple higher molecular weight forms of FOXM1 were observed, consistent with poly-SUMOylation. (c) MCF-7 cells were co-transfected with His-SUMO1, FOXM1 and Ubc9. SUMOylated proteins were purified using Ni2+-column affinity pulldown under denaturing conditions (8 M urea). Input and His-tagged proteins were probed for FOXM1, and poly-SUMOylated FOXM1 was detectable demonstrating the covalent linkage of His-SUMO1 to FOXM1. (d) IPTG-inducible recombinant His-tagged FOXM1 was expressed in E. coli and isolated using Ni2+-column affinity pulldown. Cell lysates from induced and non-induced E. coli were incubated with Ubc9 and SUMO1 in the presence and absence of Mg2+-ATP and reaction buffer, and resolved using SDS-PAGE. SUMO1 was detected by western blot analysis. (e) MCF-7 cells were transfected with constructs encoding FOXM1 fused to mouse Ubc9 (FOXM1-Ubc9) with or without eGFP-tagged SUMO1. The higher molecular weight band detected by FOXM1 and Ubc9 antibodies, which is consistent with the auto-SUMOylation of FOXM1-Ubc9, is indicated. SUMOylation was enhanced and increased in molecular weight by co-transfection of eGFP-SUMO1 (indicated by the multiple arrows). (f) MCF-7 cells were co-transfected with FOXM1, SUMO1, Ubc9, SENP-1 and the dominant-negative SENP-1 construct (SENP1-C630S), and lyssates were analysed by western blot analysis.
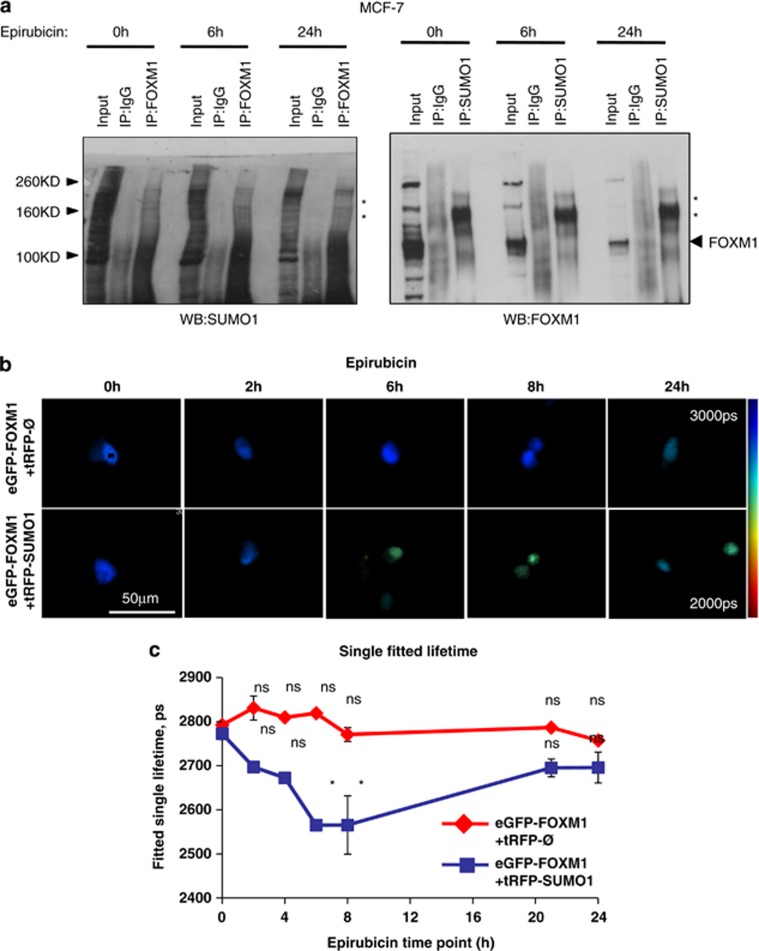
Epirubicin enhances FOXM1 SUMOylation
We next tested whether SUMOylation of FOXM1 is altered upon epirubicin treatment. Co-immunoprecipitation studies in MCF-7 cells showed that FOXM1 and SUMO1 form higher order complexes, affirming that FOXM1 is covalently modified. FOXM1 levels, but not the higher order FOXM1–SUMO1 complexes, decreased following epirubicin treatment, suggesting SUMO1 was limiting in MCF-7 cells and increased SUMOylation upon epirubicin treatment (Figure 2a and Supplementary Figures S1 and S2). Interestingly, FOXM1–SUMO1 conjugation occurred at low and stable levels in the epirubicin-resistant MCF-7EpiR cells. In agreement, endogenous FOXM1 increased its colocalization with SUMO1 in MCF-7 but not in MCF-7EpiR cells in response to epirubicin (Supplementary Figure S3). Moreover, epirubicin treatment of transfected MCF-7 cells increased the colocalization and Förster resonance energy transfer (FRET) between eGFP-tagged FOXM1 and red fluorescent protein (tRFP)-tagged SUMO1 as determined by confocal (Supplementary Figure S4) and fluorescence lifetime imaging microscopy (FLIM) (Figure 2b), respectively. Analysis of the FRET donor eGFP-FOXM1 over a 24 h period following epirubicin treatment demonstrated a significant decrease in fluorescence lifetime at 6 and 8 h, indicating increased interaction with tRFP-SUMO1 (Figure 2c). In contrast, there were no significant changes in fluorescence lifetime when the donor was expressed alone, confirming that the measured change in lifetime is due to FRET between eGFP-FOXM1 and SUMO1-tRFP in response to epirubicin treatment. Consistently, SUMO1 expression was induced by epirubicin in the MCF-7 but remained at low levels in MCF-7EpiR cells (Supplementary Fig. S5). Taken all together, these data revealed that SUMO1 expression and its interaction with FOXM1 were inducible by epirubicin in the MCF-7 cells; however, by comparison, these were occurring at relatively low and constant levels in MCF-7EpiR cells. The kinetics of FOXM1/SUMO1 expression and interaction also suggested the possibility that FOXM1-SUMOylation is associated with its downregulation in expression.
Figure 2.
FOXM1 is SUMOylated in response to epirubicin treatment. (a) MCF-7 cells were treated with epirubicin (1 μM) for 0, 6 and 24 h. Co-immunoprecipitation (co-IP) was performed with a FOXM1 antibody and was probed for SUMO1 and vice versa; inputs (1/10 of IP) and IP products with IgG and specific antibodies were resolved on western blot analysis and were probed for FOXM1 and SUMO1. High molecular weights of FOXM1- and SUMO1-containing species are highlighted ‘*'. (b) MCF-7 cells co-transfected with 0.05 μg of eGFP-FOXM1 and 0.025 μg per well of either tRFP-SUMO1 (FRET acceptor) or empty expression plasmids were treated with 0.1 μM epirubicin for 0, 2, 4, 6 and 24 h. Intensity-merged fluorescence lifetime images of doxorubicin treatment time course are shown. The reduction in donor fluorescence lifetime indicates the occurrence of FRET, implying that eGFP-FOXM1 and tRFP-SUMO1 are within 10 nm of each other. (c) Analysis of fluorescence lifetime data, showing mean lifetimes ±s.e.m. of eGFP-FOXM1 fluorescence,was determined by fitting the fluorescence decay as described. Each measurement is based on >100 cells. The results show that eGFP-FOXM1 and tRFP-SUMO1 interactions significantly increase after 6 or 8 h epirubicin treatment (Tukey's HSD test: http://www.amazon.co.uk/Biostatistics-Methodology-Sciences-Probability-Statistics/dp/0471031852: 0 versus 3, 6, 8 or 24 h; *significant P<0.05; NS, not significant).
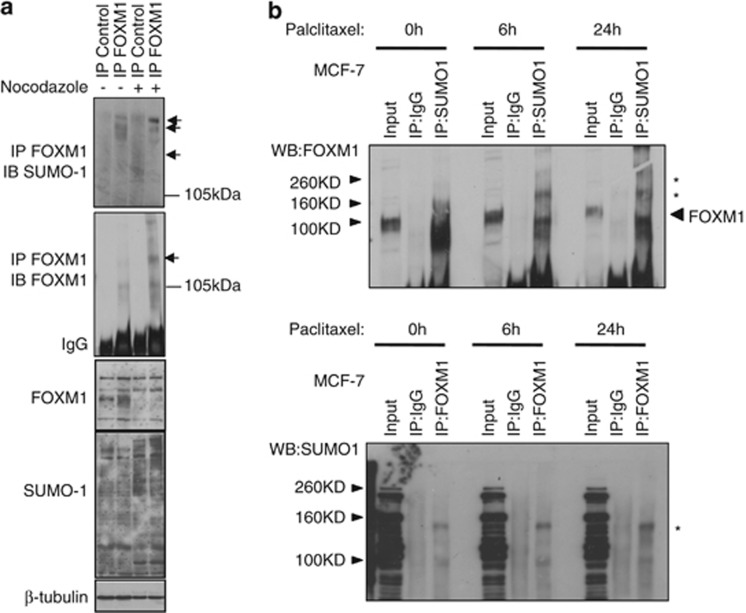
FOXM1 is SUMOylated in response to mitotic checkpoint drugs
As FOXM1 is also critical for the G2/M-phase progression,8, 21 we examined whether mitotic disruptors would also enhance FOXM1 SUMOylation. To this end, MCF-7 cells were treated with nocodazole, a spindle poison, or paclitaxel. Both compounds have previously been shown to induce a G2/M-phase delay in MCF-7 cells.8, 22 Consistent with our previous result, only low levels of SUMOylated FOXM1 were detected in untreated asynchronous cells (Figure 3a). However, this was significantly increased following treatment with nocodazole (Figure 3a), suggesting a role for SUMOylation of FOXM1 in mitotic progression. Following coexpression of FOXM1 with SUMO1, we also observed increased SUMO conjugation in response to paclitaxel treatment (Figure 3b), a response that was noticeably more pronounced than that observed with nocodazole. These data demonstrate that cytostatic drugs also enhance FOXM1 SUMOylation.
