Abstract
Prenatal alcohol exposure can lead to long-lasting changes in functional and genetic programs of the brain, which may underlie behavioral alterations seen in Fetal Alcohol Spectrum Disorder (FASD). Aberrant fetal programming during gestational alcohol exposure is a possible mechanism by which alcohol imparts teratogenic effects on the brain; however, current methods used to investigate the effects of alcohol on development often rely on either direct application of alcohol in vitro or acute high doses in vivo. In this study, we used our established moderate prenatal alcohol exposure (PAE) model, resulting in maternal blood alcohol content of approximately 20 mM, and subsequent ex vivo cell culture to assess expression of genes related to neurogenesis. Proliferating and differentiating neural progenitor cell culture conditions were established from telencephalic tissue derived from embryonic day (E) 15–17 tissue exposed to alcohol via maternal drinking throughout pregnancy. Gene expression analysis on mRNA derived in vitro was performed using a microarray, and quantitative PCR was conducted for genes to validate the microarray. Student's t tests were performed for statistical comparison of each exposure under each culture condition using a 95% confidence interval. Eleven percent of genes on the array had significantly altered mRNA expression in the prenatal alcohol-exposed neural progenitor culture under proliferating conditions. These include reduced expression of Adora2a, Cxcl1, Dlg4, Hes1, Nptx1, and Vegfa and increased expression of Fgf13, Ndn, and Sox3; bioinformatics analysis indicated that these genes are involved in cell growth and proliferation. Decreased levels of Dnmt1 and Dnmt3a were also found under proliferating conditions. Under differentiating conditions, 7.3% of genes had decreased mRNA expression; these include Cdk5rap3, Gdnf, Hey2, Heyl, Pard6b, and Ptn, which are associated with survival and differentiation as indicated by bioinformatics analysis. This study is the first to use chronic low to moderate PAE, to more accurately reflect maternal alcohol consumption, and subsequent neural progenitor cell culture to demonstrate that PAE throughout gestation alters expression of genes involved in neural development and embryonic neurogenesis.
Keywords: prenatal, alcohol, chronic, gene expression, neurogenesis, cell culture, development, epigenetic, DNA methylation
Introduction
Alcohol is a known teratogen that can impart significant damage on a developing fetus and may lead to fetal alcohol syndrome disorder (FASD) (Riley, Infante, & Warren, 2011). A collaborative effort over the past 30 years has characterized not only the more obvious outcomes of prenatal alcohol exposure (PAE) in offspring, such as microencephalopathy and speech and language pathologies, but also the molecular, structural, and cognitive consequences that persist into adulthood after PAE. Reports of alcohol's long-lasting effects on the central nervous system suggest alterations in the original molecular programming and structural elements established during gestation, also known as fetal programming. Yet, the impact of alcohol on fetal development, a particularly critical period of vulnerability, has not been fully explored.
To study the effects of alcohol on embryonic neurological development, some researchers utilize cell cultures derived from neuroepithelium, whole brain extracts, or established neuronal or glial cell lines with subsequent in vitro alcohol exposure. This reductionist approach answers highly specific questions but does not fully translate to in vivo explanations for the teratogenic effects of alcohol. Additionally, the amount of alcohol used to induce alterations, particularly in genetic programs, is relatively high, ranging from 60 to 400 mM. Further, in vitro alcohol exposures do not fully model what occurs in vivo during gestation, as the importance of the maternal environment, including the placenta, in the protection of the fetus from alcohol is ignored. However, it should be noted that studies using an in vitro application of ethanol have laid the foundation for this area of research, demonstrating that alcohol alters epigenetic programs (Veazey, Carnahan, Muller, Miranda, & Golding, 2013; Zhou et al., 2011), cell cycle dynamics (Hicks, Middleton, & Miller), cell fate (Kim et al., 2010; Miranda, Santillano, Camarillo, & Dohrman, 2008), Wnt signaling and differentiation (Vangipuram & Lyman, 2012), and transcription factors (Ogony, Malahias, Vadigepalli, & Anni, 2013) during development. Currently, in vivo exposures with subsequent cell culture characterization to derive transcriptome analysis typically deliver high doses of alcohol via intubation or intra-peritoneal injection for an acute period (Downing et al., 2012; Hashimoto-Torii, Kawasawa, Kuhn, & Rakic, 2011). These conditions may result in false positives due either to indirect effects of the high dose of alcohol used or stress of the alcohol administration. Cell culture studies have yet to model chronic alcohol consumption in a human population.
We sought to determine the effects of alcohol on genetic programs of embryonic neurogenesis using our moderate prenatal alcohol exposure (PAE) paradigm (Brady, Allan, & Caldwell, 2012). This “drinking in the dark” model results in blood alcohol content of 20 mM 4 h after consumption, similar to slightly more than one drink per day (Valenzuela, Morton, Diaz, & Topper, 2012); dams consume alcohol chronically, during preconception and gestation up to embryonic (E) days 15–17. Neurogenesis was chosen as a mechanism for investigation based on our previous publications demonstrating that alcohol elicits long-lasting effects seen in adulthood on neural progenitor cells (NPC) left over from development, including alterations in cell fate (Kajimoto, Allan, & Cunningham, 2013) and the neurogenic capacity to respond to environmental cues (Choi, Allan, & Cunningham, 2005). Ex vivo proliferating and differentiated cell cultures comprised of NPC from E15–E17 tissue exposed to alcohol in vivo were used for transcriptome analysis of neurogenesis-related genes. To our knowledge, we are the first to report that embryonic neurogenesis under both proliferating and differentiating conditions is altered by in vivo moderate alcohol exposure.
Materials and methods
Subjects and alcohol exposure
The University of New Mexico Health Sciences Center Institutional Animal Care and Use Committee approved all of the procedures and experiments described in the current studies.
Female C57BL/6 mice obtained from Jackson Labs were acclimated to the animal facility (22 °C) for one week on a reverse light/dark cycle (lights on at 2000 hours, off at 0800) in group-housing conditions (4 per cage) with ad libitum access to food and water. After acclimation, females were separated into individual housing for one week before initiation of the drinking paradigm. Prenatal exposure of mice to 10% w/v alcohol was performed using our previously published drinking in the dark method (Brady et al., 2012). Briefly, females were provided either a solution of 10% w/v alcohol sweetened with 0.066% (w/v) saccharin or a 0.066% (w/v) saccharin-only solution for 4 h starting 2 h into the dark cycle (1000–1400 hours). The concentration of alcohol was increased to 10% over the initial 4 days of the paradigm. After one week of drinking at the 10% w/v alcohol level, a single female was introduced to a singly housed male immediately after the drinking period; the female was removed from the male cage the following morning at the beginning of the dark cycle at 0800 hours. Food and water were available ad libitum during the breeding period. Females were bred for 2 days and consumed alcohol or saccharin solutions during the same 4-h period each day. One week after the last day of breeding, pregnancy was determined by monitoring weight gain. As the cell culturing method consisted of embryonic tissue (E15–E17), date of conception was not determined. Pregnant dams were sacrificed 13–15 days after the last day of mating to obtain appropriately aged tissue. Alcohol consumption was monitored before and after breeding and throughout pregnancy for several breeding rounds of mice used in these studies. Alcohol intake during pregnancy averaged 7.33 ± 0.35 g/kg body weight per day. This is similar to our previously published data (7.17 ± 0.17 g/kg body weight per day), which resulted in blood alcohol levels of 88.3 ± 11.5 mg/dL 4 h after drinking 10% w/v alcohol solution (Brady et al., 2012). This is approximately equal to 19.3 mM blood alcohol concentration.
