Abstract
Francisella tularensis is a Category A Biodefense agent that causes a fatal human disease known as tularemia. The pathogenicity of F. tularensis depends on its ability to persist inside host immune cells primarily by resisting an attack from host-generated reactive oxygen and nitrogen species (ROS/RNS). Based on the ability of F. tularensis to resist high ROS/RNS levels, we have hypothesized that additional unknown factors act in conjunction with known antioxidant defenses to render ROS resistance. By screening a transposon insertion library of F. tularensis LVS in the presence of hydrogen peroxide, we have identified an oxidant sensitive mutant in putative EmrA1 (FTL_0687) secretion protein. The results demonstrate that the emrA1 mutant is highly sensitive to oxidants and several antimicrobial agents, and exhibits diminished intramacrophage growth that can be restored to wild type F. tularensis LVS levels either by transcomplementation, inhibition of ROS generation, or infection in NADPH oxidase deficient (gp91Phox−/−) macrophages. The emrA1 mutant is attenuated for virulence, which is restored by infection in gp91Phox−/− mice. Further, EmrA1 contributes to oxidative stress resistance by affecting secretion of Francisella antioxidant enzymes SodB and KatG. This study exposes unique links between transporter activity and the antioxidant defense mechanisms of F. tularensis.
Introduction
Francisella tularensis is an important human pathogen responsible for causing tularemia in the northern hemisphere. F. tularensis has long been developed as a biological weapon due to its ability to cause severe illness upon inhalation of as few as 10 organisms and is now classified as a tier 1 category A agent by the CDC based on its possible use as a bioterror agent (Bossi and Bricaire, 2003; Dennis et al., 2001; Federal Register, 2012). The virulent strains are classified under F. tularensis subsp. tularensis (type A) and F. tularensis subsp. holarctica (type B), whereas avirulent strains belong to F. tularensis subsp. novicida or F. mediasiatica. The virulent SchuS4 strain belongs to F. tularensis subsp. tularensis while the live vaccine strain (LVS) is a derivative of F. tularensis subsp. holarctica developed in the USA from the Russian strain S15 (Wayne and Oyston, 2007). Francisella is a gram-negative, facultative intracellular pathogen that can replicate in a variety of cell types. Macrophages are considered to be the primary sites of bacterial replication. The unique intramacrophage lifestyle of F. tularensis has a transient phagosomal phase [lasting for approximately 30–60 min] followed by escape from phagosomes into the cytosol (Santic et al., 2006). The bacterial replication occurs during the cytosolic phase of bacterial residence.
Immediately upon its uptake into phagosomes by the phagocytic cells, Francisella resists its killing by potent antimicrobial host defense mechanisms such as degradative enzymes, acidic pH and reactive oxygen and nitrogen species (ROS/RNS). F. tularensis survives the attack from degradative enzymes by preventing the maturation and acidification of the phagosomes, and the phagosome-lysosome fusion (Santic et al., 2006; Santic et al., 2005). F. tularensis has evolved several strategies to inhibit the NADPH-oxidase (Phox)-dependent ROS production or to neutralize ROS within the phagosomes (Allen, 2003; Allen and McCaffrey, 2007; Mohapatra et al., 2007). The antioxidants of F. tularensis are strategically localized to protect it from macrophage derived oxidative insult. The Fe-containing superoxide dismutase (SodB) is constitutively expressed and secreted (Bakshi et al., 2006; Lee et al., 2006); while the CuZn containing SodC is located in the periplasmic space. The catalase (KatG) and alkyl hydro-peroxyreductase (AhpC) are induced in response to oxidative stress (Wehrly et al., 2009) and secreted into the macrophage cytosol (Lee et al., 2006). Moreover, the expression of these primary antioxidant enzymes starts immediately upon phagocytosis of F. tularensis and remains significantly upregulated during phagosomal and cytosolic phases (Wehrly et al., 2009), suggesting that F. tularensis experiences oxidative stress at both of these distinct intracellular locations. SodB, SodC and KatG have been shown to be required for intramacrophage survival and virulence of F. tularensis LVS in mice (Bakshi et al., 2006; Melillo et al., 2009; Melillo et al., 2010; Lindgren et al., 2007); however, KatG in F. tularensis SchuS4 is dispensable for both survival in macrophages and virulence in mice (Lindgren et al., 2007). In addition to these conventional antioxidant defenses, moxR family ATPase encoded by FTL_0200 and two additional genes present on the moxR locus of F. tularensis LVS are required for resistance against oxidative and pH stresses (Dieppedale et al., 2011). Additionally, proteins of F. novicida and F. tularensis LVS with sequence similarities to ohr gene product of Xanthomonas campestris are required for resistance to organic hydroperoxidases (Llewellyn et al., 2011). Collectively, these studies indicate that F. tularensis has evolved multitude of mechanisms to overcome oxidative stress. Based on these observations, we hypothesized that additional unknown gene products of F. tularensis may act in conjunction with known antioxidant defenses to render ROS resistance. This study has identified and characterized an additional unknown factor that contributes to the oxidant resistance of F. tularensis.
The multidrug transporters are classified either as ATP-binding cassette (ABC) transporters that use ATP as energy source or as secondary transporters that use electron gradient of protons or sodium ions as energy source for the transport of molecules across the channel. Based on their structural homology, the secondary transporters are further divided into four superfamilies including the major facilitator superfamily (MFS), the resistance-nodulation-division (RND) family, the multidrug and toxic compounds extrusion (MATE) family, and small multidrug resistance (SMR) family (Piddock, 2006). The proteins of membrane fusion protein (MFP) family serve to connect the cytoplasmic membrane transporter protein of multidrug efflux transporters to an outer membrane (OM) protein that serves as a porin. These three components in gram-negative bacteria form a contiguous channel designated as type 1 secretion system (T1SS) to pump molecules out of the cytoplasm, across both the inner and OM directly into the external milieu (Holland et al., 2005). Previously, we characterized two multidrug efflux OM proteins TolC and FtlC, and reported TolC as an important F. tularensis factor responsible for efflux of detergents, dyes, antibiotics and virulence in mice (Gil et al., 2006; Platz et al., 2010). Using an in silico approach, Atkins et al., (Atkins et al., 2006) identified 15 functional ABC systems in F. tularensis SchuS4 strain. F. tularensis is predicted to encode 31 MFS transporters. Characterization of a subset of MFS transporters revealed their role in pathogenesis and virulence of F. tularensis (Marohn et al., 2012). Bina et al., (Bina et al., 2008) identified and characterized AcrAB RND multidrug transporter. They reported that AcrB of F. tularensis LVS is required for resistance to multiple antibiotics, antimicrobial compounds, and virulence in mice (Bina et al., 2008). In a later study, it was established that both the AcrA and AcrB components of the RND-type multidrug efflux pump of F. tularensis SchuS4 are required for efflux of antibiotics, dyes, and detergents; however, they are not required for virulence in mice (Qin et al., 2008). In addition to their roles in antibiotic resistance, the multidrug transporters have also been reported to be required for oxidative stress resistance in several bacterial pathogens (Jeon et al., 2011; Santiviago et al., 2002; Chen et al., 2010).
In order to identify additional unknown factors that contribute to oxidant resistance of F. tularensis, we screened a transposon insertion library of F. tularensis LVS in the presence of hydrogen peroxide (H2O2) under in vitro culture conditions. We identified a H2O2-sensitive mutant in the FTL_0687 gene of F. tularensis LVS which encodes a MFP of MFS type multidrug efflux pump. The FTL_0687 encoded protein annotated as EmrA1 in the Francisella genome is orthologous to EmrA protein of E. coli. The E. coli Emr locus participates in the transportation of toxins and drugs from the bacterial cells. In this study, we present evidence demonstrating that the FTL_0687 gene encoded EmrA1 protein of F. tularensis LVS is required for resistance to oxidative stress, intramacrophage growth and virulence in mice. Further, EmrA1 contributes to oxidative stress resistance by affecting the secretion of Francisella antioxidant enzymes SodB and KatG. Collectively, this study describes a novel factor which contributes to oxidative stress resistance and virulence of F. tularensis.
Results
Identification of H2O2 sensitive mutants of F. tularensis LVS
It has been reported that resistance of F. tularensis to H2O2 is conferred primarily by KatG (Lindgren et al., 2007). In order to identify additional candidate genes of F. tularensis required for resistance to oxidative stress, a Tn5 transposon insertion library was screened in the presence of 250μM of H2O2 following the protocol shown in Fig. S1. Screening of about 1200 transposon insertion mutants led to the identification of five H2O2 sensitive mutants of F. tularensis LVS. The sensitive mutants were identified by comparing their OD600 values when grown for 24 hrs in Mueller Hinton (MH)-broth in the presence or absence of 250μM of H2O2. The mutants growing to an OD600 of 0.1 or above from a starting OD600 of 0.05 in the presence of H2O2 were not selected to avoid any false positive results. These mutants did not show any growth defects when grown in the absence of H2O2. Sequencing of the transposon insertion sites resulted in identification of four genes (Fig 1A). The identified mutants were characterized further for their sensitivity to H2O2, other oxidants and their ability to survive in murine alveolar macrophage cell line MH-S. Growth curves generated using large culture volumes in MH-broth revealed that a mutant in the gene FTL_0687 encoding a putative EmrA1 secretion protein and two other mutants having transposon insertions in FTL_0283 and between FTL_0283 and FTL_0284 genes encoding for an amino acid transporter failed to grow in the presence of 250μM H2O2, confirming the results from the primary screening (Fig. 1B). However, the mutants in genes FTL_0864 and FTL_0042 encoding for a Sodium decarboxylase symporter and a transposes, respectively, showed an attenuated growth in the presence of H2O2 when grown in large culture volumes (Fig. 1B). The FTL_0687 (emrA1) mutant showed enhanced sensitivity towards superoxide (O2−) generating compound pyrogallol as compared to F. tularensis LVS, FTL_0283, FTL_0283-84 and other H2O2 sensitive mutants identified in the transposon screen (Fig. 1C). Moreover, the emrA1 mutant was deficient for intramacrophage survival as compared to F. tularensis LVS and other four mutants identified in the screen when analyzed by macrophage gentamycin protection assay (Fig. 1D). Based on its extreme sensitivity to oxidants and attenuated intramacrophage growth, the emrA1 mutant was further characterized in this study to understand the additional ROS-resistance mechanisms of F. tularensis LVS.
Fig. 1. Identification and characterization of H2O2 sensitive mutants of F. tularensis LVS.
(A) H2O2 sensitive mutants identified by screening of transposon mutant library of F. tularensis LVS in the presence of 250μM of H2O2.
(B) The H2O2 sensitive mutants were grown in large culture volumes (25 ml) in the absence or presence of 250μM H2O2. The cultures were grown for 24 hrs and OD600 readings were recorded every 4 hrs.