Figure 3.
SUMOylation of FOXM1 occurs during mitotic arrest. (a) MCF-7 cells were treated with dimethyl sulfoxide (DMSO) (0.001% (v/v); 16 h) or nocodazole (0.0001 mg/ml; 16 h), and immunoprecipitation (IP) was performed with a FOXM1 antibody; inputs and IP products were resolved on western blot analysis and were probed for SUMO1. The membrane was then reprobed with a FOXM1 antibody. Arrows indicate higher molecular weight forms of FOXM1. (b) MCF-7 cells were treated with paclitaxel (100 nM) for 0, 6 and 24 h. Co-IP was performed with a FOXM1 antibody and was probed for SUMO1 and vice versa; inputs (1/10 of IP) and IP products with IgG and specific antibodies were resolved on western blot analysis and were probed for FOXM1 and SUMO1. * indicates higher molecular weight FOXM1 complex.
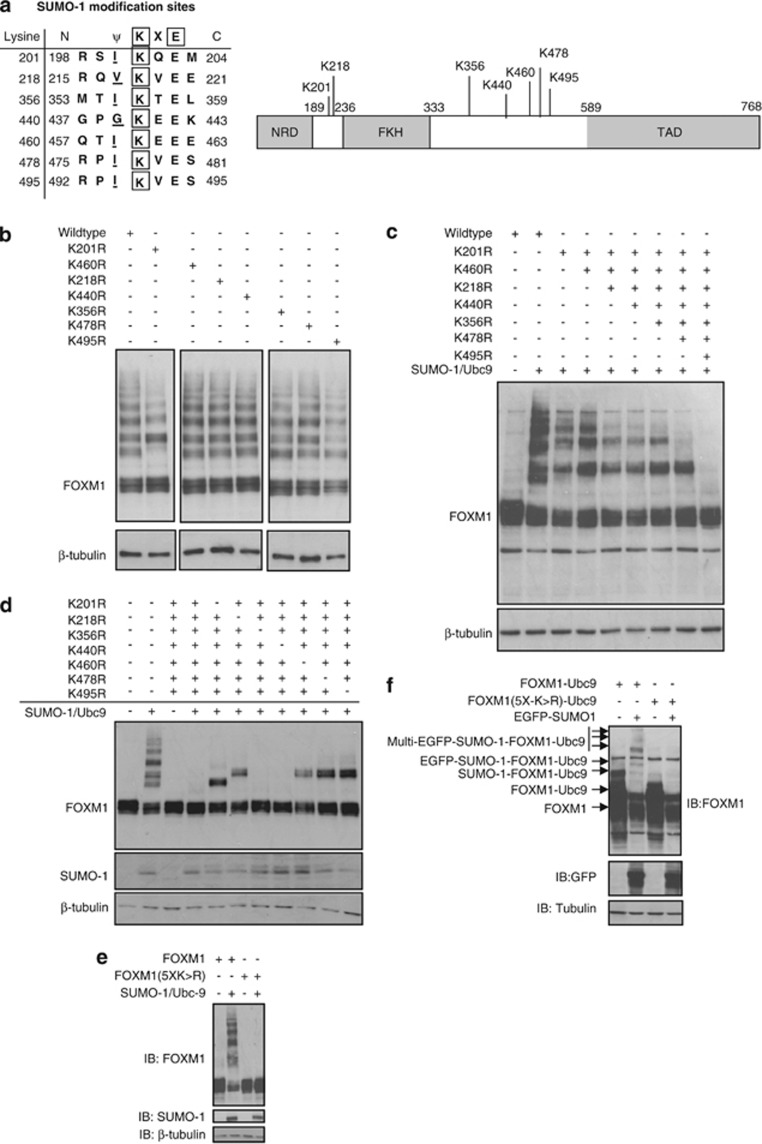
FOXM1 is modified at multiple sites
We next used in silico prediction algorithms (SUMOplot and SUMOsp) to identify potential SUMOylation sites in FOXM1 (Figure 4a). Consensus ψKXE motifs were identified throughout FOXM1 and clustered near the transactivation domain (Figure 4a). They were absent from the N-terminal repressor domain. Using site-directed mutagenesis, we generated a series of FOXM1 mutants in which individual lysines within consensus motifs were mutated to arginine (Figure 4b). With the exception of K201R, no single mutation changed the SUMOylation pattern of FOXM1. Conjugation at K201 was confirmed by immunoprecipitation with SUMO1 and immunoprobing of FOXM1 (Supplementary Figure S6; loss of SUMO-FOXM1 band highlighted). The data suggested a degree of redundancy between putative acceptor sites in FOXM1. Consequently, we simultaneously mutated multiple sites within FOXM1 to produce a SUMO-deficient mutant. Mutation of seven lysine residues (7X(K>R)-FOXM1) completely abolished SUMOylation of FOXM1 in cells overexpressing Ubc9 and SUMO1 (Figure 4c). To test for site redundancy, we then reversed individual arginine residues in FOXM17X(K>R) back to lysine and identified K440 and K356 as being redundant for FOXM1 SUMOylation (Figure 4d). Indeed, the FOXM15X(K>R) mutant, carrying five point mutations (K201R, K218R, K460R, K478R and K495R), was found to be SUMOylation-deficient following Ubc9 and SUMO1 coexpression (Figure 4e). Further, when fused to Ubc9, the FOXM15X(K>R) mutant displayed no auto-SUMOylation activity (Figure 4f).
Figure 4.
FOXM1 is SUMOylated at multiple sites. (a) Schematic showing consensus SUMOylation sites in FOXM1 identified using online computational prediction software Abgent SUMOplot (Abgent, Maidenhead, UK) and SUMOsp 2.0 (The Cuckoo Workgroup, Hefei, China). NRD, N-terminal regulatory domain; FKH, forkhead domain; TAD, transactivation domain. (b) Site-directed mutagenesis was performed using WT-FOXM1 expression vector at the indicated sites (lysine to arginine). In all cases, mutants were confirmed by sequencing. MCF-7 cells were then co-transfected with WT- or indicated mutant-FOXM1, SUMO1 and Ubc9 constructs, and 24 h later protein lysates were prepared and FOXM1 SUMOylation was determined by western blot analysis. (c) FOXM1 mutants were generated, containing multiple lysine-to-arginine mutations, from FOXM11X(K>R) (K210R) to FOXM17X(K>R) (K201R/K218R/K356R/K440R/K460R/K478R/K495R). Mutational order was based on site location. FOXM1 mutant SUMOylation was determined as above. (d) FOXM17X(K>R) was subjected to reversal of individual mutations (R>K) to examine site redundancy. Mutants were assessed for SUMOylation as above and K356R and K440R were identified as redundant mutations. (e) MCF-7 cells were transfected with WT-FOXM1 or FOXM15X(K>R) (K201R/K218R/K460R/K478R/K495R) with or without Ubc9 and SUMO1. SUMOylation of FOXM1 was determined as above. (f) MCF-7 cells were transfected with either FOXM1-Ubc9 or FOXM15X(K>R)-Ubc9 with or without eGFP-tagged SUMO1. Loss of the auto-SUMOylated form of FOXM1 was observed in FOXM15X(K>R)-Ubc9. SUMOylation of FOXM1 was determined as above.