Culture of neural progenitor cells
The neural progenitor cell proliferation and differentiation cultures were generated from telencephalic tissue derived from embryonic day (E) 15–17. Pregnant dams, either alcohol drinking (prenatal alcohol exposure; PAE) or saccharin drinking (control) were anesthetized via inhalation of isofluorane in a small chamber, and using a midline transverse abdominal incision, the gravid uterus was removed and rinsed in chilled 1X DPBS (no calcium, no magnesium; Gibco; 14190136). All embryos from the same dam were isolated and rinsed in chilled 1X DPBS solution, and the number of embryos and crown-to-rump length (mm) were recorded. Whole brains were removed and placed into chilled 1X Hank's Balanced Salt Solution (HBSS) with glucose and magnesium chloride (Cellgro; 21-023-CV). Olfactory bulbs and meninges were removed, and the telencephalons were collected into a sterile conical tube filled with cold HBSS. After complete collection, HBSS was aspirated and pre-warmed medium was added. Medium consisted of Dulbecco's Modified Eagle's Medium with Nutrient Mixture (Ham's) F-12 medium (DMEM/F12) with glutamine (Invitrogen; 11320-082) supplemented with N2 (Invitrogen; 17502-048), 20 ng/mL FGF-2 (Peprotech; 100-18B), 20 ng/mL EGF (Peprotech; AF-100-15), and 1% antibiotic-antimycotic (Invitrogen; 15240062). The tissue was mechanically dissociated and cells were filtered through a 70-μm cell strainer (VWR; 21008-952), plated on poly-D-lysine coated 6-well plates (50 μg/mL poly-D-lysine per plate; Sigma-Aldrich; P0899), and adherent monolayer proliferating cultures were established and maintained under proliferating conditions using complete medium with growth factors at 37 °C and 5% CO2. One cell culture plate consisted of 8–9 embryos from one dam; at least 6 different dams for each exposure and each culture condition were used for array assessment (n = 6). Differentiated NPC cultures were derived by removing growth factors (FGF-2, EGF) from the medium after 3 passages or 10 days in vitro (DIV). NPCs were allowed to differentiate up to 10 days with up to 2 passages if necessary (Roitbak, Thomas, Martin, Allan, & Cunningham, 2011).
Confocal Microscopy
Neural progenitor cells derived from control dams were expanded as adherent monolayer cultures under either proliferating or differentiating conditions and were prepared for immunohistochemical analysis and imaging. Cells were transferred to poly-D-lysine coated coverslips after the second passage and grown in complete medium with growth factors EGF and FGF-2. After 2 days, cells were immunostained with the neural stem cell marker Nestin (1:1000; BD Biosciences) and counterstained with DAPI (0.5 μg/mL; Sigma) (Roitbak et al., 2011). To determine appropriate culture of neurons, cells were transferred to poly-D-lysine coated plates after the second passage and grown in complete medium without growth factors. After differentiation for 7 days without growth factors, cells were immunostained for astrocytes with GFAP (1:1000; Sigma-Aldrich) and neuroblasts and immature neurons were immunostained with doublecortin (1:1500; Cell Signaling) and counterstained with DAPI (0.5 μg/mL; Sigma). Secondary antibodies used for analysis included Alexa Fluor 647 (1:250; Invitrogen; A-21236) and Cy3 (1:500; Invitrogen; A10520). Images for publication were acquired using a Zeiss LSM-510 META confocal microscope equipped with a laser diode, one argon laser, and two HeNe lasers. Maximum intensity projections of Z stacks were acquired with a 20X objective.
Microarray qPCR analysis and validation
Quantitative PCR analysis using a microarray was performed as described in our previous publication (Tyler & Allan, 2013). Briefly, total RNA was extracted from cell pellets derived from plates from either proliferating NPCs or differentiated NPCs using the RNeasy Mini Kit and a QIAshredder homogenizer (Qiagen; 74134, 79654). Each plate contained approximately 1.2 × 106 cells. The mRNA concentration was determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies); all RNA used for PCR analysis had a 260/280 absorbance ratio of ~2.0. Approximately 750 ng of isolated mRNA was converted to cDNA using the RT2 First Strand Kit (SABiosciences; 330522). PCR analysis was performed according to the manufacturer's instructions on an ABI 7300 Real-Time PCR System (Applied Biosystems) using the RT2 Profiler Mouse Neural Stem Cell and Neurogenesis PCR Array (SABiosciences; PAMM-404). Cycle threshold (CT) values for each gene were normalized to an average CT value from two housekeeping genes (Gusb and Hprt). Although Gapdh, β-actin, and Hsp90ab were also on the array and available for use as housekeeping genes, our lab has found that prenatal alcohol exposure impacts the expression of these genes (unpublished data); thus, we chose to use only average CT values from Gusb and Hprt as normalizing housekeeping genes. The comparative CT method (ΔΔCT) was used to assess changes in mRNA levels. For each gene, the fold change comparing prenatal alcohol exposure (PAE) and control conditions was determined: fold change was calculated as 2-ΔCT(PAE)/2-ΔCT(control). Relative fold-change expression was calculated as 2-ΔCT(PAE sample)/2-ΔCT(AVG control); all graphs indicate relative fold change comparing PAE to control gene expression. All control and PAE tissue was age-matched to avoid confounds. Results from the microarrays were validated using qRT-PCR in triplicate using new sets of cell cultures from different dams. RNA (1 μg) was isolated from cells as described above and stored at −80 °C until cDNA conversion. Reverse transcription was performed using a Quantitect Reverse Transcription Kit (Qiagen; 205311) and a Peltier Thermal Cycler. Quantitative PCR was performed using FastStart Universal SYBR Green (Roche; 04913850001) on an ABI 7300 Real-Time PCR System (Applied Biosystems). Primers were obtained from Qiagen: Adora2a (PPM03472F), Gdnf (PPM04315F), Hes1 (PPM05647A), Hey2 (PPM30634F), Sox3 (PPM04751A), and Vegfa (PPM03041F). Primer sets for Dnmt1, Dnmt3a, and PPIA (used as an endogenous control) were developed using PrimerBlast. Dnmt1: 5'-forward: ATCCTGTGAACGAGACCCTGT, 5'-reverse: CCGATGCGATAGGGCTCTG. Dnmt3a: 5'-forward: GAGAAGAGGAAGCCCATCCG, 5'-reverse: ATGATCTTTCCCTGGTGCCG. PPIA: 5'-forward:TGCTGGACCAAACACAAACG, 5'-reverse: AGAGAGGGGAAAGAGGCACT. Primer efficiencies were determined for each primer set and primer concentrations were optimized to achieve 94–96% efficiencies. Each reaction contained 1.25–5 mM primer and 25 ng/μL cDNA; all samples including control and PAE cDNA were run on the same plates for each primer set with the appropriate housekeeping genes. No template controls were run for each primer set for each plate; reverse transcription controls for PAE cells and control cells were also run with each primer set. The dissociated curves were evaluated. Results were assessed using the comparative CT method (ΔΔCT) and are expressed as relative fold change for each gene comparing PAE to control. At least four different cell cultures (generated from different dams, each with 8–9 embryos) were used for each group, n = 4–6.
