Abstract
Purpose
The levels and timing of expression of genes like BCLXL, HDAC1 and pluripotency marker genes namely, OCT4, SOX2, NANOG and KLF4 are known to influence preimplantation embryo development. Despite this information, precise understanding of their influence during preimplantation embryo development is lacking. The present study attempts to compare the expression of these genes in the in vivo and in vitro developed preimplantation embryos.
Methods
The in vivo and in vitro developed rabbit embryos collected at distinct developmental stages namely, pronuclear, 2 cell, 4 cell, 8 cell, 16 cell, Morula and blastocyst were compared at the transcriptional and translational levels using Real Time PCR and immunocytochemical studies respectively.
Results
The study establishes the altered levels of candidate genes at the transcriptional level and translational level with reference to the zygotic genome activation (ZGA) phase of embryo development in the in vivo and in vitro developed embryos. The expression of OCT4, KLF4, NANOG and SOX2 genes were higher in the in vitro developed embryos whereas and HDAC1 was lower. BCLXL expression had its peak at ZGA in in vivo developed embryos. Protein expression of all the candidate genes was observed in the embryos. BCLXL, KLF4 and NANOG exhibited diffused localisation whereas HDAC1, OCT4, and SOX2 exhibited nuclear localisation.
Conclusions
This study leads to conclude that BCLXL peak expression at the ZGA phase may be a requirement for embryo development. Further expression of all the candidate genes was influenced by ZGA phase of development at the transcript level, but not at the protein level.
Keywords: Pluripotency markers, Rabbit embryos, Preimplantation development, Zygotic genome activation
Introduction
The preimplantation embryo development involves synchronous or asynchronous rapid cell divisions and generation of genetic and epigenetic cues for differentiation at specific developmental stages. Insight into these processes could serve to improve the efficiency of assisted reproduction technologies, development of transgenics, and to understand nuclear reprogramming. However, the embryos developed in vivo and in vitro show differences in their developmental potential [1–3] resulting from differences in gene expression patterns [4–6]. As the genes of mature spermatozoon and oocyte are silent, and remain inactive after fertilisation, maternal transcripts form a major source for mRNA during the initial stages of embryo development; until the zygotic Genome Activation (ZGA) occurs [7, 8]. The ZGA occurs at 2-cell stage in murine, at 4-cell stage in porcine, at 4-cell to 8-cell stage in human and at 8 to 16-cell stage in bovine, rabbit and ovine embryos respectively [9–11]. The ZGA is manifested by strict regulation of genes in a temporal and stage specific manner [12, 13]. Pluripotency markers, apoptotic regulators and epigenetic regulators play significant roles in the preimplantation embryo development [13, 14].
The pluripotency markers namely OCT4, SOX2, NANOG, CMYC, LIN28 and KLF4 were used for pluripotency induction in somatic cells; and among them, OCT4, NANOG, SOX2 and KLF4 were implicated in preimplantation embryo development [15–19]. The importance of OCT4 in ZGA and cell lineage formation was reported in mouse and human preimplantation development studies [17, 20–22]. Knock-down of OCT4 expression in the mouse embryos had caused them to fail in ZGA and blastocyst formation. However, NANOG and SOX2 expressions were not altered by the OCT4 knock-down [21]. OCT4, NANOG and SOX2 were expressed in all the developmental stages from ZGA to Inner Cell Mass (ICM) of human blastocyst embryos and regulated the stemness of blastomeres [22]. In zebra fish, ZGA required the expression of NANOG, SOXB1 and OCT4 [23]. Over-expression and knock-down studies of SOX2 conducted in the mouse 2-cell embryos did not influence OCT4 and NANOG expression. However its over-expression blocked the ZGA and cleavage [24]. To bring about pluripotency in post-implantation epithelial embryonic stem cells, the expression of OCT4, SOX2 and NANOG were not sufficient and KLF4 was needed [25]. These studies have shown that the networks in Embryonic Stem Cells (ESCs) and preimplantation development could differ and be unique.
Apoptotic regulators include the BCL2 family of proteins, which have a BH domain. Among which the BAX/BCLXL ratio indicates survival or death of a cell [26, 27]. BCLXL was required for cell survival and development through the ZGA phase in murine preimplantation embryos and the micro-injection of BCLXL could rescue embryos and aid in ZGA [28, 29]. It was also shown that BCLXL has anti-proliferative effects along with anti-apoptotic properties in murine cancer cell lines [30]. BCLXL interactions for cell survival are not limited to the BCL2 family of proteins. It also interacts with VDAC1 for cell survival as shown by the studies in human cell line [31].
Among the epigenetic regulators, HDAC1 which is a class I histone deacetylase, interacts with other epigenetic regulators namely, methyl transferases and deacetylases and regulates gene expression during embryo development [32, 33]. Maternally contributed HDAC1 maintained a steady state of acetylation during ZGA and thus the developmental potential of embryo [34]. HDAC1, the primary histone deacetylase, was the most sensitive to hyperacetylation in murine preimplantation embryos.
Several other factors may also be influencing the preimplantation development. Therefore, a comparison of gene expression patterns of pluripotency markers, apoptotic regulators and epigenetic regulators in the in vivo and in vitro developed embryos in a stage wise manner, from pronuclear embryo to blastocyst, can provide deeper insights into molecular basis of embryo development. The present study was undertaken with rabbit (Oryctolagus cuniculus) oocytes and embryos to compare candidate gene expression patterns at transcriptional and translational levels, across various developmental stages of in vivo fertilized zygotes which were developed either in vivo or in vitro, with a view to understand what limits/hinders quality of preimplantation embryo development in vitro. Rabbit is a valuable experimental model for studies in regenerative medicine, reproductive biology and metabolic system studies, as it shares many biochemical and physiological properties with the human system [35, 36].
Earlier studies on rabbit embryo development were confined to the OCT4 expressions at transcriptional and translational levels [37–41] though it is very well established that BCLXL, HDAC1, SOX2, NANOG and KLF4 are also crucial for preimplantation embryo development. As more studies are needed in the context of preimplantation embryo development, this study was an attempt for the first time to monitor and compare expression of SOX2, NANOG, KLF4, HDAC1 and BCLXL simultaneously in addition to OCT4, both at the transcriptional and translational levels in rabbit preimplantation developmental stages (pronuclear, 2-cell, 4-cell, 8-cell, 16-cell, morula and blastocyst) between in vivo fertilized embryos, which were developed in vivo and in vitro. This study also tries for the first time to monitor the expression of candidate genes at transcript level and localisation of gene product simultaneously in in vivo and in vitro developed embryos.
Materials and methods
All the media and chemicals were purchased from Sigma Chemical Co. (St. Louis, MO, USA), and all plastic ware from NUNC (Rochester, NY, USA), unless mentioned otherwise. A total of 1,746 oocytes/different stages of embryos with an average of 24.6 from 71 does of 1 to 2 year old primiparous cross-bred (New Zealand white with White Giant and Soviet Chinchilla) rabbits were used for the experiments. This project was approved by the Centre for Cellular and Molecular Biology, Hyderabad, India - Institute Animal Ethics Committee vide letter No. IAEC 67/2008. Oocytes and embryos were collected in the Tissue Culture Medium (TCM199) (HEPES buffered) supplemented with heat inactivated 10 % Foetal Bovine Serum (FBS). Embryos were then cultured in TCM199 (bicarbonate buffered) supplemented with 15 % FBS. The handling and culture medium were supplemented with Penicillin (100 IU/mL) and Streptomycin (0.1 mg/mL), sterilized with 0.22 μm filter (Millipore, MA, USA) and equilibrated for at least 2 h prior to use at 38.5 °C in humidified atmosphere having 5 % CO2.
Superovulation, collection and culture of embryos
Superovulation and collection of the rabbit oocytes/embryos were carried out as described earlier [42]. Briefly, the does were superovulated with six equal doses (3 IU each) of Follicle Stimulating Hormone (FSH) administered subcutaneously at 12 h intervals. Twelve hours after the last dose of FSH, human Chorionic Gonadatrophin (hCG) (100 IU) (Chorulon, Intervet, Boxmeer, The Netherlands) was administered intravenously to induce ovulation. The oocytes were collected, 14 h post-hCG. For collection of embryos, the superovulated does were mated with a fertile buck prior to the administration of hCG. The in vivo developed pronuclear, 2-cell, 4-cell, 8-cell, 16-cell, morula and blastocyst stage embryos were collected at 18 h, 26 h, 32 h, 40 h, 48 h, 56 h and 86 h post-hCG respectively. For collection of in vitro developed embryos, the in vivo fertilized embryos were collected 14 h post-hCG and cultured at 38.5 °C in humidified atmosphere having 5 % CO2; and the embryos were collected at respective developmental stages. The in vitro embryos were collected after a 2 h delay to compensate for the delay in development of in vitro compared to in vivo developed embryos (unpublished results).