SUMOylated FOXM1 resides in the cytoplasm
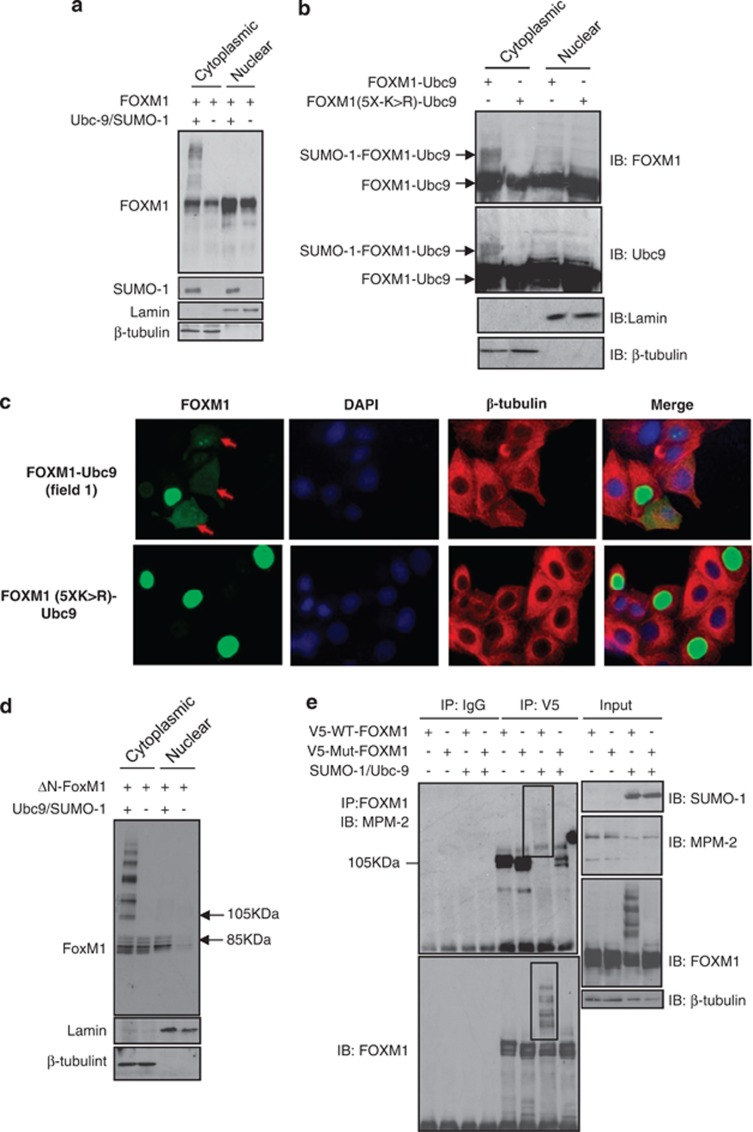
During the cell cycle, FOXM1 becomes progressively phosphorylated through G1/S- and G2/M-phase progression in an ERK- and Cdk1-dependent manner.8, 23 This leads to nuclear localization of FOXM1 and activation.8, 23 We therefore sought to determine whether SUMOylation has an impact on FOXM1 phosphorylation, nuclear accumulation and/or activity. Coexpression of SUMO1 and Ubc9 resulted in multi-SUMOylated FOXM1, which following subcellular fractionation was found to reside in the cytoplasm (Figure 5a). In agreement, the auto-SUMOylated FOXM1-Ubc9 fusion protein was also present in the cytoplasm, whereas the SUMOylation-defective FOXM15X(K>R) mutant was not (Figure 5b). Confocal microscopy further supported the notion that SUMOylation promotes cytoplasmic translocation of FOXM1. Cytoplasmic staining was present in the FOXM1-Ubc9-expressing cells, whereas it was exclusively nuclear in cells transfected with the FOXM15X(K>R)-Ubc9 mutant (Figure 5c; Supplementary Figure S7A). SUMOylation-dependent cytoplasmic translocation of FOXM1 was not dependent on modification of a specific SUMO-acceptor site (Supplementary Figure S7B), and was unaffected by deletion of the N-terminal repressor domain (ΔN-FOXM; Figure 5d). These observations suggest that binding of the N-terminal repressor domain to the trans-activating domain is in this instance not required for cytoplasmic localization of FOXM1 (Figure 5d).
Figure 5.
SUMOylation of FOXM1 promotes its cytoplasmic localization and occurs preferentially on phosphorylated FOXM1. (a) MCF-7 cells were co-transfected with FOXM1, SUMO1 and Ubc9 constructs and cytoplasmic and nuclear fractions were isolated. FOXM1 SUMOylation and cell fractionation were confirmed by western blot analysis against FOXM1, SUMO1, lamin and β-tubulin. (b) MCF-7 cells were transfected with FOXM1-Ubc9 or FOXM15X(K>R)-Ubc9 and were fractionated as above. The cytoplasmically localized auto-SUMOylation event is indicated. (c) MCF-7 cells were transfected with FOXM1-Ubc9 or FOXM15X(K>R)-Ubc9 and stained for FOXM1, β-tubulin and 4′,6-diamidino-2-phenylindole before analysis by confocal microscopy. Images are representative of three independent experiments. (d) MCF-7 cells were transfected with FOXM1 or the N-terminal truncated FOXM1 (Δ-FOXM1) and cells were fractionated as above. (e) MCF-7 cells were co-transfected with WT-FOXM1 or mutant-FOXM15X(K>R) with or without SUMO1 and Ubc9 constructs, and 24 h later protein lysates were subjected to immunoprecipitation (IP) with a FOXM1 antibody. Precipitated proteins were separated by SDS-PAGE and FOXM1 phosphorylation was detected using a phosphorylated-M-phase-associated protein-specific antibody (MPM-2). The membrane was then reprobed with a FOXM1 antibody, demonstrating that the free FOXM1 displayed no MPM-2 detectable phosphorylation following SUMOylation in the WT-FOXM1, whereas the SUMOylated form of FOXM1 was detectable by MPM-2. These are indicated by boxes.
We next determined whether there was an interdependency between SUMOylation and phosphorylation of FOXM1. No differences in basal phosphorylation were observed between the FOXM15X(K>R) mutant and wild-type (WT) FOXM1 following immunoprecipitation and immunodetection with MPM-2, an M-phase-specific antiphospho-Ser/Thr-Pro antibody that recognises FOXM1 (Figure 5e). This suggested that SUMOylation does not influence FOXM1 phosphorylation. However, coexpression of Ubc9 and SUMO1 resulted in the formation of poly-SUMO1–FOXM1 conjugates detectable with the MPM-2 antibody (Figure 5e; highlighted in panel 1). In contrast, unconjugated FOXM1 was not detectable following reprobing with MPM-2 (Figure 5e; highlighted in panel 2). Critically, upon reprobing, similar levels of unconjugated FOXM1 were detected between the control and SUMO1/Ubc9-expressing cells (Figure 5e), suggesting that phosphorylation of FOXM1 is a cue for SUMO conjugation and not vice versa. Although previous studies demonstrated that the ERK and p38 mitogen-activated protein kinases have a role in regulating the nuclear translocation and transacting activity of FOXM1,23, 24 inhibiting their activity or mutating their target sites in FOXM1 did not have any appreciable effects on FOXM1 SUMOylation (Supplementary Figure S8), arguing that other kinases are involved.
SUMOylation accelerates Cdh1-mediated degradation of FOXM1
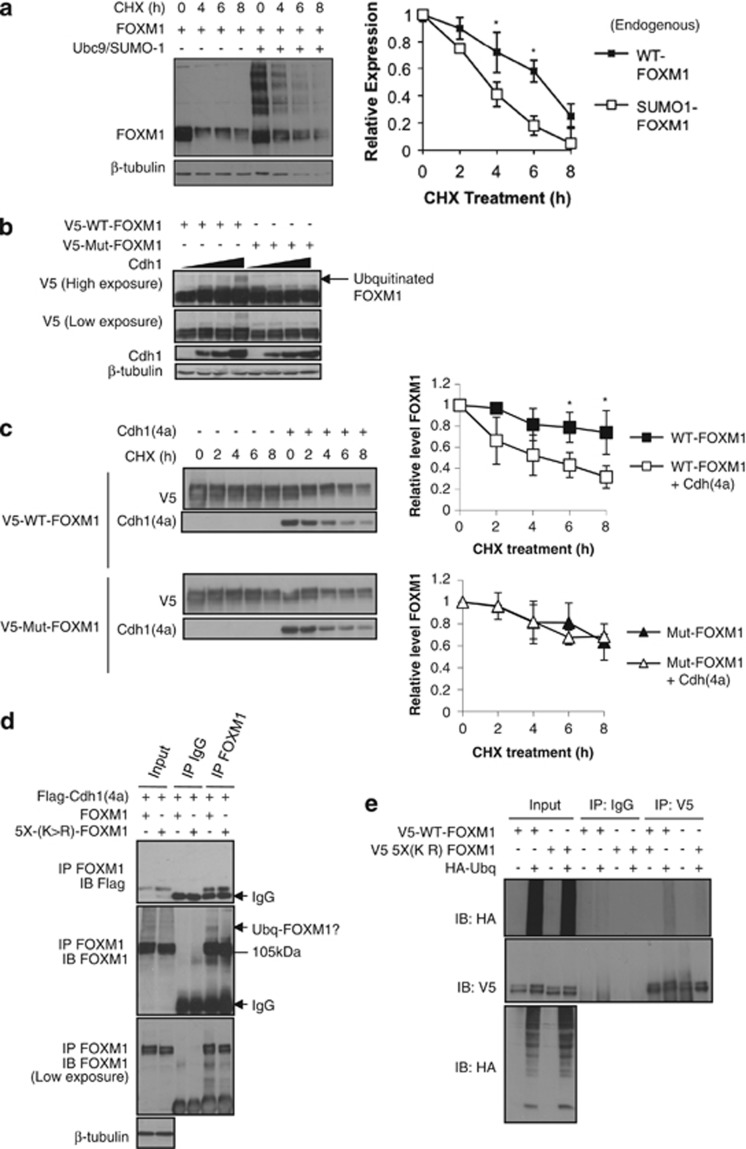
Co-immunoprecipitation experiments showed that SUMOylation is associated with a decrease in its expression levels (Figure 2a and Supplementary Figures S1, S2, S3, S4 and S5). We therefore explored the role of SUMOylation in regulating FOXM1 stability. Following cycloheximide treatment to inhibit protein translation, we observed that SUMOylated forms of FOXM1 degraded more quickly compared with unconjugated FOXM1 (Figure 6a). FOXM1 is a substrate for the APC/Cdh1 complex and is degraded during entry into anaphase to allow for mitotic progression.25 To test whether SUMOylation has an impact on FOXM1 degradation by the APC/Cdh1 complex, we transfected MCF-7 with either the WT or the SUMOylation-deficient FOXM1 and with increasing levels of an expression vector encoding Cdh1(4a), a constitutively activated form of Cdh1.26 Elevation of Cdh1 levels resulted in the appearance of a higher molecular weight form of FOXM1, consistent with ubiquitination, which was absent in the cells transfected with the SUMOylation-deficient FOXM15X(K>R) mutant (Figure 6b). This suggested that SUMO modification may earmark FOXM1 for Cdh1-mediated degradation. Consistent with this notion, we found that coexpression of Cdh1(4a) significantly shortened the half-life of WT-FOXM1 but not the FOXM15X(K>R) mutant following cycloheximide treatment. (Figure 6c). Consistently, overexpression of Cdh1(4a) decreased the half-life of the endogenous FOXM1, whereas Cdh1 depletion increased its half-life (Supplementary Figures S9 and S10). Next, we performed co-immunoprecipitation experiments in cells coexpressing Cdh1(4a) and either WT-FOXM1 or FOXM15X(K>R) mutant. Interestingly, Cdh1(4a) bound more strongly to the SUMOylation-deficient mutant (Figure 6d), suggesting that this modification is not required for interaction of FOXM1 with the APC/Cdh1 complex but has an impact on subsequent ubiquitination. To assess this possibility, we coexpressed haemagglutinin-tagged ubiquitin with either the WT-FOXM1 or FOXM1(5XK>R) and performed co-immunoprecipitation analysis (Figure 6e). Consistent with previous observations, WT-FOXM1 showed higher levels of ubiquitination compared with FOXM15X(K>R) (Figure 6e), supporting the idea that unmodified FOXM1 is more resistant to APC/Cdh1-mediated ubiquitination and degradation.