(C) Disc diffusion assay with superoxide generating compound pyrogallol (Concentration = 50μg/disc). The data are cumulative of two independent experiments. The data were analyzed by ANOVA with a Tukey-Kramer post-test, and a cut-off p value of 0.05 or less was considered significant. *, p<0.05; **, p<0.01.
(D) MHS- cells were infected with the F. tularensis (Ft) LVS, or the H2O2 sensitive mutants at 100 MOI (n=4 biological replicates). The cells were lysed at 24 hrs and plated on MH-chocolate agar plates for enumeration of bacterial CFU. The data are representative of three independent experiments conducted with identical results and were analyzed by ANOVA with a Tukey-Kramer post-test, and a cut-off p value of 0.05 or less was considered significant. Comparisons are shown with Ft LVS. **, p<0.01
EmrA1 mutant of F. tularensis
A H2O2-hypersensitive emrA1 mutant was found to contain a transposon insertion in FTL_0687 gene of F. tularensis. The FTL_0687 is the second gene of an operon that encodes for a MFS multidrug efflux pump. The FTL_0687 encodes a membrane fusion protein EmrA1 which is a component of a tri-partite MFS efflux pump. The genes flanking the FTL_0687 encode a major facilitator transporter (FTL_0688) and a TolC-like OM efflux protein (FTL_0686) designated as SilC (Huntley et al., 2007) that forms the inner and OM components of tri-partite multidrug efflux system, respectively (Fig 2A). The Emr multidrug efflux system is found in a diverse set of gram-negative pathogens in which they play a role in the secretion of proteins. In a majority of pathogens, this locus consists of core emrA and emrB genes which are separated from the OM efflux protein. The transcription of the OM efflux protein (TolC or TolC-like protein) occurs at a distant location. Genes encoding the putative Emr multidrug efflux machinery components in F. tularensis are not separated and the inner emrB (FTL_0688), MFP emrA1 (FTL_0687) and the OM component silC (FTL_0686) are tandemly positioned (Fig. 2B). The tandem positioning of these components of tri-partite multidrug efflux transporter suggested that these three genes are co-transcribed. Indeed, the RT-PCR analysis confirmed that emrB, emrA1 and silC form an operon and are co-transcribed (Fig. 2C). We also generated a transcomplemented strain of the emrA1 mutant by providing a copy of the emrA1 gene in-trans. We further investigated if the insertion of Tn5 transposon in the emrA1gene does not lead to polar effects resulting in altered transcription of genes up- and downstream of it by qRT-PCR. It was observed that the transcription of emrB and silC remained unaltered as compared to the wild type F. tularensis LVS (Fig. S2).
Fig. 2. Genomic organization of the emrA1 gene locus of F. tularensis LVS.
(A) The genomic organization of the emrA1 gene locus. Arrows on the top indicate the location of primers used for RT-PCR while the small arrows indicate location of primers used for qRT-PCR. The inverted triangle depicts the location of transposon insertion in the emrA1 gene.
(B) Unique organization of components of the Emr multidrug efflux pump of F. tularensis. Genes enclosed in dashed box indicate that all the three components of the Emr multidrug efflux pump are transcribed at the same location.
(C) RT-PCR analysis of the intergenic regions between FTL_0689 and FTL_0688 (primers A+B in Fig. 1A); FTL_0688 and FTL_0687 (C+D), and FTL_0687 and FTL_0686c (E+F). RNA (no RT) and DNA were kept as negative and positive controls, respectively.
(D) Alignment of the conserved RLS motif in EmrA1 of F. tularensis LVS, SchuS4 and other gram-negative bacterial pathogens. The numbers indicate amino acid positions. The arrows indicate the position of conserved RLS motifs.
Amino acid sequence alignment using BLAST analysis revealed 99% similarity in EmrA1 MFPs from F. tularensis SchuS4 (FTT1257) and F. novicida (FTN1276). The conserved domain search in EmrA1 protein revealed a biotin-lipoyl-like domain (amino acids 62–108, pfam13533) which anchors EmrA1 protein to the cytoplasmic major facilitator transporter, EmrA1_3 domain (amino acids 213–330, pfam 13437), and an 8a0101 efflux pump membrane protein domain (amino acids 42–337, TIGR00998). Similar to the MFP from several other gram-negative bacteria, the amino acids 146–157 of the EmrA1 protein have a conserved RxxxLxxxxxxS (RLS) motif which creates a binding interface for α-barrel tip of the OM TolC or TolC-like proteins (Fig. 2D).
EmrA1 mutant of F. tularensis is highly sensitive to oxidative stress
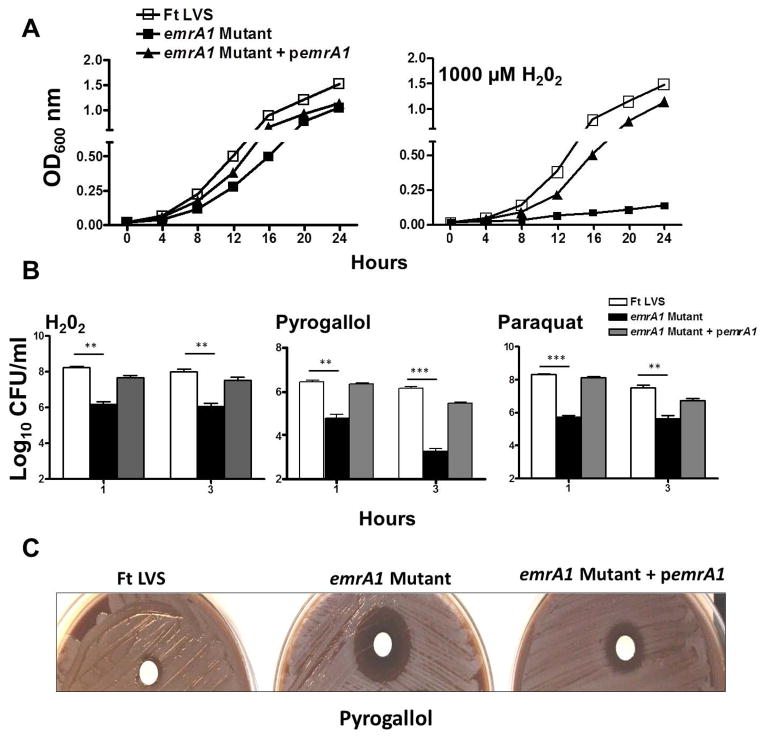
The emrA1 mutant was further characterized for its sensitivity to oxidants. Growth curves of F. tularensis LVS, the emrA1 mutant, and the transcomplemented strain were generated in the absence or presence of 1000μM H2O2. It was observed that F. tularensis LVS grew normally in the absence or presence of 1000μM concentrations of H2O2. The emrA mutant did not exhibit a growth defect as compared to F. tularensis LVS or the transcomplemented strain when grown in MH-broth in the absence of H2O2; however, it failed to grow when exposed to H2O2. Transcomplementation restored the growth of the emrA1mutant similar to wild type F. tularensis LVS when grown in the presence of 1000μM of H2O2, suggesting that the sensitivity to H2O2 is attributed to the loss of emrA1 gene (Fig. 3A). We further established the oxidant sensitivity of the emrA1 mutant by performing bacterial killing assays. In addition to H2O2, the F. tularensis LVS, the emrA1 mutant, and the transcomplemented strain were also exposed to O2− generating compounds, paraquat and pyrogallol. Significantly less numbers of emrA1 mutant as compared to the wild type F. tularensis LVS or the transcomplemented strain were recovered following 1 and 3 hrs of exposure to H2O2, paraquat and pyrogallol (Fig. 3B). A disc diffusion assay further confirmed the enhanced sensitivity of the emrA1 mutant towards pyrogallol as indicated by a larger zone of inhibition than the wild type and transcomplemented strains (Fig. 3C). Collectively, these results demonstrate that EmrA1 of F. tularensis contributes to oxidant resistance and its loss results in enhanced sensitivity to oxidative stress.
Fig. 3. The emrA1 mutant of F. tularensis is highly sensitive to oxidative stress.
(A) F. tularensis (Ft) LVS, emrA1 and emrA1+ pemrA1 transcomplemented strains were grown in MH-broth containing 1000 μM of H2O2. The cultures were grown for 24 hrs and OD600 readings were recorded every 4 hrs.
(B) The bacterial killing assay. The bacterial cultures were exposed to 1000 μM of H2O2, 1 mM Pyrogallol or 1mM Paraquat for 1 and 3 hrs (n=3 biological replicates). The cultures were diluted 10-fold and plated on MH-chocolate agar plates for bacterial enumeration. The results are expressed as Log10 CFU/ml. The data are representative of three independent experiments conducted with identical results. The data were analyzed by ANOVA with a Tukey-Kramer post-test, and a cut-off p value of 0.05 or less was considered significant. Comparisons are shown with Ft LVS. **, p<0.01; ***, p<0.001.
(D) Disc diffusion assay with superoxide generating compound pyrogallol (Concentration = 50μg/disc).
EmrA1 is required for resistance against antibiotics
EmrA1 is a membrane fusion component of MFS multidrug efflux pump of F. tularensis which is involved in actively expelling drugs from the cytosol of the bacterial cells. We next determined the role of EmrA1 in resistance against antibiotics by performing conventional disc diffusion assays. The emrA1 mutant revealed significantly enhanced sensitivity towards nalidixic acid, neomycin, streptomycin and tetracycline. Transcomplementation of the emrA1 mutant restored the wild type phenotype (Table 1). Sensitivity of the emrA1 mutant to chloramphenicol, gentamicin, oxacillin, polymyxin B and vancomycin was similar to that of wild type F. tularensis LVS. The unaltered sensitivity to polymyxin B and vancomycin suggested that the changes observed in the emrA1 mutant are not due to a general membrane defect but are specifically related to the loss of EmrA1. It was further tested if EmrA1 is also required for sensitivity to detergent and dyes by disc diffusion assays. However, the emrA1 mutant did not reveal any enhanced sensitivity to SDS or ethidium bromide indicating that EmrA1 is not required for resistance against detergents and dyes (Table 1).
Table 1.
Sensitivity of wild type F. tularensis LVS, the emrA1 mutant and the transcomplemented strain to antibiotics, detergents and dyes.
| Antibiotic | Concentration (μg/disc) | Zone of inhibition (mm) | ||
|---|---|---|---|---|
| F. tularensis LVS | emrA1 Mutant | emrA1 + pemrA1 | ||
| Nalidixic acid | 30 | 33±1 | 49±1 | 37±1 |
| Neomycin | 30 | 9±1 | 15±1 | 10±2 |
| Streptomycin | 10 | 25±1 | 32±4 | 27±2 |
| Tetracycline | 30 | 17±1 | 45±1 | 28±2 |
| Chloramphenicol | 30 | 36±4 | 40±6 | 36±2 |
| Gentamicin | 10 | 32±3 | 35±1 | 32±1 |
| Bacitracin | 10 | 6±0* | 6±0 | 6±0 |
| Oxacillin | 1 | 6±0 | 6±0 | 6±0 |
| Polymyxin B | 300 | 6±0 | 6±0 | 6±0 |
| Vancomycin | 30 | 6±0 | 10±0 | 6±0 |
| SDS | 56** | 7±0 | 7±0 | 7±0 |
| 750*** | 9±0 | 10±0 | 9±0 | |
| Ethidium Bromide | 5*** | 15±0 | 14±0 | 14±0 |
| 25** | 21±1 | 25±0 | 22±1 | |
Values represented in bold letters are significantly higher than the wild type F. tularensis LVS and emrA + pemrA transcomplemented strain (P<0.05).
