Abstract
Aromatase inhibitors (AI) are the standard endocrine therapy for postmenopausal breast cancer; however, currently used biomarkers, i.e., estrogen receptor-alpha/progesterone receptor (ERα/PR), predict only slightly more than half of the potential responders to AI treatment. To identify novel markers of AI responsiveness, a genome-wide microarray analysis was performed using primary breast tumor samples from 50 postmenopausal women (PMW) who later developed metastatic breast cancer. Sushi domain containing 3 (SUSD3) was significantly differentially expressed gene, with 3.38-fold higher mRNA levels in AI-responsive breast tumors versus non-responders (p<0.001). SUSD3 was highly expressed in ERα-positive breast tumors and treatment with estradiol increased SUSD3 expression in ERα-positive breast cancer cells. Treatment with an antiestrogen or ERα knockdown abolished basal and estradiol-dependent SUSD3 expression. Recruitment of ERα upstream of the transcription start site of SUSD3 was demonstrated by chromatin immunoprecipitation (ChIP)-PCR. Flow cytometric analysis of SUSD3 knockdown cells revealed blunted estradiol effects on progression into S and M phases. SUSD3 was localized to the plasma membrane of breast cancer cells. SUSD3 knockdown decreased the appearance of actin-rich protrusions, stress fibers and large basal focal adhesions, while increasing the presence of cortical actin concomitant with a decrease in Rho and FAK activity. SUSD3-deficient cells demonstrated diminished cell spreading, cell-cell adhesion, and motility. In conclusion, SUSD3 is a novel promoter of estrogen-dependent cell proliferation and regulator of cell-cell and cell-substrate interactions and migration in breast cancer. It may serve as a novel predictor of response to endocrine therapy and potential therapeutic target.
Keywords: Sushi domain containing 3, estrogen receptor, aromatase inhibitors, breast cancer, migration
INTRODUCTION
Breast cancer is an estrogen and progesterone-dependent disease with variable treatment responsiveness. The mitogenic role of estrogen in breast cancer is well established1,2. Both estrogen synthesis and its receptor (ERα) are targeted by endocrine therapies1,2. Aromatase inhibitors (AIs) block estrogen formation by inhibiting the enzyme aromatase, whereas the estradiol antagonist tamoxifen (TAM) targets ERα3,4. Despite clinical advances in breast cancer treatment, not all patients respond to endocrine therapy and some initial responders experience disease recurrence or progression during therapy3-13. The heterogeneous nature of the disease and the unpredictability of treatment outcomes have prompted the search for new biomarkers of responsiveness for endocrine therapies.
AIs are the most commonly used class of drugs in the long-term treatment of breast cancer3,4. Adjuvant therapy with AIs has largely replaced TAM and other anti-estrogens as the first-line endocrine treatment for postmenopausal women (PMW) with hormone receptor-positive disease3-7. There is a need to identify patients who will respond to AIs, sparing those with resistant tumors the adverse effects of ineffective therapy. Currently, biomarkers for TAM responsiveness—ERα or progesterone receptor (PR) protein immunoreactivity in breast tumors—are used as surrogate predictors for AI responsiveness8-10. Using these biomarkers, response rate to AIs is 35-70%11-13, representing a major obstacle to optimal treatment.
We studied 50 tumor RNA samples obtained between 1990-1995 from PMW with breast cancer who, after surgery and TAM treatment, experienced recurrence, progression and metastasis. Receptor status had been unknown at the time endocrine therapy was first started. Responsiveness of local and metastatic disease to AI therapy was measured by clinical benefit (complete/partial response, or stable disease) for at least 6 months of treatment14. Patients were then placed on AI, and 51% of them demonstrated clinical benefit regardless of hormone receptor status. The status of immunoreactive ERα/PR was later determined and found to have a 58% positive predictive value for clinical benefit15. The poor predictive response of ERα/PR immunoreactivity prompted the search for new markers of AI response. Here, we identify and characterize sushi domain containing-3 (SUSD3), a gene significantly overexpressed in AI responders in a microarray analysis of these tumor samples. We also demonstrate its role in breast cancer cell proliferation as well as cell-cell and cell-substrate adhesion and migration through Rho-focal adhesion kinase (FAK) signaling.
RESULTS
Microarray Gene Expression Analysis of Primary Breast Cancer Tumors
Of the 50 patients, 27 demonstrated clinical benefit from AI treatment for at least 6 months, and 23 did not. The top 50 differentially expressed genes are listed based on the best p-values (Table 1). SUSD3 is highly expressed in AI responders compared with non-responders (3.04-fold), with the highest significance (p=0.0000319). Given the lack of scientific knowledge regarding SUSD3, we focused our efforts on its characterization. Interestingly, ESR1 which encodes ERα ranked number 9, whereas PR encoding PGR was not in top 50 (Table 1).
Table 1.
Top 50 differentially expressed genes between AI responders (n=27) and AI non-responders (n=23).
| Gene | P-value | Fold Change |
|---|---|---|
| SUSD3 | 3.19E-05 | 3.039 |
| PRRT2 | 0.000348 | 2.182 |
| DISP1 | 0.000453 | 1.508 |
| GPR6 | 0.000533 | 1.695 |
| RHBDL1 | 0.00058 | 2.038 |
| KCNE4 | 0.000745 | 5.650 |
| ANXA9 | 0.000781 | 2.479 |
| MYL5 | 0.000867 | 1.703 |
| ESR1 | 0.00103 | 2.007 |
| GON4 | 0.001169 | 1.568 |
| HLXB9 | 0.001196 | 1.702 |
| LRRC50 | 0.001247 | 2.578 |
| PTGER3 | 0.001607 | 1.662 |
| PDE4A | 0.001618 | 1.753 |
| C4A|C4B | 0.001707 | 1.926 |
| ARTN | 0.001974 | 2.031 |
| SELENBP1 | 0.002145 | 1.652 |
| SMYD3 | 0.002828 | 1.560 |
| MAPT | 0.002896 | 1.770 |
| ABAT | 0.002919 | 2.277 |
| RUNDC1 | 0.003055 | 1.515 |
| GREB1 | 0.003213 | 1.742 |
| SEMA3C | 0.003595 | 1.859 |
| KRT19 | 0.004185 | 1.788 |
| GDF15 | 0.004888 | 2.758 |
| SIAH2 | 0.005041 | 1.707 |
| SLC39A6 | 0.005194 | 1.615 |
| SYTL4 | 0.005432 | 1.692 |
| SMYD3 | 0.006022 | 1.588 |
| CELSR1 | 0.006073 | 1.557 |
| KCNK15 | 0.006503 | 1.674 |
| LTC4S | 0.006598 | 1.524 |
| HPN | 0.006685 | 1.679 |
| KRTHA7 | 0.007158 | 1.531 |
| ATP7B | 0.007346 | 1.675 |
| TNFRSF10C | 0.007629 | 2.156 |
| ATP6V1G2 | 0.00852 | 1.640 |
| GJA1 | 0.008552 | 1.698 |
| GRP | 0.00857 | 1.690 |
| CA12 | 0.008711 | 1.824 |
| ANKRD42 | 0.009261 | 1.645 |
| ARRB1 | 0.009329 | 1.578 |
| LRG1 | 0.009972 | 2.082 |
| PLA2G10 | 0.011227 | 2.758 |
| HOXA10 | 0.011789 | 1.735 |
| BAI2 | 0.011838 | 5.770 |
| MAP3K12 | 0.012113 | 1.539 |
| ATHL1 | 0.012313 | 2.594 |
| ELA2B | 0.013659 | 1.519 |
| PCGF2 | 0.01403 | 2.031 |
SUSD3 qRT-PCR Validation
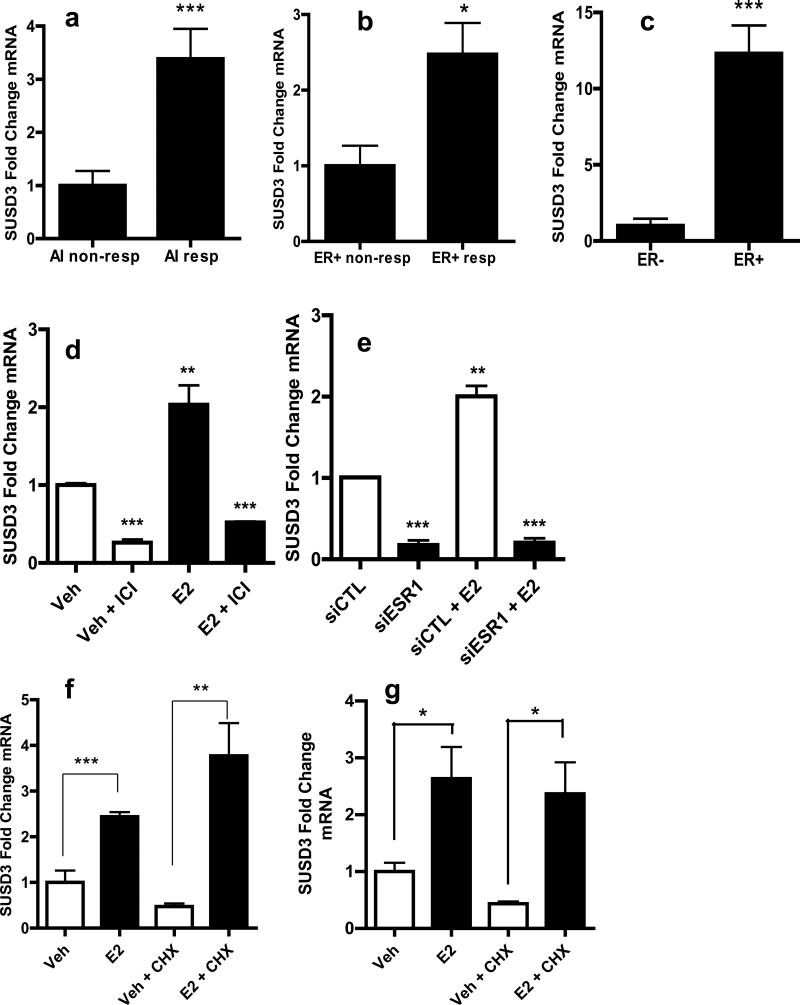
Forty-nine high-quality mRNA samples were available for validation (27 responders and 22 non-responders). SUSD3 mRNA levels were 3.38-fold higher in responders (p=0.0004; Figure 1A). In clinical practice, a tumor's ERα-positive status is often used as a predictive marker of AI responsiveness8-10; therefore, we assessed the correlation between SUSD3 and ERα expression. Among ERα-positive tumors, AI-responders (n=27) had 2.48-fold higher levels of SUSD3 mRNA compared with non-responders (n=15; p=0.0118; Figure 1B). In analyzed tumors, SUSD3 mRNA was 12.3-fold higher in ERα-positive (n=42) compared to ERα-negative (n=7) tumors (p=0.0009; Figure 1C). In vivo, there was a robust positive correlation between ERα and SUSD3 expression (R=0.82), suggesting strong association (Supplemental Figure 1A). Expression of SUSD3 was assessed in MCF7, T47D, MDA-MB231, and SKBR3 breast cancer cell lines and Ishikawa endometrial adenocarcinoma cell line. Highest SUSD3 expression was observed in ERα/PR-positive cell lines (Ishikawa, T47D, MCF7), whereas the two ERα/PR-negative cells (MDA-MB231, SKBR3) contained the lowest levels (Supplemental Figure 1B).