Figure 6.
SUMOylation of FOXM1 promotes its degradation and a SUMOylation mutant is resistant to Cdh1-mediated degradation. (a) MCF-7 cells were transfected with FOXM1 with or without Ubc9 and SUMO1, and 16 h later cells were treated with cycloheximide (CHX) or vehicle (0.001% (v/v) dimethyl sulfoxide (DMSO)) and protein lysates were prepared from 0 to 8 h following CHX treatment. Densitometry was used to quantify the unconjugated FOXM1 relative to the SUMOylated-FOXM1, for which independent background readings were taken. Western blot analysis was representative of three independent experiments; densitometry is mean±s.e.m. Densitometry was used to quantify FOXM1 levels and were normalized to β-tubulin. Western blot analysis was representative of three independent experiments; densitometry is mean±s.e.m. (t-test: *significant P<0.05; otherwise not significant). (b) Asynchronous MCF-7 cells were co-transfected with FOXM-1 or FOXM15X(K>R) with or without increasing levels of Cdh1(4a). At 24 h after transfection, protein lysates were prepared and FOXM1 was examined by western blot analysis. The arrow indicates the higher molecular weight form of FOXM1 suggesting protein ubiquitination. (c) MCF-7 cells were transfected with FOXM1 or FOXM1(5XK>R) with or without Cdh1(4a), and 16 h later cells were treated with cycloheximide or vehicle (0.001% (v/v) DMSO) and protein lysates were prepared from 0 to 8 h following cycloheximide treatment. Densitometry was used to quantify FOXM1 levels and were normalized to β-tubulin. Western blot analysis was representative of three independent experiments; densitometry is mean±s.e.m. (t-test: *significant P<0.05; otherwise not significant). (d) MCF-7 cells were co-transfected with FOXM1 or FOXM15X(K>R) with Flag-Cdh1(4a), and 24 h later protein lysates were subjected to co-immunoprecipitation (co-IP) with a FOXM1 antibody. Precipitated proteins were separated by SDS–PAGE and Flag-Cdh1(4a) binding was detected using an anti-Flag antibody. The membrane was then reprobed with a FOXM1 antibody. Ubiquitinated-FOXM1 (Ubq-FOXM1) and IgG are indicated by arrows. (e) MCF-7 cells were co-transfected with FOXM1 or FOXM15X(K>R) with or without haemagglutinin-ubiquitin (HA-Ubq). After 24 h, protein lysates were subjected to IP with a FOXM1 antibody. Precipitated proteins were separated by SDS–PAGE and ubiquitination of FOXM1 was detected by anti-HA antibody. The membrane was then reprobed with a FOXM1 antibody.
FOXM1 SUMOylation inhibits proliferation and delays mitotic progression
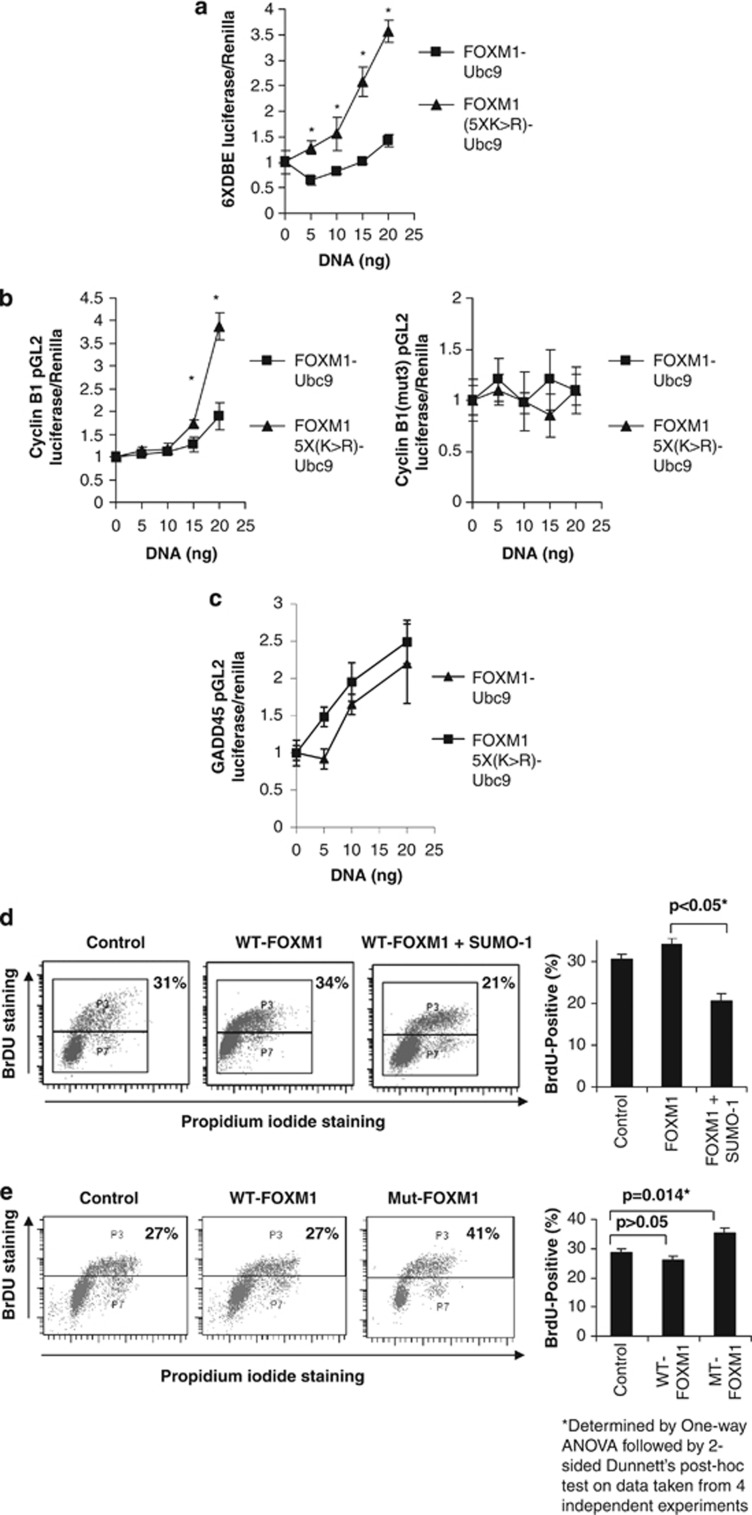
As SUMOylation promoted cytoplasmic accumulation and degradation of FOXM1, we hypothesized that the SUMOylation-deficient mutant would exert stronger transcriptional activity and promote cell proliferation. In agreement, the transcriptional activity of the FOXM15X(K>R)-Ubc9 was found to be consistently higher than that of FOXM1-Ubc9 using a variety of FOXM1-responsive promoter–reporter systems, including 6 × DNA-binding element, cyclin B1 and GADD45 luciferase reporter assays (Figures 7a–c and Supplementary Figure S11). Moreover, this activity was also dependent on the presence of forkhead response elements, as demonstrated by the cyclin B1-mut3 reporter, which contains mutations in the forkhead response element (Figure 7b). We next determined the effect of FOXM1 SUMOylation on cell proliferation using bromodeoxyuridine (BrdU) staining. Transfection of WT-FOXM1 did not significantly alter cell proliferation. However, transfection of SUMO1 with or without FOXM1 markedly reduced cell proliferation (Figure 7d and Supplementary Figure S12). Conversely, expression of the FOXM1(5XK>R) mutant significantly increased the percentage of BrdU-positive MCF-7 cells (Figure 7e). Furthermore, we also found that the SUMOylation-deficient mutant FOXM1 is more effective than WT-FOXM1 in overcoming the antiproliferative effects of epirubicin (Supplementary Figure S13). Taken together, the data indicate that epirubicin treatment promotes SUMOylation of FOXM1, which downregulates FOXM1 expression and restrains cell proliferation.