RNA isolation and RT-PCR
Prior to the RNA isolation using RNeasy mini kit (Qiagen GmbH, Hilden, Germany) different embryonic stages and oocytes in respective groups of 25 were collected randomly from the does and were denuded. The RNA isolation was repeated twice for each embryonic stage. The isolated total RNA was quantified in a NanoDrop spectrophotometer (Thermo scientific, Wilmington, DE, USA) and treated with Dnase I (Roche Applied Science, Mannheim, Germany) prior to reverse transcription. Reverse transcription was carried out using the Sensiscript RT kit (Qiagen GmbH, Hilden, Germany) using three replicates of RNA in each group. Each 20 μl cDNA synthesis reaction used a volume of RNA equivalent to five embryos/oocytes as the case may be. Oligo (dT) primers (Invitrogen, CA, USA) were used for priming the reaction. The cDNAs were then stored in aliquots at −20 °C.
Semi-quantitative PCR
Gene specific primers for candidate genes and reference genes (Table 1) were designed based on the sequences available in NCBI-GenBank nucleotide database using Fast PCR software [43]. For KLF4, NANOG and SOX2, the primers were designed from conserved regions identified by alignment of murine, human and bovine gene sequences in ClustalW2 [26]. Specific binding of the primers was confirmed based on expected amplicon sizes in agarose gel electrophoresis and by sequencing the amplicons in 3730 DNA Analyzer (Applied Biosystems, CA, USA).
Table 1.
Gene specific primers used in the study
| Gene | Accession number | Primer sequence | Amplicon size (bp) |
|---|---|---|---|
| BCLXL | NM_001082135.1 | 5′aggagatggaggtattggtgagtc3′ | 122 |
| 5′gttgccgtagagttccacaaac3′ | |||
| HDAC1 | XM_002720715.1 | 5′gctgaggagatgaccaagtacc3′ | 173 |
| 5′acagagccaccagtagacag3′ | |||
| H2A | AF030235.1 | 5′caaggcagtgtctcgctcacag3′ | 187 |
| 5′gagatccttggaagcgttacctgc3′ | |||
| HPRT1 | EF062857.1 | 5′ataagttctttgctgacctgctg3′ | 134 |
| 5′tacttttatgtcccctgttgactg3′ | |||
| KLF4 a | 5′ctcaaggcacacctgcgaac3′ | 193 | |
| 5′tcttcatgtgtaaggcgaggt3′ | |||
| NANOG a | 5′cagccctgattcttctaccagtcc3′ | 196 | |
| 5′ggagagttcttgcatctgctggag3′ | |||
| OCT4 | EF062856.1 | 5′ggtgttcagccaaaccaccatctg3′ | 352 |
| 5′ttgggaacagtcactgcttgatcg3′ | |||
| SOX2 a | 5′cgggtgctccttcatgtgcag3′ | 120 | |
| 5′cacaactcggagatcagcaagc3′ | |||
| YWHAZ | ENSOCUG00000000734 | 5′ggtctggcccttaacttctctgtgttcta3′ | 142 |
| 5′gcgtgctgtctttgtatgattcttcactt3′ |
aPrimers designed from conserved regions of human, murine and bovine sequences
Cyanine dye based real time amplification detection was done using the SYBR® GreenER qPCR Supermix (Invitrogen, CA, USA) in ABI PRISM® 7900HT Sequence Detection System (Applied Biosystems, CA, USA). Biological replicates of in vivo and in vitro developed embryos were used for the semi-quantitative PCR. The PCR programme used was: 50 °C for 2 min, initial hold at 95 °C for 10 min, followed by 40 cycles of denaturation (95 °C for 15 s), annealing (55–60 °C for 30 s) and extension (60 °C for 30 s). One-tenth equivalent template of the embryo or oocyte was used in each real time PCR experiment of 10 μl volume and was repeated thrice. Melting curve analysis was done to confirm the amplicon specificity. The oocyte (Metaphase II arrested) derived cDNA was used for evaluating amplification efficiency of all primer pairs using standard curve method. Amplification efficiency was calculated using the following equation from the slope of respective standard curves.
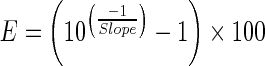 |
Statistical analysis of qPCR data
Reference genes were selected based on reports demonstrating that H2A, HPRT1 and YWHAZ were expressed stably throughout the rabbit preimplantation embryo development [37]. Data analysis was done based on the geometric averaging and normalisation method described as in geNorm [44]. Briefly, the mean quantification cycle value (Cq) of triplicates was transformed to quantities following comparative Cq method, and the highest relative quantity was set to one. Geometric mean of relative quantities of the three reference genes at a developmental stage was used as normalisation factor for calculating relative quantity of a candidate gene for that developmental stage. Statistical significance of the data within a developmental condition (in vivo or in vitro) was calculated using modified t-Test (Tukey’s test) at p < 0.05 and significance of the data between developmental conditions (in vivo vs in vitro) at different developmental stages was calculated using Student’s t-Test at p < 0.05.
Immunocytochemical studies
Oocytes/embryos were washed in Phosphate Buffered Saline (PBS) and then fixed in the freshly prepared 4 % paraformaldehyde. The membrane permeabilization was achieved by treating with 1 % TritonX100 in PBS for 15 min and the non-specific binding was blocked by treating with 1 % bovine serum albumin in PBS for 1 h. Subsequently, oocytes/embryos in groups of six or more were incubated with the mouse monoclonal antibodies (1:200 dilution) (Abcam, Cambridge, UK), raised against BCLXL (ab77571), HDAC1 (ab51846), KLF4 (ab75486), NANOG (ab62734), OCT4 (ab91194) and SOX2 (ab75485) at 4 °C overnight. The negative controls were prepared by omission of primary antibody. Samples were then incubated with FITC conjugated goat polyclonal antibody (1:500 dilution) (Abcam, Cambridge, UK) raised against the mouse IgG, for an hour at room temperature in dark. The oocytes and embryos were counterstained with 300 nM DAPI for 5 min at room temperature in dark and then mounted on a clean glass cover slip using VECTASHIELD mounting medium (Vector Labs, CA, USA) over concavity glass slides. The fluorescence emission spectra were documented using Leica TCS SP5 confocal system (Leica Microsystems CMS GmbH, Mannheim, Germany). The Leica Application Suite – Advanced Flourescence (LAS-AF) was used for scanning, capturing and analysing images. The total emission was quantified using LAS-AF. The regions of interest were designated and the difference of their intensities from background intensity was averaged and taken as the mean total intensity. The significance of intensities within a developmental (in vivo or in vitro) condition was calculated using Tukey’s test at P < 0.05 and the significance of data between developmental conditions (in vivo vs in vitro) at different developmental stages was calculated using Student’s t-Test at p < 0.05.
Results
All embryos used in this study were fertilized in vivo to rule out the variations resulting from in vitro fertilization on embryo quality. The percentage of in vitro blastocyst development for embryos cultured in TCM199 (bicarbonate buffered) was 80. The in vitro and in vivo developed rabbit preimplantation embryos from pronuclear stage to blastocyst were compared for expression levels of candidate genes. Multiple reference genes namely, H2A, HPRT1 and YWHAZ were used to reduce the bias in normalization [44]. Relative expression levels of candidate genes were normalised with respective normalisation factors derived from the reference gene expressions.