Diameter of the filter paper disc.
acrB mutant is sensitive to these concentrations of SDS and ethidium bromide (Qin et al., 2008).
ΔtolC mutant is sensitive to these concentrations of SDS and ethidium bromide (Gil et al., 2006).
SDS = Sodium Dodecyl Sulphate
Intramacrophage killing of emrA1 mutant is ROS-dependent
Since F. tularensis is an intracellular pathogen, we next determined the role of EmrA1 in intramacrophage survival by performing a conventional gentamicin protection assay in alveolar macrophage cell line MH-S, and primary bone marrow derived macrophages (BMDMs) from C57BL/6 mice. The emrA1 mutant did not reveal any invasion defect as bacterial numbers similar to that of F. tularensis LVS and the transcomplemented strain were taken up by macrophages 4 hrs post-infection. However, significantly less numbers of emrA1 mutant were recovered at 24 hrs post-infection as compared to the wild type and transcomplemented strains (Fig. 4A and D). Further, inhibition of ROS production by treating macrophages with ROS inhibitors apocynin or N-acetyl cysteine (NAC) restored the replication of the emrA1 mutant to wild type F. tularensis LVS levels (Fig. 4B and C). Similar results were obtained when experiments were conducted using gp91Phox−/− macrophages which fail to generate Phox-dependent ROS production (Fig. 4E). Collectively, these results demonstrate that the emrA1 mutant of F. tularensis is killed by macrophages in a ROS-dependent fashion and that EmrA1 plays an essential role in resisting oxidative stress inside the macrophages.
Fig. 4. The emrA1 mutant of F. tularensis is deficient for intramacrophage survival and its killing by macrophages is ROS-dependent.
MHS- cells (A, B and C) or primary BMDMs derived from wild type C57BL/6 (D) or gp91Phox−/− mice (E) were infected with the F. tularensis (Ft) LVS, the emrA1 mutant or the transcomplemented strain (emrA1 + pemrA1) at 100 MOI (n=4 biological replicates). The cells were lysed at 4 and 24 hrs and plated on MH-chocolate agar plates for enumeration of bacterial CFU. The MH-S cells were treated with ROS inhibitors apocynin (B) or N- Acetyl Cysteine (NAC) (C) prior to infection. The data are representative of three independent experiments conducted with identical results. The data are expressed as Log10 CFU/ml. The data were analyzed by ANOVA with a Tukey-Kramer post-test, and a cut-off p value of 0.05 or less was considered significant. Comparisons are shown with Ft LVS. **, p<0.01.
EmrA1 mutant of F. tularensis is attenuated for virulence and its virulence is restored in gp91Phox−/− mice
We next investigated the requirement of EmrA1 for virulence in mice. C57BL/6 mice were inoculated intranasally with 1×105 or 1×106 CFUs of F. tularensis LVS or the emrA1 mutant and observed for morbidity and mortality for a period of 21 days. Mice infected with 1×105 or 1×106 CFU of F. tularensis LVS started exhibiting the signs of illness by day 6–7 post-infection and succumbed to infection by days 9–12 post-infection, respectively. However, 100% of mice challenged with similar doses of the emrA1 mutant survived the infection (Fig. 5A and B). When a similar experiment was performed in gp91Phox−/− mice, the onset of symptoms in gp91Phox−/− mice inoculated with 1×105 CFU of the emrA1 mutant occurred by day 13 post-infection and 66% mortality was observed when the experiment was terminated at day 30 post-infection. On the other hand, mice inoculated with 1×106 CFU of the emrA1 mutant started showing the symptoms of sickness as early as day 6 post-infection and all mice died by day 17 post-infection. Control wild type C57BL/6 mice inoculated with similar doses of the emrA1 mutant survived the infection (Fig. 5C and D). Collectively, these results demonstrate that the emrA1 mutant of F. tularensis is attenuated for virulence in mice and similar to that in macrophages, its clearance from mice is dependent upon Phox-generated ROS.
Fig. 5. EmrA1 mutant of F. tularensis is attenuated for virulence and its virulence is restored in gp91Phox−/− mice.
Survival of C57BL/6 mice (n = 5) infected intranasally with 1×105 (A) and 1×106 CFU (B) of F. tularensis LVS or the emrA1 mutant and observed for mortality.
C57BL/6 wild type mice or gp91Phox−/− mice (n=3) were infected intranasally with 1×105 (C) and 1×106 CFU (D) of the emrA1 mutant. The data from a single experiment are shown. The mice were observed for the indicated periods for morbidity and mortality. The results are expressed as Kaplan-Meier survival curves and the P values were determined using Log-rank test.
EmrA1 mutant fails to secrete antioxidant enzymes SodB and KatG
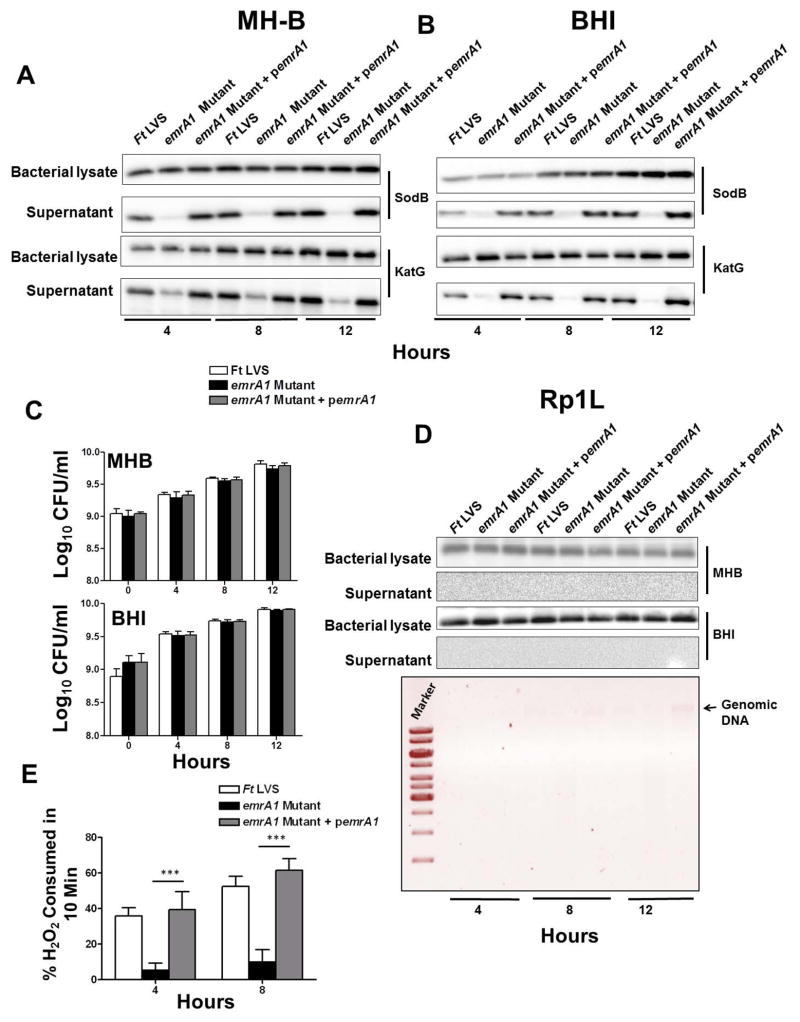
Our previous results showed that the emrA1 mutant of F. tularensis is sensitive to oxidants, attenuated for intramacrophage growth and virulence in mice, and that Phox-dependent ROS is required for its in vitro or in vivo killing. We next investigated how EmrA1 renders resistance to oxidants. Since EmrA1 is a component of MFS multidrug efflux pump involved in transport function and the antioxidant enzymes SodB and KatG are secreted by F. tularensis (Lee et al., 2006), we hypothesized that EmrA1 may have role in secretion of these antioxidant enzymes and this is how it contributes to the resistance to oxidative stress. The cultures of F. tularensis LVS, the emrA1 mutant and the transcomplemented strain were grown in MH-broth for shorter durations (4, 8 and 12 hrs) to minimize the amount of proteins released into the culture filtrates due to bacterial autolysis. The culture supernatants were filtered through a 0.22μ filter and concentrated. Equal quantities of proteins from bacterial lysates and concentrated culture filtrates were resolved separately on SDS gels, transferred onto PVDF membranes and blotted against anti-sodB and anti-KatG antibodies. Unlike F. tularensis LVS and the emrA1 transcomplemented strain, we consistently failed to detect both the SodB and KatG in culture filtrates from the emrA1 mutant. To rule out that the absence of SodB or KatG in the culture filtrates of the emrA1 mutant was due to reduced quantities of these proteins being generated in the emrA1 mutant, we analyzed the bacterial cell lysates by western blot analysis. It was observed that nearly equal amounts of these proteins were present in the lysates from all three bacterial strains tested (Fig. 6A).
Fig. 6. EmrA1 mutant fails to secrete antioxidant enzymes SodB and KatG.
The cultures of F. tularensis (Ft) LVS, the emrA1 mutant or the transcomplemented strain (emrA1+ pemrA1) were grown in MH- (A) or BHI-broth (B). The culture filtrates or the lysates of the bacterial pellet were analyzed at the indicated times by western blot analysis using anti-SodB and anti-KatG antibodies.
(C) Aliquots of the bacterial strain grown in MHB or BHI were collected at the indicated time points, diluted 10-fold and plated on MH-chocolate agar plates to enumerate bacterial numbers (n=3).
(D) The western blots of the MH-B or BHI-grown culture filtrates probed with anti-SodB antibodies were stripped and re-probed with antibodies against 50S ribosomal protein Rp1L at the indicated times (top panel). Agarose gel electrophoresis of the genomic DNA isolated from culture filtrates from the BHI-grown bacteria at indicated time points (lower panel).
(E) Determination of catalase activity of secreted KatG by amplex red assay. The amount of H2O2 consumed by culture filtrates of Ft LVS, the emrA1 mutant and the transcomplemented strain collected at the indicated times. The data are representative of at least 3–5 independent experiments conducted with identical results. The data were analyzed by ANOVA with a Tukey-Kramer post-test, and a cut-off p value of 0.05 or less was considered significant. Comparisons are shown with Ft LVS. ***, p<0.001.