Figure 7.
SUMOylation of FOXM1 represses its transcriptional activity and a SUMOylation-deficient FOXM1 mutant can enhance cell proliferation. MCF-7 cells were co-transfected with FOXM1-Ubc9 or FOXM1(5XK>R)-Ubc9 (0–20 ng) and luciferase reporters were driven by (a) the FOXM1 6X-DNA-binding element (6XDBE), (b) the wild-type cyclin B1 promoter (cyclin B1-pGL2) or the cyclin B1 promoter containing mutations in the three consensus forkhead binding sites (cyclin B1-mut3-pGL2), or (c) the GADD45 promoter (GADD45-pGL2). After 8 h, cells were placed in 0.5% fetal calf serum for 24 h before the luciferase assay was performed. All DNA concentrations were normalized using empty vector. Reporter gene activity was expressed as a ratio of firefly luciferase activity to control Renilla luciferase activity. (d) MCF-7 cells were transfected with FOXM1 with or without SUMO1. After 20 h, cells were treated with BrdU (10 μmol/l) for 4 h and then harvested and stained with propidium iodide and fluorescein isothiocyanate-conjugated anti-BrdU antibody and analysed by fluorescence-activated cell sorter (FACS) (20 000 gated events were counted; data are representative of three independent experiments; graph shows mean of three experiments±s.e.m.). (e) MCF-7 cells were transfected with empty vector, FOXM1 or FOXM15X(K>R), and 20 h later they were treated with BrdU for 4 h before being harvested and stained with propidium iodide- and fluorescein isothiocyanate-conjugated anti-BrdU. Fluoresence was determined by FACS analysis (20 000 gated events were counted; data are representative of three independent experiments; graph shows mean of three experiment ±s.e.m.) (t-test: *significant P<0.05; otherwise not significant).
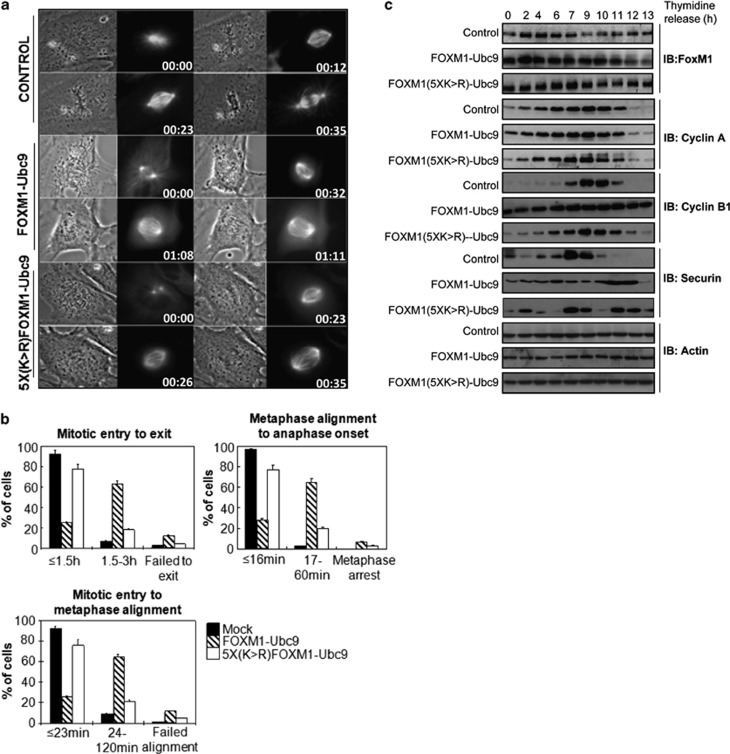
We next sought to determine whether the SUMOylation of FOXM1 would perturb mitotic progression under otherwise normal conditions. To test this hypothesis, we co-transfected FOXM1-Ubc9 or 5X(K>R)-FOXM1-Ubc9 with a pmCherry expression vector into RPEhTert-GFP-α-tubulin cells (retinal pigmented epithelial cell line stably transfected with human telomerase reverse transcriptase and GFP-α-tubulin).27 Progression through mitosis following synchronisation by double thymidine block was monitored using time-lapse phase-contrast and fluorescent microscopy (Figure 8a and Supplementary Videos VS1 and VS2). The FOXM15X(K>R)-Ubc9 mutant had no discernible effects on time from mitotic entry to exit (Figure 8a), suggesting that SUMOylation of FOXM1 is not required for normal progression of cells through mitosis. However, the FOXM1-Ubc9 fusion protein markedly increased the time needed for mitotic entry to metaphase alignment, and also from metaphase alignment to anaphase onset (Figure 8b). To further examine the mitotic delay in response to persistent FOXM1 SUMOylation, we examined the expression of cell cycle regulatory proteins following double thymidine block and release in HeLa cells transfected with a control vector or expression vectors encoding FOXM1-Ubc9 or 5X(K>R)-FOXM1-Ubc9. Intriguingly, expression of FOXM1-Ubc9, but not 5X(K>R)-FOXM1-Ubc9, caused persistent cyclin B1 expression (Figure 8c). This suggested that the observed mitotic delay may be due to inadequate cyclin B1 degradation, thus delaying anaphase entry.28 Collectively, these data also demonstrate that the SUMOylation of FOXM1 inhibits cell cycle transition by delaying mitotic progression.
Figure 8.
SUMOylation of FOXM1 delays mitotic progression and deregulates cyclin B1 expression. (a) RPE-hTERT-GFP-α-tubulin cells were transfected with empty vector, pmCherry plus FOXM1-Ubc9 or pmCherry plus FOXM15X(K>R)-Ubc9 and synchronized in the G1 phase by double thymidine block. Following removal of thymidine, progression through mitosis was monitored by time-lapse phase-contrast and fluorescent microscopy. (b) Graphical representation of mitotic progression. Time from mitotic entry to exit, mitotic entry to metaphase alignment and metaphase alignment to anaphase onset were calculated on a total of 200 cells per condition from three independent experiments (mean±s.e.m.). Only cells that were transfected with the pmCherry-tagged FOXM1 constructs were included. (c) HeLa cells were transfected with empty vector, FOXM1-Ubc9 or FOXM15X(K>R)-Ubc9, and 24 h later they were synchronised by double thymidine block. Following release from thymidine block, protein lysates were prepared from 0 to 13 h; lysates were resolved by gel electrophoresis and western blot analysis was performed for mitotic and S-phase-associated cyclins and checkpoint protein. β-Actin was used as a loading control; data are representative of three independent experiments.
Discussion
Overexpression of FOXM1 is a hallmark of various aggressive solid carcinomas.29 It has a crucial role in the transformation of cells, in the response of malignant cells to chemotherapeutic agents, and in the development of drug resistance.1, 4, 8, 10 We now provide evidence that FOXM1 activity is controlled by SUMO1 conjugation and deconjugation. We demonstrated that SUMOylation of FOXM1 promotes its degradation in the cytoplasm by the APC/Cdh1 complex, leading to loss of FOXM1 activity and, consequently, mitotic delay. We also provide evidence that enhanced FOXM1 SUMOylation is integral to the cellular response to epirubicin and taxanes.