Comparison of BCLXL expression levels
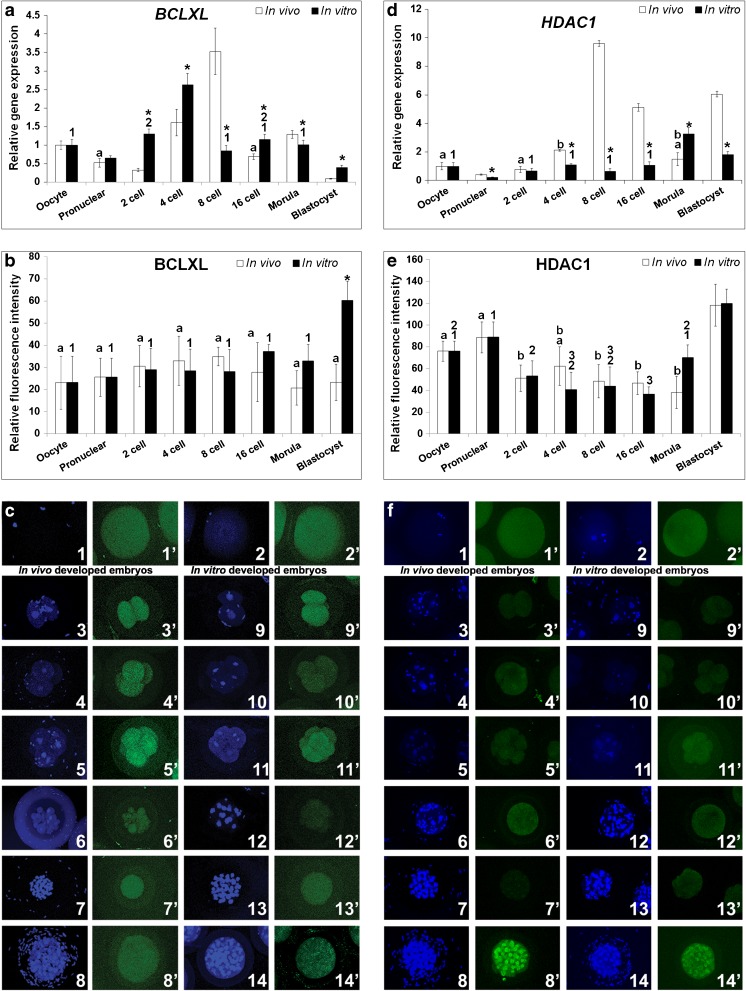
This is the first report to compare BCLXL expression patterns at transcriptional and translational level in a sequential manner in the in vitro and in vivo developed rabbit preimplantation embryos. Compared to the matured oocyte, BCLXL expression levels in the in vivo developed pronuclear and 2-cell embryos were found to be reduced (Fig. 1a). In the successive developmental stages, mRNA levels had increased and peaked at 8-cell stage. The expression was ten-times higher than that of 2-cell embryo. BCLXL expression then sharply reduced to the lowest level in blastocyst. The overall trend in transcription of BCLXL indicated an increase in expression towards zygotic genome activation (ZGA), followed by a decline. BCLXL expression in the in vitro developed embryos also followed a similar pattern, except that the increase in expression started earlier, i.e., at 2-cell embryo and peak expression was observed at the 4-cell stage, prior to ZGA. The in vitro developed embryos showed significantly higher BCLXL expression than in vivo embryos, except for 8-cell stage and morula (Fig. 1a). BCLXL protein was detected from all the developmental stages including oocytes, implying its maternal contribution (Fig. 1b). Fluorescence intensity did not change significantly in the in vivo embryos or in vitro embryos, except for the in vitro blastocyst showing a higher BCLXL expression (Fig. 1b). BCLXL showed a diffused localisation in all the embryo developmental stages and oocytes (Fig. 1c).
Fig. 1.
Relative expression levels of BCLXL and HDAC1 in the in vivo and in vitro developed rabbit embryos. a & d Relative mRNA expression. b & e Relative protein expression. c & f Protein localization by immunocytochemistry. In c & f (1 & 1′) oocyte, (2 & 2′) pronuclear embryo, (3 & 3′ and 9 & 9′) 2-cell embryo, (4 & 4′ and 10 &10′) 4-cell embryo, (5 & 5′ and 11 & 11′) 8-cell embryo, (6 & 6′ and 12 & 12′) 16-cell embryo, (7 & 7′ and 13 &13′) Morula and (8 & 8′ and 14 &14′) Blastocyst. Figures from 3 to 8 are in vivo developed embryos and figures from 9 to 14 are in vitro developed embryos. DAPI (1–14) and FITC (1′–14′) emission signals for respective developmental stages are shown side by side. Asterisk (*) indicates significant difference between in vitro and in vivo developed embryos in respective developmental stages (P < 0.05). Identical superscripts (alphabets for in vivo developed embryos and numerals for in vitro developed embryos) indicate similar levels of expression between developmental stages (P < 0.05)
Comparison of HDAC1 expression levels
HDAC1 expression was observed in all of the preimplantation developmental stages (Fig. 1d). In the in vivo embryos, HDAC1 expression increased gradually and exhibited maximum expression in 8-cell embryo coinciding with ZGA; after which it declined in the 16-cell embryo and morula to increase again in blastocyst. In contrast, the in vitro embryos showed peak expression in morula and declined subsequently in the blastocyst. In both the embryos, a phase of reduced expression had followed a surge in expression. Significant increase in expression observed prior to the ZGA in in vivo was absent in in vitro developed embryos. Except for morulae, the expression of HDAC1 in in vivo embryos was significantly higher compared to the respective in vitro embryos (Fig. 1d). The HDAC1 expression was similar between the developmental conditions. There was a gradual reduction in the fluorescent intensity until 16-cell embryo and was highest in the blastocyst stage for both the types of embryos (Fig. 1e). Even though HDAC1 showed a diffused localization in the early stages, the localization was distinctly nuclear from morula onwards (Fig. 1f). In fact, the quantitative data showed significant increase in intensity in blastocysts over ZGA phase, irrespective of the developmental conditions.
Comparison of pluripotency marker expression levels
This is the first study to compare the expression patterns of four pluripotent markers namely, KLF4, NANOG, OCT4 and SOX2 at the transcriptional and translational levels, simultaneously in in vivo and in vitro developed rabbit preimplantation embryos.
KLF4
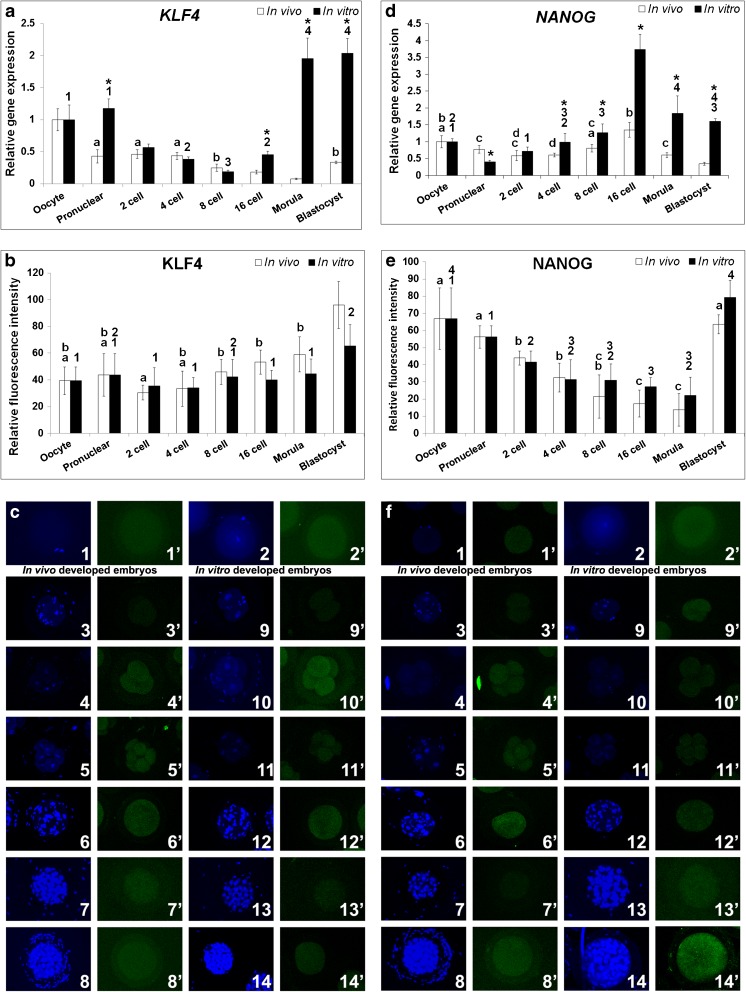
The transcript and protein of KLF4 were expressed in all stages of rabbit preimplantation embryo development (Fig. 2a–c). In sharp contrast to the in vitro embryos, in vivo embryos showed a delayed embryonic transcription of KLF4 i.e., at the blastocyst; while the in vitro embryos showed embryonic expression from the 16-cell stage onwards. It was also observed that at the pronuclear stage KLF4 expression was significantly higher in in vitro embryos than in in vivo embryos and was similar to the oocyte. The expression of KLF4 in in vitro embryos was significantly higher than that in in vivo embryos at the post ZGA stages (Fig. 2a). KLF4 was detected in oocytes and all the developmental stages of both embryo types (Fig. 2b). The fluorescence intensity was observed to be similar at various developmental stages. The intensity increased after ZGA and was the highest in blastocyst stage. The KLF4 localization was diffused in cytoplasm in all the embryonic stages (Fig. 2c).