Figure 1.
SUSD3 expression is up-regulated in AI non-responders and regulated by ERα. (a) Real-time PCR was performed on a total of 49 patient tumor samples. SUSD3 mRNA expression in AI responders (n=27) vs. non-responders (n=22). (b) SUSD3 mRNA expression in ERα positive/AI responders (n=27) vs. ERα positive/AI non-responders (n=15). (c) SUSD3 mRNA expression in ERα positive (n=42) vs. ERα negative tumors (n=7). Results are expressed as the means ± SD from triplicate experiments. (d) MCF7 cells were treated with vehicle control (EtOH), 100-nM E2, 10-μM ICI 182780, or both. SUSD mRNA was quantified by RT-PCR. Cells were pretreated with ICI 182780 1h prior to the addition of E2. (e) siRNA-mediated knockdown of ERα followed by treatment with vehicle control or 100-nM E2. *, p< 0.05; **, p< 0.01; ***, p< 0.001. (f) SUSD3 mRNA expression in MCF7 cells and (g) T47D cells after treatment with vehicle control, 100-nM E2, 5-μM cycloheximide (CHX), or both. (h) ChIP assay performed with ERα antibody in MCF7 cells treated with vehicle (EtOH) or 100-nM E2 for 30min. Amplicons 10 kb upstream of SUSD3 exon 1 are shown. Mock ChIP was performed with IgG as a control. Error bars represent mean ± SD. TFF1 was used as a positive control and its mRNA expression after E2 treatment was compared to control. ***, p< 0.001. All experiments were performed in triplicate.
ERα Directly Regulates SUSD3 Expression
Given the strong correlation observed between ERα and SUSD3 expression, we investigated whether ERα and its ligand E2 regulate SUSD3. E2 induced SUSD3 mRNA in MCF7 cells in a time- and dose-dependent fashion, with the highest levels seen after a 24h treatment (Supplemental Figure 1C,D). The antiestrogen ICI 182780 inhibited the stimulatory effect of E2 on SUSD3 expression (p< 0.0001; Figure 1D). ERα knockdown led to the significant down-regulation of SUSD3 mRNA that could not be restored with E2 treatment (Figure 1E). The addition of translational inhibitor cycloheximide (CHX) had no effect on E2-stimulated SUSD3 expression in MCF7 and T47D cells, suggesting that induction of SUSD3 mRNA is a direct effect of ERα and does not require new protein synthesis (Figure 1F,G).
We further examined the role of ERα in SUSD3 regulation using ChIP assays. SUSD3 transcription start site in MCF7 cells was found using 5′ RACE, likely transcription start site (TSS) being in a region -21bp upstream of Exon 1 of isoform 1. The mapped SUSD3 TSS was consistent with its annotated TSS16. After 30min of E2 treatment, ERα is recruited to a distinct region approximately 5,000bp upstream of the SUSD3 TSS (Figure 1H). Transcription Element Search Software (TESS) identified four half ERE sequences in this region17. Together, our data strongly suggest that ERα regulates SUSD3 expression by interacting directly with its regulatory region in an E2-dependent manner.
Effect of SUSD3 Knockdown on Breast Cancer Cells Growth
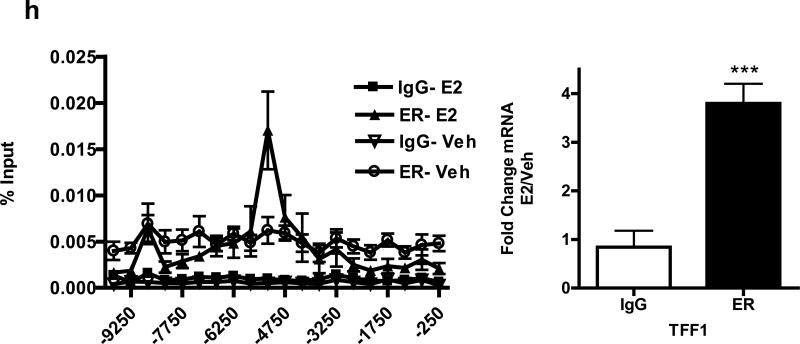
We transfected MCF7 and T47D cells with control or selected SUSD3 siRNAs; qRT-PCR and western blot demonstrated that two distinct siRNAs (oligo4 and 5) robustly ablated SUSD3 expression (Supplemental Figures 2A-D). The ORF-targeted oligo4 was used for all work done, whereas the 3’UTR targeted oligo5 was used to confirm all the major findings (see Supplemental figures). SUSD3 knockdown led to dramatic cell growth arrest after siRNA transfection of MCF7 and T47D cells (Figure 2A, Supplemental 3A). This effect was not due to an increase in cell death, as apoptosis levels, measured by TUNEL staining and nuclear morphology changes, remained the same in control and SUSD3-ablated MCF7 and T47D cells (Supplemental Figure 3B). To assess effects of SUSD3 on the cell cycle, we used a DNA binding dye to resolve cells into three groups (G0/G1; S; G2/M) and chromatin staining with phosphorylated histone H3, which specifically determines the percentage of cells in M phase. In control cells, E2 treatment led to a significant decrease in the percentage of cells in G0/G1 phase with a concomitant increase in the percentage of cells in S, G2 and M phases (Figures 2B-E). SUSD3 knockdown eliminated the effect of E2 in all phases of cell cycle (Figures 2B-E).
Figure 2.
SUSD3-knockdown MCF7 cells show cell cycle defects. (a) Cell counts of control (siCTL) or SUSD3 siRNA-transfected (oligo 4) MCF7 cells (siSUSD3) were performed at 24, 48, and 72 hours post-transfection using a hemocytometer. **, p< 0.01. (b) MCF7 cells were transfected with control or SUSD3 siRNA (oligo 4) for 72 hours and the percent of cells in the indicated phases of the cell cycle were determined by FACS. E2 treatment led to a decrease in the percentage of cells in the G0/G1 phase (83.37% vs. 80.97, vehicle vs. E2, p=0.0533). (c) E2 treatment caused a significant increase in control cells entering S phase (6.09% vs. 13.8%, vehicle vs. E2, p<0.001). siRNA knockdown of SUSD3 abrogated the effect of E2 treatment (6.9% vs. 7.89%; vehicle vs. E2, p=0.36). (d) E2 significantly increased the number of control cells in G2 phase, an effect that was blunted after SUSD3 knockdown. (e) E2 robustly increased the M-phase fraction, whereas SUSD3 knockdown significantly decreased the fraction of cells in the M phase, from 0.6% to 0.35% (p=0.012). The percentage of E2-treated SUSD3-ablated cells entering M phase was significantly lower than E2-treated control cells (2.13% vs. 0.55%; p<0.0001). Results are reported as mean ± SD from triplicate experiments.
SUSD3 Localizes to the Plasma Membrane and Promotes Cell-Cell Adhesion
To study functions of SUSD3 we investigated its cellular localization. Confocal imaging with E-cadherin shows GFP-tagged SUSD3 expressed in MCF7 cells and prominently localized to cell-cell borders (Figures 3A,B). This raised the possibility that SUSD3 may play a role in cell-cell adhesion.
Figure 3.
SUSD3 localizes to the MCF7 cell plasma membrane. (a) Control GFP-empty vector was transfected into MCF7 cells. Overlay shows localization of nuclear DAPI and E-cadherin to the plasma membrane (stained in red). The control GFP-construct is shown to be diffusely distributed in the cell. (b) A GFP-tagged ORF of human SUSD3 was transfected into MCF7 cells. Co-localization was performed with E-cadherin (stained in red). Nuclei were stained with DAPI. SUSD3 appears in green. The overlay shows the localization of nuclear DAPI and co-localization of SUSD3 and E-cadherin on the cell membrane.
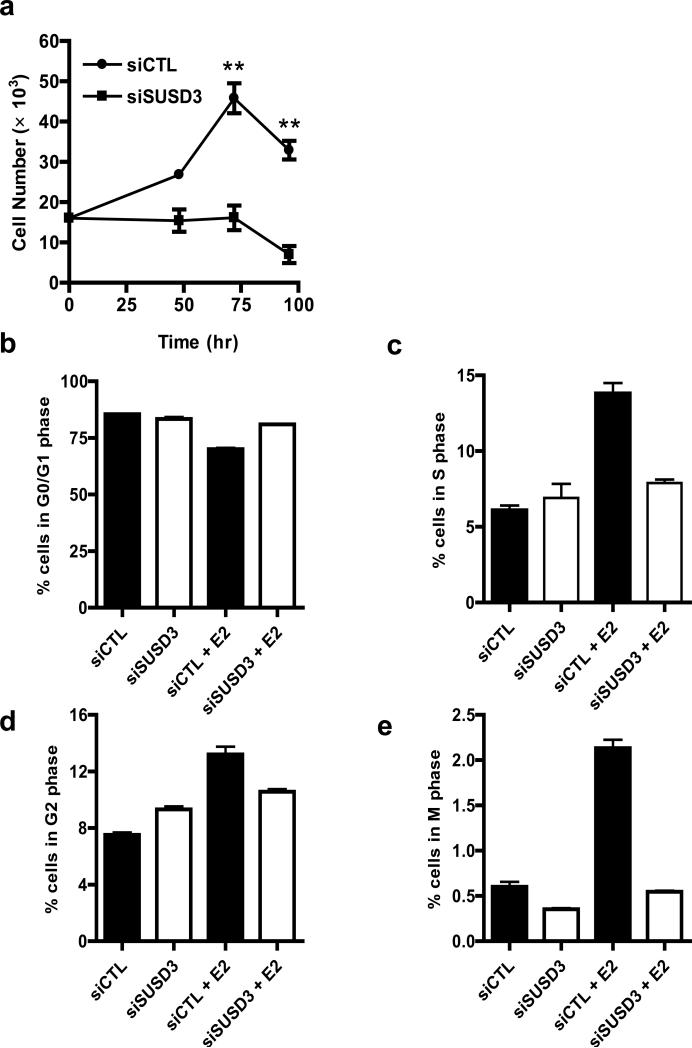
We tested cell-cell adhesion strength in control and siSUSD3 MCF7 cells using dispase assay. Whereas dispase treated monolayers of control cells were moderately disturbed upon mechanical stress producing on average 30 fragments, SUSD3-ablated MCF7 monolayers completely disintegrated, producing over 400 fragments in each experiment (Figures 4A,B). This finding indicated that SUSD3 is critical for promoting cell-cell adhesion in MCF7 breast cancer cells.