Our observations are in agreement with the broad and general function of SUMOylation as an inhibitory post-translational modification for many transcription factors. Further, the interplay between SUMOylation and ubiquitination has been shown to control the subcellular localisation and stability of a number of transcription factors, including PPARs,30 RXR,31 PGC-1,32 IRF3,33 Sp134 and SERCA2a.35 In most cases, SUMO-targeting E3 ubiquitin ligases (STUbLs), such as RNF4, are recruited to SUMO-conjugated target proteins, resulting in ubiquitination and subsequent degradation. We demonstrate here that the Cdh1 subunit of the APC complex mediates ubiquitination and degradation of FOXM1. The N-terminal region of FOXM1 contains both destruction box (D box) and KEN box sequences that are recognized and targeted by the E3 ubiquitin ligase Cdh1 during mitotic exit.25 However, we found that, although SUMOylation of FOXM1 is required for its ubiquitination and degradation by the APC/Cdh1 complex, it does not mediate the interaction of FOXM1 with APC/Cdh1, suggesting other E3 ubiquitin ligases, including the SUMOylation-dependent. This suggests that other E3 ubiquitin ligases, including STUbLs, enzymes with SUMO-targeted ubiquitin ligation activity (e.g. RNF4, RNF8 and BRCA1),36, 37, 38 may be involved in recognizing SUMOylated FOXM1. It is also possible that FOXM1 undergoes further ubiquitination mediated by the mitosis-specific E3 ligase Cdh1 following delays in M phase in response to DNA damage. In support of this notion, STUbLs as well as Cdh1 have been shown to be involved in mediating the DNA damage response.39, 40 However, STUbLs are E3 ubiquitin ligases that bind preferentially to poly-SUMO2/3 chains but poorly to monomers.38, 41, 42 However, there is evidence that certain target proteins modified by SUMO1, such as PML-RARα,38, 43, 44 can recruit STUbLs and be targeted for ubiquitination. It is possible that clustering of multiple SUMOylation sites as found in FOXM1 and other proteins may function in a similar manner as poly-SUMO2/3 chains in providing the interface for binding to SUMO-interacting motifs of STUbLs.
FOXM1 has a central part in the response to the anticancer genotoxic agent epirubicin. Accordingly, epirubicin causes a decline in FOXM1 expression accompanied by cell death in the sensitive MCF-7 breast cancer cells.1, 10, 11 We also observed here that FOXM1-SUMOylation increases in response to epirubicin in the MCF-7 cells, suggesting that FOXM1-SUMOylation negatively regulates its expression and activity. FOXM1 regulates the expression of genes important for DNA damage repair, cell cycle control and cell survival, which are also involved in DNA damage response.29 SUMOylation of FOXM1 mediates its ubiquitination, degradation and cytoplasmic relocalization, which will culminate in a decrease in its transcriptional activity. Accordingly, our results show that the SUMOylation-deficient FOXM1 has a better transactivation activity compared with WT-FOXM1. In consequence, FOXM1-SUMOylation has a key role in DNA damage response and genotoxic drug sensitivity through negatively regulating FOXM1-dependent transcription. Consistently, the E3 ligases PIAS1 and -4, which mediate SUMOylation in response to DNA damage, have been shown to be essential for the subsequent DNA damage response.12 Collectively, these findings underscore the importance of SUMOylation in regulating FOXM1 expression and activity. As such, FOXM1-SUMOylation may function like a molecular switch that ultimately controls DNA damage response and genotoxic drug resistance.
Methods and materials
Plasmid constructs, mutagenesis and antibodies
The FOXM1 and ΔN-FOXM1 plasmids have previously been described.45 The SUMO1, SUMO2, SUMO3 and Ubc-9 plasmids have previously been described41, 46 and were generous gifts from RT Hay (University of Dundee, Dundee, UK). The SENP-1 and mutant SENP-1-C630S plasmids47 were gifts from P O'Hare (Marie Curie Research Institute, Oxted, UK). FOXM1 SUMOylation sites were predicted using computational methods by combining SUMOSp (http://bioinformatics.lcd-ustc.org/sumosp/)48 with analysis of lysine residue conservation across FOXM1 orthologues using Bioedit and ClustalW (http://www.ebi.ac.uk/clustalw/) software. FOXM1 SUMOylation mutants were generated using the QuickChange Site-Directed mutagenesis kit (Stratagene, Leicester, UK), as per the manufacturer's instructions, to generate lysine-to-arginine mutations. The FOXM1-Ubc9 and FOXM1(5XK>R)-Ubc9 fusion proteins were generated by cloning full-length WT/mutant-FOXM1 into the BamHI and EcoRI sites of the pcDNA3-MCS-Ubc9 vector, which was generated by cloning full-length mouse Ubc9 (mUbc9) into the EcoRI and XbaI sites of pcDNA3 (Invitrogen, Paisley, UK). The pcDNA3-MCS-Ubc9 vector20 was a kind gift from R Niedenthal (Institut für Physiologische Chemie, Hanover, Germany). For the mitosis studies the FOXM1-Ubc9 and the FOXM1-mut-Ubc9 plasmids were subcloned into pmCherry plasmid (Invitrogen). All plasmids were sequenced. Antibodies used in this study were polyclonal and from Santa Cruz Biotechnology unless otherwise stated: FOXM1 (c-20; 1:1000), β-tubulin (H-235; 1:5000), cyclin B1 (GNS1; 1:1000), cyclin A1 (c-19; 1:2000), Securin (DCS-280; NeoMarkers, Fremont, CA, USA), SUMO1 (21C7; 1:1000; monoclonal; Zymed, San Francisco, CA, USA), SUMO2/3 (N18; 1:1000; monoclonal; Cell Signaling Technology, New England Biolabs, Hitchin, UK), acetylated lysine (7F8; 1:1000), phospho-ERK1/2 (1:1000), total ERK1/2 (1:1000) (Cell Signaling Technology), monoclonal ubiquitin (1:500) and monoclonal MPM-2 (Upstate, New York, NY, USA; 1:1000). The V5 mouse monoclonal antibody (R960-25) detects recombinant proteins containing the 14 amino-acid V5 epitope and was purchased from Life Technologies (Paisley, UK). The eGFP-FOXM1 expression plasmid was generated by subcloning the full-length FOXM1 into the peGFP-C1 vector (Clonetech, Mountain View, CA, USA), and the tRFP-SUMO1expression plasmid has been described13 and was a kind gift from Dr J Morris and Professor T Ng (University of Birmingham and King's College London, London, UK, respectively).
Cell culture and synchronization
Cell lines used in this study originated from the American Type Culture Collection (LGC standards, Middlesex, UK). MCF-7 cells were cultured in Dulbecco's modified Eagle's medium (Sigma, Poole, UK) supplemented with 10% (v/v) fetal calf serum and 2 mM glutamine at 37 °C. For early S-phase block, HeLa cells were exposed to double thymidine block (2 mM thymidine for 18 h, fresh culture media for 10 h, 2 mM thymidine for 18 h, and release into fresh media), and for mitotic block MCF-7 cells were exposed to nocodazole (100 ng/ml; 16 h).
Cell fractionation, immunoprecipitation and western blotting
For western blot analysis, cells were lysed as described previously49 with the addition of N-ethyl-amide (10 mM) (Sigma UK, Poole, UK) to the lysis buffer. For immunoprecipitation and cell fractionation, see Supplementary Materials and methods. Proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE), transferred to Hybond-C membranes and immunoblotted with the indicated antibodies.
Cell cycle and BrdU analysis
Propidium iodide staining was conducted as described previously.50 For staining with BrdU, see Supplementary Materials and methods. The analysis was conducted using a FACSCanto analyser (Becton Dickinson, Oxford, UK); the cell cycle profile was analysed using BD FACSDiva software (Becton Dickinson).
In vitro SUMOylation assay
Human FOXM1 was cloned into an IPTG-inducible bacterial vector with a C-terminal His-tag. Escherichia coli bacteria were transformed and grown in the presence and absence of IPTG, and recombinant FOXM1 was purified by nickel column under denaturing conditions and used for an in vitro SUMOylation assay (Biomol, Enzo Life Sciences, Exeter, UK) with or without Mg-ATP, as per the manufacturer's instructions. RanGAP was used as a positive control.
Confocal microscopy
MCF-7 cells were transiently transfected with the eGFP-FOXM1 and tRFP-SUMO1 plasmids, treated with epirubicin and fixed as described. MCF-7 cells were washed with room temperature phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde solution for 10 min and washed again with PBS. Cells were then permeabilized with 0.1% Triton in 0.2% bovine serum albumin (BSA)-PBS for 10 min, washed with PBS and nonspecific protein binding blocked with 2% BSA-PBS for 30 min. The blocking solution was removed and the specified antibody was added in 1% BSA-PBS for 45 min, followed by washing with PBS. The process was repeated when staining with two antibodies. The secondary antibodies were added (anti-rabbit 488 nM; anti-mouse 594 nM; Molecular Probes, Life Technologies) in 1% BSA-PBS for 45 min before a repeat wash with PBS. Mounting medium containing 4′,6-diamidino-2-phenylindole (DAKO, Ely, UK) was added and a coverslip was applied. Confocal microscopy was performed using a Zeiss confocal microscope with LSM meta 510 software (Carl Zeiss, Cambridge, UK). All experiments included a non-primary control and were repeated with three independent experiments.
Luciferase assays
MCF-7 cells were transfected with the indicated plasmids using Fugene-6 (Qiagen, West Sussex, UK) as per the manufacturer's instructions. Luciferase activity was determined 24 h after transfection using the Perkin-Elmer Luclite assay kit (Perkin-Elmer, Cambridge, UK). Renilla luciferase was used for normalization. Luminescence was read using the 9904 Topcount Perkin-Elmer plate reader and relative luciferase activity was expressed as a percentage of induction of the 6 × Forkhead response element promoter8 and cyclin B1 promoter,51 where basal promoter activity was set as 100%.