Previous studies have reported that Francisella grown in MH-broth is prone to bacterial lysis resulting in a release of bacterial contents into the culture supernatants; while Brain-Heart Infusion (BHI)-broth grown Francisella mimics host-adapted phenotype with minimal in vitro bacterial autolysis (Singh et al., 2013). To ensure that the presence of SodB and KatG in culture filtrates of MH-broth grown F. tularensis LVS and the transcomplemented strain was not due to bacterial lysis and release of cytosolic contents, we repeated the immunoblots using culture filtrates and bacterial cell lysates from BHI-broth grown bacteria. The results obtained were similar to those observed for MH-broth grown bacteria showing the absence of both the SodB and KatG only in culture filtrates from the emrA1 mutant (Fig. 6B). We further investigated if the absence of antioxidant enzymes SodB and KatG in emrA1 culture filtrates was due to the differences in growth rates between the emrA1 mutant and the wild type F. tularensis LVS or the transcomplemented strain. Aliquots of MH- or BHI-broth grown bacteria collected at 0, 4, 8 and 12 hrs were diluted ten-fold and plated on MH-chocolate agar plates to quantitate the bacterial numbers. The bacterial numbers did not differ amongst the three bacterial strains at all-time points examined (Fig. 6C). We further confirmed if the release of SodB and KatG in F. tularensis LVS and the transcomplemented strain in the BHI grown medium was not due to excessive bacterial autolysis. Blots probed with the anti-SodB antibodies were stripped and re-probed with antibodies against the cytosolic 50S ribosomal protein Rp1L (FTL_1745) of F. tularensis LVS (Savitt et al., 2009) (a kind gift from Dr. Ann Savitt, SUNY Stony Brook). As shown in Fig. 7D, the Rp1L protein was detected in bacterial cell lysates, but not in the culture filtrates from any of the bacterial strains tested. We also attempted to isolate genomic DNA from the culture filtrates to detect bacterial cell lysis; however, we did not detect genomic DNA in any of the filtrates tested except a small quantity in culture filtrates from the transcomplemented strain after 12 hrs of growth (Fig. 6D).
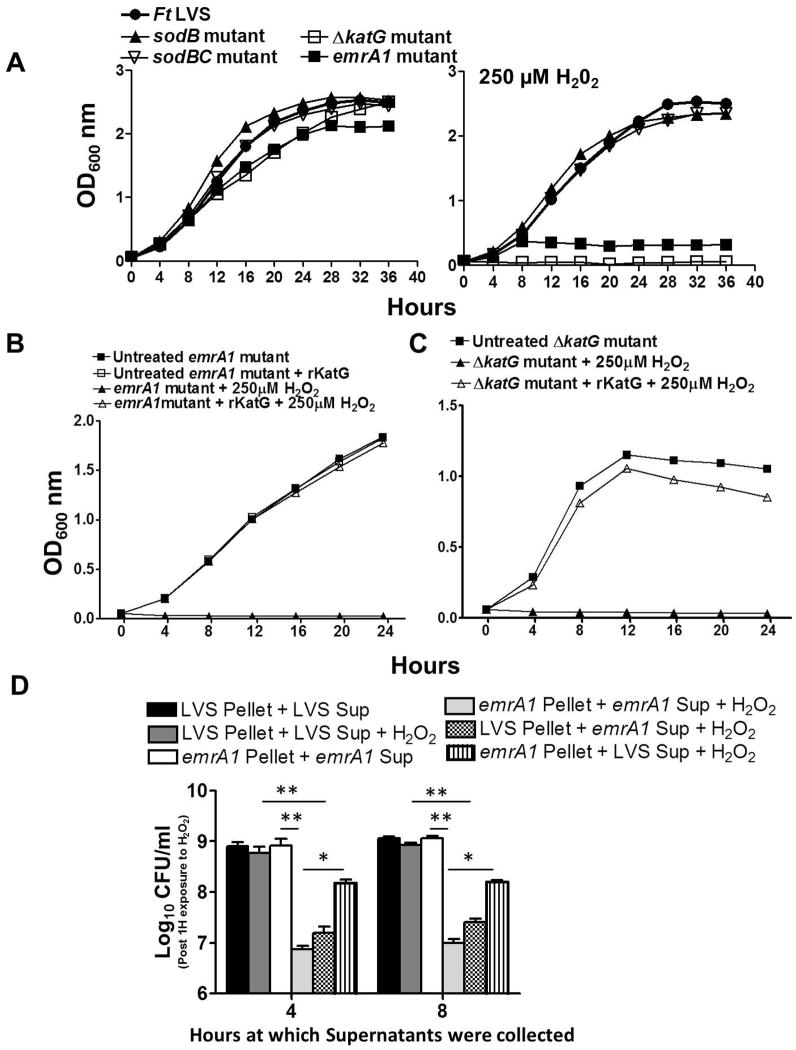
Fig. 7. EmrA1 contributes to the secretion of KatG and extracellular KatG is essential to overcome H2O2 toxicity.
(A) Growth curves of F. tularensis (Ft) LVS and the sodB, sodBC and ΔkatG mutant in the absence or presence of 250μM of H2O2. The cultures were grown for 24 hrs and OD600 readings were recorded every 4 hrs.
Growth curves of F. tularensis (Ft) LVS and the emrA1 mutant (B) and ΔkatG mutant (C) in the absence or presence of 250μM of H2O2 and with or without the addition of recombinant F. tularensis KatG (rKatG). The cultures were grown for 24 hrs and OD600 readings were recorded every 4 hrs.
(D) F. tularensis LVS (LVS) and the emrA1 mutant were grown in MH-broth for 4 and 8 hrs. The bacteria were pelleted and the supernatants were collected and filtered through a 0.22μ filter to remove the bacteria. The F. tularensis LVS pellets were then either resuspended in its own culture supernatants or those from the emrA1 mutant. Similarly, the emrA1 pellets were either resuspended in its own culture supernatants or those from F. tularensis LVS. The resuspended bacteria were either left untreated or exposed to 500μM of H2O2 for 1 hr. The bacteria were diluted 10-fold and plated on MH-chocolate-agar plates to enumerate the viable bacteria. The data are representative of 3 independent experiments conducted with identical results. The data were analyzed by ANOVA with a Tukey-Kramer post-test, and a cut-off p value of 0.05 or less was considered significant. Comparisons are shown with Ft LVS. *, p<0.05, **, p<0.01.
We further investigated if KatG secreted by the F. tularensis LVS and transcomplemented strain is enzymatically active by performing amplex red assay. This assay enabled us to determine the amount of H2O2 consumed by the catalase activity of KatG present in the culture filtrates over a period of 10 minutes. It was observed that the culture filtrates from the F. tularensis LVS and transcomplemented strain collected at 4 hrs of growth consumed about 30–40% H2O2; while about 50–60% of H2O2 was consumed by the filtrates collected after 8 hrs of growth. In contrast, the culture filtrates from the emrA1 mutant failed to neutralize any substantial amount of H2O2 (Fig. 6E). Collectively, these results demonstrate that EmrA1 is required for the secretion of antioxidant enzymes SodB and KatG and the absence of these antioxidant enzymes in culture filtrates from the emrA1 mutant is not due to altered growth rate or bacterial autolysis. Moreover, the secreted KatG is enzymatically active and can effectively neutralize extracellular H2O2.
EmrA1-dependent secretion of KatG is required for resistance against H2O2
Our preceding results indicated a role of EmrA1 in the secretion of antioxidant enzymes SodB and KatG. We next investigated if lack of secretion of these antioxidants is indeed responsible for the enhanced sensitivity of the emrA1 mutant to H2O2. SodB is required for the dismutation of O2− to H2O2, while KatG neutralizes H2O2 to water. We first compared the H2O2 sensitivity of the emrA1 mutant with sodB, sodBC and ΔkatG; the mutants deficient in known antioxidants of F. tularensis LVS by growing them in the presence of 250μM of H2O2. It was observed that all the mutants grew normally in the absence of H2O2 however, only emrA1 and the ΔkatG mutants failed to grow in the presence of H2O2 (Fig. 7A) indicating that KatG is essential for resistance against H2O2. Since the emrA1 mutant can produce but fails to secrete KatG in the extracellular milieu, we next investigated if addition of recombinant F. tularensis KatG (rKatG) in culture medium can restore its growth in the presence of H2O2. Growth curves of the emrA1mutant were generated in the presence or absence of 250μM of H2O2 with or without the addition of F. tularensis LVS rKatG. Indeed, the addition of rKatG to the culture medium restored the growth of the emrA1 mutant when grown in the presence of 250μM of H2O2 (Fig. 7B). Similarly, the growth of the ΔkatG mutant was also restored to its untreated control levels by addition of rKatG to the culture medium (Fig. 7C). To confirm that the addition of rKatG does not have a non-specific effect on growth in the presence of 250μM of H2O2, we repeated experiments using equal concentration of recombinant bacterioferritin (rBfr), a F. tularensis LVS cytosolic protein. Both the emrA1 and ΔkatG mutants failed to grow in presence of 250μM of H2O2 when rBfr was added to the culture medium (Fig. S3). Taken together, these results demonstrate that failure to secrete KatG results in enhanced H2O2 sensitivity of the emrA1 mutant.
To further establish that extracellular KatG is required for resistance against H2O2, wild type F. tularensis LVS and the emrA1 mutant were grown in MH-broth for 4 and 8 hrs. The bacteria were pelleted and the supernatants were collected and filtered through a 0.22μ filter to remove the bacteria. The F. tularensis LVS pellets were then either resuspended in its own culture supernatants or those from the emrA1 mutant. Similarly, the emrA1 pellets were either resuspended in its own culture supernatants or those from F. tularensis LVS. The resuspended bacteria were either left untreated or exposed to 500μM of H2O2 for 1 hr. The bacteria were diluted 10-fold and plated on MH chocolate-agar plates to enumerate the viable bacteria. No differences were observed in the numbers of viable bacteria recovered either from untreated emrA1 and F. tularensis LVS resuspended in their respective supernatants or from F. tularensis LVS pellets resuspended in F. tularensis LVS culture supernatants and exposed to 500μM H2O2 for 1 hr. On the contrary, significantly less numbers of viable F. tularensis LVS were recovered when the LVS pellets were resuspended in emrA1culture supernatants. On the other hand, significantly higher numbers of viable emrA1 mutant bacteria were recovered when the emrA1 pellets were resuspended in F. tularensis LVS culture supernatants as compared to the emrA1 pellets resupended in its own culture supernatants and exposed to H2O2 (Fig. 7D). Collectively, these results provide evidence that EmrA1 contributes to the secretion of KatG and that extracellular KatG is essential to overcome H2O2 toxicity. Further, the loss of EmrA1 results in enhanced sensitivity to H2O2.