Figure 4.
SUSD3 knockdown impairs cell-cell adhesion in MCF7 cells. (a) Cell-cell adhesion (dispase) assay. Number of cell fragments from control (siCTL) or SUSD3-deficient (siSUSD3 oligo 4) cultures counted with a dissecting scope after 30min incubation with 2.4 U/mL dispase. ***, p < 0.001. (b) Representative light microscopy image of cell fragments after the dispase assay (siCTL vs. siSUSD3). (c) Hanging drop aggregation assay. Number of fragments observed in siCTL vs. siSUSD3 oligo 4 prior to mechanical stress. **, p < 0.01. (d) Number of cells in clusters of 1-20, 21-100, >100 in hanging drops of non-triturated cells and (e) triturated cells. The percentage of cells in clusters is shown from experiments performed in triplicate.
To study cell-cell adhesion independent of interactions with an underlying substrate, we performed an adhesion assay of cells in suspension, followed by exposure of aggregates to shear stress. SUSD3-deficient cells had diminished ability to form adhesive clusters compared to control cells (Figures 4C,D). After applying shear stress, >25% of control cells remained in large clusters (>100 cells), which were virtually absent in the SUSD3-deficient cells (Figure 4E). Approximately 70% of SUSD3-deficient cells were individual or in very small (<20 cells) clusters, indicating that SUSD3 expression increases cell-cell adhesive strength and resistance of MCF7 breast cancer cells to shear stress independent of cell-substrate interactions.
SUSD3 Deficiency Alters MCF7 Breast Cancer Cell Morphology
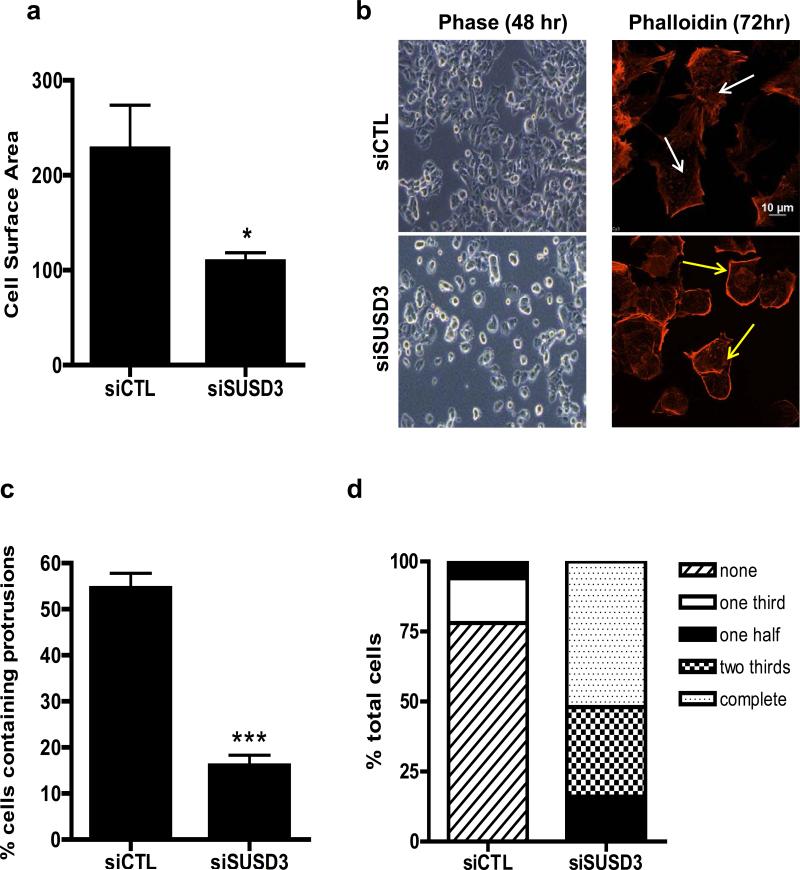
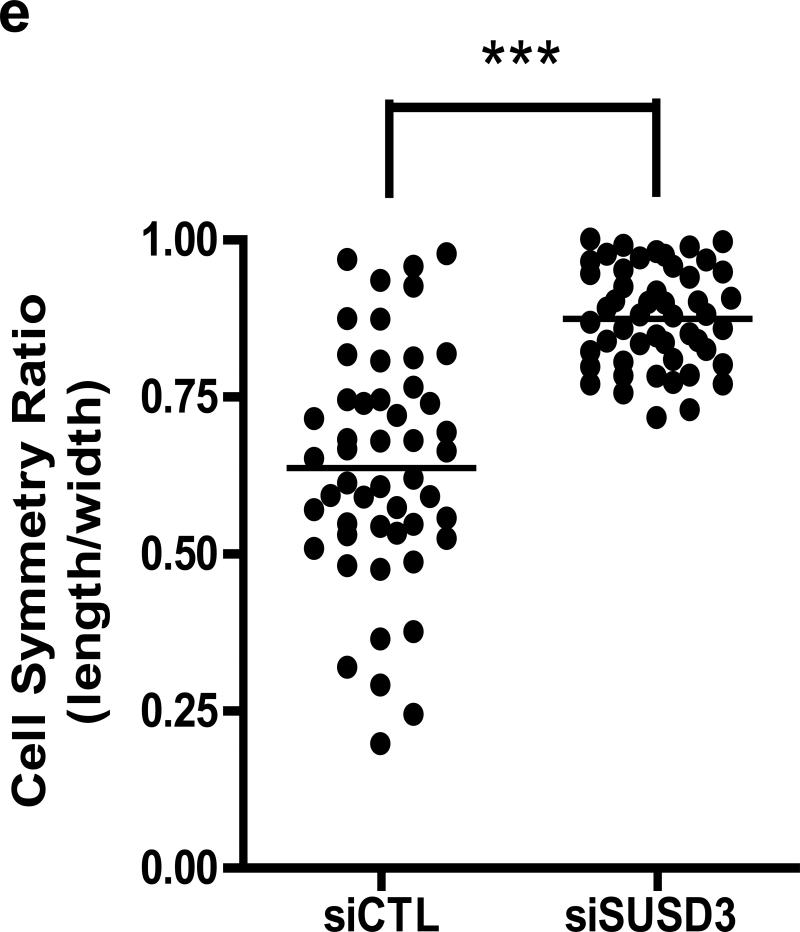
Studies have demonstrated the role of E2 and ERα in the formation of actin-rich protrusions such as filopodia and lamellipodia in breast cancer cells18-20. We therefore assessed the effect of SUSD3-knockdown on MCF7 cell morphology. Phase contrast and confocal imaging revealed that, compared with controls, SUSD3-deficient cells had a significantly lower surface area (Figure 5A). Knockdown cells displayed a smaller and rounder phenotype, prompting us to speculate that they may have spreading defects (Figure 5B, Supplemental Figure 4A). SUSD3-ablated cells demonstrated over 3-fold decrease in actin-rich protrusions and stress fibers (Figure 5B,C; Supplemental Figure 4B). Whereas all of the knockdown cells exhibited thickened cortical actin on at least 50% of their cell borders, almost none of the control cells did, with 78% of them having no cortical acting thickening at all (Figures 5B,D, Supplemental Figure 4B). Cell symmetry measurements indicated a prominent shift towards circular phenotype of SUSD3-ablated cells while control cells displayed a varied morphology (Figure 5E).
Figure 5.
SUSD3-knockdown alters MCF7 cell morphology. (a) siSUSD3 (oligo 4) cells displayed lower cell surface area compared to control cells (109.08 ± 16.25 vs. 227.99 ± 45.93, siSUSD3 vs. siCTL, p<0.05). (b) Early morphological changes in MCF7 cells observed via phase contrast microscopy 48h after SUSD3 siRNA oligo 4 transfection compared to control (left panels). Confocal images were taken of SUSD3-knockdown and control MCF7 cells at 72h post-transfection (right panels). Immunofluorescent staining of actin with phalloidin was performed to assess stress fiber formations (white arrows). Thickened cortical actin formations (yellow arrows) were noted on SUSD3-knockdown cells. (c) A significantly lower fraction of siSUSD3 cells (16%) contained protrusions compared with siCTL cells (54.5%; p< 0.0002). Results are reported as mean percentage ± SD of triplicate experiments. ***, p< 0.001. (d) Diagram showing the percentage of MCF7 cells (n≥50) treated with control or SUSD3 siRNA with cortical actin staining absent (no ring), or present over one third (one third), one half (one half), two thirds (two thirds) or whole cell border (complete). (e) Cell symmetry ratio measurements of control (siCTL) and SUSD3-knockdown (siSUSD3 oligo 4) MCF7 cells. A total of 50 cells were measured in each population. The closer the ratio to 1, the more circular or cuboidal the cell. ***, p < 0.001.