Statistical analyses
Statistical analyses were performed using GraphPad Prism 5 (San Diego, CA). All studies used at least three different breeding rounds, with the reported n corresponding to the number of dams. Gene expression data was analyzed using the Student's 2-tailed t test; n = 6 per group for microarray analysis and for qRT-PCR validation, with separate cohorts of animals used for each analysis. Statistical significance was set at a 95% confidence interval.
Results
Ex vivo cell culture following moderate alcohol exposure in vivo
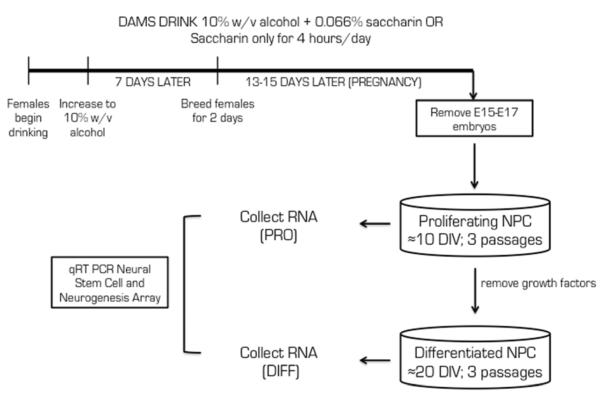
Fig. 1 depicts the moderate alcohol exposure paradigm (in vivo) and the cell culture model used from ex vivo derived neural progenitor cells used in these studies. Animals were exposed to a moderate dose (10% w/v) of alcohol in utero for up to 17 days (E15–E17) via a limited access model of alcohol consumption by the dam (Brady et al., 2012). Telencephalonic cells derived from E15-E17 tissue were cultured (ex vivo) with growth factors to obtain a proliferating neural progenitor cell (NPC) culture. The age of extraction for each set of PAE and control tissue was matched for each culture condition and determined via crown-to-rump length. After 3 passages (approximately 10+ days in vitro), growth factors were removed for differentiation of NPCs either into neural or glial lineage; NPCs typically differentiate into neurons between 7–10 days after growth factor removal. The differentiated cell culture contained a heterogeneous mixture of neurons and glial cells. RNA was extracted from two culture conditions and used for analysis in a microarray of genes associated with neurogenesis.
Figure 1. Prenatal alcohol exposure paradigm and ex vivo cell culture.
Female C57BL/6 mice, aged 2 months, were acclimated to drinking 10% w/v alcohol with 0.066% (w/v) saccharin or 0.066% (w/v) saccharin only for 4 days. Females continued to drink for 1 week and were then mated for 2 days; 13–15 days after the last day of mating, embryonic tissue (E15–E17) was extracted from pregnant females and the telencephalons from all embryos from one dam were cultured under proliferating conditions with growth factors. After 10 days in vitro (DIV) and 3 passages, RNA was collected from the proliferating NPC cultures. Other NPC cultures had the growth factors removed from their media and were permitted to differentiate for approximately 7–10 DIV for a total of 20 DIV and 3 passages. RNA was collected from the differentiated NPCs at 20 DIV for analysis.
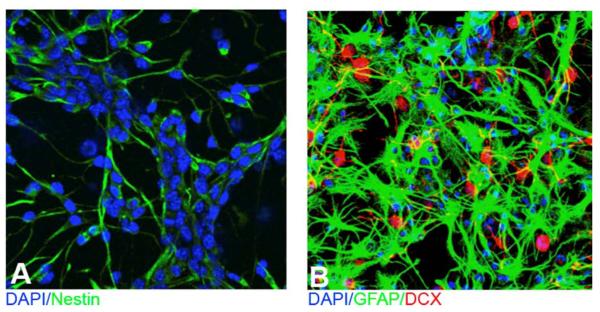
Microscopy analysis was performed to ensure that cell cultures contained appreciable numbers of proliferating and differentiated neural progenitor cells. Fig. 2A shows a 20X image of the proliferating NPCs from control mice expanded as adherent monolayer culture in the presence of growth factors (EGF/FGF) using immunostaining with DAPI for the nuclear marker and Nestin for the NPC marker. Fig. 2B shows a 20X image of cultured NPCs 10 days after the removal of growth factors from the media using immunostaining DAPI for the nuclear marker, doublecortin (DCX) for the neuroblasts and immature neuron marker, and GFAP for the astrocytic marker. These images demonstrate that culture conditions were appropriate for proliferating NPCs (Fig. 2A) and differentiated NPCs (Fig. 2B).
Figure 2. Representative images of cell culture conditions.
Neural progenitor cells were collected from E15–E17 tissue without exposure to alcohol and expanded as adherent monolayer cultures in the presence of growth factors (EGF, FGF-2) for immunohistochemistry. Fig. 2A is a 20X representative image of the proliferating NPC culture conditions; Nestin, which marks neural progenitor cells, is in green with a DAPI nuclear stain in blue. Fig. 2B is a 20X representative image of the differentiated NPC culture conditions; GFAP, which marks astrocytes, is in green with a DAPI nuclear stain in blue and the immature neuronal marker doublecortin (DCX) in red.
Prenatal alcohol exposure alters neurogenesis-related genes in proliferating and differentiated NPC cultures
Microarray analysis of the proliferating and differentiated NPCs for genes related to neural stem cell development and neurogenesis revealed significant effects of in vivo developmental exposure to alcohol. Alcohol significantly altered mRNA expression (fold change ≤ 0.8 or ≥ 1.5) of 9 genes in the proliferating NPC cultures and 6 genes in the differentiated NPC cultures out of 82 possible targets. Figs. 3A and 4A show gene expression, and associated Figs. 3B and 4B show relative gene expression compared to controls for proliferating and differentiating NPCs, respectively.
Figure 3. Exposure to alcohol in utero induces alterations in gene expression in proliferating NPC in vitro.
A) A microarray containing probes for genes related to neurogenesis and neural stem cell development was used to assess RNA expression under proliferating conditions. Fold change and p values are provided for each gene significantly altered by PAE. Data presented were acquired from control proliferating NPCs and prenatal alcohol-exposed (PAE) proliferating NPCs (n = 6 dams for each condition).
B) Relative gene expression of the most significant changes in gene expression from the array data using RNA derived from the proliferating NPC culture conditions. *p < .05,**p < .01
Figure 4. Exposure to alcohol in utero induces alterations in gene expression in differentiated NPC in vitro.
A) A microarray containing probes for genes related to neurogenesis and neural stem cell development was used to assess RNA expression in both culture conditions. Data presented here were acquired from control differentiated NPCs and PAE differentiated NPCs (n = 6 dams for each condition).
