Abstract
A series of bifunctional compounds was prepared consisting of 17β estradiol linked to a DNA damaging N,N-bis(2-chloroethyl)-aniline. The objective of our studies was to detem1ine the characteristics of the linker that permitted both reaction with DNA and binding of the resultant covalent adducts to the estrogen receptor. Linker characteristics were pivotal determinants underlying the ability of the compounds to kill selectively breast cancer cells that express the estrogen receptor.
1. Introduction
Many antitumor drugs produce their cytotoxic effect through covalent damage to DNA.1;2 Repair enzymes that remove such covalent lesions, restoring the integrity of genetic information, limit the effectiveness of these drugs. Inhibition of DNA repair in cancer cells is an attractive yet not significantly explored strategy to potentiate the therapeutic effects of alkylating chemotherapeutic drugs.3;4 We have previously described the synthesis and biological activity of novel bifunctional compounds that form covalent DNA adducts with high affinity for the estrogen receptor (ER)5;6 The compounds were designed to test the hypothesis that adduct-ER complexes would be concealed from DNA repair proteins and therefore be refractory to repair. The compounds would thus exhibit greater toxicity in cancer cells that over express the ER protein.
The original series of compounds consisted of an N,N-bis-chloroethylaniline cormected to a 2-phenylindole (2PI) group by alkyl-amino-carbamate linkers of various lengths5. Investigations with these derivatives identified several molecular characteristics of the linker that were important for the compound to react with DNA and bind to the ER. A compound with greater affinity for the ER was obtained by replacing the 2-PI group with that of 17β-estradiol c01mected via the 7α position.6 Both the 2PI- and estradiol-based compounds showed increased toxicity toward breast cancer cells that express the ER. Evidence was presented that the differential toxicity of the DNA damaging agents was not due to an anti-honnonal mechanism.
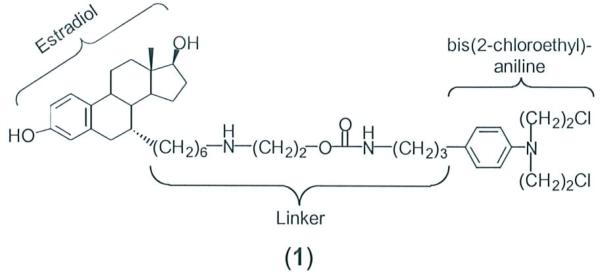
We investigated the molecular features of the linker that are essential for the biological activities of the estradiol-linked compound 1 (Fig. 1) by varying the structure of the linker c01mecting the N,N-bis-(2-chloroethyl)aniline and estradiol moiety. The roles of the amino and carbamate groups in the linker were of particular interest because their substitution with different chemical groups might lead to a less complex synthetic process and increase overall yields. The modified linkers retained the six carbon alkyl chain that appears, based on our earlier work, to be essential for the attached ligand to fit into the estradiol binding site of the ER.5 Amino-, ami do- and guanidino- groups were incorporated into the region connecting the hexanyl-substituted estradiol and the aniline group. We report here the synthesis and physical properties of these molecules and their cytotoxic effects toward breast cancer cells.
Figure 1.
Structure and molecular features of lead compound.