Signal IP analysis revealed that the SodB protein of F. tularensis lacks a classical signal sequence required for protein secretion. On the other hand, an N-terminal 24 amino acid long signal peptide with a putative cleavage site between amino acids 23 and 24 is present in the KatG protein. We further investigated the requirement of this signal sequence for secretion of KatG. We deleted the putative signal sequence of katG gene (ΔSkatG), cloned it in the pMP822 transcomplementation vector (LoVullo et al., 2006) and electroporated in ΔkatG mutant of F. tularensis LVS. The ΔkatG+pΔSkatG transcomplemented strain was analyzed for the expression of KatG by western blot analysis. Surprisingly, no expression of KatG was observed in the ΔkatG+pΔSkatG transcomplemented strain. However, as reported earlier (Lee et al., 2006), KatG expression was observed when E. coli were transformed with pΔSkatG plasmid (data not shown). Further, consistent with its lack of expression, the ΔkatG+pΔSkatG transcomplemented strain similar to the ΔkatG mutant failed to grow in the presence of 250μM of H2O2. These results indicate that the putative KatG signal sequence is required for expression of KatG protein (Fig. S4).
Secretion of antioxidant enzymes SodB and KatG is independent of tolC and Sec-dependent secretion pathways
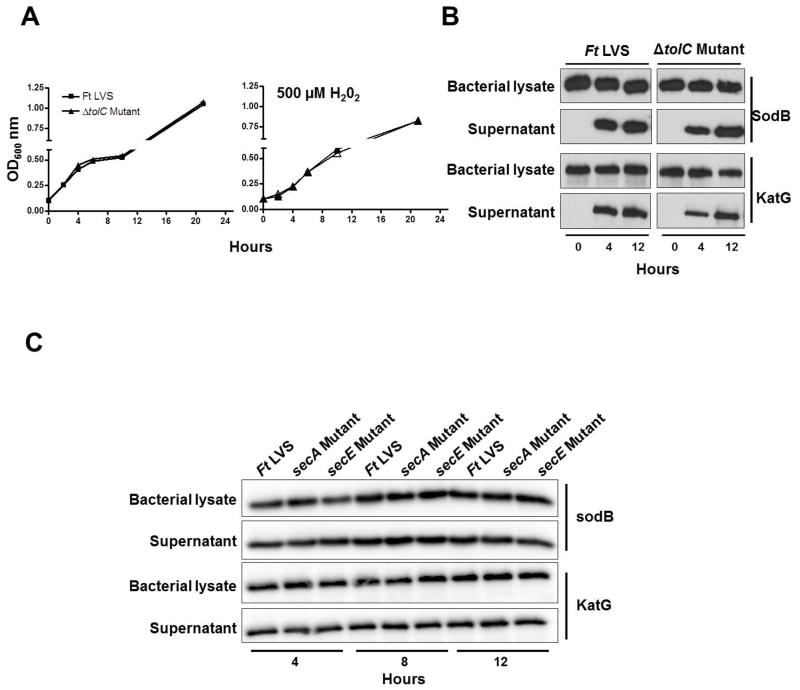
In several other bacterial systems, it has been shown that the Acr RND and Emr multidrug efflux systems work together with a common OM protein TolC for the efflux of a range of antibiotics and harmful substances. We further investigated if the loss of OM protein TolC results in an oxidant-sensitive phenotype similar to that observed for the emrA1 mutant. Growth curves of the TolC deletion mutant (ΔtolC) were generated in the presence of 500μM of H2O2 and compared with those of F. tularensis LVS. It was observed that the ΔtolC mutant did not exhibit any growth defect when grown in the presence of H2O2 (Fig. 8A). Further, both SodB and KatG were detected in the culture supernatants of the ΔtolC mutant by western blot analysis indicating that TolC is not required for secretion of these antioxidant enzymes (Fig. 8B).
Fig. 8. Secretion of SodB and katG is independent of TolC and Sec-dependent secretion pathway.
(A) Growth curves of F. tularensis (Ft) LVS and the ΔtolC mutant in the absence or presence of 500μM of H2O2.
(B) The culture filtrates or the lysates of the bacterial pellet of F. tularensis (Ft) LVS or the ΔtolC mutant grown in BHI-broth were analyzed at the indicated times by western blot analysis using anti-SodB and anti-KatG antibodies. The data are representative of at least 2–3 independent experiments conducted with identical results.
(C) Wild type F. tularensis (Ft) LVS and the secA and secE mutants of F. tularensis LVS were grown in BHI-broth for 4, 8 and 12 hrs. The culture filtrates or the lysates of the bacterial pellet were analyzed at the indicated times by western blot analysis using anti-SodB and anti-KatG antibodies. The data are representative of 2 independent experiments conducted with identical results.
A SecA-dependent mechanism has been shown to be involved in the secretion of an Fe-containing SodA and KatG in M. tuberculosis and it has been reported that considerably less quantities of these antioxidants are released by the secA2 mutant in the extracellular milieu (Braunstein et al., 2003). Since the results obtained with the emrA1 mutant of F. tularensis LVS mirror the phenotype reported for secA2 mutant of M. tuberculosis (Braunstein et al., 2003), we investigated if SecA and SecE proteins of F. tularensis are also involved in the secretion of these antioxidant enzymes. Analysis of the KatG signal sequence failed to exhibit any features characteristic of the signal peptides of Sec-dependent pathway. The secA and secE mutants of F. tularensis LVS were grown in BHI-broth for 4, 8 and 12 hrs. The bacterial pellets and the culture supernatants were tested for the presence of SodB and KatG. The western blot analysis of secA and secE mutants with anti-SodB and KatG antibodies revealed the presence of both the SodB and KatG in culture filtrates as well in the cell lysates indicating that the secretion of both these antioxidant enzymes by F. tularensis LVS is independent of SecA and SecE-dependent secretion systems (Fig. 8C).
Discussion
This study focused on the investigation of antioxidant defense mechanisms of F. tularensis presents unique mechanism found to be involved in this process. Upon phagocytosis of bacterial pathogens by macrophages, Phox initiates ROS production by reducing molecular oxygen to highly reactive O2− radicals. The O2− rapidly dismutate into H2O2 which may further get converted into highly microbicidal hydroxyl radicals via iron-mediated Fenton chemistry (Bedard and Krause, 2007). These ROS are highly reactive microbicidal agents and kill engulfed pathogens by oxidizing their proteins, lipids, carbohydrates and nucleic acids. Deficiencies in the Phox-encoding genes result in a none to minimal level of ROS production, thereby enhancing the host’s susceptibility towards bacterial, fungal and yeast infections (Bedard and Krause, 2007; Bustamante et al., 2007). The significance of ROS in host defense against F. tularensis has been well established (Lindgren et al., 2004; Lindgren et al., 2005). Even though studies have demonstrated that ROS are required for killing of F. tularensis, it has been shown that this bacterium can resist killing by oxidative stress in macrophages (Allen, 2003; McCaffrey and Allen, 2006; Mohapatra et al., 2010). F. tularensis genes showing similarities with genes known to be involved in ROS resistance in different bacterial species have been identified (Larsson et al., 2005). Previous studies including ours have shown that F. tularensis antioxidant enzymes KatG, SodB and SodC are required for oxidant resistance, survival in macrophages, and virulence in mice (Bakshi et al., 2006; Melillo et al., 2009; Lindgren et al., 2007). The SodB and KatG are secreted; however, the mechanism of secretion of these Francisella antioxidant enzymes is not known (Lee et al., 2006). The Secretome software analysis also predicts both the SodB and KatG as secreted proteins (data not shown). Further, the transcriptional analysis has shown that many of these genes are induced immediately upon Francisella’s entry into phagosomes and escape into the cytosol (Wehrly et al., 2009). These findings suggest that Francisella is continuously primed for oxidative stress and has evolved multitude of mechanisms to overcome oxidative stress in order to survive in macrophages.
In this study, by screening 1200 F. tularensis LVS transposon mutants for H2O2 sensitivity, five mutants were identified. The low recovery of the mutants could have been due to higher stringency for selection of the sensitive mutants and a low dose of H2O2 used for screening. A 250μM concentration of H2O2 was chosen for screening of transposon mutants to eliminate the identification of mutants in known antioxidant genes of F. tularensis. It has been reported that sodB, sodC, sodBC and moxR gene mutants of F. tularensis LVS exhibit enhanced susceptibility when exposed to 1–10mM of H2O2 (Bakshi et al., 2008; Melillo et al., 2009; Dieppedale et al., 2011), whereas; the ohr gene mutant remains resistant to 3% of H2O2 (Llewellyn et al., 2011). As a result, these mutants were not recovered in our screen. Only the ΔkatG mutant of F. tularensis LVS is susceptible to lower concentrations (100μM) of H2O2 (Lindgren et al., 2007). However, despite the sensitivity of the ΔkatG mutant to the concentration of H2O2 used for screening, our screen did not identify katG mutant. The reason could be a non-saturating number of transposon mutants used for screening that did not cover the entire genome of F. tularensis LVS that contains 1754 open reading frames (Zvi et al., 2011). Interestingly, four of the five H2O2 sensitive mutants identified in our screens were found to be involved in transporter functions. We identified two H2O2 sensitive mutants in gene FTL_0283 and FTL_0283-284 encoding an amino acid transporter, indicating its important role in resistance to oxidative stress. Another identified mutant in FTL_0687 gene of F. tularensis which encodes for EmrA1 protein is a component of MFS type multidrug efflux pump. We also identified a mutant in sodium decarboxylase symporter encoded by FTL_0684 gene and FTL_0042 encoding a transposes in our primary screen, however, these two mutants revealed only an attenuated growth in the presence of H2O2 when grown in large culture volumes. Both the aromatic amino acid transporter mutants and the emrA1 mutant were equally sensitive to H2O2. However, only emrA1 mutant was found to be more sensitive to pyrogallol which generates O2− extracellularly and highly attenuated for intramacrophage growth as compared to the aromatic amino acid transporter mutant.