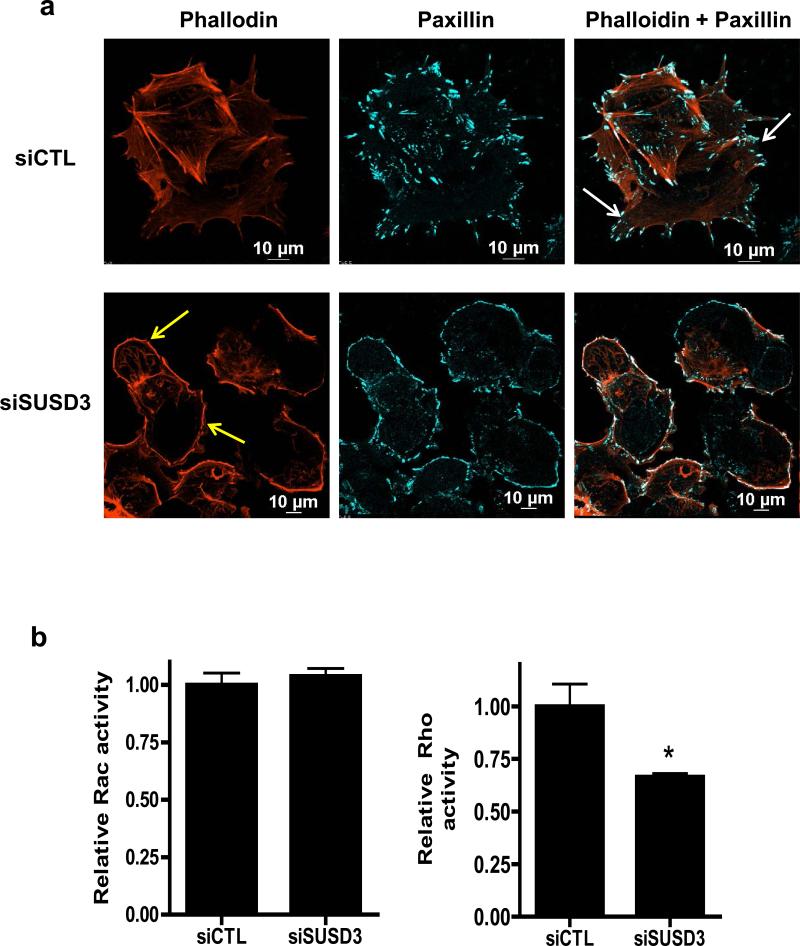
We also examined focal adhesions in control and knockdown cells. Paxillin-containing focal adhesions were observed at edges of SUSD3-ablated cells coinciding with thickened cortical actin (Figure 6A, Supplemental Figure 4B). Moreover, large pointed focal adhesions anchoring stress fibers, prominent in control cells, were almost completely absent from SUSD3-knockdown cells21 (Figure 6A). Rescue experiment demonstrated knockdown specificity with co-expression of a siRNA-resistant construct. SUSD3-GFP expressing cells were resistant to SUSD3-siRNA induced morphological changes (Supplemental Figure 4C).
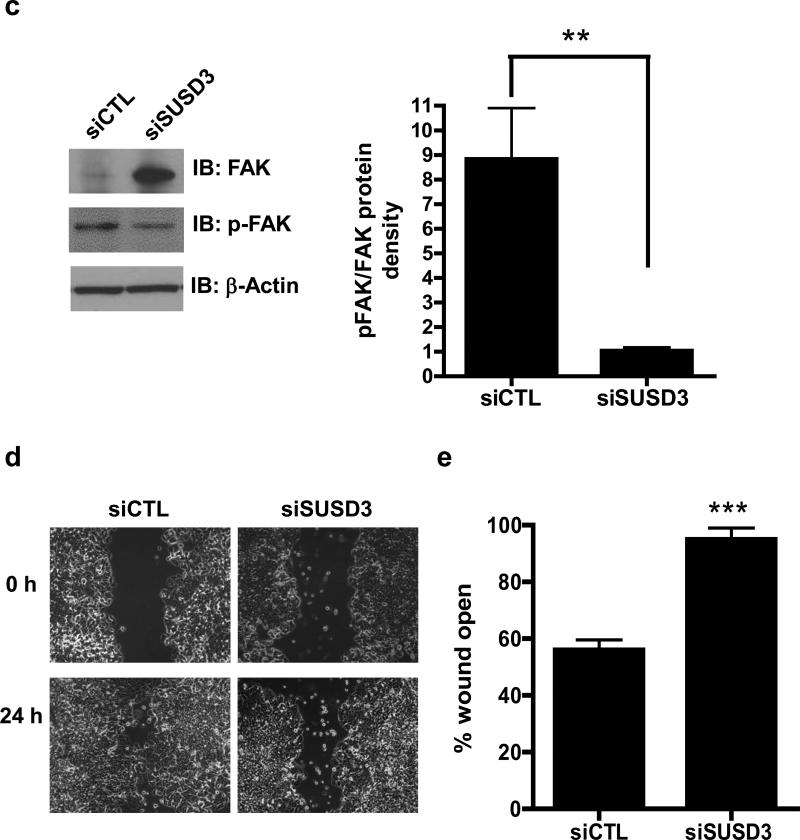
Figure 6.
SUSD3 knockdown deregulates FAK/Rho-mediated focal adhesion dynamics. (a) Immunofluorescent staining of control (siCTL) and SUSD3-knockdown (siSUSD3 oligo 4) MCF7 cells was performed after a 72h transfection with Alexa-568 phalloidin-actin and Alexa-647 paxillin. White arrows in siCTL cells point to focal adhesions anchoring to stress fibers. Yellow arrows in siSUSD3 cells point to thickened cortical actin. (b) Rac and Rho-activation assays. (c) Immunoblot analysis of FAK and phosphorylated (activated) FAK. Control (Lane 1) vs. siSUSD3 oligo 4 (Lane 2) transfected MCF7 cells. β-actin was used as internal control. Ratio of phosphorylated (activated) FAK and total FAK in control (siCTL) and SUSD3-knockdown (siSUSD3) cells. Protein densities were normalized to β-actin. **, p< 0.01. (d) Scratch wound healing assay. MCF7 cells were imaged at time 0 and 24h. (e) 24h post-wound creation, 56% of the wound remained open in MCF7 CTL cells vs. 95% in MCF7 siSUSD3 oligo 4 cells (p=0.0002). Experiments in panels (b and c) was performed in triplicate, with graph results reported as mean percentage ± SD. *, p<0.05, **, p < 0.01. Experiments performed in (d) were replicated 6 times, with representative images shown.
Changes observed in stress fibers and focal adhesions of SUSD3-knockdown cells were suggestive of changes in Rho GTPase signaling, which has been implicated in the establishment of cell-cell contacts and cell matrix interactions22,23. Therefore, we investigated levels of active GTP-bound Rho and Rac in control and SUSD3-knockdown cells. Interestingly, no significant differences in Rac activity were observed; however, Rho activity was significantly lower in SUSD3-knockdown cells compared to controls (Figure 6B). Since Rho activity is required for formation of stress fibers and focal adhesions24, the decrease in its activity could be linked to the observed disruption of stress fibers and ventral focal adhesions in SUSD3-ablated cells.
We next analyzed focal adhesion kinase (FAK) activity in SUSD3-knockdown cells. FAK is a cytoplasmic protein tyrosine kinase whose activity plays an important role in integrin-mediated signal transduction pathways and has been shown to be important in breast cancer progression, invasion, and dynamic turnover of focal adhesions25,26. Expression levels of FAK were higher in SUSD3-knockdown cells than in controls; however, phosphorylated (active) FAK levels were lower than in controls, suggesting a large decrease in the portion of activated FAK in SUSD3-knockdown cells (Figure 6C). SUSD3-ablated cells demonstrated >8-fold decrease in activated FAK (Figure 6C). Decreased FAK activity has been demonstrated in cells displaying a thickened cortical actin phenotype21. Taken together, these data suggest that loss of SUSD3 may interfere with normal FAK/Rho-mediated focal adhesion dynamics in MCF7 and T47D breast cancer cells27.
Loss of SUSD3 Impedes Breast Cancer Cell Motility
Cell migration is a critical step in cancer invasion and metastasis, and FAK, paxillin, and Rho GTPases have been implicated in metastasis23,25,26,28-31. Localization of SUSD3 to the plasma membrane and its effects on actin-rich cell protrusions, focal adhesions, and Rho and FAK activity point to a role of SUSD3 in cell migration. We therefore performed wound healing experiments with MCF7 cells transfected with control or SUSD3 siRNA (Figure 6D). To prevent the confounding effect of cell proliferation, mitomycin C, a potent inhibitor of DNA synthesis, was added to the media. Measured as percentage of wound left open after 24h, SUSD3-knockdown cells showed 2-fold lower motility compared with control cells (Figure 6E, Supplemental Figure 5A). A continued deficiency in motility in SUSD3-knockdown cells was observed up to 72h after the scratch wound (Supplemental Figure 5B). Results were replicated in T47D cells (Supplemental Figure 5C).
DISCUSSION
AIs occupy a central role in the endocrine treatment of breast cancer by blocking the metastatic spread of hormone-dependent breast cancer1-7,32-34. Despite demonstrated superiority over TAM, response rates of AIs remain about 50% in advanced breast cancer3,4,12-14. Genome-wide searches using clinical samples from patients treated with AIs offer hope in identifying new markers that may better predict responsiveness to therapy35,36. To date, however, a biochemically and functionally defined in vivo marker for AI responsiveness has not been reported8-10. Using a similar genome-wide approach, our lab profiled the mRNA from tumors of PMW who developed breast cancer and were subsequently treated with an AI for recurrent and metastatic disease. Hormone receptor status was unknown at time of treatment, but 10-year survival data after AI treatment was available to assess responsiveness. We found that SUSD3 expression was 3.38-fold higher in patients who responded to AI therapy.
SUSD3 is located on chromosome 9 and has three isoforms37. Literature regarding SUSD3 is limited and its function is unknown. SUSD3 expression has been reported in ERα-positive breast tumors38 with decreased expression reported in aggressive malignant tumors (triple-negative status, endocrine insensitivity, and short-term survival)39. Recent literature has linked its homolog, SUSD2, to increased invasion of breast cancer cells. SUSD2 contains a transmembrane domain and functional domains inherent to adhesions molecules40.