2. Chemistry
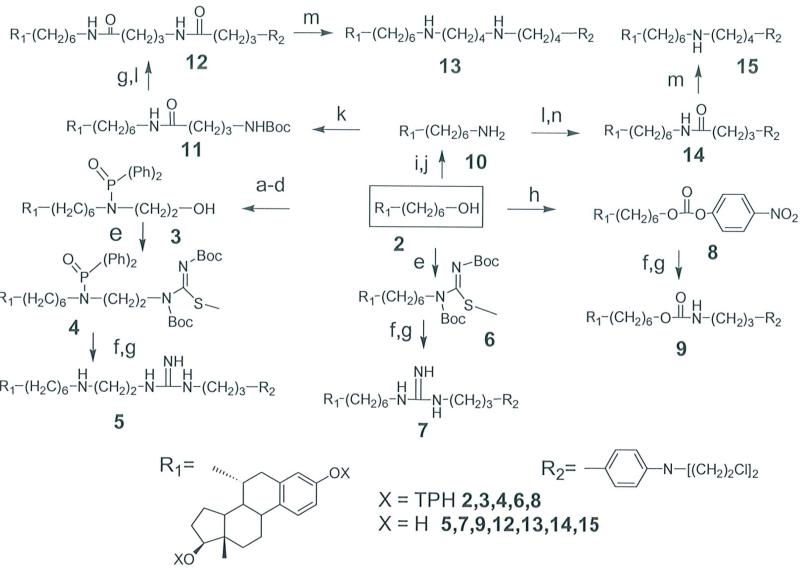
The synthesis of compound 1 was described in our previous report.6 The syntheses utilized 3,17β-bis(2-tetrahydropyranyloxy)-7α-(6-hydroxyhexan-1-yl)-estra-1,3,5(1O)triene 2 as the starting compound (see box in Scheme 1); its preparation has also been described.6 Construction of linkers proceeded by linear additions to 2 with final addition of the N,N-bis(2-chloroethyl)aniline moiety. Compound 5 was prepared by conversion alcohol 2 to the bromide, which was allowed to react with a protected ethanolamine providing 3 as described in reference 10. The Mitsunobo reaction7 then was used to couple 1,3-bis(tert-butoxycarbonyl)-2-methyl-2-thiopseudourea with 3. Reaction of the resulting product 4 with excess (N,N-bis-2-chloroethylaminophenyl)-propylamine followed by acid deprotection produced 5. Procedures described by Litmey et al.7 were applied to incorporate an N,N-disubstituted guanidine moiety into the linker. The preparation of 7 proceeded with the initial reaction of 2 with 1,3-bis(tert-butoxycarbonyl)-2-methyl-2-thiopseudourea under Mitsunobu conditions.7 The resulting product 6 was then allowed to react with an excess of (N,N-bis-2-chloroethylaminophenyl)-propylamine followed by acid deprotection to furnish 7. Compound 9 was prepared by conversion of 2 to the p-nitrophenyl carbonate 8, which was then allowed to react with (N,N-bis-2-chloroethylaminophenyl)-propylamine.6 Removal ofTHP groups under acidic conditions produced 9. Compound 10, the starting material for compounds 12-15, was prepared by hydrazinolysis of the phthalimide of 2 that was formed under Mitsunobu conditions. 8 Compound 12 containing two amide groups was synthesized by first reacting 10 with the NHS ester of 4(tert-butoxycarbonylamino)butyric acid. Following removal of the THP and Boc groups the terminal amino group was allowed to react with the NHS ester of chlorambucil producing 12. Compound 13 in which the linker contains two amino groups was produced by reduction of 12 with borane dimethylsulfide complex.9 The preparation of 14 was also accomplished by conversion of 2 to the phthalimide via Mitsunobo conditions with subsequent hydrazinolysis to obtain the amine 10. The NHS ester of chlorambucil was then allowed to react with the terminal amine, producing 14. Compound 15, containing a secondary amino group in the linker, was prepared by reduction of the amide in 14 using borane dimethylsulfide complex.9
Scheme 1.
Synthesis of estradiol linked aniline mustards. The starting compound 2 is shown in the box (center). Reagents and conditions: (a) methanesulfonyl chloride, DIEA, THF; (b) LiBr, TEA, DMF, 60°C; (c) Ph2P(O)NHCH2CH20TBDMS, NaH, TBAB, Ph-H, 60°C; (d) TBAF, THF; (e) Boc-NHC(=N-Boc)SCH3, PPh3, DIP AD, THF; (f) 4-(N,N-bis-2-chloroethylamino-phenyl)-propylamine, THF/H20(90: 10), reflux; (g) HCl/dioxane, CH2Cl2; (h) p-nitrophenylchloroformate, DIEA, THF; (i) phthalimide, DIP AD, PPh3, THF; (j) Hydrazine, EtOH, reflux; (k) 4-(N-Boc)n-butyl(N-hydroxysuccinimide)ester, TEA, DMF; (1) 4-(N,N-bis-chloroethylamino-phenyl)butyl(N -hydroxysuccinimide )ester, TEA, DMF; (m) BH3S(CH3)2 THF, HCl; (n) HCl, THF. DIEA= Diisopropylethyl amine; TEA= Triethyl amine; TBAB= Tetrabutylammonium bromide;TBAF= Tetrabutylammonium fluoride; DIP AD= Diisopropylethyl azadicarboxylate; DMF= Dimethylformamide; THF= Tetrahydrofuran
3. Biology
In the initial characterization of the biochemical properties of new compounds 5, 7, 9, 12- 15 we evaluated their affinities for the ER. A radiometric competitive binding assay10 with the rabbit uterine ER was used to determine the relative binding affinity (RBA) of each compound for the ERas compared with estradiol; RBA=100. All of the compounds exhibited some affinity for the ER. The data in Table 1 show that the new compounds have RBA values for the ER ranging from 6 to 40. Among the new compounds, 15 containing a single amino group in the linker had an RBA of 40 which is comparable to 1.