Relative gene expression of the most significant changes in gene expression from the array data using RNA derived from the differentiated NPC cell culture conditions. *p < .05, **p < .01
Decreased mRNA levels of the following genes were found in the PAE proliferating NPC cultures: the adenosine receptor 2A (Adora2a, p = .02), which encodes the A2A receptor; chemokine ligand 1 (Cxcl1, p = .04), involved in development of the spinal cord; PSD-95 (Dlg4, p = .02), important for synapse formation and maintenance; hairy enhancer of split 1 (Hes1, p = .01), a Notch pathway signaling effector; neuronal pentraxin 1 (Nptx1, p = .03), which mediates the uptake of degraded synaptic material and is involved in immunological responses; and vascular endothelial growth factor A (Vegfa, p = .01), crucial for maintaining the proliferating pool of progenitors (Fournier, Lee, Banasr, Elsayed, & Duman, 2012) and responsible for directly stimulating neurogenesis (Schänzer et al., 2004). PAE also reduced mRNA expression of DNA methyltransferase 1 (Dnmt1, p = .02) and DNA methyltransferase 3a (Dnmt3a, p = .04), both of which are critical for maintenance and de novo methylation of genes involved in fetal programming. Increased expression of the following factors was found after PAE in the proliferating NPC cultures: fibroblast growth factor 13 (Fgf13, p = .02), important for microtubule stabilization; necdin (Ndn, p = .004), an imprinted gene that regulates quiescence; and SRY-box containing gene 3 (Sox3, p = .02), which regulates NPC maintenance and proliferation and acts to inhibit differentiation.
Decreased mRNA levels of the following genes were found after PAE in differentiated NPC cultures: CDK5 regulatory subunit protein 3 (Cdk5rap3, p = .04), which governs cell cycle progression, particularly neuronal differentiation; glial derived neurotrophic factor (Gdnf, p = .004), a neurotrophic factor that promotes survival of neurons; hairy enhancer of split (Hey2, p = .01, and Heyl, p = .03), both of which are important for vascularization in the embryo during development (Fischer, Schumacher, Maier, Sendtner, & Gessler, 2004); Par-6 family cell polarity regulator beta (Pard6b, p = .004), which encodes for a protein with a PDZ domain and is involved in cell division and polarization; and pleiotropin (Ptn, p = .05), which promotes neurite outgrowth during development. PAE did not induce increased mRNA levels in any genes tested in the differentiated NPC cultures.
Functional annotation analysis of genes affected by prenatal alcohol exposure
Functional clustering using the Ingenuity Pathway Analysis (IPA) software was performed to determine common processes (within the subset of neurogenesis) affected by moderate PAE. All genes altered by PAE for each set of culture conditions were analyzed together. Tables 1 and 2 show results from the analysis for the proliferating and differentiated culture conditions, respectively, indicating the most significant gene ontology (GO) categories and functional annotations within those categories altered by PAE. Gene clusters derived from genes altered by PAE indicate that alcohol influences nervous system development and function, cell death and survival, cellular movement, and embryonic development, cellular function and maintenance, and behavior. The functional annotations within the GO categories for genes altered under proliferating conditions are particularly interesting: they include the quantity and migration of neuroglia and neurons, neuritogenesis and outgrowth of neurites, long-term potentiation, neuronal apoptosis, behavior, blood vessel development, proliferation, and organization of the cytoskeleton, all with p <.001 as seen in Table 1. These categories mostly include Adora2a and Vegfa. Functional annotations for genes altered under differentiated conditions include survival of neurons; differentiation of NPCs, neurons, and cells; neurogenesis; and proliferation of microglia, all with p <.001 as seen in Table 2. These categories are likely predominantly driven by the alterations in Gdnf and Hey2.
Table 1.
Significant Gene Ontology (GO) categories and molecular functions annotations for genes differentially expressed after prenatal alcohol exposure in ex vivo cell culture of proliferating NPCs
| GO CATEGORY | FUNCTIONS ANNOTATION | p VALUE | MOLECULES |
|---|---|---|---|
| Nervous System Development and Function | quantity of neuroglia | 8.64E-08 | CXCL1,FGF13,HES1,VEGFA |
| development of central nervous system | 2.66E-07 | ADORA2A,HES1,NDN,NPTX1,SOX3,VEGFA | |
| migration of neuroglia | 3.78E-06 | CXCL1,NDN,VEGFA | |
| quantity of neurons | 9.77E-06 | ADORA2A,FGF13,HES1,VEGFA | |
| migration of oligodendrocyte precursor cells | 1.20E-05 | CXCL1,VEGFA | |
| neurological function | 2.60E-05 | ADORA2A,VEGFA | |
| proliferation of neuroglia | 2.71E-05 | CXCL1,HES1,VEGFA | |
| outgrowth of neurites | 5.18E-05 | ADORA2A,NDN,NPTX1,VEGFA | |
| development of neurons | 9.01E-05 | HES1,NDN,VEGFA | |
| neuritogenesis | 9.51E-05 | ADORA2A,DLG4,FGF13,NDN | |
| development of forebrain | 2.01E-04 | HES1,SOX3,VEGFA | |
| long-term potentiation | 2.52E-04 | ADORA2A,DLG4,VEGFA | |
| proliferation of neural precursor cells | 2.74E-04 | HES1,VEGFA | |
| synaptic transmission of cells | 2.83E-04 | ADORA2A,DLG4,NPTX1 | |
| morphogenesis of neurites | 6.77E-04 | ADORA2A,DLG4,FGF13 | |
| abnormal morphology of neurons | 1.01E-03 | HES1,NDN,VEGFA | |
|
| |||
| Cell Death and Survival | cell death of brain | 2.14E-07 | ADORA2A,CXCL1,DLG4,NPTX1,VEGFA |
| cell death of cerebral cortex cells | 3.49E-06 | ADORA2A,CXCL1,NPTX1,VEGFA | |
| cell death of medium spiny neurons | 9.78E-06 | ADORA2A,DLG4 | |
| neuronal cell death | 1.84E-05 | ADORA2A,CXCL1,DLG4,NPTX1,VEGFA | |
| apoptosis of neurons | 5.62E-05 | ADORA2A,DLG4,NPTX1,VEGFA | |
| apoptosis | 2.01E-04 | ADORA2A,CXCL1,DLG4,HES1,NDN,NPTX1,VEGFA | |
|
| |||
| Embryonic Development | development of sensory organ | 5.39E-06 | ADORA2A,FGF13,HES1,SOX3,VEGFA |
| development of head | 5.79E-05 | ADORA2A,FGF13,HES1,SOX3,VEGFA | |
| eye development | 6.19E-05 | ADORA2A,FGF13,HES1,VEGFA | |
| development of pituitary gland | 1.77E-04 | HES1,SOX3 | |
|
| |||
| Behavior | behavior | 1.04E-04 | ADORA2A,DLG4,NDN,NPTX1,VEGFA |
| hypoactivity of mice | 5.51E-05 | ADORA2A,DLG4,VEGFA | |
| emotional behavior | 2.33E-04 | ADORA2A,DLG4,VEGFA | |
|
| |||
| Organismal Development | proliferation of vascular endothelial cells | 3.04E-05 | CXCL1,FGF13,VEGFA |
| development of blood vessels | 7.77E-04 | ADORA2A,CXCL1,FGF13,VEGFA | |
|
| |||
| Cardiovascular Development and Function | angiogenesis | 1.83E-05 | ADORA2A,CXCL1,FGF13,NPTX1,VEGFA |
|
| |||
| Cellular Assembly and Organization | organization of cytoskeleton | 2.21E-05 | ADORA2A,CXCL1,DLG4,FGF13,NDN,VEGFA |
|
| |||
| Cellular Development | differentiation of cells | 3.49E-03 | ADORA2A,HES1,NDN,SOX3,VEGFA |
|
| |||
| Cellular Movement | migration of cells | 2.54E-03 | ADORA2A,CXCL1,FGF13,NDN,VEGFA |
|
| |||
| Post-Translational Modification | assembly of protein-protein complex | 6.47E-05 | DLG4,HES1,VEGFA |
|
| |||
| Tissue Morphology | quantity of cells | 1.70E-04 | ADORA2A,CXCL1,FGF13,HES1,SOX3,VEGFA |
Ingenuity Pathway Analysis (IPA) was used to determine the gene ontology categories and functions annotations for genes altered (both increased and decreased) in response to PAE in proliferating NPC conditions.