Fig. 2.
Relative expression levels of KLF4 and NANOG in the in vivo and in vitro developed rabbit embryos. a & d Relative mRNA expression. b & e Relative protein expression. c & f Protein localization by immunocytochemistry. In c & f (1 & 1′) oocyte, (2 & 2′) pronuclear embryo, (3 & 3′ and 9 & 9′) 2-cell embryo, (4 & 4′ and 10 &10′) 4-cell embryo, (5 & 5′ and 11 & 11′) 8-cell embryo, (6 & 6′ and 12 & 12′) 16-cell embryo, (7 & 7′ and 13 &13′) Morula and (8 & 8′ and 14 &14′) Blastocyst. Figures from 3 to 8 are in vivo developed embryos and figures from 9 to 14 are in vitro developed embryos. DAPI (1–14) and FITC (1′–14′) emission signals for respective developmental stages are shown side by side. Asterisk (*) indicates significant difference between in vitro and in vivo developed embryos in respective developmental stages (P < 0.05). Identical superscripts (alphabets for in vivo developed embryos and numerals for in vitro developed embryos) indicate similar levels of expression between developmental stages (P < 0.05)
NANOG
In vivo developed embryos showed similar NANOG expression levels till the 8-cell stage and later attained a peak expression in the 16-cell stage, after which it declined in morula and blastocyst stages (Fig. 2d). Even though the in vitro and in vivo embryos showed a similar expression trend, in vitro developed embryos had a lower expression level at the pronuclear stage and higher expression levels during and after the ZGA (8-cell, 16-cell, morula and blastocyst). The results also indicate a basal level of transcription of NANOG from early stages of development. The fluorescence intensity of NANOG decreased continually from pronuclear stage to morula and then increased in the blastocyst; and both the embryo types followed this trend faithfully (Fig. 2e). Further, the intensity levels in oocyte and blastocyst were similar and the expression was diffused in all the developmental stages studied (Fig. 2f).
OCT4
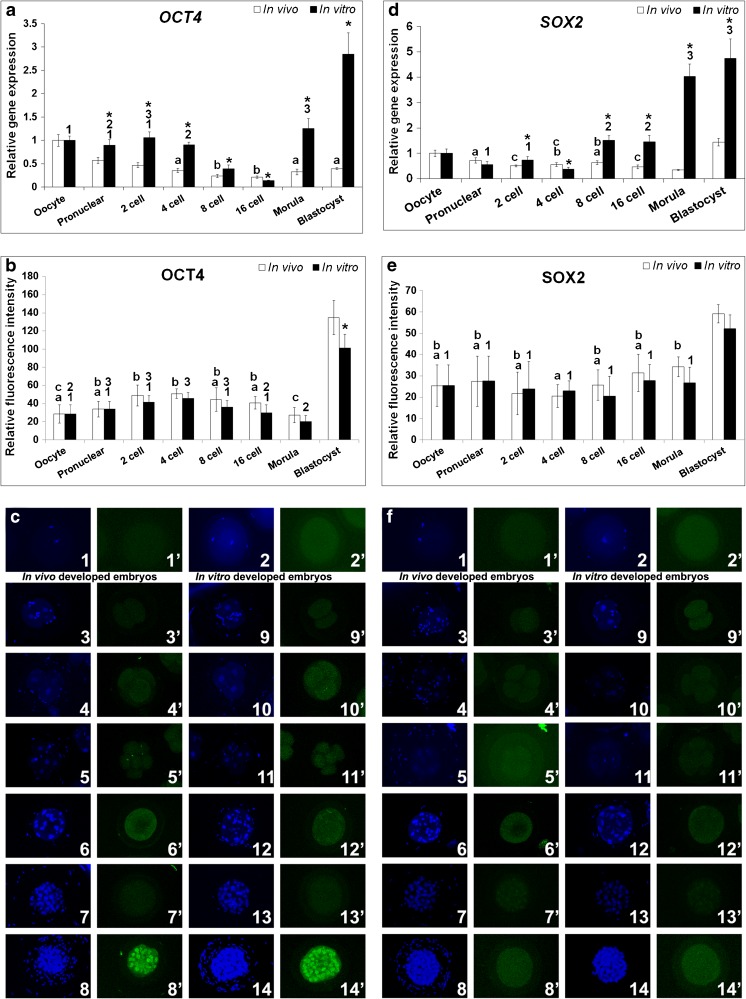
OCT4 expression pattern in the in vivo and in vitro embryos decreased gradually till 16-cell stage, after which it increased in the morula (Fig. 3a). The in vitro embryos had a significantly higher gene expression level than in vivo embryos at all stages of development, except for 16-cell stage. In the in vitro morula and blastocyst stages, expression levels had increased substantially. The fluorescence intensity was similar in both types of embryos up to morulae, after which it increased substantially (Fig. 3b). OCT4 was detected in all the stages of preimplantation development and showed a diffused localisation pattern in the earlier developmental stages. However, OCT4 was nuclear localised in the blastocyst (Fig. 3c).
Fig. 3.
Relative expression levels of OCT4 and SOX2 in the in vivo and in vitro developed rabbit embryos. a & d Relative mRNA expression. b & e Relative protein expression. c & f Protein localization by immunocytochemistry. In c & f (1 & 1′) oocyte, (2 & 2′) pronuclear embryo, (3 & 3′ and 9 & 9′) 2-cell embryo, (4 & 4′ and 10 &10′) 4-cell embryo, (5 & 5′ and 11 & 11′) 8-cell embryo, (6 & 6′ and 12 & 12′) 16-cell embryo, (7 & 7′ and 13 &13′) Morula and (8 & 8′ and 14 &14′) Blastocyst. Figures from 3 to 8 are in vivo developed embryos and figures from 9 to 14 are in vitro developed embryos. DAPI (1–14) and FITC (1′–14′) emission signals for respective developmental stages are shown side by side. Asterisk (*) indicates significant difference between in vitro and in vivo developed embryos in respective developmental stages (P < 0.05). Identical superscripts (alphabets for in vivo developed embryos and numerals for in vitro developed embryos) indicate similar levels of expression between developmental stages (P < 0.05)
SOX2
SOX2 expression in the in vivo embryos decreased gradually till morula stage and later increased in the blastocyst (Fig. 3d). In contrast, the in vitro embryos showed gradual increase in expression from 8-cell stage and exhibited peak expression in the morula and blastocyst. Except for the pronuclear embryos, all the embryos developed in vitro had higher expression levels than those embryos developed in vivo. The in vitro developed morula and blastocyst showed a comparable level of expression. SOX2 protein was localised diffusely in all the stages of preimplantation development and the fluorescence intensity indicated a similar trend in both types of embryos (Fig. 3f). The intensity did not vary significantly up to morula but it increased in both the in vivo and in vitro developed blastocysts (Fig. 3e).
Discussion
In this study, a comparison was made between the in vivo developed embryos and in vitro developed embryos (in vivo fertilized) with respect to expression levels and patterns of candidate genes and their proteins at distinct developmental stages during rabbit preimplantation embryo development. Even though a higher percentage of in vitro blastocyst development was reported using the B2 medium in rabbit [39], the 80 % in vitro blastocyst development achieved in this study was in agreement with earlier reports in cattle and rabbit [2, 45]. The expression of candidate genes namely, OCT4, SOX2, NANOG, KLF4, HDAC1 and BCLXL were normalised with endogenous expression levels of reference genes namely, H2AFZ, HPRT1 and YWHAZ. These genes showed stable expression levels in all the developmental stages and thus agreeing with previous reports on rabbit preimplantation development [37].