Here, we demonstrated that SUSD3 is regulated by E2 and ERα in MCF7 breast cancer cells, and that siRNA knockdown of SUSD3 abolishes the mitogenic effects of E2 on these cells. It is possible that SUSD3 mediates some of the mitogenic effects of E2 via enhancing the progression of malignant cells from the G2 into the S and M phases. Cancer cells often have alterations in activity or expression levels of genes that control cell cycle, which has been proposed as a mechanism for resistance to endocrine therapy41.
We found that SUSD3 is located at cell-cell borders, and that its loss leads to changes in cell-cell adhesion, cell morphology, and migration. Studies have demonstrated the role of estrogens and ERα in stimulating breast cancer cell migration via activation of various signaling pathways regulating cell morphology and motility18,19,42,43. E2 treatment of breast cancer cells leads to formation of focal adhesion complexes, filopodia, lamellipodia, and pseudopodia42,43. Migrating cells must coordinate extension of the leading edge of the cell and retraction of the back edge which require turnover of focal adhesion complexes29,30,44. During the process of cell spreading, integrin-containing filopodia form initial adhesion sites with subsequent recruitment of FAK and paxillin leading to formation of mature focal adhesions29,30,44. Phosphorylated FAK is required for organization of the leading edge of migrating cells and for focal adhesion disassembly at the trailing edge of the cell25,28,30. FAK is important in breast cancer progression, invasion, and focal adhesions26,27.
While our data demonstrated an increase in total FAK expression, phosphorylated FAK was significantly decreased in SUSD3-knockdown cells, suggesting a defect in FAK activation in these cells. The ratio of active to inactive FAK is greatly reduced in SUSD3 deficient cells indicating a functional loss of FAK signaling in these cells. It is possible that the increase of total FAK is an attempt of cells to compensate for the loss of FAK activity due to SUSD3 loss. In wound healing experiments and morphological studies, SUSD3-knockdown showed decreased cell motility, reduced formation of actin-rich cell protrusions, paxillin containing focal adhesions, stress fibers, and a reduction in overall cell surface area. Together, the data suggest that SUSD3 deficiency leads to a defect in focal adhesion and stress fiber formation; thereby inhibiting cell spreading and subsequent migration22.
The breakdown of cell adhesion is directly implicated in carcinogenesis, whereas deregulation of intercellular adhesion has been linked to the onset of breast cancers among other solid tumors45-47. For example, E-cadherin loss and inappropriate expression of non-epithelial cadherins have been implicated in invasion and metastasis47. Here we demonstrated that SUSD3 is essential for cell-cell adhesive strength. In SUSD3-deficient MCF7 cells, mechanical stress resulted in complete disintegration of cell-cell adhesions. This result was further validated in our hanging drop test which revealed the inability of SUSD3-deficient cells to form large cell clusters.
The Rho GTPases are key regulators of actin assembly and control the formation of stress fibers, filopodia, and lamellipodia. Moreover, they regulate focal adhesion assembly, cell motility, polarity, and cell cycle progression. Alterations in Rho GTPase signaling have been implicated in cancer cell invasion22,24,31,32. In this study, confocal imaging revealed thickened cortical actin coinciding with paxillin-containing focal adhesions at the edges of the SUSD3-knockdown cells. Furthermore, the diminished number of both stress fibers and large basal focal adhesions suggested disruption in Rho GTPase signaling compared to control cells. GLISA confirmed significantly lower Rho GTPase activity in SUSD3-deficient cells compared with controls. Taken together with the decrease in FAK activation discussed above, our data indicate deregulation of FAK/Rho-mediated focal adhesion dynamics in SUSD3-deficient cells.
In conclusion, we report a novel estrogen-mediated ERα-regulated gene, SUSD3, which plays an important role in E2-mediated breast cancer cell proliferation, adhesion and migration. We suspect that in ERα-positive tumors, high levels of SUSD3 induced by E2 support breast cancer cell motility. In the presence of an AI, estrogen levels are significantly decreased, leading to reduced levels of SUSD3 and thereby diminished motility. Multiple signaling pathways that facilitate the invasion of extra-mammary tissues and underlie the metastatic nature of breast cancer cells have been investigated to help aid in the development of treatments for breast cancer invasion. As cell migration is a key first step in the metastatic process, further experiments need to be performed to better delineate the interplay between SUSD3, Rho GTPases, and numerous signaling molecules, including paxillin and FAK, which are involved in the complex steps of cell migration, morphology, and cytoskeleton dynamics. SUSD3 may serve as a future diagnostic and therapeutic target in the treatment of breast cancer. Its subcellular localization to the cell surface makes it an attractive therapeutic target.
MATERIALS AND METHODS
Cells and Tissues
RNA samples from 50 primary tumors from PMW who developed breast cancer in 1990-1995 were obtained from Royal Marsden Hospital, UK. Patients underwent surgery and were treated with adjuvant TAM. All patients subsequently developed recurrent locally advanced or metastatic disease and were then placed on either anastrozole or letrozole. Hormone receptor status of the tumors had not been determined before administration of adjuvant therapy because this treatment was experimental at that time15. ER/PR status of these tumors were determined by immunohistochemistry after treatment was completed.
Human breast cancer cells, MCF7, T47D, endometrial adenocarcinoma Ishikawa cancer cells (all ERα/PR+), MD-MBA231 (ERα/PR−), and SKBR3 (ERα/PR−, HER2+), were obtained from ATCC and cultured as described48-50.
For experiments evaluating estradiol-β (E2), ICI 182780 (anti-estradiol), and cycloheximide (CHX) responsiveness, cells were cultured in charcoal-stripped FBS for 48h followed by overnight serum starvation prior to treatment with E2 (100nM) for 24h. For ICI and CHX experiments, cells were treated 1h with either compound (10μM) prior to the addition of E2. Time course and dose response experiments were performed to optimize gene expression.
Microarray and Data Analysis
mRNA microarray analysis was performed using an ABI 1700 chemiluminescent microarray analyzer (Life Technologies, Grand Island, NY) and data was normalized by the quantile normalization procedure using the bioconductor package Affy (Affymetrix, Santa Clara, CA). Gene expression data sets containing information on fold change and p-values from two sample t-tests comparing Al responders to non-responders were determined. We analyzed differentially expressed genes with fold change >1.5 and p-values <0.05. 32,878 probes were used in the microarray. Two criteria, flag counts ≤10, and coefficients of variation >0.07 were used to select probes whose transcript levels were likely to be above background noise and vary across tumor samples. This filtering procedure resulted in 5197 probes for data analysis. We observed 524 genes with p-value <0.05 and false discovery rate of 37%, which is more than expected by chance alone51 Genes were verified by quantitative real-time PCR (qRT-PCR).
RNA Extraction and qRT-PCR
Total RNA was extracted using TRIzol (Life Technologies). cDNA was prepared with qScript cDNA SuperMix (Quanta Biosciences, Gaithersburg, MD). Expression of mRNA was measured by qRT-PCR using ABI 7900 Sequence Detection and SYBR green (Life Technologies) in triplicate. Gene expression was normalized to GAPDH (Supplemental Table 1). SUSD3 primers were obtained from Qiagen (Valencia, CA).
siRNA Transfection
MCF7 and T47D cells were transfected with ERα, SUSD3, and non-targeting control siRNA (Thermo Scientific, Waltham, MA) using Lipofectamine RNAiMax reagent (Life Technologies). SUSD3 siRNA target sequences employed are shown in Supplemental Table 1.
Flow Cytometry
Control and siRNA transfected SUSD3 cells were serum starved prior to vehicle or E2 treatment for 24h. Cells were prepared as previously described52,53, with minor modifications. Cells were fixed in 1% formaldehyde, permeabilized with methanol, and immunostained with phosphorylated histone H3 Alexa Fluor 647 antibody (Beckman Coulter, Brea, CA). DNA was counterstained with propidium iodide solution and the mitotic index was determined by flow cytometry as percentage of phosphorylated histone H3 positive cells in the population.
Generation and purification of SUSD3 antibody
A custom rabbit polyclonal antibody was generated against SUSD3 (AA 151 to 170). The peptide sequence, CKDEDLETVQAAYLGLKHFNK, was custom made by Princeton BioMolecules (Langhorne, PA). A cysteine residue was added to the N-terminus for conjugation purposes. Animal immunization, serum collection, and affinity purification were performed by Covance (Princeton, NJ). ELISA was performed to monitor titers and confirm animals’ responses to the antigen. Specificity of the affinity purified SUSD3 antibody was confirmed by immunoblotting.
Rescue Experiment
The silencing resistant SUSD3-GFP expression construct was generated by introducing three silent mutations within the siRNA targeting sequence of pCMV6-AC-SUSD3-GFP. Site directed mutagenesis was performed using QuikChange Lightning Site Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA). Non-silenceable SUSD3 was subcloned as a C-terminal EGFP fusion protein into the retroviral expression vector pBABE-puro. Sequences of all constructs were verified by DNA sequencing. VSV-G pseudotyped retrovirus was produced in 293T/17 cells and used to transduce MCF7 cells as described elsewhere54. Stable pools of MCF7 cells expressing SUSD3-EGFP or EGFP alone were selected using 2μg/ml puromycin beginning two days post-transduction. For rescue experiments, the siRNA resistant pCMV6-AC-SUSD3-GFP construct was co-transfected into MCF7 cells together with the oligonucleotides used shown in Supplemental Table 1. Alternately, GFP-only and SUSD3-GFP stable cell lines were transfected with SUSD3 and control-siRNA.
Immunoblotting
Immunoblotting was performed as described previously48 with samples resolved on 4-12% Ready Gel precise Gels (BioRad, Hercules, CA), transferred onto PVDF membranes, and probed using primary antibodies: anti-phosphorylated FAK (Millipore, Billerica, MA), anti-FAK (Cell Signaling, Danvers, MA), and anti-SUSD3. Equal loading was confirmed using anti-β-actin (Sigma Aldrich, St. Louis, MO). Anti-mouse and rabbit IgG secondary antibodies were used (Cell Signaling). Western blots were developed using Amersham ECL Plus (GE Healthcare, San Francisco, CA) and SuperSignal West Femto Chemiluminescent substrate (Thermo Scientific). Quantification was performed using Image J (NIH, Bethesda, MD).
5′-Rapid Amplification of Complementary DNA Ends (5′-RACE)
The SUSD3 transcription start site (TSS) was determined by RNA ligase-mediated 5’ end cDNA amplification with First-Choice RLM-RACE (Life Technologies) following manufacturer's protocols. RACE products were cloned into the pCR-TOPO TA vector and sequenced.