Table 2.
Significant Gene Ontology categories and molecular functions annotations for genes differentially expressed after prenatal alcohol exposure in ex vivo cell culture of differentiated NPC
| GO CATEGORY | FUNCTIONS ANNOTATION | p VALUE | MOLECULES |
|---|---|---|---|
| Nervous System Development and Function | survival of spinal neuron | 7.07E-06 | GDNF,PTN |
| survival of dopaminergic neurons | 2.94E-05 | GDNF,PTN | |
| differentiation of neuronal progenitor cells | 6.01E-05 | GDNF,HEYL | |
| differentiation of neurons | 2.79E-04 | GDNF,HEYL,PTN | |
| neuritogenesis | 5.26E-04 | GDNF,PARD6B,PTN | |
| proliferation of neuroglia | 7.13E-04 | GDNF,PTN | |
|
| |||
| Embryonic Development | development of mesenchymal cells | 3.26E-06 | HEY2,HEYL |
| differentiation of neural crest cells | 8.25E-06 | HEY2,HEYL | |
| morphogenesis of embryonic tissue | 9.37E-06 | GDNF,HEY2,HEYL | |
| morphogenesis of ventricular septum | 2.29E-05 | HEY2,HEYL | |
|
| |||
| Cellular Development | development of neurons | 2.20E-05 | GDNF,HEY2,PTN |
| differentiation of cells | 2.37E-04 | CDK5RAP3,GDNF,HEY2,HEYL,PTN | |
|
| |||
| Gene Expression | activation of DNA endogenous promoter | 2.76E-04 | CDK5RAP3,GDNF,HEY2,HEYL |
| transcription of RNA | 2.92E-03 | CDK5RAP3,GDNF,HEY2,HEYL | |
|
| |||
| Cell Death and Survival | cell viability of brain cells | 3.75E-04 | GDNF,PTN |
| apoptosis | 1.33E-02 | CDK5RAP3,GDNF,HEYL,PTN | |
|
| |||
| Cellular Function and Maintenance | microtubule dynamics | 3.31E-04 | CDK5RAP3,GDNF,PARD6B,PTN |
| Cellular Growth and Proliferation | proliferation of cells | 4.05E-02 | GDNF,HEY2,HEYL,PTN |
| Cellular Movement | cell movement | 4.58E-03 | GDNF,HEY2,PARD6B,PTN |
Ingenuity Pathway Analysis (IPA) was used to determine the gene ontology categories and functions annotations for genes altered (both increased and decreased) in response to PAE in differentiated NPC conditions.
The validity of the microarray results was confirmed with selected genes from each data set chosen based on genes on either large positive or negative fold changes or highly significant p values. Real-time results of selected genes are shown in Table 3. For the proliferating culture conditions, decreased expression of Adora2a (p = .05), Hes1 (p = .01), Vegfa (p = .01), and increased expression of Sox3 (p = .02) were confirmed. For differentiated culture conditions, the reduced expression of Gdnf (p = .04) and Hey2 (p = .01) were also confirmed.
Table 3.
Validated Genes Significantly Altered After Prenatal Exposure to Alcohol in both sets of culture conditions (E15–E17)
| Gene Name | Gene Symbol | Fold Change | p value | Culture conditions |
|---|---|---|---|---|
| Adenosine A2a receptor | Adora2a | 0.62 | 0.05 | Proliferation |
| Hairy and enhancer of split 1 | Hesl | 0.47 | 0.002 | |
| SRY-box containing gene 3 | Sox3 | 1.53 | 0.05 | |
| Vascular endothelial growth factor A | Vegfa | 0.29 | 0.007 | |
|
| ||||
| Glial cell line derived neurotrophic factor | Gdnf | 0.76 | 0.04 | Differentiation |
| Hairy/enhancer-of-split related with YRPW motif 2 | Hey2 | 0.28 | 0.008 | |
Relative gene expression from the genes that were validated using qRT-PCR in the differentiated NPC culture condition, (n = 4–6) *p < .05, **p < .01
Discussion
The current paradigms used to investigate the teratogenic effects of alcohol on neurodevelopmental processes typically involve the direct application of alcohol on stem or neural progenitor cells derived in vivo from unexposed animals or in vitro from cell cultures of neural or glial origin. Without the influence of the placenta, maternal metabolism, and the environment of the womb, it is difficult to extrapolate results from these in vitro studies on the effects of alcohol exposure in vivo. However, research using in vitro alcohol exposure on neural stem cells has demonstrated reduced neurogenesis either via increased length of the cell cycle (Hicks et al., 2010; Jacobs & Miller, 2001) or alterations of transcription factors that provide the balance between maintenance of the progenitor pool and differentiation (Ogony et al., 2013). Based on these findings, we sought to investigate the effects of in vivo alcohol on expression of neurogenesis-related genes using two sets of ex vivo culture conditions: proliferating neural progenitor cells derived from the telencephalons of alcohol-exposed and control embryonic tissue and the differentiation of those cells into a mixed cell culture of neurons and astrocytes (Figs. 1 & 2).