The peak expression of anti-apoptotic factor BCLXL coincided with start of ZGA in in vivo developed rabbit embryo similar to the reports in mouse, human and cattle embryos [27, 29, 46, 47]. Housekeeping genes and apoptotic regulators were reported to peak at the ZGA in bovine and murine embryos [48, 49]. The early BCLXL expression peak in in vitro embryos indicates either a compensative measure for enhanced protein degradation or a higher pro-apoptotic signal being balanced for the embryo survival. BAX/BCL ratio could give further insight into importance of the early expression peak in vitro. BCLXL protein showed cytoplasmic localization in all the preimplantation stages of rabbit embryo and had similar levels of expression, except for the in vitro blastocyst; indicating that BCLXL translated from the maternal mRNA was continually present in embryos until newly translated proteins appear. Similar observation was also made in the human embryos [27]. The maintenance of BCLXL titre could be regulated through degradation of the maternal protein and replacement with newly translated protein as the expression level of BCLXL remained stable even after the ZGA. Hence the peak expression of transcripts at ZGA could be a requirement for embryo development. Unlike the reports on other BCL2 family members forming a perinuclear ring, BCLXL in the rabbit embryos showed diffused cytoplasmic localisation as observed in human embryos [27]. The absence of perinuclear ring implies that cells were less apoptotic in those embryos [50].
Studies on human cancer cell lines have provided evidence for the regulation of BCL2 and BCLXL by HDAC1 [51]. It was demonstrated that, when the expression of HDAC1 is inhibited, expression of BCLXL goes down and results in apoptosis [51]. Studies conducted on mouse preimplantation embryos have also demonstrated the crucial role of BCLXL in protecting the embryos to enter ZGA phase [28, 29]. Even though it is yet to be established in rabbit embryos, it is safe to assume that the embryo relies on BCLXL and HDAC1 expression to achieve ZGA. However, the contrast in their expression trend may imply a requirement for early onset of BCLXL transcription in in vitro embryos than their in vivo counterparts. HDAC1 peak expression in the in vitro rabbit embryos was delayed compared to that of in vivo embryos implying a delay in chromatin remodelling during the course of in vitro development. This could in turn reflect in the expression of other genes. The peak expression at ZGA phase in this study shows a positive correlation to the lowest acetylation levels in rabbit embryo reported earlier [40]. In cattle, HDAC1 expression in the in vitro embryos was low until 8-cell stage, but increased eight to ten times at the blastocyst stage [52]. The presence of its protein in all the stages studied in both in vitro and in vivo embryos implies maternal contribution of HDAC1 prior to ZGA and its embryonic contribution post ZGA. With respect to HDAC1 localisation, similar to the mouse embryos, HDAC1 was localized to the nucleoplasm during later stages of rabbit preimplantation embryo development [34]. The discordance between mRNA and protein levels of HDAC1 was observed during both in vivo and in vitro embryo development. This may be attributed to the significant decrease of HDAC1 expression post ZGA and lineage specification [40]. However in this study, it is difficult to rule out the effect of in vitro conditions. The disagreement between HDAC1 transcript and protein levels was also reported in in vitro mouse blastocyst embryo, which had lower protein expression than morula even though the transcript levels were steadily increasing in each successive stage of development [34]. The higher expression levels of HDAC1 in rabbit in vivo blastocyst may imply a role in pluripotency maintenance, as the cells start to differentiate in the later stages of preimplantation development. HDAC1 expression was reported to be critical for pluripotency, since the pluripotent markers namely, OCT4, SOX2, KLF4 and NANOG were targets for transcription regulation by HDAC1 [53]. The lack or delay in increase of HDAC1 in in vitro embryos may reduce their developmental potential. It has been shown that knockdown of HDAC1 releases the regulation and the pluripotent markers get overexpressed [53]. The dynamics of HDAC1 in this study hints at its role in pluripotency maintenance.
The delay in decrease in KLF4 observed in in vitro pronuclear stage could be ascribed to the incubation of the zygote for 4 h under in vitro conditions unlike the in vivo pronuclear stage which was directly obtained from the tract. However an increased expression of KLF4 was observed post ZGA, during rabbit preimplantation development. The extent of increase in transcription was higher in in vitro developed embryos; however this did not get translated into higher levels of protein. Earlier reports on murine embryos have demonstrated that the regulation of KLF4 expression was not influenced by ZGA, rather it was being influenced by the Leukaemia inhibitory factor/STAT3 signalling [54]. This implies that KLF4 was probably influenced by the ZGA either through LIF/STAT3 or by some other gene. The role of HDAC1 regulation of KLF4 also could have an influence in these high levels of KLF4 expression. KLF4 expression was also reported to increase at the 8-cell embryo in bovine embryos [49]. This study showed that KLF4 localisation in both the in vitro and in vivo developed rabbit preimplantation embryos was diffused. As a transcription factor, KLF4 is expected to have a nuclear localisation as reported in the Macaque morulae and blastocysts [3]. The diffused localisation observed in the present study raises more questions related to delay in transport even though it possesses a nuclear localisation signal [55]. It is likely that when KLF4 localisation is compromised, genes like KLF2 and KLF5 function to maintain the pluripotent state as shown in mouse ESCs [56]. The human ICM showed a higher expression of KLF2 than KLF4, which may imply its compensatory role [22].
Expression of NANOG in the preimplantation embryos was reported to be varying with species. In mouse, the NANOG expression was observed from compact morula onwards [57], in human from pronuclear stage onwards [58], in bovine embryos from 8-cell onwards [59], in caprine embryos from 8- to 16-cell onwards [60], and in porcine from 4-cell embryo onwards [61, 62]. In this study, transcripts of NANOG were detected in all embryonic stages similar to the observation in human embryos. Post ZGA, as reported in bovine embryos, NANOG expression had steadily declined from the peak expression at ZGA and showed similar expression trend in both in vivo and in vitro developed rabbit embryos [59]. This may indicate its critical role in the lineage specification, post ZGA for pluripotency maintenance. The decrease in NANOG expression levels in rabbit blastocysts, similar to caprine and human blastocysts, could be attributed to a reduction in number of cells expressing NANOG which gets limited to the inner cell mass (ICM) [58, 60]. Rabbit blastocysts showed a diffused NANOG protein expression similar to that observed in the bovine embryos prior to blastocyst stage [59]. In the caprine and bovine blastocysts, NANOG was expressed in both ICM and trophectoderm [60]. In this study, we found that the expression of NANOG transcripts and protein starts prior to ZGA and peaked after ZGA, which is an indication of its consequential role in ZGA. NANOG expression in embryo development is crucial and probably depends on its ability to regulate the expression of OCT4 and SOX2, which have been implicated in transcription regulation of a number of genes in the pluripotency network [20, 63]. It was also demonstrated that HDAC1 mediated suppression of OCT4 and SOX2 results in suppression of NANOG [64]. It has been suggested that HDAC1, OCT4 and NANOG form the NODE complex responsible for regulation of gene expression and for maintaining pluripotency [65]. Thus OCT4/NANOG ratio form a quantifier for the maintenance of pluripotency where the reduction in NANOG will cause differentiation [66]. The expression levels observed for NANOG and OCT4 in rabbit preimplantation shows a similarly maintained pattern and at blastocyst the OCT4 level was much higher than NANOG. This change could be attributed to the initiation of lineage specification as reported earlier [41].
OCT4 has also been implicated in regulation of transcription of a number of genes in the pluripotency network [20, 63] namely, UTF1, FGF4, FBXO15 and LEFTY1 in mouse [67]. In the current study, OCT4 transcripts were present in all stages of preimplantation development. The level of OCT4 declined in early stages of preimplantation development and increased post ZGA, which was in agreement with the earlier observations on OCT4 or POU5F1 expression in in vitro developed rabbit preimplantation embryos [37] and bovine preimplantation embryos [68]. But this observation was in discordance with what was observed in the porcine preimplantation development, where ZGA coincided with the peak expression of OCT4 [62]. In the human preimplantation development studies, an increase in expression of OCT4 post ZGA was clearly demonstrated [22]. Lower expression levels of OCT4 in the in vivo blastocysts against in vitro embryos, as observed in this study was in accordance with reports on porcine and bovine embryos, which showed an increased expression in morulae and blastocysts [59, 62]. However, lower expression levels in the in vivo blastocysts may be a consequence of higher HDAC1 activity and differentiation [69]. In murine and human expanded blastocysts, expression of OCT4 was limited to the ICM [20, 22, 70, 71] whereas in bovine and porcine blastocysts, OCT4 was expressed in both the trophectoderm and ICM [72, 73]. Localization of OCT4 was diffused in the early stages of rabbit preimplantation embryos; while nuclear localization was evident in the blastocyst. In the bovine blastocysts, a similar OCT4 localization was reported [59, 73]. A nuclear localisation in the early stages of rabbit development and a decline in protein levels recovering after ZGA were reported earlier [40]. The difference in these observations needs to be addressed further.