Chromatin Immunoprecipitation-PCR (ChIP)
ChIP was conducted as described previously, with minor modifications55. Sonicated samples were incubated overnight with ERα antibody (Santa Cruz Biotechnology, Santa Cruz, CA) and immunoprecipitated with Dynal magnetic beads (Life Technologies). Mock precipitation with pre-immune IgG was performed. For quantitative detection of retained DNA, qRT-PCR was performed in triplicate. SUSD3 gene primers were designed that span 10kb upstream of exon 1. TFF1 was used as a positive control (Supplemental Table 1).
Confocal Imaging
SUSD3 localization experiments were performed in MCF7 and T47D cells plated on glass coverslips. After allowing the cells to attach overnight, they were transfected with a GFP-tagged ORF clone of SUSD3 (OriGene, Rockville, MD) using FuGENE HD transfection reagent (Promega, Madison, WI) according to manufacturer's protocol. Images were taken with the Zeiss LSM 510 Meta laser scanning confocal microscope and analyzed with LSM Image Browser 4.2 (Carl Zeiss). siRNA transfected cells were fixed with formaldehyde, treated with 0.05% IGEPAL CA-630 (Sigma), washed, and sequentially overlaid with primary ((anti-phosphorylated paxillin (Millipore), rhodamine phalloidin, and DAPI nuclear stain (Life Technologies)), E-cadherin (gift from M. Takeichi and O. Abe, Kobe, Japan) and secondary antibodies (Alexa 568 goat anti-mouse and Alexa-647 goat anti-rabbit IgG (Life Technologies)). Images were taken with a Nikon A1 laser scanning confocal microscope and analyzed with NIS-Elements software (Nikon).
Cell Surface Area and Cell Symmetry Ratio Measurement
Subconfluent control or SUSD3-depleted MCF7 cells were imaged 72h post-transfection. Surface area of individual cells was determined using Image J lasso tool. A minimum of 50 cells per experiment were measured and the average surface area was calculated. Cell symmetry ratio was determined by taking the ratio of two perpendicular diameters of each cell, with higher value used as the numerator.
Mechanical Strength Assay
Confluent control or SUSD3 siRNA-transfected MCF7 cells in 6-well plates were washed with PBS and incubated with 2.4U/ml dispase (Roche) for 30min at 37°C. Released monolayers were fixed by formalin and fragments were counted using an MZ6 dissecting scope (Leica, Germany) as described previously56 imaged with a Hamamatsu Orca digital camera and analyzed using MetaVue imaging software (Universal Imaging, Downingtown, PA). Under experimental conditions where fragmentation was excessive, a maximum of 400 fragments was counted.
Hanging Drop Aggregation Assay
Aggregation assay was performed as described previously57, with modifications56 20-μl drops of cell suspensions (4000 cells) were seeded onto inner surfaces of 35-mm culture dish lids and cultured for 20h. To examine the ability of cells to form aggregates, culture dish lids were inverted, and hanging drops were flattened with coverslips for imaging. To examine the adhesive strength of cellular aggregates, parallel cultures were triturated 10 times through a 20-μl pipette tip. Five random fields of phase-contrast images from each drop were acquired using a Zeiss Axiovert 200 inverted microscope with a Zeiss Axiocam camera and Zeiss Axiovision software. Total number of cells in clusters of 1-20, 21-100, or >100 cells was counted from triplicate hanging drops; percentage of cells in the clusters and total number of fragments were determined.
Rho/Rac Activation Assays
RhoA- and Rac-GTP levels were detected using colorimetric GLISA activation assays (Cytoskeleton, Denver, CO), according to manufacturer's protocol. Signal produced by the detection reagent, proportional to the amount of Rho- or Rac-GTP, was detected by measuring absorbance at 490nm using a Synergy 2 plate reader (BioTek, Winooski, VT). Constitutively active Rho and Rac were used as positive controls.
Scratch Wound Assay
MCF7 and T47D cells were transfected for 72h with control or SUSD3 siRNAs in 6-well plates. Cell monolayers were wounded with a 20-μl pipette tip, washed and submerged in media with or without 40μg/ml mytomycin C to prevent cell division. Cells were imaged immediately, 24, 48, and 72h after wounding. Percentage wound closure was determined using Image J software.
TUNEL Assay
For TUNEL assay, MCF7 and T47D cells were cultured on glass coverslips. Cells were fixed, permeabilized and TUNEL assay was performed using the ApopTag Red detection kit (Chemicon, Inc.) following manufacturer's protocol. Samples were counterstained with DAPI before mounting. Cells were visualized using fluorescent microscopy.
Statistical Methods
Cell culture experiments were carried out in minimum 3 replicates. Values are given as mean with error bars indicating standard deviations. Statistical analyses were conducted using GraphPad Prism software (San Diego, USA). All statistical tests were 2-sided. Student's t-test and Wilcoxon rank sum test for line and bar graph comparisons were performed. Pearson correlation analysis was performed for ERα and SUSD3 expression levels in breast cancer tumor samples. p<0.05 was considered statistically significant.
Supplementary Material
Supplemental Figure 1. (a) Pearson correlation analysis demonstrates a strong positive correlation between ERα and SUSD3. (b) SUSD3 mRNA expression is higher in ERα/PR-positive cell lines. Expression profile in MCF7, T47D, MDA-MB231, SKBR3, and Ishikawa cell lines. (c) Treatment of MCF7 cells with 100-nM E2 demonstrated robust induction of SUSD3 mRNA expression. (d) Time course of MCF7 cells treated with 100-nM E2 demonstrated the highest induction of SUSD3 mRNA expression at 24h. All experiments were performed in triplicate. Results reported as mean percentage ± SD compared to vehicle control (EtOH). *, p<0.05; **, p<0.01; ***, p<0.001.
Supplemental Figure 2. (a) RT-PCR of SUSD3 expression in MCF7 cells transfected with control (siCTL) or SUSD3 siRNA (siSUSD3) in the presence or absence of E2 (100-nM). (b) Trial of 5 different SUSD3 siRNA target sequences. Sequences labeled siSUSD3 oligo 4 and siSUSD3 oligo 5 were most effective in knockdown as verified by RT-PCR. (c) RT-PCR of SUSD3 expression in MCF7 and T47D cells after siSUSD3 oligo 4 and 5 knockdown. Experiments in panel (a), (b), (c) were all performed in triplicate. Results reported as mean percentage ± SD for triplicate experiments. *, p<0.05; **, p<0.01; ***, p<0.001. (d) Immunoblot analysis of MCF7 CTL vs. SUSD3 siRNA oligo 4 demonstrating effective SUSD3 knockdown. Lanes 1-3 were loaded with siCTL in the amount of 18, 14, and 10μg of protein respectively. Lanes 4 and 5 were loaded with siSUSD3 oligo 4 and 5 samples respectively (18μg of protein). Custom SUSD3 antibody was utilized.
Supplemental Figure 3. (a) Cell counts of control (siCTL) or SUSD3 siRNA-transfected (oligo 4 and 5) T47D cells (siSUSD3) were performed at 72 hours post-transfection using a hemocytometer. (b) TUNEL assay demonstrating similar apoptotic levels in control and SUSD3-ablated MCF7 and T47D cells. TUNEL reaction in control and UV-B treated MCF7 and T47D cells are shown in the left two columns. MCF7 and T47D cells treated with siSUSD3 oligo 4 and 5 are shown in the right 2 columns. TUNEL staining appears red. DAPI nuclear stain appears blue. RT-PCR of SUSD3 expression in MCF7 and T47D cells after siSUSD3 oligo 4 and 5 knockdown.
Supplemental Figure 4. SUSD3-knockdown with siSUSD3 oligo 5 alters MCF7 cell morphology. (a) Early morphological changes in MCF7 cells observed via phase contrast microscopy 48h after SUSD3 siRNA transfection compared to control. Western blot of MCF7 cells demonstrating effective SUSD3 knockdown utilizing oligo 5 is shown. (b) Immunofluorescent staining of control (siCTL) and SUSD3-knockdown (siSUSD3) MCF7 cells was performed after a 72h transfection with Alexa-568 phalloidin-actin and Alexa-647 paxillin. (c) Rescue experiment utilizing GFP-only and SUSD3-GFP stably transfected MCF7 cells demonstrated that SUSD3-GFP expressing cells were resistant to SUSD3-siRNA induced morphological changes. Both cell lines were treated with control and SUSD3-siRNA. Phallodin-actin, GFP, and merged confocal pictures were taken.
Supplemental Figure 5. SUSD3 ablation led to decreased MCF7 and T47D breast cancer cell motility. (a) Percentage wound closure was determined and compared between control (siCTL) and SUSD3-knockdown (siSUSD3, oligo 5) MCF7 cells 24h after scratch test. Results are reported as means ± SD from 5 replicate experiments. ***, p< 0.001. Western blot of MCF7 cells demonstrating effective SUSD3 knockdown utilizing oligo 5 is shown. (b) Percentage wound closure in MCF7 control and SUSD3-knockdown cells (oligo 4) from time 0 to 72h after scratch test. (c) Percentage wound closure in T47D control and SUSD3-knockdown cells from time 0 to 72h after scratch test. Western blot of T47D cells demonstrating effective SUSD3 knockdown utilizing oligo 5 is shown. Results are reported as means ± SD from triplicate experiments. *, p< 0.05; **, p< 0.01.
ACKNOWLEDGMENTS
This work was supported by grants from the AVON Foundation (to S.E.B.), a Northwestern Memorial Foundation Dixon Priority Initiative Grant (S.E.B.) and National Cancer Institute (CA122151, to K.J.G.). The imaging work performed at Northwestern University Cell Imaging Facility was supported by NCI CCSG P30 CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center. We thank Satya Khuon for her assistance in our confocal work. We thank Mitch Dowsett, Helen Cotterill, Roger A'Hern, Ben Haynes, and Ian Smith from the Royal Marsden Hospital in London, England, for making the patient samples available.
Financial support: Avon Foundation, Northwestern Memorial Foundation Dixon Priority Initiative Grant, NIH CA 122151
Footnotes
Conflicts of Interest: The authors have nothing to disclose.