Using our limited-access, moderate prenatal alcohol exposure (PAE) paradigm (Brady et al., 2012), we found that alcohol significantly (p < .05) decreased expression of several neurogenic factors involved in development and function of the central nervous system, particularly mechanisms associated with cell growth and death (Table 1). These include Adora2a, Cxcl1, Dlg4 (PSD-95), Hes1, Nptx1, and Vegfa (Fig. 3). Several of these genes have been studied in the context of alcohol exposure; for example, Adora2a, which encodes the A2A receptor and is highly expressed in the adult striatum, has been implicated in alcohol use disorders and other addictive behaviors (Hack & Christie, 2003; Nam, Bruner, & Choi, 2013; Nam, Hinton, et al., 2013). Transient expression of A2A during development of the brain aids in neuritogenesis and proliferation (Sun et al., 2010; Weaver, 1993); however, the role of A2A in development after PAE has not been studied extensively. Additionally, A2A activation leads to upregulation of VEGF expression, suggesting that the decreased mRNA levels of both of these genes after PAE may be connected in the proliferating NPC culture conditions (Escudero et al., 2013). Further, gestational alcohol exposure alters cortical vascularization in the neonatal brain resulting in reduced expression of VEGF receptors (Jégou et al., 2012). Vascularization, neural development, cell growth, and proliferation are dependent on the oscillatory signaling of Notch pathway components; as such, reduced expression of Hes1 after PAE may alter proliferation of the NPC culture (Barton & Fendrik, 2013; Fischer et al., 2004; Shimojo, Ohtsuka, & Kageyama, 2011). Other studies have demonstrated the sensitivity of Notch signaling to alcohol using several different exposure methods: these include application of 100 mM alcohol on human neural stem cells in vitro (Hashimoto-Torii et al., 2011; Melendez, McGinty, Kalivas, & Becker, 2012) and chronic intermittent (Melendez et al., 2012) or acute in vivo exposure to high doses of alcohol (Rubert, Miñana, Pascual, & Guerri, 2006). Maternal alcohol consumption from E7–E21 in the rat model (25% v/v alcohol) has been shown to reduce proliferation and induce rapid differentiation via down-regulation of Notch pathway factors (Kim et al., 2010). The increase in levels of Ndn may exacerbate these anti-proliferative effects of PAE, as Ndn facilitates cell cycle arrest, acts as a growth suppressor, and has been shown to suppress NPC proliferation in the embryonic neocortex (Minamide, Fujiwara, Hasegawa, & Yoshikawa, 2014). While we were unable to find other studies demonstrating a connection between Ndn and alcohol exposure, it is likely that Ndn is impacted by alcohol. Ndn is an imprinted gene and since alcohol has been demonstrated to deplete methyl donors, lack of DNA methylation due to alcohol exposure may result in increased expression of this particular gene, similar to alcohol's effect on Sox3 methylation (Haycock, 2009; Zhou et al., 2011). Alterations in these genes suggest that PAE results in perturbed cell growth; however, it is possible that the observed increase in expression of Fgf13 and Sox3 could be a compensatory mechanism to shift the emphasis back to self-renewal and stem cell maintenance programs. Sox3, part of the SoxB family of transcription factors expressed in self-renewing progenitor cells, is important for stem cell maintenance and inhibits neuronal differentiation (Bylund, Andersson, Novitch, & Muhr, 2003). Thus, we may conclude that PAE is altering components of proliferation and cell growth, with possibly compensatory mechanisms increased in response to the presence of alcohol, as seen in other studies (Pickering, Wicher, Rosendahl, Schiöth, & Fex-Svenningsen, 2010; Rubert et al., 2006).
In the differentiated NPC culture, alcohol decreased the mRNA levels of genes involved in survival, differentiation, and neuritogenesis (Table 2); these include Cdk5rap3, Gdnf, Hey2, Heyl, Pard6b, and Ptn. Of particular interest are the Notch components Hey2 and Heyl, which are transcription factor effectors of the Notch pathway. Hey2 particularly acts to inhibit transcription of proneural factors in association with histone deacetylase 2, to allow for stem cell maintenance and retention of pluripotency (Wen, Li, & Liu, 2009: Zhou, Kumari, Xiao, & Tan, 2010). Thus, PAE may induce rapid differentiation of NPCs by reducing these Notch effectors; however, reduced Gdnf, which has been demonstrated to increase differentiation when applied to cell culture, does not support this assessment (Roussa & Krieglstein, 2004). Indeed, gene ontology analysis suggests that decreased Gdnf and Ptn would result in less survival and differentiation (Table 2). GDNF protects against alcohol-mediated cell death in culture but is reduced in response to alcohol along with neurite outgrowth (Barak, Carnicella, Yowell, & Ron, 2011; Carnicella, Kharazia, Jeanblanc, Janak, & Ron, 2008; Chen & Charness, 2012). Interestingly, GDNF has been used to prevent relapse of alcohol consumption (Carnicella et al., 2008). Thus, it is unclear if PAE is preventing or enhancing the differentiation program; these mixed results may derive from the fact that differentiation occurred in vitro from NPCs derived in vivo after PAE. Indeed, alcohol increases differentiation at the expense of proliferation under some conditions (Kim et al., 2010); as such, the effects we have observed from differentiated NPC cultures may occur due to a combined effect of PAE and culture conditions.
Alterations in gene expression presented here, combined with studies demonstrating alcohol's effects on methyl donors (Haycock, 2009), suggest that alcohol may impact transcriptional regulation, particularly DNA methylation. A recent study using 88 mM alcohol on cultured neural stem cells revealed that alcohol inhibits the methylation of Sox genes possibly resulting in upregulation of this transcription factor (Zhou et al., 2011); this is similar to increased mRNA expression of Sox3 after PAE presented here. Indeed, PAE also decreased mRNA levels of DNA methyltransferase 1 (Dnmt1) and 3a (Dnmt3a) in the proliferating culture of NPCs (Fig. 3A). DNMT1, known as the maintenance methyltransferase, is important for proper gene expression during neurogenesis (Ma et al., 2010), while DNMT3a is required for de novo methylation and is primarily localized to neural precursor cells during development (Feng, Chang, Li, & Fan, 2005). Conditional knockout of Dnmt1 and Dnmt3a results in hypomethylation of neuronal DNA resulting in loss of synaptic function (Feng et al., 2010). While we did observe an increase in Sox3 and Ndn expression (an imprinted gene requiring hypermethylation on one allele), other epigenetic or transcription factors may play a role in mediating the effects of PAE, as mRNA expression of other genes is decreased in both culture conditions. However, decreased expression of DNMT1 and DNMT3a after prenatal alcohol exposure has been demonstrated at postnatal day 21 (PD21), suggesting that this effect is persistent (Perkins, Lehmann, Lawrence, & Kelly, 2013). Studies that have investigated the effect of alcohol on DNA methylation indicate that alcohol impairs normal DNA methylation programming particularly in progenitor cells leading to decreased growth and development (Chen, Ozturk, & Zhou, 2013; Zhou, 2012).
Results from several studies investigating long-lasting outcomes after prenatal alcohol exposure point to aberrant fetal programming, particularly in the development of the brain, as a mechanism for alcohol's effects (Kleiber, Mantha, Stringer, & Singh, 2013; Zhou, 2012). However, to date, embryonic neurogenesis has not been fully explored in vivo. Cell culture models used for investigating the effects of alcohol during development, while elucidating important findings for the field, lack the ability to recapitulate gestational alcohol exposure. Investigating the pathogenesis of fetal alcohol syndrome requires modeling the phenotype of the disease in the closest approximation possible, which includes the method of alcohol exposure; however, the use of cell culture does allow for more flexible and direct characterization of distinct activities specifically within proliferating neural progenitor systems that are difficult to isolate in vivo. Thus, PAE followed by ex vivo culture is a more physiologically relevant model of alcohol exposure, which allows for assessment of embryonic neurogenesis on particular cellular phenotypes. While acute in vivo alcohol exposure and subsequent gene expression analysis has been performed in either whole embryo or whole brain extracts using relatively high doses of alcohol (Aoto, Shikata, Higashiyama, Shiota, & Motoyama, 2008; Chen et al., 2013; Da Lee et al., 2004; Du & Hamre, 2003; Green et al., 2007; Hard, Abdolell, Robinson, & Koren, 2005; Zhou, 2012; Zhou et al., 2011), we present findings using a chronic moderate alcohol exposure on neural progenitor cells. Our results suggest that prenatal alcohol exposure imparts significant alterations in genes responsible for embryonic neurogenesis; thus, this ex vivo model may be useful in the transition from in vitro to in vivo studies for assessing fetal programming.