This study characterized and investigated role of FTL_0687 encoded EmrA1 protein in resistance to oxidant stress, intramacrophage survival and virulence in mice. The orthologs of FTL_0687 in F. novicida (FTN1276) and its upstream OM component FTN1277 have been identified in the negative selection screen of F. novicida transposon mutants amongst several other genes required for in vivo growth and survival in mouse model of infection (Weiss et al., 2007). The FTL_0687 mutant was also identified as deficient for intramacrophage growth when the transposon mutant library of F. tularensis was screened in MH-S cells, an alveolar macrophage cell line (our unpublished data). F. tularensis has been predicted to encode 31 MFS transporters. A subset of MFS transporters belonging to Francisella phagosomal transporter family have been characterized and shown to play an important role in the pathogenesis of F. tularensis (Marohn et al., 2012). Additionally, based on software analysis, F. tularensis has been predicted to contain about 15 functional ABC transport systems (Atkins et al., 2006); however, none of these have been functionally characterized for their role in efflux of substrates, intracellular survival, and virulence. The functional roles of the inner and membrane fusion components of the RND type AcrA/B multidrug efflux system of F. tularensis have been characterized (Bina et al., 2008; Qin et al., 2008). It has been shown that AcrB of F. tularensis LVS which is a functional equivalent of the EmrA1 protein of the MFS transporters is required for resistance to multiple antibiotics, antimicrobial compounds and virulence in mice (Bina et al., 2008). Contrary to this, it has been established that although AcrA or AcrB components of the RND-type efflux pump of F. tularensis SchuS4 are required for efflux of antibiotics, dyes and detergents, they are not required for virulence in mice (Qin et al., 2008). Additionally, two multidrug efflux OM proteins TolC and FtlC of F. tularensis have been characterized for their role in efflux of detergents, dyes, antibiotics, and virulence in mice (Gil et al., 2006; Platz et al., 2010). Similar to several other bacterial systems, it has been suggested that the AcrA/B RND multidrug efflux system works either with TolC or FtlC OM components for the efflux of a range of antibiotics, detergents, and drugs (Qin et al., 2008; Gil et al., 2006); however, their functional association has not been established. In several other bacterial systems, it has been shown that the Acr RND and Emr MFS multidrug efflux systems work together with a common OM protein TolC for the efflux of a range of antibiotics and harmful substances. In F. tularensis LVS loss of OM protein TolC neither result in oxidant-sensitive phenotype similar to that observed for the emrA1 mutant nor required for secretion of antioxidant enzymes SodB and KatG (Fig. 8A, B).
The results from this study indicate that both the AcrA/B RND and Emr MFS multidrug efflux systems are designed to serve specific roles in F. tularensis and that these systems differ from one another in many respects. Firstly, the genomic organization of the Emr locus of F. tularensis is unique and all the three components of Emr MFS multidrug efflux pump are co-transcribed as an operon (Fig. 2B and C). On the other hand, only the inner and membrane fusion components (AcrA and AcrB) of the RND type multidrug efflux transporter are co-transcribed whereas the OM component, the TolC/FtlC like protein is transcribed at a distant location. Secondly, unlike emrA1 mutant, the acrB or tolC mutants are neither sensitive to oxidants nor restricted in secretion of SodB and KatG in culture filtrates (Fig. 8A, B and S5A, B, D); however, they are sensitive towards detergent and dyes (Gil et al., 2006; Qin et al., 2008; Bina et al., 2008). It was observed that mutation of emrA1 enhanced the sensitivity of F. tularensis towards antibiotics such as nalidixic acid, neomycin, streptomycin, and tetracycline but not against SDS and ethidium bromide (Table 1). This selective specificity indicates that AcrA/B RND rather than EmrA1 MFS multidrug efflux may have a major role in the efflux of detergents and dyes in F. tularensis LVS. Thirdly, neither AcrB (Fig. S5C) nor TolC are required for intramacrophage survival and AcrB is dispensable for virulence in mice (Qin et al., 2008; Gil et al., 2006). On the other hand, the results from this study demonstrate that functional EmrA1 is required for both intramacrophage survival and virulence in mice. Further, this study demonstrates that failure of the emrA1 mutant to survive in macrophages is primarily due to its inability to resist ROS generated consequent to the infection. When ROS generation was abolished either by chemical inhibitors such as apocynin and NAC or using macrophages deficient in gp91Phox, the replication defect of the emrA1 mutant in macrophages was restored to its wild type levels. Consistent with its inability to grow in macrophages, the emrA1 mutant was highly attenuated for virulence in mice. Mice challenged intranasally with 10–100LD100 dose survived the infection and required gp91phox-dependent ROS generation for clearance of the emrA1 mutant. Collectively, these observations suggests that the EmrA1 of F. tularensis may have a dual role and function in both the Emr drug efflux pathway as well as participate in a separate type 1 protein secretion system.
A novel and very interesting finding from this study was the enhanced sensitivity of the emrA1 mutant towards H2O2. Further, the emrA mutation affected secretion of antioxidant enzymes SodB and KatG. Several recent studies have addressed the role of bacterial efflux pumps in rendering resistance to oxidative stress. The multidrug efflux transporters of Campylobacter jejuni (Jeon et al., 2011), the SmvA efflux pump of Salmonella Typhimurium (Santiviago et al., 2002), MexGHI-OmpD multidrug efflux pump of Pesudomonas aeruginosa (Chen et al., 2010), and an MFS-type efflux pump of Mycobacterium tuberculosis are all been shown to contribute to defense against oxidative stress (Ramon-Garcia et al., 2009). It has been speculated that multidrug efflux pumps provide protection against oxidative stress by detoxification occurring due to the efflux of oxidants. Another possible explanation for the increased sensitivity to oxidants could be due to indirect effects resulting from the loss of multidrug efflux system. However, this study instead provides evidence that MFS transporters contribute to oxidant resistance probably by secreting antioxidant enzymes in the extracellular milieu. The presence of catalytically active KatG in the culture filtrate further supports the notion that MFS multidrug efflux mediated oxidant resistance may in fact involve neutralization of oxidants outside of the bacterial cells by pumping out the antioxidant enzymes. A previous study that has demonstrated that both SodB and KatG are secreted during infection of the host cells and in culture filtrates by actively growing F. tularensis (Lee et al., 2006). Our results demonstrating that addition of rKatG restored the growth of the emrA1 mutant in the presence of H2O2 further support the notion that extracellular KatG is essential for overcoming the oxidant stress. These characteristic features of Emr MFS multidrug efflux pump suggest that it serves to provide oxidant resistance in addition to serving as a conventional MFS multidrug efflux system.
The mechanism of recognition and secretion of both the SodB and KatG by the Emr multidrug efflux system remain undetermined in this study. Signal IP analysis revealed that the SodB protein of F. tularensis lacks a classical signal sequence required for protein secretion. On the other hand, an N-terminal 24 amino acid long signal peptide with a putative cleavage site between amino acids 23 and 24 is present in the KatG protein. Our results demonstrate that the signal sequence of KatG is required for expression in addition to its potential role in localization. An iron-containing SodA and KatG in M. tuberculosis are secreted by a SecA-dependent mechanism and considerably less quantities of these antioxidants are released by the secA2 mutant in the extracellular milieu (Braunstein et al., 2003). However, we observed that the secretion of SodB and KatG proteins is independent of both SecA and SecE-dependent secretion systems in F. tularensis LVS. Further, unlike the M. tuberculosis secA2 mutant, intramacrophage growth and virulence of the emrA1 mutant was restored in gp91phox−/− macrophages and mice or by chemical inhibition of ROS in wild type macrophages. Finally, we detected both SodB and KatG in the culture filtrates of F. tularensis LVS, the emrA1 transcomplemented strain, and the acrB or tolC mutants irrespective of the growth conditions (MHB or BHI) which contradicts the findings from an earlier study showing that BHI-broth grown F. tularensis fails to secrete these antioxidant enzymes (Zarrella et al., 2011).
In this study SodB and KatG proteins that are secreted in an EmrA1-dependent fashion were identified using an antibody-probe targeted approach. However, additional proteins that are secreted via this secretion machinery remain unidentified in this study. We were unsuccessful in detecting additional proteins in the supernatants by conventional SDS-PAGE and staining. Further, immunoblotting using F. tularensis mouse hyperimmune serum detected only a single immunogenic protein in culture supernatants from F. tularensis LVS that corresponds to KatG based on its molecular weight. This protein band was not detected in supernatants from the emrA1 mutant (data not shown). The mechanism of secretion of SodB and KatG, and identification of additional proteins secreted by the EmrA1 MFS type multidrug efflux pump remains the focus of future investigations in our laboratory. Finally, based on results from previous studies that mutants of F. tularensis LVS and SchuS4 in transporter proteins exhibit similar phenotypes (Atkins et al., 2006; Qin et al., 2008), we speculate that given a 99% identity of nucleotide and amino acid sequences between F. tularensis LVS and SchuS4, the emrA1mutant of F. tularensis SchuS4 may have a phenotype similar to that observed for the emrA1 mutant of F. tularensis LVS. However, a more rigorous assessment of the role of emrA1 in resistance to oxidative stress and virulence would require generation of a mutant in the F. tularensis SchuS4 background.
To conclude, this study identified five H2O2 sensitive mutants, including a mutant in emrA1gene encoding MFS-type MFP. We demonstrate that the lack of functional EmrA1 results in extreme oxidant sensitivity, enhanced ROS-dependent killing in macrophages, and attenuated virulence in mice. Most importantly, the loss of EmrA1 resulted in a defect in secretion of antioxidant enzymes SodB and KatG. This study provides evidence that the MFS type multidrug efflux pump of F. tularensis probably acts in concert with known antioxidant defenses to resist oxidative stress. The identification of functional component required for antioxidant enzyme secretion allows us to describe for the first time a novel antioxidant defense mechanism in F. tularensis. A better understanding of ROS resistance mechanisms of F. tularensis will contribute to the development of effective anti-tularemia prophylactics and therapeutics.
Experimental procedures
Bacterial strains and media
The bacterial strains, plasmids, and primers used in this study are listed in Table S1. The F. tularensis subsp. holarctica live vaccine strain (F. tularensis LVS) (ATCC 29684; American Type Culture Collection, Rockville, MD) was used in this study. The F. tularensis cultures were grown on Mueller-Hinton (MH) chocolate agar plates (BD Biosciences, San Jose, CA) supplemented with IsoVitaleX at 37°C with 5% CO2; MH-broth (BD Biosciences, San Jose, CA) supplemented with ferric pyrophosphate and IsoVitaleX (BD Biosciences, San Jose, CA); or in Brain-Heart Infusion (BHI)-broth at 37°C with shaking (160 rpm). Active mid-log phase bacteria grown in MH- or BHI broth were harvested and stored at −80°C; 1ml aliquots were thawed periodically for use. Escherichia coli strain DH5α was used for routine cloning. E. coli cultures were grown in Luria-Bertani (LB) broth or on LB agar plates. When necessary, kanamycin (10 μg/ml), or hygromycin (100 μg/ml) was included in broth and agar media for selection purposes.
F. tularensis mutant library and transcomplementation
A stable mini transposon Tn5 insertion library of Francisella tularensis LVS was constructed using the EZ::TN transposon system as described previously (Kawula et al., 2004). These mutants were screened in vitro for sensitivity to oxidative stress in 96-well plates. Briefly, the transposon insertion mutants were grown in MH-broth in the presence of 250μM of hydrogen peroxide (H2O2) in a 96-well plate for 24 hrs. The mutant strains showing retarded/no growth in the presence of H2O2 in comparison to the untreated controls were identified. The transposon insertion sites in the H2O2 sensitive mutants were further identified by DNA sequencing the transposon flanking regions using outwardly designed primers within the transposon. The overall screening protocol is shown in Fig. S1. A mutant identified in the gene locus FTL_0687 was selected for further characterization. For transcomplementation of the FTL_0687 (emrA1) mutant, a complementation construct containing FTL_0687 coding sequence was PCR amplified with primers shown in Table S1. The PCR amplified FTL_0687 gene was digested with BamHI and PstI restriction enzymes and cloned into the E. coli-Francisella shuttle vector pMP822 (LoVullo et al., 2006) (A kind gift from Dr. Martin Pavelka, University of Rochester, NY). The resulting plasmid, pMM001 was verified by DNA sequencing and electroporated into the FTL_0687 mutant. The transcomplementation was confirmed by PCR and qRT-PCR for FTL_0687 transcripts.