REFERENCES
- 1.Anderson WF, Chatterjee N, Ershler WB, Brawley O. Estrogen receptor breast cancer phenotypes in the Surveillance, Epidemiology, and End Results Database. Breast Cancer Res Treat. 2002;76:26–36. doi: 10.1023/a:1020299707510. [DOI] [PubMed] [Google Scholar]
- 2.Osborne CK, McGuire WL. Hormone receptors in primary and advanced breast cancer. Clin Endocrinol Metab. 1980;9:361–368. doi: 10.1016/s0300-595x(80)80038-7. [DOI] [PubMed] [Google Scholar]
- 3.Saranya C, Howes T, Bao T, Sabnis G, Brodie A. Aromatase, aromatase inhibitors, and breast cancer. J Steroid Biochem Mol Biol. 2011;125(1-2):13–22. doi: 10.1016/j.jsbmb.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller WR. Aromatase and the breast: Regulation and clinical aspects. Maturitas. 2006;54(4):335–341. doi: 10.1016/j.maturitas.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 5.Goldhirsch A, Ingle JN, Gelber RD, Coates AS, Thurlimann B, Senn HJ. Thresholds for therapies: highlights of the St Gallen International Expert Consensus on the primary therapy of early breast cancer 2009. Ann Oncol. 2009;20(8):1319–1329. doi: 10.1093/annonc/mdp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith IE, Dowsett M, Yap YS, Walsh G, Lonning PE, Santen RJ, et al. Adjuvant aromatase inhibitors for early breast cancer after chemotherapy-induced amenorrhoea: caution and suggested guidelines. J Clin Oncol. 2006;24(16):2444–2447. doi: 10.1200/JCO.2005.05.3694. [DOI] [PubMed] [Google Scholar]
- 7.Burstein HJ, Prestrud AA, Seidenfeld J, Anderson H, Buchholz T, Davidson N, et al. American Society of Clinical Oncology Clinical Practice Guideline: Update on Adjuvant Endocrine Therapy for Women with Hormone-Receptor-Positive Breast Cancer. J Clin Oncol. 2010;28(23):3784–3796. doi: 10.1200/JCO.2009.26.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson H, Bulun SE, Smith I, Dowsett M. Predictors of response to aromatase inhibitors. J Steroid Biochem Mol Biol. 2007;106(1-5):49–54. doi: 10.1016/j.jsbmb.2007.05.024. [DOI] [PubMed] [Google Scholar]
- 9.Stuart-Harris R, Shadbolt B, Palmquist C, Chaudri Ross HA. The prognostic significance of single hormone receptor positive metastatic breast cancer: an analysis of three randomized phase III trials of aromatase inhibitors. Breast. 2009;18:351–355. doi: 10.1016/j.breast.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Anderson H, Hills M, Zabaglo L, A'hern R, Leary AF, Haynes BP, et al. Relationship between estrogen receptor, progesterone receptor, HER-2 and Ki67 expression and efficacy of aromatase inhibitors in advanced breast cancer. Ann Oncol. 2011;22(8):1770–6. doi: 10.1093/annonc/mdq700. [DOI] [PubMed] [Google Scholar]
- 11.Eiermann W, Paepke S, Appfelstaedt J, Llombart-Cussac A, Eremin J, Vinholes J, et al. Preoperative treatment of postmenopausal breast cancer patients with letrozole: a randomized double-blind multicenter study. Ann Oncol. 2001;12:1527–1532. doi: 10.1023/a:1013128213451. [DOI] [PubMed] [Google Scholar]
- 12.Smith IE, Dowsett M, Ebbs SR, Dixon JM, Skene A, Blohmer JU, et al. Neoadjuvant treatment of postmenopausal breast cancer with anastrozole, tamoxifen, or both in combination: the Immediate Preoperative Anastrozole, Tamoxifen, or Combined with Tamoxifen (IMPACT) multicenter double-blind randomized trial. J Clin Oncol. 2005;23:5108–5116. doi: 10.1200/JCO.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Miller WR. Aromatase inhibitors: prediction of response and nature of resistance. Expert Opin Pharmacother. 2010;(11):1873–1887. doi: 10.1517/14656566.2010.487863. [DOI] [PubMed] [Google Scholar]
- 14.Bernhard J, Thurlimann B, Schimtz SFH, Castiglione-Gertsch M, Cavalli F, Morant R, et al. Defining clinical benefit in postmenopausal patients with breast cancer under second-line endocrine treatment: Does quality of life matter? J Clin Oncol. 1999;17:1672–1679. doi: 10.1200/JCO.1999.17.6.1672. [DOI] [PubMed] [Google Scholar]
- 15.Moy I, Lin ZH, Rademaker F, Reierstad S, Khan S, Bulun S. Expression of estrogen-related gene markers in breast cancer tissue predicts aromatase inhibitor responsiveness. PLoS One. doi: 10.1371/journal.pone.0077543. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim JM, Lee KH, Jeon YJ, Oh JH, Jeong SY, Song IS, et al. Identification of genes related to Parkinson's disease using expressed sequence tags. DNA Res. 2006;13(6):275–286. doi: 10.1093/dnares/dsl016. [DOI] [PubMed] [Google Scholar]
- 17.Schug J. Curr Protoc Bioinformatics. J. Wiley and Sons; Malden, MA: 2008. Using TESS to Predict Transcription Factor Binding Sites in DNA Sequence. Chapter 2: Unit 2.6. [DOI] [PubMed] [Google Scholar]
- 18.Li Y, Wang JP, Santen FJ, Kim TH, Park Y, Fan P, et al. Estrogen stimulation of cell migration involves multiple signaling pathway interactions. Endocrinology. 2010;151(11):5146–5156. doi: 10.1210/en.2009-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chakravarty D, Nair SS, Santhamma B, Nair BC, Wang L, Bandyopadhyay A, et al. Extranuclear functions of ER impact invasive migration and metastasis by breast cancer cells. Cancer Res. 2010;70(10):4092–4101. doi: 10.1158/0008-5472.CAN-09-3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanchez AM, Flamini MI, Baldacci C, Goglia L, Genazzani AR, Simoncini T. Estrogen Receptor α promotes breast cancer cell motility and invasion via focal adhesion kinase and N-WASP. Mol Endocrinol. 2010;24(11):2114–2125. doi: 10.1210/me.2010-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sieg DJ, Hauck CR, Schlaepfer DD. Required role of focal adhesion kinase (FAK) for integrin-stimulated cell migration. J Cell Sci. 1999;112(Pt 16):2677–2691. doi: 10.1242/jcs.112.16.2677. [DOI] [PubMed] [Google Scholar]
- 22.Hall Alan. RhoGTPase and actin cytoskeleton. Science. 1998;279:509–513. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 23.Spiering D, Hodgson L. Dynamics of the Rho-family small GTPases in actin regulation and motility. Cell Adh Migr. 2011;5(2):170–180. doi: 10.4161/cam.5.2.14403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chrzanowska-Wodnicka M, Burridge K. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J Cell Biol. 1996;133(6):1403–1315. doi: 10.1083/jcb.133.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan KT, Cortesio CL, Huttenlocher A. FAK alters invadopodia and focal adhesion composition and dynamics to regulate breast cancer invasion. J Cell Biol. 2009;185(2):357–70. doi: 10.1083/jcb.200809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Provenzao PP, Keely PJ. The role of focal adhesion kinase in tumor initiation and progression. Cell Adh Migr. 2009;3(4):347–350. doi: 10.4161/cam.3.4.9458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomar A, Schlaepfer DD. Focal adhesion kinase: switching between GAPS and GEFs in the regulation of cell motility. Curr Opin Cell Biol. 2009;21(5):676–683. doi: 10.1016/j.ceb.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Webb E, Donais K, Whitmore LA, Thomas SM, Turner CE, Parsons JT, et al. FAK-Src signaling through paxillin, ERK, and MLCK regulates adhesion disassembly. Nat Cell Biol. 2004;6(2):154–161. doi: 10.1038/ncb1094. [DOI] [PubMed] [Google Scholar]
- 29.Dermaudt TB, Dujardin D, Hamadi A, Noulet F, Kolli K, De Mey J, et al. FAK phosphorylation at Tyr-925 regulates cross-talk between focal adhesion turnover and cell protrusion. Mol Biol Cell. 2011;22(7):964–975. doi: 10.1091/mbc.E10-08-0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burbelo P, Wellstein A, Pestell RG. Altered RhoGTPase signaling pathways in breast cancer cells. Breast Cancer Res Treat. 2004;84:43–48. doi: 10.1023/B:BREA.0000018422.02237.f9. [DOI] [PubMed] [Google Scholar]
- 31.Tang Y, Olufemi L, Wang MT, Nie D. Role of Rho GTPases in breast cancer. Front Biosci. 2008;13:759–776. doi: 10.2741/2718. [DOI] [PubMed] [Google Scholar]
- 32.Arimidex Tamoxifen, Alone or in Combination (ATAC) Trialists’ Group. Forbes JF, Cuzick J, Buzdar A, Howell A, Tobias JS, et al. Effect of anastrozole and tamoxifen as adjuvant treatment for early breast cancer: 100 month analysis of the ATAC trial. Lancet Oncol. 2008;9:45–53. doi: 10.1016/S1470-2045(07)70385-6. [DOI] [PubMed] [Google Scholar]
- 33.Coombes RC, Kilburn LS, Snowdon CF, Paridaens R, Coleman RE, Jones SE. Survival and safety of exemestane versus tamoxifen after 2-3 years’ tamoxifen treatment (Intergroup Exemestane Study): a randomized controlled trial. Lancet. 2007;(9561):559–570. doi: 10.1016/S0140-6736(07)60200-1. [DOI] [PubMed] [Google Scholar]
- 34.The Breast International Group 1-98 Collaborative Group A comparison of letrozole and tamoxifen in postmenopausal women with early breast cancer. N Engl J Med. 2005;353(26):2747–2757. doi: 10.1056/NEJMoa052258. [DOI] [PubMed] [Google Scholar]
- 35.Galanina N, Bossuyt V, Harris LN. Molecular predictors of response to therapy for breast cancer. Cancer J. 2011;17(20):96–103. doi: 10.1097/PPO.0b013e318212dee3. [DOI] [PubMed] [Google Scholar]
- 36.Weigel MT, Dowsett M. Current and emerging biomarkers in breast cancer: prognosis and prediction. Endocr Relat Cancer. 2010;17(4):R245–262. doi: 10.1677/ERC-10-0136. [DOI] [PubMed] [Google Scholar]