Supplementary Material
Acknowledgments
The authors would like to provide special thanks to Samantha Goggin, Matthew Labrecque, and Xun Guo for technical assistance and to Dr. Kevin Caldwell and Samantha Goggin for critical reading of the manuscript. This work was funded by National Institutes of Health grant, 1R01AA0174499 (A. M. Allan) and the UNM institutional NIAAA training grant, T32-AA014127 (C. F. Valenzuela).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aoto K, Shikata Y, Higashiyama D, Shiota K, Motoyama J. Fetal ethanol exposure activates protein kinase A and impairs Shh expression in prechordal mesendoderm cells in the pathogenesis of holoprosencephaly. Birth Defects Research. Part A, Clinical and Molecular Teratology. 2008;82:224–231. doi: 10.1002/bdra.20447. [DOI] [PubMed] [Google Scholar]
- Barak S, Carnicella S, Yowell QV, Ron D. Glial cell line-derived neurotrophic factor reverses alcohol-induced allostasis of the mesolimbic dopaminergic system: implications for alcohol reward and seeking. The Journal of Neuroscience. 2011;31:9885–9894. doi: 10.1523/JNEUROSCI.1750-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton A, Fendrik AJ. Sustained vs. oscillating expressions of Ngn2, Dll1 and Hes1: a model of neural differentiation of embryonic telencephalon. Journal of Theoretical Biology. 2013;328:1–8. doi: 10.1016/j.jtbi.2013.03.004. [DOI] [PubMed] [Google Scholar]
- Brady ML, Allan AM, Caldwell KK. A limited access mouse model of prenatal alcohol exposure that produces long-lasting deficits in hippocampal-dependent learning and memory. Alcoholism: Clinical and Experimental Research. 2012;36:457–466. doi: 10.1111/j.1530-0277.2011.01644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylund M, Andersson E, Novitch BG, Muhr J. Vertebrate neurogenesis is counteracted by Sox1–3 activity. Nature Neuroscience. 2003;6:1162–1168. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- Carnicella S, Kharazia V, Jeanblanc J, Janak PH, Ron D. GDNF is a fast-acting potent inhibitor of alcohol consumption and relapse. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:8114–8119. doi: 10.1073/pnas.0711755105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Charness ME. Ethanol disrupts axon outgrowth stimulated by netrin-1, GDNF, and L1 by blocking their convergent activation of Src family kinase signaling. Journal of Neurochemisty. 2012;123:602–612. doi: 10.1111/j.1471-4159.2012.07954.x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chen Y, Ozturk NC, Zhou FC. DNA methylation program in developing hippocampus and its alteration by alcohol. PLoS One. 2013;8:e60503. doi: 10.1371/journal.pone.0060503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi IY, Allan AM, Cunningham LA. Moderate fetal alcohol exposure impairs the neurogenic response to an enriched environment in adult mice. Alcoholism: Clinical and Experimental Research. 2005;29:2053–2062. doi: 10.1097/01.alc.0000187037.02670.59. [DOI] [PubMed] [Google Scholar]
- Da Lee R, Rhee GS, An SM, Kim SS, Kwack SJ, Seok JH, et al. Differential gene profiles in developing embryo and fetus after in utero exposure to ethanol. Journal of Toxicology and Environmental Health. Part A. 2004;67:2073–2084. doi: 10.1080/15287390490515001. [DOI] [PubMed] [Google Scholar]
- Downing C, Flink S, Florez-McClure ML, Johnson TE, Tabakoff B, Kechris KJ. Gene expression changes in C57BL/6J and DBA/2J mice following prenatal alcohol exposure. Alcoholism: Clinical and Experimental Research. 2012;36:1519–1529. doi: 10.1111/j.1530-0277.2012.01757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Hamre K. Identity and neuroanatomical localization of messenger RNAs that change expression in the neural tube of mouse embryos within 1 h after ethanol exposure. Brain Research. Developmental Brain Research. 2003;144:9–23. doi: 10.1016/s0165-3806(03)00135-4. [DOI] [PubMed] [Google Scholar]
- Escudero C, Bertoglia P, Hernadez M, Celis C, Gonzalez M, Aguayo C, et al. Impaired A2A adenosine receptor/nitric oxide/VEGF signaling pathway in fetal endothelium during late- and early-onset preeclampsia. Purinergic Signalling. 2013;9:215–226. doi: 10.1007/s11302-012-9341-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. Journal of Neuroscience Research. 2005;79:734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nature Neuroscience. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Schumacher N, Maier M, Sendtner M, Gessler M. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes & Development. 2004;18:901–911. doi: 10.1101/gad.291004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier NM, Lee B, Banasr M, Elsayed M, Duman RS. Vascular endothelial growth factor regulates adult hippocampal cell proliferation through MEK/ERK- and PI3K/Akt-dependent signaling. Neuropharmacology. 2012;63:642–652. doi: 10.1016/j.neuropharm.2012.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green ML, Singh AV, Zhang Y, Nemeth KA, Sulik KK, Knudsen TB. Reprogramming of genetic networks during initiation of the Fetal Alcohol Syndrome. Developmental Dynamics. 2007;236:613–631. doi: 10.1002/dvdy.21048. [DOI] [PubMed] [Google Scholar]
- Hack SP, Christie MJ. Adaptations in adenosine signaling in drug dependence: therapeutic implications. Critical Reviews in Neurobiology. 2003;15:235–274. doi: 10.1615/critrevneurobiol.v15.i34.30. [DOI] [PubMed] [Google Scholar]
- Hard ML, Abdolell M, Robinson BH, Koren G. Gene-expression analysis after alcohol exposure in the developing mouse. The Journal of Laboratory and Clinical Medicine. 2005;145:47–54. doi: 10.1016/j.lab.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Hashimoto-Torii K, Kawasawa YI, Kuhn A, Rakic P. Combined transcriptome analysis of fetal human and mouse cerebral cortex exposed to alcohol. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4212–4217. doi: 10.1073/pnas.1100903108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haycock PC. Fetal alcohol spectrum disorders: the epigenetic perspective. Biology of Reproduction. 2009;81:607–617. doi: 10.1095/biolreprod.108.074690. [DOI] [PubMed] [Google Scholar]
- Hicks SD, Middleton FA, Miller MW. Ethanol-induced methylation of cell cycle genes in neural stem cells. Journal of Neurochemistry. 2010;114:1767–1780. doi: 10.1111/j.1471-4159.2010.06886.x. [DOI] [PubMed] [Google Scholar]
- Jacobs JS, Miller MW. Proliferation and death of cultured fetal neocortical neurons: effects of ethanol on the dynamics of cell growth. Journal of Neurocytology. 2001;30:391–401. doi: 10.1023/a:1015013609424. [DOI] [PubMed] [Google Scholar]
- Jégou S, El Ghazi F, de Lendeu PK, Marret S, Laudenbach V, Uguen A, et al. Prenatal alcohol exposure affects vasculature development in the neonatal brain. Annals of Neurology. 2012;72:952–960. doi: 10.1002/ana.23699. [DOI] [PubMed] [Google Scholar]