SOX2 forms a complex with the OCT4 and performs self-regulation and regulation of other developmentally important genes like FGF4, UTF1, FBX15 and NANOG. The murine blastocysts, like rabbit blastocysts showed a peak expression of SOX2, implying a central role for SOX2 in trophectoderm formation and maintenance of pluripotency [67, 74–76]. The stable level of SOX2 expression observed even after ZGA (8-cell to morula) in the in vivo embryos indicated a need for SOX2 along with other partners in the pluripotency gene regulatory network. In contrast to in vivo rabbit embryos, the in vitro embryos showed SOX2 gene activation from 8-cell embryo onwards and transcript levels remained higher than that of the in vivo embryos. The in vitro developed bovine preimplantation embryos also showed a higher level of SOX2 expression than in vivo embryos [77]. This deviation in the expression pattern from that of in vivo embryos could be a result of stress brought about by culture conditions. In later stages of rabbit embryo development, SOX2 was localised in the nuclei, similar to the reports in mouse preimplantation development [78]. SOX2 had been reported to have detrimental effects on embryo development when over expressed in early stages of embryo and also influence maternal to embryo transition [24]. Thus the expression trends of SOX2 transcripts and protein in this study helps to safely conclude that the maternal contribution and stable protein expression level were crucial to the regulation of differentiation and rapid cell proliferation in developing embryos.
Conclusion
The comparisons made between in vivo and in vitro developed rabbit embryos in this study showed that OCT4, SOX2, NANOG, KLF4, HDAC1 and BCLXL transcript levels varied; possibly due to the difference in developmental conditions. As a consequence of variation observed in the expression of above genes, chromatin remodelling and generation of differentiation cues were altered in the in vitro and in vivo developed embryos. For instance, HDAC1 showed lower level of expression, while KLF4, NANOG, OCT4 and SOX2 showed higher levels of expression under in vitro conditions compared to the in vivo developed embryos post ZGA. However, the levels of protein expression and localisation for these genes did not differ between in vivo and in vitro development. In case of BCLXL, the levels of transcript followed an early but identical expression pattern in in vitro developed embryos. The BCLXL transcript and protein expression levels and protein localisation were observed to be similar to the earlier reports in mouse and human embryos [27, 29].
Our study indicates a maternal protein contribution for all the candidate genes. The lower acetylation levels reported from ZGA till blastocyst, when HDAC1 reaches peak expression implies a requirement for HDAC1 to tide over ZGA. The requirement of BCLXL for ZGA was also evident from the expression pattern observed [51]. The lower levels of HDAC1 under in vitro condition may indicate a comparatively poor reprogramming of genome. Further, the lower levels of OCT4 expression at ZGA under in vitro conditions may also hinder the developmental potential and reprogramming of embryos. The stable levels for OCT4, SOX2 and NANOG in this study further emphasise the importance of OCT4/NANOG ratio in maintaining pluripotency and also lineage specification with the association of SOX2 [66]. In fact, the differential SOX2 transcript levels between in vivo and in vitro developed embryos could be an indication of a reduced embryo development potential during in vitro development. Increased levels of KLF4 in the later stages of preimplantation development indicated the influence of ZGA on LIF/STAT3 pathway which triggers the KLF4 expression. Higher expression levels of KLF4, NANOG and SOX2 in the in vitro embryos compared to in vivo developed embryos observed in our study warrants further deliberation on their role in preimplantation development. The role of HDAC1 regulation of these genes may also be a reason for this observation.
More detailed studies on expression patterns of the pluripotency genes and apoptotic markers in preimplantation embryos which fail to develop or slow-down in development, would give further information on the factors that play a pivotal role in embryo development.
Acknowledgments
The authors are thankful to the Council of Scientific and Industrial Research, India, CSIR-Centre for Cellular and Molecular Biology, India and Department of Biotechnology, Govt. of India.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Capsule The gene expression of OCT4, KLF4, NANOG, SOX2, HDAC1 and BCLXL was altered at transcriptional and translational levels with reference to zygotic genome activation of rabbit embryos developed in vivo and in vitro.
References
- 1.Dattena M, Ptak G, Loi P, Cappai P. Survival and viability of vitrified in vitro and in vivo produced ovine blastocysts. Theriogenology. 2000;53(8):1511–9. doi: 10.1016/S0093-691X(00)00293-4. [DOI] [PubMed] [Google Scholar]
- 2.Rizos D, Ward F, Duffy P, Boland MP, Lonergan P. Consequences of bovine oocyte maturation, fertilization or early embryo development in vitro versus in vivo: implications for blastocyst yield and blastocyst quality. Mol Reprod Dev. 2002;61(2):234–48. doi: 10.1002/mrd.1153. [DOI] [PubMed] [Google Scholar]
- 3.Rizos D, Clemente M, Bermejo-Alvarez P, De La Fuente J, Lonergan P, Gutiérrez-Adán A. Consequences of in vitro culture conditions on embryo development and quality. Reprod Domest Anim. 2008;43:44–50. doi: 10.1111/j.1439-0531.2008.01230.x. [DOI] [PubMed] [Google Scholar]
- 4.Brownell MS, Warner CM. Ped gene expression by embryos cultured in vitro. Biol Reprod. 1988;39(4):806–11. doi: 10.1095/biolreprod39.4.806. [DOI] [PubMed] [Google Scholar]
- 5.Lonergan P. State-of-the-art embryo technologies in cattle. Soc Reprod Fertil Suppl. 2007;64:315–25. doi: 10.5661/rdr-vi-315. [DOI] [PubMed] [Google Scholar]
- 6.Lonergan P, Fair T, Corcoran D, Evans ACO. Effect of culture environment on gene expression and developmental characteristics in IVF-derived embryos. Theriogenology. 2006;65(1):137–52. doi: 10.1016/j.theriogenology.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 7.DePamphilis ML, Kaneko KJ, Vassilev A. Activation of zygotic gene expression in mammals. In: Melvin LD, editor. Advances in developmental biology and biochemistry. Netherlands: Elsevier; 2002. pp. 55–84. [Google Scholar]
- 8.Latham KE, Garrels JI, Chang C, Solter D. Quantitative analysis of protein synthesis in mouse embryos. I. Extensive reprogramming at the one- and two-cell stages. Development. 1991;112(4):921–32. doi: 10.1242/dev.112.4.921. [DOI] [PubMed] [Google Scholar]
- 9.Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136:3033–42. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- 10.Brunet-simon A, Henrion G, Renard JP, Duranthon V. Onset of zygotic transcription and maternal transcript legacy in the rabbit embryo. Mol Reprod Dev. 2001;58:127–36. doi: 10.1002/1098-2795(200102)58:2<127::AID-MRD1>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 11.Braude P, Bolton V, Moore S. Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature. 1988;332(6163):459–61. doi: 10.1038/332459a0. [DOI] [PubMed] [Google Scholar]
- 12.Howe CC, Solter D. Cytoplasmic and nuclear protein synthesis in preimplantation mouse embryos. J Embryol Exp Morpholog. 1979;52:209–25. [PubMed] [Google Scholar]
- 13.Marlow F. Maternal control of development in vertebrates: My mother made me do it! San Rafael (CA): Morgan & Claypool Life Sciences; 2010. Available from: http://www.ncbi.nlm.nih.gov/books/NBK53194/ [PubMed]