- 37.The UniProt Consortium. Ongoing and future developments in the Universal Protein Resource. Nucleic Acids Res. 2011;39:D214–D219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abba MC, Hu Y, Sun H, Drake JA, Gaddis S, Baggerly K, et al. Gene expression signature of estrogen receptor alpha status in breast cancer. BMC Genomics. 2005;6(1):37. doi: 10.1186/1471-2164-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parris TZ, Danielsson A, Nemes S, Kovacs A, Delle U, Fallenius G, et al. Clinical implications of gene dosage and gene expression patterns in diploid breast carcinoma. Clin Cancer Res. 2010;16(15):3860–3874. doi: 10.1158/1078-0432.CCR-10-0889. [DOI] [PubMed] [Google Scholar]
- 40.Watson AP, Evans RL, Egland KA. Multiple functions of sushi domain containing 2 (SUSD2) in breast tumorigenesis. Mol Cancer Res. Jan. 2013;11(1):74–85. doi: 10.1158/1541-7786.MCR-12-0501-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Butt AJ, McNeil CM, Musgrove EA, Sutherland RL. Downstream targets of growth factor and oestrogen signaling and endocrine resistance: the potential roles of c-Myc, cyclin D1 and cyclin E. Endocr Relat Cancer. 2005;12(Suppl. 1):S47–59. doi: 10.1677/erc.1.00993. [DOI] [PubMed] [Google Scholar]
- 42.Sanchez AM, Flamini MI, Baldacci C, Goglia L, Genazzani AR, Simoncini T. Estrogen Receptor-alpha promotes breast cancer cell motility and invasion via focal adhesion kinase and N-WASP. Mol Endocrinol. 2010;24(11):2114–2125. doi: 10.1210/me.2010-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song RX, McPherson RA, Adam L, Bao Y, Shupnik M, Kumar R, et al. Linkage of rapid estrogen action to MAPK activation by ERα-Shc association and Shc pathway activation. Mol Endocrinol. 2002;16:116–127. doi: 10.1210/mend.16.1.0748. [DOI] [PubMed] [Google Scholar]
- 44.Tilghman RW, Slack-Davis JK, Sergina N, Martin KH, Iwanicki M, Hershey ED. Focal adhesion kinase is required for the spatial organization of the leading edge in migrating cells. J Cell Sci. 2005;118:2613–2623. doi: 10.1242/jcs.02380. [DOI] [PubMed] [Google Scholar]
- 45.Makrilia N, Kollias A, Manolopoulos L, Syrigos K. Cell adhesion molecules: role and clinical significance in cancer. Cancer Invest. 2009;27:1023–1037. doi: 10.3109/07357900902769749. [DOI] [PubMed] [Google Scholar]
- 46.Lombaerts M, van Wezel T, Philippo K, Dierssen JW, Zimmerman RM, Oosting J, et al. E-cadherin transcriptional downregulation by promoter methylation but not mutation is related to epithelial-to-mesenchymal transition in breast cancer cell lines. Br J Cancer. 2006;94:661–67. doi: 10.1038/sj.bjc.6602996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hajra KM, Fearon ER. Cadherin and catenin alterations in human cancer. Genes Chromosomes Cancer. 2002;34(3):255–68. doi: 10.1002/gcc.10083. [DOI] [PubMed] [Google Scholar]
- 48.Lin Z, Yin P, Reierstad S, O'Halloran M, Coon VJ, Pearson EK, et al. Adenosine A1 receptor, a target and regulator of estrogen receptor alpha action, mediates the proliferative effects of estradiol in breast cancer. Oncogene. 2010;29(8):1114–1122. doi: 10.1038/onc.2009.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng YH, Utsunomiya H, Pavone ME, Yin P, Bulun SE. Retinoic acid inhibits endometrial cancer cell growth via multiple genomic mechanisms. J Mol Endocrinol. 2011;46(2):139–153. doi: 10.1530/JME-10-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahn RW, Chen F, Chen H, Stern ST, Clogston JD, Patri AK, et al. A novel nanoparticulate formulation of arsenic trioxide with enhanced therapeutic efficacy in a murine model of breast cancer. Clin Cancer Res. 2010;16(14):3607–3617. doi: 10.1158/1078-0432.CCR-10-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Storey J. A direct approach to false discovery rates. JRSS. 2002;64(3):479–498. [Google Scholar]
- 52.Jacobberger JW. Flow cytometric analysis of intracellular protein epitopes. In: Stewart CC, Nicholson JKA, editors. Immunophenotypic Cytometric Cellular Analysis. John Wiley & Sons; New York: 2000. pp. 361–406. [Google Scholar]
- 53.Juan G, Darzynkiewicz Z. Current Protocols in Cytometry. John Wiley & Sons; New York: 2004. Detection of Mitotic Cells. Unit 7.24.1-7.24.7. [DOI] [PubMed] [Google Scholar]
- 54.Parker JB, Palchaudhuri S, Yin HW, Wei J, Chakravarti D. A transcriptional regulatory role of the THAP11-HCF1 complex in colon cancer cell function. Mol Cell Biol. 2012;32(9):1654–1670. doi: 10.1128/MCB.06033-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein localization. Nature Protocols. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hudson TY, Fontao L, Godsel LM, Choi HJ, Huen AC, Borradori L, et al. In vitro methods for investigating desmoplakin-intermediate filament interactions and their role in adhesive strength. Methods Cell Biol. 2004;78:757–786. doi: 10.1016/s0091-679x(04)78026-7. [DOI] [PubMed] [Google Scholar]
- 57.Kim JB, Islam S, Kim KJ, Prudoff RS, Sass KM, et al. N-cadherin extracellular repeat 4 mediates epithelial to mesenchymal transition and increased motility. J Cell Biol. 2000;151(6):1193–1206. doi: 10.1083/jcb.151.6.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. (a) Pearson correlation analysis demonstrates a strong positive correlation between ERα and SUSD3. (b) SUSD3 mRNA expression is higher in ERα/PR-positive cell lines. Expression profile in MCF7, T47D, MDA-MB231, SKBR3, and Ishikawa cell lines. (c) Treatment of MCF7 cells with 100-nM E2 demonstrated robust induction of SUSD3 mRNA expression. (d) Time course of MCF7 cells treated with 100-nM E2 demonstrated the highest induction of SUSD3 mRNA expression at 24h. All experiments were performed in triplicate. Results reported as mean percentage ± SD compared to vehicle control (EtOH). *, p<0.05; **, p<0.01; ***, p<0.001.
Supplemental Figure 2. (a) RT-PCR of SUSD3 expression in MCF7 cells transfected with control (siCTL) or SUSD3 siRNA (siSUSD3) in the presence or absence of E2 (100-nM). (b) Trial of 5 different SUSD3 siRNA target sequences. Sequences labeled siSUSD3 oligo 4 and siSUSD3 oligo 5 were most effective in knockdown as verified by RT-PCR. (c) RT-PCR of SUSD3 expression in MCF7 and T47D cells after siSUSD3 oligo 4 and 5 knockdown. Experiments in panel (a), (b), (c) were all performed in triplicate. Results reported as mean percentage ± SD for triplicate experiments. *, p<0.05; **, p<0.01; ***, p<0.001. (d) Immunoblot analysis of MCF7 CTL vs. SUSD3 siRNA oligo 4 demonstrating effective SUSD3 knockdown. Lanes 1-3 were loaded with siCTL in the amount of 18, 14, and 10μg of protein respectively. Lanes 4 and 5 were loaded with siSUSD3 oligo 4 and 5 samples respectively (18μg of protein). Custom SUSD3 antibody was utilized.
Supplemental Figure 3. (a) Cell counts of control (siCTL) or SUSD3 siRNA-transfected (oligo 4 and 5) T47D cells (siSUSD3) were performed at 72 hours post-transfection using a hemocytometer. (b) TUNEL assay demonstrating similar apoptotic levels in control and SUSD3-ablated MCF7 and T47D cells. TUNEL reaction in control and UV-B treated MCF7 and T47D cells are shown in the left two columns. MCF7 and T47D cells treated with siSUSD3 oligo 4 and 5 are shown in the right 2 columns. TUNEL staining appears red. DAPI nuclear stain appears blue. RT-PCR of SUSD3 expression in MCF7 and T47D cells after siSUSD3 oligo 4 and 5 knockdown.
Supplemental Figure 4. SUSD3-knockdown with siSUSD3 oligo 5 alters MCF7 cell morphology. (a) Early morphological changes in MCF7 cells observed via phase contrast microscopy 48h after SUSD3 siRNA transfection compared to control. Western blot of MCF7 cells demonstrating effective SUSD3 knockdown utilizing oligo 5 is shown. (b) Immunofluorescent staining of control (siCTL) and SUSD3-knockdown (siSUSD3) MCF7 cells was performed after a 72h transfection with Alexa-568 phalloidin-actin and Alexa-647 paxillin. (c) Rescue experiment utilizing GFP-only and SUSD3-GFP stably transfected MCF7 cells demonstrated that SUSD3-GFP expressing cells were resistant to SUSD3-siRNA induced morphological changes. Both cell lines were treated with control and SUSD3-siRNA. Phallodin-actin, GFP, and merged confocal pictures were taken.
Supplemental Figure 5. SUSD3 ablation led to decreased MCF7 and T47D breast cancer cell motility. (a) Percentage wound closure was determined and compared between control (siCTL) and SUSD3-knockdown (siSUSD3, oligo 5) MCF7 cells 24h after scratch test. Results are reported as means ± SD from 5 replicate experiments. ***, p< 0.001. Western blot of MCF7 cells demonstrating effective SUSD3 knockdown utilizing oligo 5 is shown. (b) Percentage wound closure in MCF7 control and SUSD3-knockdown cells (oligo 4) from time 0 to 72h after scratch test. (c) Percentage wound closure in T47D control and SUSD3-knockdown cells from time 0 to 72h after scratch test. Western blot of T47D cells demonstrating effective SUSD3 knockdown utilizing oligo 5 is shown. Results are reported as means ± SD from triplicate experiments. *, p< 0.05; **, p< 0.01.