Table 1.
| Compound | RBA | % Oligo Modified | % Oligo Shifted | logP | logD | ED30 (uM) | |
|---|---|---|---|---|---|---|---|
| MCF-7 | MDA-MB231 | ||||||
| 1 | 46 | 45 | 93 | 5.32 | 2.22 | 5.1 | 9.6 |
| 5 | 10 | 29 | 93 | 6.27 | 3.12 | >20 | >20 |
| 7 | 28 | 58 | 38 | 5.67 | 2.52 | >20 | >20 |
| 9 | 6 | 3 | ND | 5.61 | 5.61 | >20 | >20 |
| 12 | 29 | 39 | ND | 4.85 | 4.85 | >20 | >20 |
| 13 | 29 | 79 | 38 | 5.32 | 2.22 | 5.1 | 7.9 |
| 14 | 13 | 14 | ND | 5.07 | 5.07 | >20 | >20 |
| 15 | 40 | 44 | 67 | 5.96 | 2.87 | 5.1 | 8.6 |
RBA = relative binding affinity for the rabbit uterine ER as compared to estradiol (RBA = 100); %01igo modified = percent of 16-mer cleaved by piperidine after treatment with test compound for 24 h at 37°C; %01igo shifted = percent of covalently modified 16-mer that formed a slowly migrating complex with the ER-LBD under electrophoresis; ED30 = concentration of compound that resulted in 30% clonal survival of cells exposed for 2 h.
Although it is apparent that the original combination of the positively charged secondary amine with the neutral carbamyl group (compound 1) results in a bifunctional compound with excellent affinity for the rabbit uterine ER, compounds 7, 12 and 13 also have good affinities. These molecules were viewed as valuable assets as we move ahead toward probing structure-activity relationships and the biochemical mechanisms underlying the biological activity of 1. It is likely that our 7α-link:ed estradiol compounds adopt a binding mode similar to that identified for the 7α-undecylamide estradiol analog ICI 164,384.11 The positioning and orientation ofthe estradiol moiety ofiCI-164,384 within the hydrophobic binding cavity of the ER is directed by its 7α side chain, which protrudes out of a hydrophobic channel extending from the binding pocket. At the surface ofthe LBD, a 90° flexion of the undecyl chain enables the remainder of the linker to track closely with the surface contours of the LBD. 11 The low RBAs of compounds 5, 9 and 14 may result from surface interactions adopted by the linkers in these molecules that do not permit optimal alig1m1ent of the estradiol moiety within the binding cavity.