Determination of susceptibility of emrA1 mutant to ROS
The wild type F. tularensis LVS, the emrA1 mutant, the transcomplemented strain, acrB (FTL_1672), tolC, sodB, SodBC and ΔkatG mutants were tested for their susceptibility to ROS by generating growth curves in the presence of H2O2 as described previously (Bakshi et al., 2006; Melillo et al., 2009). Wherever indicated recombinant F. tularensis KatG (rkatG) or recombinant bacterioferritin protein (rBfr) (1μg/ml) [kindly provided by Dr Daniel Clemens, University of California, Los Angeles, CA] were added to the culture medium. For growth curves with the recombinant rKatG and rBFr proteins the emrA1 and ΔkatG mutants were grown in the presence of kanamycin (10μg/ml) or ampicillin (100μg/ml), respectively, to avoid contamination due to addition of recombinant proteins. Susceptibility of the FTL_0687 mutant to H2O2 and superoxide generating compounds, paraquat (1mM) and pyrogallol (500μM) was also investigated by bacterial killing assays. The numbers of viable bacteria were determined after 1 and 3 hrs of incubation and plating serial dilutions on MH-chocolate agar plates. Bacterial colonies were counted after 48 hrs and expressed as log10 CFU/ml and compared with those obtained for the wild type F. tularensis LVS or the transcomplemented strain. For disc diffusion assay, bacteria were spread on MH chocolate agar plates. Sterile filter paper discs were impregnated with 50μg of pyrogallol in 5μl of sterile PBS. The plates were incubated for 48 hrs and zones of inhibition around the discs were measured.
Macrophage invasion assay
Previously described gentamicin protection assay (Mahawar et al., 2012) was performed to determine the role of FTL_0687 and FTL_1672 mutant in intramacrophage survival. Briefly, the murine alveolar macrophage cell line, MH-S (ATCC CRL-2019) or the bone marrow derived macrophages (BMDMs) isolated from wild type or gp91Phox−/− C57BL/6 mice were infected with F. tularensis LVS, the FTL_0687 mutant, or the transcomplemented strain at a multiplicity of infection (MOI) of 100. Two hrs after infection, the macrophages were treated with gentamicin (100μg/ml) for 2 hrs to kill the extracellular bacteria. Medium containing gentamicin was then replaced with medium without antibiotics, followed by incubation at 37°C in 5% CO2. The macrophages were collected and lysed at 4 and 24 hrs post-infection with 0.1% sodium deoxycholate. The macrophage cell lysates were serially diluted in sterile PBS and plated on MH chocolate agar plates for bacterial enumeration. Gentamicin protection assays were also performed by treating macrophages with ROS inhibitors, apocynin (250μM) (Arcos Organics, Morris Plains, NJ) or 1mM of N-acetyl cysteine (NAC) (Sigma Aldrich, St. Louis, MO). The results were expressed as log10CFU/ml.
Antibiotic and detergent sensitivity assays
F. tularensis LVS, the emrA1 mutant, or the transcomplemented strain grown on MH chocolate agar plates were scraped and suspended in MH broth to achieve an OD600 of 1.0. The bacterial suspensions were spread with a sterile cotton swab onto MH chocolate agar plates (supplemented with 10 μg/ml kanamycin for the FTL_0687 mutant and 100μg/ml hygromycin for the transcomplemented strain). Sterile antibiotic discs (Becton Dickinson, San Jose, CA) or plain filter paper discs soaked with 56 and 750 μg of SDS, and 5 and 25 μg of ethidium bromide (Qin et al., 2008; Gil et al., 2006) were placed on the plates. The plates were incubated at 37°C in the presence of 5% CO2. After three days of incubation, the zones of growth inhibition around the disks were measured. The experiments were repeated at least three times for each drug and strain.
Mouse studies
All mice experiments were performed according to the protocols approved by IACUC at New York Medical College. Six to eight weeks old wild type C57BL/6 or gp91Phox−/− mice (Jackson Laboratories) were used in the study. Prior to intranasal inoculations, the mice were deeply anesthetized using a ketamine/xylazine cocktail. Mice were inoculated with wild type F. tularensis LVS or the FTL_0687 mutant and observed for morbidity and mortality for a period of 21–30 days. The results were expressed as Kaplan-Meier survival curves and the statistical significance was determined by Log-rank test.
Western blot analysis
F. tularensis strains were grown at 37°C with shaking in MHB or BHI-broth overnight and adjusted to OD600 of 0.2. Aliquots were collected at 4, 8 and 12 hrs of growth and centrifuged at 10,000 rpm for 10 min. The supernatants were collected and filtered through 0.22μ filters to remove any bacteria. The culture filtrates were concentrated to 1/10th their original volume. The bacterial cell pellets were resuspended in 200μl lysis buffer [200mM Tris –HCl (pH 8.0), 320mM (NH4)2SO4, 5mM MgCl2,10mM EDTA, 10mM EGTA, 20% glycerol, 1 mM dithiothreitol (DTT), protease and phosphatase inhibitors]. The protein concentrations of both the culture filtrates and cell lysates were determined using a Pierce BCA Protein Assay kit (Thermoscientific). Five micrograms of protein from each sample was run on a 12% SDS-PAGE gel, transferred to polyvinylidene difluoride membranes (Millipore) and probed with anti-KatG (1:20000) or anti-SodB antibodies (1:20000) (kindly provided by Dr. Karsten Hazlett, Albany Medical College, Albany NY) or anti-F. tularensis hyperimmune serum (1:40) and secondary monoclonal antibodies (anti-rabbit or anti-mouse immunoglobulin, IgG,1:5000) conjugated to horseradish peroxidase (Amersham). The protein bands on the membrane were visualized using Supersignal West Pico chemiluminescent substrate (Thermoscientific) on a Chemidoc XRS system (BioRad). The membranes were stripped and re-probed with antibodies against the 50S-ribosomal subunit (Rp1L) to determine the extent of bacterial lysis.
RNA isolation, RT- PCR and qRT-PCR analysis
The qRT-PCR was performed to determine if emrA1 constitutes a part of an operon and to determine that the insertion of transposon in the FTL_0687 gene does not affect the transcription of upstream and downstream genes. The F. tularensis LVS, the emrA1 mutant, and the transcomplemented strains were grown to an OD600 of 0.7. Bacterial cells were pelleted and total RNA was purified using Purelink RNA Mini Kit (Ambion). The contaminating DNA from RNA preparations was removed using on-Column Purelink DNase treatment. cDNA was synthesized using iScript cDNA Synthesis Kit (Ambion). The primer sequences used for RT-PCR are shown in Table S1. Genomic DNA was isolated from the culture filtrates using a genomic DNA isolation kit (Invitrogen) to ascertain the extent of bacterial lysis. The isolated DNA was run on 0.8% agarose gel and stained with ethidium bromide to visualize the DNA bands.
Determination of catalase activity in culture filtrates
F. tularensis LVS, the emrA1 mutant, and the transcomplemented strain were grown overnight in BHI broth. The OD600 of the cultures was adjusted to 0.2. The cultures were allowed to grow for 4 and 8 hrs in a shaker incubator. The filtrates were collected and filtered through a 0.22μ filter. The catalase activity in the culture filtrates was determined by measuring the consumption of H2O2 over a period of 10 min. using Amplex Red Hydrogen peroxide/Peroxidase assay kit (Invitrogen) according to the manufacturer’s instructions.
Statistical analysis
Results were expressed as mean ± standard error of mean (SEM) or standard deviation (SD). The comparisons between multiple groups were made using One-way Analysis of Variance (ANOVA) with Tukey-Kramer Multiple Comparisons post-test. Differences between the experimental groups were considered statistically significant at a P < 0.05 level. The survival data were analyzed using Logrank test and P-values were determined. Differences in survival between the experimental groups were considered statistically significant at a P < 0.005 level.
Supplementary Material
Acknowledgments
This work was supported in whole or in part, by National Institutes of Health Grants R15AI107698 (M. Malik), 2P01AI056320 (CSB) and startup funds from the Albany College of Pharmacy and Health Sciences (M. Malik) and New York Medical College (CSB). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
The authors have no conflict of interest to declare.