Mitotic progression analysis
RPE-hTert cells expressing GFP-tagged α-tubulin (RPE-hTERT-GFP-α-tubulin cells) (Clonetech)27 were injected with empty vector, pmCherry-FOXM1-Ubc9 or pmCherry-FOXM15X(K>R)-Ubc9 and synchronized in the G1 phase by double thymidine block as described.52 Following removal of thymidine, progression through mitosis was monitored by phase-contrast and fluorescent microscopy. Time from mitotic entry to exit, mitotic entry to metaphase alignment and metaphase alignment to anaphase onset was calculated on a total of 200 cells per condition from three independent experiments. Only cells that were transfected with the pmCherry-tagged FOXM1 constructs were included.
Microinjection and live cell microscopy
Mitotic progression was detected with live cell microscopy in RPE-hTert cells expressing GFP-tagged α-tubulin followed by microinjection of the following expression plasmids: control pmCherry-C1, pmCherry-C1 and FOXM1-Ubc9, pmCherry-C1 and FOXM15X(K>R)-Ubc9. The expression plasmids were coinjected at a maximum needle concentration of 300 ng/μl. Live cell microscopy was started 6 h after microinjection of cells on the stage of a Zeiss Axiovert microscope (Preston, UK). Images were acquired at 12-min intervals using a Hamamatsu Orca camera (Hamamatsu Photonics, Hamamatsu, Japan) for a total of 5 h. Times taken from mitotic entry (starting of chromosome condensation) to exit (chromosome decondensation in daughter cells), from mitotic entry to metaphase alignment and from metaphase alignment to anaphase onset were calculated. These mitotic phase analyses were performed only on mCherry-positive cells (n=30 per group). Uninjected cells were used as internal control to ensure that delay in mitotic timing is not due to the procedure. MetaMorph imaging software (Molecular Devices, Wokingham, UK) was used as platforms for acquisition and assembly of the acquired time lapse images.
Cell synchronization and western blotting
HeLa cells were transfected with empty vector, FOXM1-Ubc9 or FOXM1(5XK>R)-Ubc9 and synchronized by double thymidine block as described. Briefly, HeLa cells were blocked in thymidine (2 mM) for 20 h, removed for 6 h and blocked for another 16 h. Cells entered mitosis about 6–8 h after removing the second thymidine block. Progression of cells through mitosis was monitored by detection of cyclin A, cyclin B1 and securin. Proteins were separated by SDS–PAGE gel electrophoresis and immunoblotted with the indicated antibodies.
FLIM-FRET
MCF-7 cells were seeded on a 96-well plate and transfected 24 h later with 0.1 μg per well of eGFP-FOXM1 (FRET donor) and 0.025 μg per well of either tRFP-SUMO1 or tRFP empty vector (FRET acceptor) using XtremeGene HP (Roche, Hertfordshire, UK). Treatment time course with 0.1 μM epirubicin commenced 24 h later. Upon time-course completion, cells were washed and immersed in imaging medium (Hank's balanced salt solution+glucose+L-glutamine) and imaged on a fully automated 96-well plate reading FLIM microscope similar to that described previously.53 Configured for wide-field time-gated lifetime imaging, the instrument acquired FLIM and acceptor intensity images for 420 fields of view in <95 min. Lifetime decays were fitted to a monoexponential model, accounting for instrument response and background fluorescence, using FLIMfit (Imperial College London, London, UK).54
Statistical analysis
All statistical analyses, unless otherwise specified, were carried out using SPSS 15.0 (SPSS inc, Chicago, IL, USA) and Windows XP (Microsoft, Redmond, WA, USA). Where appropriate, a two-tailed independent sample t-test was performed or where more than two conditions were present an analysis of variance followed by Dunnett's post hoc test for data sets showing significant variance (P<0.05). The FLIM-FRET data were analysed using Tukey's HSD (honestly significant difference) test.
Acknowledgments
This work was funded by grants from Breast Cancer Campaign and from Cancer Research UK. MK and PK are recipients of studentships from the Royal Thai Government, and UK is supported by BBSRC. ARG is a post-doctoral fellow supported by Cancer Research UK, and SZ is a post-doc funded by Breast Cancer Campaign. DJK is supported by a studentship from EPSRC. J.J.B was supported by the Biomedical Research Unit in Reproductive Health, University Hospital Coventry and Warwickshire, UK.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Monteiro LJ, Khongkow P, Kongsema M, Morris JR, Man C, Weekes D, et al. The Forkhead Box M1 protein regulates BRIP1 expression and DNA damage repair in epirubicin treatment. Oncogene. 2012;32:4634–4645. doi: 10.1038/onc.2012.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Wu X, Yang L, Xiao F, Zhang H, Zhou A, et al. FoxM1 inhibition sensitizes resistant glioblastoma cells to temozolomide by downregulating the expression of DNA-repair gene Rad51. Clin Cancer Res. 2012;18:5961–5971. doi: 10.1158/1078-0432.CCR-12-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YY, Jung SY, Jennings NB, Rodriguez-Aguayo C, Peng G, Lee SR, et al. FOXM1 mediates Dox resistance in breast cancer by enhancing DNA repair. Carcinogenesis. 2012;33:1843–1853. doi: 10.1093/carcin/bgs167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr JR, Park HJ, Wang Z, Kiefer MM, Raychaudhuri P. FoxM1 mediates resistance to herceptin and paclitaxel. Cancer Res. 2010;70:5054–5063. doi: 10.1158/0008-5472.CAN-10-0545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien AJ, Moasser MM.Cellular mechanisms of resistance to anthracyclines and taxanes in cancer: intrinsic and acquired Semin Oncol 200835S1–S14.quiz S39. [DOI] [PubMed] [Google Scholar]
- Alvarez-Fernandez M, Halim VA, Krenning L, Aprelia M, Mohammed S, Heck AJ, et al. Recovery from a DNA-damage-induced G2 arrest requires Cdk-dependent activation of FoxM1. EMBO Rep. 2010;11:452–458. doi: 10.1038/embor.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darieva Z, Bulmer R, Pic-Taylor A, Doris KS, Geymonat M, Sedgwick SG, et al. Polo kinase controls cell-cycle-dependent transcription by targeting a coactivator protein. Nature. 2006;444:494–498. doi: 10.1038/nature05339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, et al. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- Kim IM, Ramakrishna S, Gusarova GA, Yoder HM, Costa RH, Kalinichenko VV. The forkhead box m1 transcription factor is essential for embryonic development of pulmonary vasculature. J Biol Chem. 2005;280:22278–22286. doi: 10.1074/jbc.M500936200. [DOI] [PubMed] [Google Scholar]
- Millour J, de Olano N, Horimoto Y, Monteiro LJ, Langer JK, Aligue R, et al. ATM and p53 regulate FOXM1 expression via E2F in breast cancer epirubicin treatment and resistance. Mol Cancer Ther. 2011;10:1046–1058. doi: 10.1158/1535-7163.MCT-11-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Olano N, Koo CY, Monteiro LJ, Pinto PH, Gomes AR, Aligue R, et al. The p38 MAPK-MK2 axis regulates E2F1 and FOXM1 expression after epirubicin treatment. Mol Cancer Res. 2012;10:1189–1202. doi: 10.1158/1541-7786.MCR-11-0559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009;462:935–939. doi: 10.1038/nature08657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A, et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462:886–890. doi: 10.1038/nature08593. [DOI] [PubMed] [Google Scholar]
- Werner A, Flotho A, Melchior F. The RanBP2/RanGAP1*SUMO1/Ubc9 complex is a multisubunit SUMO E3 ligase. Mol Cell. 2012;46:287–298. doi: 10.1016/j.molcel.2012.02.017. [DOI] [PubMed] [Google Scholar]
- Leisner C, Kammerer D, Denoth A, Britschi M, Barral Y, Liakopoulos D. Regulation of mitotic spindle asymmetry by SUMO and the spindle-assembly checkpoint in yeast. Curr Biol. 2008;18:1249–1255. doi: 10.1016/j.cub.2008.07.091. [DOI] [PubMed] [Google Scholar]
- Zhang XD, Goeres J, Zhang H, Yen TJ, Porter AC, Matunis MJ. SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Mol Cell. 2008;29:729–741. doi: 10.1016/j.molcel.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y, Arnaoutov A, Dasso M. SUMO-2/3 regulates topoisomerase II in mitosis. J Cell Biol. 2003;163:477–487. doi: 10.1083/jcb.200304088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannoun Z, Greenhough S, Jaffray E, Hay RT, Hay DC. Post-translational modification by SUMO. Toxicology. 2010;278:288–293. doi: 10.1016/j.tox.2010.07.013. [DOI] [PubMed] [Google Scholar]