The reactivity of each compound with DNA was assessed by its ability to produce piperidine labile sites in the self complementary deoxyoligonucleotide 5’-d(AA TATTGGCCAA TATT). The results in Table 1 (column 3) indicate the percent ofthe oligomer that was cleaved by piperidine. Compound 9 in which the alkyl linker contains a single carbamyl group produced the lowest level of modification (i.e., 3% cleaved by piperidine). Compound 14 containing an amido instead of the carbamyl group produced approximately five times the number ofDNA adducts (14% cleaved by piperidine). High levels of reactivity towards DNA were observed with compounds with linkers containing secondary amino groups. The combination of the amino and carbamyl groups in the linker of 1 resulted in a 10-fold increase in reactivity over 9 in which the linker contains only the carbamyl group. The reactivity of 1 was similar to that of 15 in which the linker contains a single secondary amine suggesting that the charged amino group is the major determinant of reaction rate. Compound 13 in which the linker contains a diamine -NH-(CH2)NH-CH2- was the most reactive (79% cleaved by piperidine). The same is likely the case for molecules 5 and 7 in which the strongly basic guanidino groups would be cationic under assay conditions. It is likely that the cationic nature of these molecules gives them a high reactivity with DNA by localizing the reactive alkylating group in the vicinity of nucleophilic atoms. A similar result has been repmied for a conjugate of chlorambucil with the polyamine spetmidine.12
Using an electrophoretic gel mobility shift assay6, we observed that covalent DNA adducts of 1, 5, 7, 13 and 15 form complexes with the portion of the ER containing the ligand binding domain (ER-LBD) (Table 1, column 4). Under conditions that allowed complex formation, addition of the ER to the modified DNAs resulted in the appearance of a slowly migrating band by electrophoresis that was eliminated by addition of excess competitor, estradiol (data not shown). The results in Table 1 (column 4) indicate that the extent of complex formation for 1, 7, 13 and 15 were colTelated with the RBAs of the unreacted compounds. The exception was compound 5 in which the linker contained both amino and guanidino groups. In this case, despite its low RBA, virtually all of the modified oligonucleotide fanned a slowly migrating band. We do not know the basis for this unexpected finding.
LogP and logD values can be predictive of aqueous solubility, absorbtion and permeability.13 The lipophilicities of 1, 5, 7, 9, 12- 15 were assessed using an HPLC method to estimate the logP of the neutral fmm of each compound. LogD values at pH 7.4 were estimated using an equation derived by Horvath et al.14 for basic compounds. The logD values in Table 1 indicated that the aqueous solubilities of the eight compounds span approximately a 2,500-fold range under physiological conditions. The compounds with logD values >5 (compounds 9 and 14) had both low affinities for the ER and low reactivity with DNA. Compounds containing charged groups with calculated logD values <3 generally had the highest affinities for the ER along with the greatest reactivities towards DNA. These relationships, however, did not prove to be reliable predictors of biological activities in cytotoxicity assays against breast cancer cells.
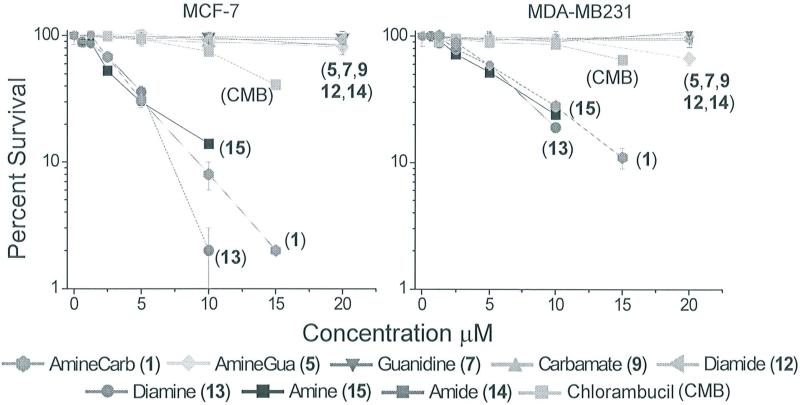
The lethal effects of our new compounds were investigated in the MCF-7 (ER+) and MDAMB231 (ER-) breast cancer cell lines. The data shown in Figure 2 and Table 1 indicate that most but not all of the modifications that were introduced in the linker resulted in decreased toxicity towards both cell lines. The low toxicity of 5 and 7, which contain guanindinium groups, may be related to either their poor uptake by cells or their rapid excretion once absorbed.15 Despite showing reactivity towards DNA in vitro, neither compound showed significant toxicity at the highest dose; i.e., 20 μM. Lack of uptake may also be responsible for the low toxicity of 9,12 and 14, which have high logD values that are not predictive of good absorption.13 Further work is warranted to determine if cellular uptake is indeed limiting for these compounds.
Figure 2.
Survival ofMCF-7 (ER+) and MDA-MB231 (ER−) breast cancer cells after 2 hr exposure to estradiol-linked toxicants.