References
- Federal Register. Biennial review, Final rule. 194. Vol. 77. Federal Register; 2012. Possession, use and transfer of select agents and toxins; pp. 10–5. [PubMed] [Google Scholar]
- Allen LA. Mechanisms of pathogenesis: evasion of killing by polymorphonuclear leukocytes. Microbes Infect. 2003;5:1329–1335. doi: 10.1016/j.micinf.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Allen LA, McCaffrey RL. To activate or not to activate: distinct strategies used by Helicobacter pylori and Francisella tularensis to modulate the NADPH oxidase and survive in human neutrophils. Immunol Rev. 2007;219:103–117. doi: 10.1111/j.1600-065X.2007.00544.x. [DOI] [PubMed] [Google Scholar]
- Atkins HS, Dassa E, Walker NJ, Griffin KF, Harland DN, Taylor RR, et al. The identification and evaluation of ATP binding cassette systems in the intracellular bacterium Francisella tularensis. Res Microbiol. 2006;157:593–604. doi: 10.1016/j.resmic.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Bakshi CS, Malik M, Mahawar M, Kirimanjeswara GS, Hazlett KR, Palmer LE, et al. An improved vaccine for prevention of respiratory tularemia caused by Francisella tularensis SchuS4 strain. Vaccine. 2008;26:5276–5288. doi: 10.1016/j.vaccine.2008.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi CS, Malik M, Regan K, Melendez JA, Metzger DW, Pavlov VM, Sellati TJ. Superoxide dismutase B gene (sodB)-deficient mutants of Francisella tularensis demonstrate hypersensitivity to oxidative stress and attenuated virulence. J Bacteriol. 2006;188:6443–6448. doi: 10.1128/JB.00266-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev. 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- Bina XR, Lavine CL, Miller MA, Bina JE. The AcrAB RND efflux system from the live vaccine strain of Francisella tularensis is a multiple drug efflux system that is required for virulence in mice. FEMS Microbiol Lett. 2008;279:226–233. doi: 10.1111/j.1574-6968.2007.01033.x. [DOI] [PubMed] [Google Scholar]
- Bossi P, Bricaire F. Tularemia, a potential bioterrorism weapon. Presse Med. 2003;32:1126–1130. [PubMed] [Google Scholar]
- Braunstein M, Espinosa BJ, Chan J, Belisle JT, Jacobs WR., Jr SecA2 functions in the secretion of superoxide dismutase A and in the virulence of Mycobacterium tuberculosis. Mol Microbiol. 2003;48:453–464. doi: 10.1046/j.1365-2958.2003.03438.x. [DOI] [PubMed] [Google Scholar]
- Bustamante J, Aksu G, Vogt G, de BL, Genel F, Chapgier A, et al. BCG-osis and tuberculosis in a child with chronic granulomatous disease. J Allergy Clin Immunol. 2007;120:32–38. doi: 10.1016/j.jaci.2007.04.034. [DOI] [PubMed] [Google Scholar]
- Chen H, Yi C, Zhang J, Zhang W, Ge Z, Yang CG, He C. Structural insight into the oxidation-sensing mechanism of the antibiotic resistance of regulator MexR. EMBO Rep. 2010;11:685–690. doi: 10.1038/embor.2010.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis DT, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, et al. Tularemia as a biological weapon: medical and public health management. JAMA. 2001;285:2763–2773. doi: 10.1001/jama.285.21.2763. [DOI] [PubMed] [Google Scholar]
- Dieppedale J, Sobral D, Dupuis M, Dubail I, Klimentova J, Stulik J, et al. Identification of a putative chaperone involved in stress resistance and virulence in Francisella tularensis. Infect Immun. 2011;79:1428–1439. doi: 10.1128/IAI.01012-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil H, Platz GJ, Forestal CA, Monfett M, Bakshi CS, Sellati TJ, et al. Deletion of TolC orthologs in Francisella tularensis identifies roles in multidrug resistance and virulence. Proc Natl Acad Sci U S A. 2006;103:12897–12902. doi: 10.1073/pnas.0602582103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland IB, Schmitt L, Young J. Type 1 protein secretion in bacteria, the ABC-transporter dependent pathway (review) Mol Membr Biol. 2005;22:29–39. doi: 10.1080/09687860500042013. [DOI] [PubMed] [Google Scholar]
- Huntley JF, Conley PG, Hagman KE, Norgard MV. Characterization of Francisella tularensis outer membrane proteins. J Bacteriol. 2007;189:561–574. doi: 10.1128/JB.01505-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon B, Wang Y, Hao H, Barton YW, Zhang Q. Contribution of CmeG to antibiotic and oxidative stress resistance in Campylobacter jejuni. J Antimicrob Chemother. 2011;66:79–85. doi: 10.1093/jac/dkq418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawula TH, Hall JD, Fuller JR, Craven RR. Use of transposon-transposase complexes to create stable insertion mutant strains of Francisella tularensis LVS. Appl Environ Microbiol. 2004;70:6901–6904. doi: 10.1128/AEM.70.11.6901-6904.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson P, Oyston PC, Chain P, Chu MC, Duffield M, Fuxelius HH, et al. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat Genet. 2005;37:153–159. doi: 10.1038/ng1499. [DOI] [PubMed] [Google Scholar]
- Lee BY, Horwitz MA, Clemens DL. Identification, recombinant expression, immunolocalization in macrophages, and T-cell responsiveness of the major extracellular proteins of Francisella tularensis. Infect Immun. 2006;74:4002–4013. doi: 10.1128/IAI.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren H, Shen H, Zingmark C, Golovliov I, Conlan W, Sjostedt A. Resistance of Francisella tularensis strains against reactive nitrogen and oxygen species with special reference to the role of KatG. Infect Immun. 2007;75:1303–1309. doi: 10.1128/IAI.01717-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren H, Stenmark S, Chen W, Tarnvik A, Sjostedt A. Distinct roles of reactive nitrogen and oxygen species to control infection with the facultative intracellular bacterium Francisella tularensis. Infect Immun. 2004;72:7172–7182. doi: 10.1128/IAI.72.12.7172-7182.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren H, Stenman L, Tarnvik A, Sjostedt A. The contribution of reactive nitrogen and oxygen species to the killing of Francisella tularensis LVS by murine macrophages. Microbes and Infection. 2005;7:467–475. doi: 10.1016/j.micinf.2004.11.020. [DOI] [PubMed] [Google Scholar]
- Llewellyn AC, Jones CL, Napier BA, Bina JE, Weiss DS. Macrophage replication screen identifies a novel Francisella hydroperoxide resistance protein involved in virulence. PLoS ONE. 2011;6:e24201. doi: 10.1371/journal.pone.0024201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoVullo ED, Sherrill LA, Perez LL, Pavelka MS., Jr Genetic tools for highly pathogenic Francisella tularensis subsp. tularensis. Microbiology. 2006;152:3425–3435. doi: 10.1099/mic.0.29121-0. [DOI] [PubMed] [Google Scholar]
- Mahawar M, Atianand MK, Dotson RJ, Mora V, Rabadi SM, Metzger DW, et al. Identification of a novel Francisella tularensis factor required for intramacrophage survival and subversion of innate immune response. J Biol Chem. 2012;287:25216–25229. doi: 10.1074/jbc.M112.367672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marohn ME, Santiago AE, Shirey KA, Lipsky M, Vogel SN, Barry EM. Members of the Francisella tularensis phagosomal transporter subfamily of major facilitator superfamily transporters are critical for pathogenesis. Infect Immun. 2012;80:2390–2401. doi: 10.1128/IAI.00144-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey RL, Allen LA. Francisella tularensis LVS evades killing by human neutrophils via inhibition of the respiratory burst and phagosome escape. J Leukoc Biol. 2006;80:1224–1230. doi: 10.1189/jlb.0406287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melillo AA, Bakshi CS, Melendez JA. Francisella tularensis antioxidants harness reactive oxygen species to restrict macrophage signaling and cytokine production. J Biol Chem. 2010;285:27553–27560. doi: 10.1074/jbc.M110.144394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melillo AA, Mahawar M, Sellati TJ, Malik M, Metzger DW, Melendez JA, Bakshi CS. Identification of Francisella tularensis live vaccine strain CuZn superoxide dismutase as critical for resistance to extracellularly generated reactive oxygen species. J Bacteriol. 2009;191:6447–6456. doi: 10.1128/JB.00534-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra NP, Balagopal A, Soni S, Schlesinger LS, Gunn JS. AcpA is a Francisella acid phosphatase that affects intramacrophage survival and virulence. Infect Immun. 2007;75:390–396. doi: 10.1128/IAI.01226-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra NP, Soni S, Rajaram MV, Dang PM, Reilly TJ, El-Benna J, et al. Francisella acid phosphatases inactivate the NADPH oxidase in human phagocytes. J Immunol. 2010;184:5141–5150. doi: 10.4049/jimmunol.0903413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddock LJ. Multidrug-resistance efflux pumps - not just for resistance. Nat Rev Microbiol. 2006;4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- Platz GJ, Bublitz DC, Mena P, Benach JL, Furie MB, Thanassi DG. A tolC mutant of Francisella tularensis is hypercytotoxic compared to the wild type and elicits increased proinflammatory responses from host cells. Infect Immun. 2010;78:1022–1031. doi: 10.1128/IAI.00992-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin A, Scott DW, Mann BJ. Francisella tularensis subsp. tularensis Schu S4 disulfide bond formation protein B, but not an RND-type efflux pump, is required for virulence. Infect Immun. 2008;76:3086–3092. doi: 10.1128/IAI.00363-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon-Garcia S, Martin C, Thompson CJ, Ainsa JA. Role of the Mycobacterium tuberculosis P55 efflux pump in intrinsic drug resistance, oxidative stress responses, and growth. Antimicrob Agents Chemother. 2009;53:3675–3682. doi: 10.1128/AAC.00550-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santic M, Molmeret M, Abu KY. Modulation of biogenesis of the Francisella tularensis subsp. novicida-containing phagosome in quiescent human macrophages and its maturation into a phagolysosome upon activation by IFN-gamma. Cell Microbiol. 2005;7:957–967. doi: 10.1111/j.1462-5822.2005.00529.x. [DOI] [PubMed] [Google Scholar]
- Santic M, Molmeret M, Klose KE, Abu KY. Francisella tularensis travels a novel, twisted road within macrophages. Trends Microbiol. 2006;14:37–44. doi: 10.1016/j.tim.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Santiviago CA, Fuentes JA, Bueno SM, Trombert AN, Hildago AA, Socias LT, et al. The Salmonella enterica sv. Typhimurium smvA, yddG and ompD (porin) genes are required for the efficient efflux of methyl viologen. Mol Microbiol. 2002;46:687–698. doi: 10.1046/j.1365-2958.2002.03204.x. [DOI] [PubMed] [Google Scholar]
- Savitt AG, Mena-Taboada P, Monsalve G, Benach JL. Francisella tularensis infection-derived monoclonal antibodies provide detection, protection, and therapy. Clin Vaccine Immunol. 2009;16:414–422. doi: 10.1128/CVI.00362-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Rahman T, Malik M, Hickey AJ, Leifer CA, Hazlett KR, Sellati TJ. Discordant results obtained with Francisella tularensis during in vitro and in vivo immunological studies are attributable to compromised bacterial structural integrity. PLoS ONE. 2013;8:e58513. doi: 10.1371/journal.pone.0058513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Yang J, Zhao D, Kawula TH, Banas JA, Zhang JR. Genome-wide identification of Francisella tularensis virulence determinants. Infect Immun. 2007;75:3089–3101. doi: 10.1128/IAI.01865-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne CJ, Oyston PC. Vaccines against Francisella tularensis. Ann N Y Acad Sci. 2007;1105:325–350. doi: 10.1196/annals.1409.012. [DOI] [PubMed] [Google Scholar]
- Wehrly TD, Chong A, Virtaneva K, Sturdevant DE, Child R, Edwards JA, et al. Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages. Cell Microbiol. 2009;11:1128–50. doi: 10.1111/j.1462-5822.2009.01316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss DS, Brotcke A, Henry T, Margolis JJ, Chan K, Monack DM. In vivo negative selection screen identifies genes required for Francisella virulence. Proc Natl Acad Sci U S A. 2007;104:6037–6042. doi: 10.1073/pnas.0609675104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrella TM, Singh A, Bitsaktsis C, Rahman T, Sahay B, Feustel PJ, et al. Host-adaptation of Francisella tularensis alters the bacterium’s surface-carbohydrates to hinder effectors of innate and adaptive immunity. PLoS ONE. 2011;6:e22335. doi: 10.1371/journal.pone.0022335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvi A, Rotem S, Bar-Haim E, Cohen O, Shafferman A. Whole-genome immunoinformatic analysis of F. tularensis: predicted CTL epitopes clustered in hotspots are prone to elicit a T-cell response. PLoS ONE. 2011;6:e20050. doi: 10.1371/journal.pone.0020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.