- 14.Vassena R, Boué S, González-Roca E, Aran B, Auer H, Veiga A, et al. Waves of early transcriptional activation and pluripotency program initiation during human preimplantation development. Development. 2011;138(17):3699–709. doi: 10.1242/dev.064741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 16.Boiani M, Eckardt S, Schöler HR, McLaughlin KJ. Oct4 distribution and level in mouse clones: consequences for pluripotency. Gene Dev. 2002;16(10):1209–19. doi: 10.1101/gad.966002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pesce M, Schöler HR. Oct-4: gatekeeper in the beginnings of mammalian development. Stem Cells. 2001;19(4):271–8. doi: 10.1634/stemcells.19-4-271. [DOI] [PubMed] [Google Scholar]
- 18.Haase A, Olmer R, Schwanke K, Wunderlich S, Merkert S, Hess C, et al. Generation of induced pluripotent stem cells from human cord blood. Cell Stem Cell. 2009;5(4):434–41. doi: 10.1016/j.stem.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 19.Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, et al. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138(4):722–37. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ovitt CE, Schöler HR. The molecular biology of Oct-4 in the early mouse embryo. Mol Hum Reprod. 1998;4(11):1021–31. doi: 10.1093/molehr/4.11.1021. [DOI] [PubMed] [Google Scholar]
- 21.Foygel K, Choi B, Jun S, Leong DE, Lee A, Wong CC, et al. A novel and critical role for Oct4 as a regulator of the maternal-embryonic transition. PLoS One. 2008;3(12):e4109. doi: 10.1371/journal.pone.0004109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galán A, Montaner D, Póo ME, Valbuena D, Ruiz V, Aguilar C, et al. Functional genomics of 5- to 8-cell stage human embryos by blastomere single-cell cDNA analysis. PLoS ONE. 2010;5(10):e13615. doi: 10.1371/journal.pone.0013615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee MT, Bonneau AR, Takacs CM, Bazzini AA, DiVito KR, Fleming ES, et al. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature. 2013;503(7476):360–4. doi: 10.1038/nature12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan H, Schultz RM. Sox2 modulates reprogramming of gene expression in two-cell mouse embryos. Biol Reprod. 2011;85(2):409–16. doi: 10.1095/biolreprod.111.090886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo G, Yang J, Nichols J, Hall JS, Eyres I, Mansfield W, et al. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136(7):1063–9. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu FT, Goff LK, Hao JH, Newland AC, Jia L. Increase in the ratio of mitochondrial Bax/Bcl-XL induces Bax activation in human leukemic K562 cell line. Apoptosis. 2004;9(3):377–84. doi: 10.1023/B:APPT.0000025815.78761.5c. [DOI] [PubMed] [Google Scholar]
- 27.Metcalfe AD, Hunter HR, Bloor DJ, Lieberman BA, Picton HM, Leese HJ, et al. Expression of 11 members of the BCL-2 family of apoptosis regulatory molecules during human preimplantation embryo development and fragmentation. Mol Reprod Dev. 2004;68(1):35–50. doi: 10.1002/mrd.20055. [DOI] [PubMed] [Google Scholar]
- 28.Liu X, Fernandes R, Gertsenstein M, Perumalsamy A, Lai I, Chi M, et al. Automated microinjection of recombinant BCL-X into mouse zygotes enhances embryo development. PLoS One. 2011;6(7):e21687. doi: 10.1371/journal.pone.0021687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jurisicova A, Latham KE, Casper RF, Varmuza SL. Expression and regulation of genes associated with cell death during murine preimplantation embryo development. Mol Reprod Dev. 1998;51:243–53. doi: 10.1002/(SICI)1098-2795(199811)51:3<243::AID-MRD3>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 30.Janumyan YM, Sansam CG, Chattopadhyay A, Cheng N, Soucie EL, Penn LZ, et al. Bcl-xL/Bcl-2 coordinately regulates apoptosis, cell cycle arrest and cell cycle entry. EMBO J. 2003;22(20):5459–70. doi: 10.1093/emboj/cdg533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arbel N, Ben-Hail D, Shoshan-Barmatz V. Mediation of the antiapoptotic activity of Bcl-xL protein upon interaction with VDAC1 protein. J Biol Chem. 2012;287(27):23152–61. doi: 10.1074/jbc.M112.345918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brunmeir R, Lagger S, Seiser C. Histone deacetylase HDAC1/HDAC2-controlled embryonic development and cell differentiation. Int J Dev Biol. 2009;53(2–3):275–89. doi: 10.1387/ijdb.082649rb. [DOI] [PubMed] [Google Scholar]
- 33.Dufourcq P, Victor M, Gay F, Calvo D, Hodgkin J, Shi Y. Functional requirement for Histone deacetylase 1 in Caenorhabditis elegans gonadogenesis. Mol Cell Biol. 2002;22(9):3024–34. doi: 10.1128/MCB.22.9.3024-3034.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma P, Schultz RM. Histone deacetylase 1 (HDAC1) regulates histone acetylation, development, and gene expression in preimplantation mouse embryos. Dev Biol. 2008;319:110–20. doi: 10.1016/j.ydbio.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan J, Watanabe T. Transgenic rabbits as therapeutic protein bioreactors and human disease models. Pharmacol Ther. 2003;99(3):261–82. doi: 10.1016/S0163-7258(03)00069-X. [DOI] [PubMed] [Google Scholar]
- 36.Ogonuki N, Inoue K, Miki H, Mochida K, Hatori M, Okada H, et al. Differential development of rabbit embryos following microinsemination with sperm and spermatids. Mol Reprod Dev. 2005;72(3):411–7. doi: 10.1002/mrd.20363. [DOI] [PubMed] [Google Scholar]
- 37.Mamo S, Baji Gal A, Polgar Z, Dinnyes A. Expression profiles of the pluripotency marker gene POU5F1 and validation of reference genes in rabbit oocytes and preimplantation stage embryos. BMC Mol Biol. 2008;9(1):67. doi: 10.1186/1471-2199-9-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kobolak J, Kiss K, Polgar Z, Mamo S, Rogel Gaillard C, Tancos Z, et al. Promoter analysis of the rabbit POU5F1 gene and its expression in preimplantation stage embryos. BMC Mol Biol. 2009;10:88. doi: 10.1186/1471-2199-10-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen CH, Du F, Xu J, Chang WF, Liu CC, Su HY, et al. Synergistic effect of trichostatin A and scriptaid on the development of cloned rabbit embryos. Theriogenology. 2013;79(9):1284–93. doi: 10.1016/j.theriogenology.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen CH, Chang WF, Liu CC, Su HY, Shyue SK, Cheng WT, et al. Spatial and temporal distribution of Oct-4 and acetylated H4K5 in rabbit embryos. Reprod BioMed Online. 2012;24(4):433–42. doi: 10.1016/j.rbmo.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen CH, Xu J, Chang WF, Liu CC, Su HY, Chen YE, et al. Dynamic profiles of Oct-4, Cdx-2 and acetylated H4K5 in in-vivo-derived rabbit embryos. Reprod BioMed Online. 2012;25(4):358–70. doi: 10.1016/j.rbmo.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naik BR, Rao BS, Vagdevi R, Gnanprakash M, Amarnath D, Rao VH. Conventional slow freezing, vitrification and open pulled straw (OPS) vitrification of rabbit embryos. Anim Reprod Sci. 2005;86:329–38. doi: 10.1016/j.anireprosci.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 43.Kalendar R, Lee D, Schulman A. Fast PCR software for PCR, in silico PCR, and oligonucleotide assembly and analysis. In: Valla S, Lale R, editors. DNA cloning and assembly methods. Methods in molecular biology. New York: Humana Press; 2014. p. 271–302. [DOI] [PubMed]
- 44.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):1–12. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maurer RR, Beier HM. Uterine proteins and development in vitro of rabbit preimplantation embryos. J Reprod Fertil. 1976;48(1):33–41. doi: 10.1530/jrf.0.0480033. [DOI] [PubMed] [Google Scholar]
- 46.Jurisicova A, Antenos M, Varmuza S, Tilly JL, Casper RF. Expression of apoptosis-related genes during human preimplantation embryo development: potential roles for the Harakiri gene product and Caspase-3 in blastomere fragmentation. Mol Hum Reprod. 2003;9(3):133–41. doi: 10.1093/molehr/gag016. [DOI] [PubMed] [Google Scholar]