As previously reported, 1 was significantly more toxic toward MCF-7 cells than MDA-MB231 cells.6 Compounds 13 and 15 containing amino groups showed toxicity similar to that of l . Both of these compounds also showed greater toxicity toward the ER-positive MCF-7 cells than towards the ER-negative MDA-MB231 cells. This result was consistent with our intended mechanisms, since the RBAs and reactivities with DNA of 13 and 15 imply greater toxicity on ER-positive cells. It is interesting that the results of the electrophoretic mobility shift assay indicate that DNA adducts of 1 have the greatest affinity for the ER-LBD; compound 1 also shows the largest differential toxicity between the two cell lines.
4. Conclusion
We designed and synthesized a series of estradiol-aniline mustard-linked bifunctional molecules that differ in their relative affinities for the ER and their capacities to covalently modify DNA. The selective cytotoxic effects of3 of these compounds (1, 13, 15) towards ER+ breast cancer cells correlated with their ability to react with DNA and their affinity for the ER, as well as favorable solubility. We are directing research to determine if the interaction ofthe ER with DNA adducts formed by these compounds is responsible for their selective effects.
Supplementary Material
Acknowledgements
Research was supported by NIH grant CA77743, the Natural Sciences and Engineering Research Council of Canada (PDF-242490-2001 Fellowship to AND), an Award from the Susan B. Komen Foundation, and NIH grant 1SS10RR13386-01 for support of the NMR and the mass spectrometry facility at MIT.
References and Notes
- 1.Teicher BA. Antitumor alkylating agents. In: DeVita VT, Hellman S, Rosenberg SA, editors. Cancer Principles and Practice. Vol. 97. Lippincott-Raven; New York: pp. 405–417. [Google Scholar]
- 2.Hurley LH. Nature Rev. Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- 3.Barret JM, Hill BT. Anticancer Drugs. 1998;9:105–123. doi: 10.1097/00001813-199802000-00001. [DOI] [PubMed] [Google Scholar]
- 4.Dolan ME. Adv.Drug Del.Rev. 1997;26:105–118. doi: 10.1016/s0169-409x(97)00028-8. [DOI] [PubMed] [Google Scholar]
- 5.Rink SM, Yarema KJ, Solomon MS, Paige LA, Tadayoni-Rebek BM, Essigmam1 JM, Croy RG. Proc.Natl.Acad.Sci.U.S.A. 93:15063–15068. doi: 10.1073/pnas.93.26.15063. 12-24-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitra K, Marquis JC, Hillier SM, Rye PT, Zayas B, Lee AS, Essigmmm JM, Croy RG. JAmer.Chem.Soc. 2002;124:1862–1863. doi: 10.1021/ja017344p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linney ID, Buck IM, Harper EA, Kalindjian B, Pether M. j., Shankley NP, Watt GF, Wright PT. J. Med.Chem. 2000;43:2362–2370. doi: 10.1021/jm990952j. [DOI] [PubMed] [Google Scholar]
- 8.Hughes DL. Organic Reactions. John Wiley and Sons; New York: 1992. pp. 335–656. [Google Scholar]
- 9.Brown HC, Heim PJ. Org. Chem. 1973;38:912–916. [Google Scholar]
- 10.Korenman SG. Endocrinology. 1970;87:1119–1123. [Google Scholar]
- 11.Pike ACW, Brzozowski AM, Walton J, Hubbard RE, Thorsell A-G, Li Y-L, Gustafsson J-Å, Carlquist M. Structure. 2001;9:145–153. doi: 10.1016/s0969-2126(01)00568-8. [DOI] [PubMed] [Google Scholar]
- 12.Holley JL, Mather A, Wheelhouse RT, Cullis PM, Hartley JA, Bingham JP, Cohen GM. Cancer Res. 1992;52:4190–4195. [PubMed] [Google Scholar]
- 13.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Adv.Drug Del.Rev. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 14.Horváth Cs., Melánder W, Molnár I. Anal.Chem. 1977;49:865. [Google Scholar]
- 15.Zhang L, Brett CM, Giacomini KM. Annu.Rev.Pharmacol.Toxicol. 1998;38:431–460. doi: 10.1146/annurev.pharmtox.38.1.431. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.