- Kajimoto K, Allan A, Cunningham LA. Fate analysis of adult hippocampal progenitors in a murine model of fetal alcohol spectrum disorder (FASD) PLoS One. 2013;8:e73788. doi: 10.1371/journal.pone.0073788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KC, Go HS, Bak HR, Choi CS, Choi I, Kim P, et al. Prenatal exposure of ethanol induces increased glutamatergic neuronal differentiation of neural progenitor cells. Journal of Biomedical Science. 2010;17:85. doi: 10.1186/1423-0127-17-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiber ML, Mantha K, Stringer RL, Singh SM. Neurodevelopmental alcohol exposure elicits long-term changes to gene expression that alter distinct molecular pathways dependent on timing of exposure. Journal of Neurodevelopmental Disorders. 2013;5:6. doi: 10.1186/1866-1955-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Marchetto MC, Guo JU, Ming GL, Gage FH, Song H. Epigenetic choreographers of neurogenesis in the adult mammalian brain. Nature Neuroscience. 2010;13:1338–1344. doi: 10.1038/nn.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendez RI, McGinty JF, Kalivas PW, Becker HC. Brain region-specific gene expression changes after chronic intermittent ethanol exposure and early withdrawal in C57BL/6J mice. Addiction Biology. 2012;17:351–364. doi: 10.1111/j.1369-1600.2011.00357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamide R, Fujiwara K, Hasegawa K, Yoshikawa K. Antagonistic interplay between necdin and Bmi1 controls proliferation of neural precursor cells in the embryonic mouse neocortex. PLoS One. 2014;9:e84460. doi: 10.1371/journal.pone.0084460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda RC, Santillano DR, Camarillo C, Dohrman D. Modeling the impact of alcohol on cortical development in a dish: strategies from mapping neural stem cell fate. Methods in Molecular Biology. 2008;447:151–168. doi: 10.1007/978-1-59745-242-7_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam HW, Bruner RC, Choi DS. Adenosine signaling in striatal circuits and alcohol use disorders. Molecules and Cells. 2013;36:195–202. doi: 10.1007/s10059-013-0192-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam HW, Hinton DJ, Kang NY, Kim T, Lee MR, Oliveros A, et al. Adenosine transporter ENT1 regulates the acquisition of goal-directed behavior and ethanol drinking through A2A receptor in the dorsomedial striatum. The Journal of Neuroscience. 2013;33:4329–4338. doi: 10.1523/JNEUROSCI.3094-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogony JW, Malahias E, Vadigepalli R, Anni H. Ethanol alters the balance of Sox2, Oct4, and Nanog expression in distinct subpopulations during differentiation of embryonic stem cells. Stem Cells and Development. 2013;22:2196–2210. doi: 10.1089/scd.2012.0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins A, Lehmann C, Lawrence RC, Kelly SJ. Alcohol exposure during development: Impact on the epigenome. International Journal of Developmental Neuroscience. 2013;31:391–397. doi: 10.1016/j.ijdevneu.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering C, Wicher G, Rosendahl S, Schiöth HB, Fex-Svenningsen A. A low ethanol dose affects all types of cells in mixed long-term embryonic cultures of the cerebellum. Basic & Clinical Pharmacology & Toxicology. 2010;106:472–478. doi: 10.1111/j.1742-7843.2009.00528.x. [DOI] [PubMed] [Google Scholar]
- Riley EP, Infante MA, Warren KR. Fetal alcohol spectrum disorders: an overview. Neuropsychology Review. 2011;21:73–80. doi: 10.1007/s11065-011-9166-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitbak T, Thomas K, Martin A, Allan A, Cunningham LA. Moderate fetal alcohol exposure impairs neurogenic capacity of murine neural stem cells isolated from the adult subventricular zone. Experimental Neurology. 2011;229:522–525. doi: 10.1016/j.expneurol.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussa E, Krieglstein K. GDNF promotes neuronal differentiation and dopaminergic development of mouse mesencephalic neurospheres. Neuroscience Letters. 2004;361:52–55. doi: 10.1016/j.neulet.2003.12.106. [DOI] [PubMed] [Google Scholar]
- Rubert G, Miñana R, Pascual M, Guerri C. Ethanol exposure during embryogenesis decreases the radial glial progenitorpool and affects the generation of neurons and astrocytes. Journal of Neuroscience Research. 2006;84:483–496. doi: 10.1002/jnr.20963. [DOI] [PubMed] [Google Scholar]
- Schänzer A, Wachs FP, Wilhelm D, Acker T, Cooper-Kuhn C, Beck H, et al. Direct stimulation of adult neural stem cells in vitro and neurogenesis in vivo by vascular endothelial growth factor. Brain Pathology. 2004;14:237–248. doi: 10.1111/j.1750-3639.2004.tb00060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojo H, Ohtsuka T, Kageyama R. Dynamic expression of notch signaling genes in neural stem/progenitor cells. Frontiers in Neuroscience. 2011;5:78. doi: 10.3389/fnins.2011.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun CN, Chuang HC, Wang JY, Chen SY, Cheng YY, Lee CF, et al. The A2A adenosine receptor rescues neuritogenesis impaired by p53 blockage via KIF2A, a kinesin family member. Developmental Neurobiology. 2010;70:604–621. doi: 10.1002/dneu.20802. [DOI] [PubMed] [Google Scholar]
- Tyler CR, Allan AM. Adult hippocampal neurogenesis and mRNA expression are altered by perinatal arsenic exposure in mice and restored by brief exposure to enrichment. PLoS One. 2013;8:e73720. doi: 10.1371/journal.pone.0073720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela CF, Morton RA, Diaz MR, Topper L. Does moderate drinking harm the fetal brain? Insights from animal models. Trends in Neurosciences. 2012;35:284–292. doi: 10.1016/j.tins.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vangipuram SD, Lyman WD. Ethanol affects differentiation-related pathways and suppresses Wnt signaling protein expression in human neural stem cells. Alcoholism: Clinical and Experimental Research. 2012;36:788–797. doi: 10.1111/j.1530-0277.2011.01682.x. [DOI] [PubMed] [Google Scholar]
- Veazey KJ, Carnahan MN, Muller D, Miranda RC, Golding MC. Alcohol-induced epigenetic alterations to developmentally crucial genes regulating neural stemness and differentiation. Alcoholism: Clinical and Experimental Research. 2013;37:1111–1122. doi: 10.1111/acer.12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver DR. A2a adenosine receptor gene expression in developing rat brain. Brain Research. Molecular Brain Research. 1993;20:313–327. doi: 10.1016/0169-328x(93)90058-w. [DOI] [PubMed] [Google Scholar]
- Wen S, Li H, Liu J. Dynamic signaling for neural stem cell fate determination. Cell Adhesion & Migration. 2009;3:107–117. doi: 10.4161/cam.3.1.7602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou FC. DNA methylation program during development. Frontiers in Biology. 2012;7:485–494. doi: 10.1007/s11515-012-9246-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou FC, Balaraman Y, Teng M, Liu Y, Singh RP, Nephew KP. Alcohol alters DNA methylation patterns and inhibits neural stem cell differentiation. Alcoholism: Clinical and Experimental Research. 2011;35:735–746. doi: 10.1111/j.1530-0277.2010.01391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZD, Kumari U, Xiao ZC, Tan EK. Notch as a molecular switch in neural stem cells. IUBMB Life. 2010;62:618–623. doi: 10.1002/iub.362. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.