- Gill G. Post-translational modification by the small ubiquitin-related modifier SUMO has big effects on transcription factor activity. Curr Opin Genet Dev. 2003;13:108–113. doi: 10.1016/s0959-437x(03)00021-2. [DOI] [PubMed] [Google Scholar]
- Jakobs A, Koehnke J, Himstedt F, Funk M, Korn B, Gaestel M, et al. Ubc9 fusion-directed SUMOylation (UFDS): a method to analyze function of protein SUMOylation. Nat Methods. 2007;4:245–250. doi: 10.1038/nmeth1006. [DOI] [PubMed] [Google Scholar]
- Down CF, Millour J, Lam EW, Watson RJ. Binding of FoxM1 to G2/M gene promoters is dependent upon B-Myb. Biochim Biophys Acta. 2012;1819:855–862. doi: 10.1016/j.bbagrm.2012.03.008. [DOI] [PubMed] [Google Scholar]
- Sunters A, Fernandez de Mattos S, Stahl M, Brosens JJ, Zoumpoulidou G, Saunders CA, et al. FoxO3a transcriptional regulation of Bim controls apoptosis in paclitaxel-treated breast cancer cell lines. J Biol Chem. 2003;278:49795–49805. doi: 10.1074/jbc.M309523200. [DOI] [PubMed] [Google Scholar]
- Ma RY, Tong TH, Cheung AM, Tsang AC, Leung WY, Yao KM. Raf/MEK/MAPK signaling stimulates the nuclear translocation and transactivating activity of FOXM1c. J Cell Sci. 2005;118:795–806. doi: 10.1242/jcs.01657. [DOI] [PubMed] [Google Scholar]
- Behren A, Muhlen S, Acuna Sanhueza GA, Schwager C, Plinkert PK, Huber PE, et al. Phenotype-assisted transcriptome analysis identifies FOXM1 downstream from Ras-MKK3-p38 to regulate in vitro cellular invasion. Oncogene. 2010;29:1519–1530. doi: 10.1038/onc.2009.436. [DOI] [PubMed] [Google Scholar]
- Laoukili J, Alvarez-Fernandez M, Stahl M, Medema RH. FoxM1 is degraded at mitotic exit in a Cdh1-dependent manner. Cell Cycle. 2008;7:2720–2726. doi: 10.4161/cc.7.17.6580. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Ching YP, Chun AC, Jin DY. Nuclear localization of the cell cycle regulator CDH1 and its regulation by phosphorylation. J Biol Chem. 2003;278:12530–12536. doi: 10.1074/jbc.M212853200. [DOI] [PubMed] [Google Scholar]
- Morales CP, Holt SE, Ouellette M, Kaur KJ, Yan Y, Wilson KS, et al. Absence of cancer-associated changes in human fibroblasts immortalized with telomerase. Nat Genet. 1999;21:115–118. doi: 10.1038/5063. [DOI] [PubMed] [Google Scholar]
- Chang DC, Xu NH, Luo KQ. Degradation of cyclin B is required for the onset of anaphase in mammalian cells. J Biol Chem. 2003;278:37865–37873. doi: 10.1074/jbc.M306376200. [DOI] [PubMed] [Google Scholar]
- Koo CY, Muir KW, Lam EW. FOXM1: from cancer initiation to progression and treatment. Biochim Biophys Acta. 2012;1819:28–37. doi: 10.1016/j.bbagrm.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Wadosky KM, Willis MS. The story so far: post-translational regulation of peroxisome proliferator-activated receptors by ubiquitination and SUMOylation. Am J Physiol Heart Circ Physiol. 2012;302:H515–H526. doi: 10.1152/ajpheart.00703.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrage PS, Schmucker AC, Ren Y, Sporn MB, Brinckerhoff CE. Retinoid X receptor and peroxisome proliferator-activated receptor-gamma agonists cooperate to inhibit matrix metalloproteinase gene expression. Arthritis Res Ther. 2008;10:R139. doi: 10.1186/ar2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rytinki MM, Palvimo JJ. SUMOylation attenuates the function of PGC-1alpha. J Biol Chem. 2009;284:26184–26193. doi: 10.1074/jbc.M109.038943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran Y, Liu TT, Zhou Q, Li S, Mao AP, Li Y, et al. SENP2 negatively regulates cellular antiviral response by deSUMOylating IRF3 and conditioning it for ubiquitination and degradation. J Mol Cell Biol. 2011;3:283–292. doi: 10.1093/jmcb/mjr020. [DOI] [PubMed] [Google Scholar]
- Spengler ML, Brattain MG. Sumoylation inhibits cleavage of Sp1 N-terminal negative regulatory domain and inhibits Sp1-dependent transcription. J Biol Chem. 2006;281:5567–5574. doi: 10.1074/jbc.M600035200. [DOI] [PubMed] [Google Scholar]
- Kho C, Lee A, Jeong D, Oh JG, Chaanine AH, Kizana E, et al. SUMO1-dependent modulation of SERCA2a in heart failure. Nature. 2011;477:601–605. doi: 10.1038/nature10407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Leverson JD, Hunter T. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 2007;26:4102–4112. doi: 10.1038/sj.emboj.7601839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Seifert A, Chua JS, Maure JF, Golebiowski F, Hay RT. SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev. 2012;26:1196–1208. doi: 10.1101/gad.189274.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- Heideker J, Perry JJ, Boddy MN. Genome stability roles of SUMO-targeted ubiquitin ligases. DNA Repair (Amst) 2009;8:517–524. doi: 10.1016/j.dnarep.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJ, Tainer JA, et al. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 2007;26:4089–4101. doi: 10.1038/sj.emboj.7601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, et al. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem. 2001;276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- Knipscheer P, van Dijk WJ, Olsen JV, Mann M, Sixma TK. Noncovalent interaction between Ubc9 and SUMO promotes SUMO chain formation. EMBO J. 2007;26:2797–2807. doi: 10.1038/sj.emboj.7601711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakli M, Karvonen U, Janne OA, Palvimo JJ. SUMO-1 promotes association of SNURF (RNF4) with PML nuclear bodies. Exp Cell Res. 2005;304:224–233. doi: 10.1016/j.yexcr.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, et al. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol. 2008;10:547–555. doi: 10.1038/ncb1717. [DOI] [PubMed] [Google Scholar]
- Kwok JM, Myatt SS, Marson CM, Coombes RC, Constantinidou D, Lam EW. Thiostrepton selectively targets breast cancer cells through inhibition of forkhead box M1 expression. Mol Cancer Ther. 2008;7:2022–2032. doi: 10.1158/1535-7163.MCT-08-0188. [DOI] [PubMed] [Google Scholar]
- Jones MC, Fusi L, Higham JH, Abdel-Hafiz H, Horwitz KB, Lam EW, et al. Regulation of the SUMO pathway sensitizes differentiating human endometrial stromal cells to progesterone. Proc Natl Acad Sci USA. 2006;103:16272–16277. doi: 10.1073/pnas.0603002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey D, O'Hare P. Characterization of the localization and proteolytic activity of the SUMO-specific protease, SENP1. J Biol Chem. 2004;279:692–703. doi: 10.1074/jbc.M306195200. [DOI] [PubMed] [Google Scholar]
- Xue Y, Zhou F, Fu C, Xu Y, Yao X. SUMOsp: a web server for sumoylation site prediction. Nucleic Acids Res. 2006;34:W254–W257. doi: 10.1093/nar/gkl207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui RC, Gomes AR, Constantinidou D, Costa JR, Karadedou CT, Fernandez de Mattos S, et al. The forkhead transcription factor FOXO3a increases phosphoinositide-3 kinase/Akt activity in drug-resistant leukemic cells through induction of PIK3CA expression. Mol Cell Biol. 2008;28:5886–5898. doi: 10.1128/MCB.01265-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M, Medema RH, Garcia-Cao I, Dubuisson ML, Barradas M, Glassford J, et al. Inhibition of the phosphoinositide 3-kinase pathway induces a senescence-like arrest mediated by p27Kip1. J Biol Chem. 2000;275:21960–21968. doi: 10.1074/jbc.M000759200. [DOI] [PubMed] [Google Scholar]
- Leung TW, Lin SS, Tsang AC, Tong CS, Ching JC, Leung WY, et al. Over-expression of FoxM1 stimulates cyclin B1 expression. FEBS Lett. 2001;507:59–66. doi: 10.1016/s0014-5793(01)02915-5. [DOI] [PubMed] [Google Scholar]
- Lam EW, Watson RJ. An E2F-binding site mediates cell-cycle regulated repression of mouse B-myb transcription. EMBO J. 1993;12:2705–2713. doi: 10.1002/j.1460-2075.1993.tb05932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alibhai D, Kelly DJ, Warren S, Kumar S, Margineau A, Serwa RA, et al. Automated fluorescence lifetime imaging plate reader and its application to Forster resonant energy transfer readout of Gag protein aggregation. J Biophotonics. 2013;6:398–408. doi: 10.1002/jbio.201200185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren SC, Margineanu A, Alibhai D, Kelly DJ, Talbot C, Alexandrov Y, et al. Rapid global fitting of large fluorescence lifetime imaging microscopy datasets. PLoS One. 2013;8:e70687. doi: 10.1371/journal.pone.0070687. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.