- 47.Zaraza J, Oropeza A, Velazquez MA, Korsawe K, Herrmann D, Carnwath JW, et al. Developmental competence and mRNA expression of preimplantation in vitro–produced embryos from prepubertal and postpubertal cattle and their relationship with apoptosis after intraovarian administration of IGF-1. Theriogenology. 2010;74(1):75–89. doi: 10.1016/j.theriogenology.2009.11.033. [DOI] [PubMed] [Google Scholar]
- 48.Wang Q, Latham KE. Requirement for protein synthesis during embryonic genome activation in mice. Mol Reprod Dev. 1997;47(3):265–70. doi: 10.1002/(SICI)1098-2795(199707)47:3<265::AID-MRD5>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 49.Vigneault C, Gravel C, Vallée M, McGraw S, Sirard M-A. Unveiling the bovine embryo transcriptome during the maternal-to-embryonic transition. Reproduction. 2009;137(2):245–57. doi: 10.1530/REP-08-0079. [DOI] [PubMed] [Google Scholar]
- 50.Portier BP, Taglialatela G. Bcl-2 localized at the nuclear compartment induces apoptosis after transient overexpression. J Biol Chem. 2006;281(52):40493–502. doi: 10.1074/jbc.M606181200. [DOI] [PubMed] [Google Scholar]
- 51.Chen S, Dai Y, Pei X-Y, Grant S. Bim upregulation by histone deacetylase inhibitors mediates interactions with the Bcl-2 antagonist ABT-737: evidence for distinct roles for Bcl-2, Bcl-xL, and Mcl-1. Mol Cell Biol. 2009;29(23):6149–69. doi: 10.1128/MCB.01481-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McGraw S, Robert C, Massicotte L, Sirard M-A. Quantification of histone acetyl transferase and histone deacetylase transcripts during early bovine embryo development. Biol Reprod. 2003;68(2):383–9. doi: 10.1095/biolreprod.102.005991. [DOI] [PubMed] [Google Scholar]
- 53.Kidder BL, Palmer S. HDAC1 regulates pluripotency and lineage specific transcriptional networks in embryonic and trophoblast stem cells. Nucleic Acids Res. 2012;40(7):2925–39. doi: 10.1093/nar/gkr1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang P, Andrianakos R, Yang Y, Liu C, Lu W. Kruppel-like Factor 4 (Klf4) prevents embryonic stem (ES) cell differentiation by regulating Nanog gene expression. J Biol Chem. 2010;285(12):9180–9. doi: 10.1074/jbc.M109.077958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vangapandu H, Ai W. Kruppel like factor 4 (KLF4): a transcription factor with diverse context-dependent functions. Gene Ther Mol Biol. 2009;13:194–204. [Google Scholar]
- 56.Parisi S, Cozzuto L, Tarantino C, Passaro F, Ciriello S, Aloia L, et al. Direct targets of Klf5 transcription factor contribute to the maintenance of mouse embryonic stem cell undifferentiated state. BMC Biol. 2010;8(1):128. doi: 10.1186/1741-7007-8-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113(5):643–55. doi: 10.1016/S0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 58.Kimber SJ, Sneddon SF, Bloor DJ, El-Bareg AM, Hawkhead JA, Metcalfe AD, et al. Expression of genes involved in early cell fate decisions in human embryos and their regulation by growth factors. Reproduction. 2008;135(5):635–47. doi: 10.1530/REP-07-0359. [DOI] [PubMed] [Google Scholar]
- 59.Khan DR, Dubé D, Gall L, Peynot N, Ruffini S, Laffont L, et al. Expression of pluripotency master regulators during two key developmental transitions: EGA and early lineage specification in the bovine embryo. PLoS One. 2012;7(3):e34110. doi: 10.1371/journal.pone.0034110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He S, Pant D, Schiffmacher A, Bischoff S, Melican D, Gavin W, et al. Developmental expression of pluripotency determining factors in caprine embryos: Novel pattern of NANOG protein localization in the nucleolus. Mol Reprod Dev. 2006;73(12):1512–22. doi: 10.1002/mrd.20525. [DOI] [PubMed] [Google Scholar]
- 61.Magnani L, Cabot RA. In vitro and in vivo derived porcine embryos possess similar, but not identical, patterns of Oct4, Nanog, and Sox2 mRNA expression during cleavage development. Mol Reprod Dev. 2008;75(12):1726–35. doi: 10.1002/mrd.20915. [DOI] [PubMed] [Google Scholar]
- 62.Xing X, Magnani L, Lee K, Wang C, Cabot RA, Machaty Z. Gene expression and development of early pig embryos produced by serial nuclear transfer. Mol Reprod Dev. 2009;76(6):555–63. doi: 10.1002/mrd.20974. [DOI] [PubMed] [Google Scholar]
- 63.Schöler HR, Ruppert S, Suzuki N, Chowdhury K, Gruss P. New type of POU domain in germ line-specific protein Oct-4. Nature. 1990;344(6265):435–9. doi: 10.1038/344435a0. [DOI] [PubMed] [Google Scholar]
- 64.You JS, Kang JK, Seo D-W, Park JH, Park JW, Lee JC, et al. Depletion of embryonic stem cell signature by histone deacetylase inhibitor in NCCIT cells: involvement of Nanog suppression. Cancer Res. 2009;69(14):5716–25. doi: 10.1158/0008-5472.CAN-08-4953. [DOI] [PubMed] [Google Scholar]
- 65.Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY, et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol. 2008;10(6):731–9. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- 66.Muñoz Descalzo S, RuÉ P, Garcia-Ojalvo J, Arias AM. Correlations between the levels of Oct4 and Nanog as a signature for naïve pluripotency in mouse embryonic stem cells. Stem Cells. 2012;30(12):2683–91. doi: 10.1002/stem.1230. [DOI] [PubMed] [Google Scholar]
- 67.Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9(6):625–35. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- 68.Kurosaka S, Eckardt S, McLaughlin KJ. Pluripotent lineage definition in bovine embryos by Oct4 transcript localization. Biol Reprod. 2004;71(5):1578–82. doi: 10.1095/biolreprod.104.029322. [DOI] [PubMed] [Google Scholar]
- 69.de Juano MD S, Peñaranda DS, Marco-Jiménez F, Llobat L, Vicente JS. Differential mRNA expression in rabbit in vivo Pre-implantatory embryos. Reprod Domest Anim. 2011;46(4):567–72. doi: 10.1111/j.1439-0531.2010.01702.x. [DOI] [PubMed] [Google Scholar]
- 70.Pesce M, Wang X, Wolgemuth DJ, Schöler HR. Differential expression of the Oct-4 transcription factor during mouse germ cell differentiation. Mech Dev. 1998;71(1–2):89–98. doi: 10.1016/S0925-4773(98)00002-1. [DOI] [PubMed] [Google Scholar]
- 71.Hansis C, Grifo JA, Krey LC. Oct-4 expression in inner cell mass and trophectoderm of human blastocysts. Mol Hum Reprod. 2000;6(11):999–1004. doi: 10.1093/molehr/6.11.999. [DOI] [PubMed] [Google Scholar]
- 72.van Eijk MJT, van Rooijen MA, Modina S, Scesi L, Folkers G, van Tol HTA, et al. Molecular cloning, genetic mapping, and developmental expression of bovine POU5F1. Biol Reprod. 1999;60(5):1093–103. doi: 10.1095/biolreprod60.5.1093. [DOI] [PubMed] [Google Scholar]
- 73.Kirchhof N, Carnwath JW, Lemme E, Anastassiadis K, Schöler H, Niemann H. Expression pattern of Oct-4 in preimplantation embryos of different species. Biol Reprod. 2000;63(6):1698–705. doi: 10.1095/biolreprod63.6.1698. [DOI] [PubMed] [Google Scholar]
- 74.Wang QT, Piotrowska K, Ciemerych MA, Milenkovic L, Scott MP, Davis RW, et al. A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev Cell. 2004;6(1):133–44. doi: 10.1016/S1534-5807(03)00404-0. [DOI] [PubMed] [Google Scholar]
- 75.Li X, Kato Y, Tsunoda Y. Comparative analysis of development-related gene expression in mouse preimplantation embryos with different developmental potential. Mol Reprod Dev. 2005;72(2):152–60. doi: 10.1002/mrd.20346. [DOI] [PubMed] [Google Scholar]
- 76.Keramari M, Razavi J, Ingman KA, Patsch C, Edenhofer F, Ward CM, et al. Sox2 is essential for formation of trophectoderm in the preimplantation embryo. PLoS One. 2010;5(11):e13952. doi: 10.1371/journal.pone.0013952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Balasubramanian S, Son WJ, Kumar BM, Ock SA, Yoo JG, Im GS, et al. Expression pattern of oxygen and stress-responsive gene transcripts at various developmental stages of in vitro and in vivo preimplantation bovine embryos. Theriogenology. 2007;68(2):265–75. doi: 10.1016/j.theriogenology.2007.05.044. [DOI] [PubMed] [Google Scholar]
- 78.Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Gene Dev. 2003;17(1):126–40. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]