Abstract
Background & Aim: The aim of this study was the detection of OqxAB efflux pumps, OmpK35 and OmpK36 porins among extended-spectrum-β-lactamase-producing Klebsiella pneumoniae isolates from Iran.
Materials and Methods: This study was conducted with 83 K. pneumoniae isolates from two hospitals in Tehran, Iran. Antibiotic susceptibility tests were performed by Kirby-Bauer disc diffusion and Broth Microdilution methods according to CLSI guidelines. The OqxAB , blaTEM, blaSHV and blaCTX-M genes were detected by PCR and sequencing methods.The outer membrane porins OmpK35 and OmpK36 were analyzed by SDS-PAGE, PCR and sequencing methods.
Results: Among the 83 K. pneumoniae strains, 48 (57.5%) were ESBL positive. The existence of blaTEM, blaSHV and blaCTX-M was detected in 24 (50%), 30 (62.5%) and 28 (58.33%) ESBL-producing isolates respectively. The prevalence of both oqxA and oqxB detected in K. pneumoniae was high: 50 (60.2%) and 50 (60.2%), respectively. OmpK35 was detected in 30 (62.5%) while OmpK36 was found in 35 (72.91%) out of 48 ESBL-producing isolates. In this study, fosfomycin and tigecycline were more active than other antibiotics.
Conclusions: The prevalence of beta-lactamase-producing K. pneumoniae detected in this study is of great concern and highlights the need of infection control measures including antibacterial management and prompt identification of beta-lactamase-producing isolates.
Keywords: Klebsiella pneumoniae, OqxAB, β-Lactamases, outer membrane porins
Introduction
Multidrug resistance in bacteria is a significant issue in the treatment of infectious diseases1. Extended spectrum beta-lactamases (ESBLs) are a rapidly evolving group of β-lactamase enzymes produced by bacteria. These enzymes have the ability to hydrolyze aztreonam and cephalosporins but are inhibited by beta-lactamase inhibitors such as clavulanic acid. ESBLs are often located on plasmids and many of them derived from mutations in SHV (Sulphydryl variable) and TEM (Temoneira) genes determined by amino acid substitutions around the active site. Apart from SHV and TEM ESBL types, K. pneumoniae isolates may additionally produce CTX-M (Cefotaximase-Munchen) enzymes. CTX-M β-lactamases are more active against ceftriaxone and cefotaxime than against ceftazidime, even though point mutations can increase their activity against ceftazidime as well2. Rapid and adequate ESBL detection is crucial for infection control measures and for the choice of appropriate antibacterial therapy. In 1998, plasmid-mediated quinolone resistance (PMQR) was detected. The qnrA, qnrB, qnrC, qnrD, and qnrS genes have been identified as major groups of qnr. Two additional PMQR determinants, AAC(6′)-Ib-Cr and quinolone extrusion by OqxA or OqxB have been detected3 as well as other non-specific mechanisms including decreased intracellular antibiotic accumulation by up-regulation of efflux pumps and decreased permeability related to porin loss4. OqxAB is one of the first plasmid-borne efflux pumps of the RND family. It is encoded by the OqxA and OqxB genes, located on a 52 kb conjugative plasmid, designated pOLA52, and confers resistance to multiple agents, including fluoroquinolones such as nalidixic acid, norfloxacin and ciprofloxacin, as well as biocides such as chlorhexidine and triclosan5. Furthermore, these pumps confer resistance to ethidium bromide and chloramphenicol6,7. K. pneumoniae produces two major porins, OmpK35 and OmpK36. However, most ESBL-expressing K. pneumoniae clinical isolates produce only the OmpK36 porin7. OmpK35 and OmpK36 provide a channel that allows a wide range of antibiotics to penetrate into the periplasmic space7. The aim of this study was to analyse the presence of OqxAB efflux pumps, OmpK35 and OmpK36 porins among extended-spectrum β-lactamase (ESBL)-producing K. pneumoniae strains.
Materials and Methods
From October 2011 to May 2012, 83 non-duplicate non-consecutive K. pneumoniae that were isolated from males 27 (32.53%), females 14 (16.86%) and infants 42 (50.60%) were collected from hospitalized patients in Mofid Children and Taleghani Hospitals, Tehran, Iran. The susceptibility to 19 antibiotics was measured by disk diffusion and broth microdilution methods and an ESBL production test was performed according to the guidelines provided by the CLSI8. Plasmids were prepared by Plasmid Mini Extraction Kit (Bioneer Company, Korea). The genes Ompk35, Ompk36, blaTEM, blaSHV and blaCTX-M were detected by PCR and Sequencing using described primers9-11. The primers used for OqxA and OqxB were as follows: OqxA-F (5’-GGCAACAGCCAAAACGCAGG-3’) and oqxA-R (5’-GGGGCGGTCACTTTGGTGAA-3’) for OqxA; and OqxB-F (5’-ATGCACTTCCCGATCTCGAC-3’) and OqxB-R (5’– TGGCGATATCTTCCACGCTC-3 ‘) for OqxB.Amplification was carried out with the following thermal cycling conditions: 5 min at 94°C and 36 cycles of amplification consisting of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C, with 5 min at 72°C for the final extension. DNA fragments were analysed by electrophoresis in a 1% agarose gel at 95 V for 45 min in 1X TBE containing ethidium bromide. K. pneumoniae ATCC700603 was used as the control strain. Outer-membrane proteins (OMPs) were analysed by SDS-PAGE using standard methods. Briefly, isolates were grown in Mueller-Hinton broth, sonicated and centrifuged. Cell membranes were obtained following centrifugation at 12,000 g, and extracted with 2 % sodium N-lauroyl sarcosinate. SDS-PAGE was performed with a 11% acrylamide gel, which was then stained with Coomassie blue10.
Results
Antimicrobial drug–resistance patterns of 83 K. pneumoniae isolates are shown in Table 1 and Table 2. The Combination Disk Diffusion Test (CDDT) was applied for the phenotypic detection of ESBLs in 83 K. pneumoniae isolates using ceftazidime and cefotaxime alone and in combination with clavulanic acid. Forty eight (57.5%) isolates were positive for ESBL production. Outer Membrane Porin, OmpK35, was detected in 30 (62.5%) out of 48 ESBL-producing isolates while OmpK36 was found in 35 (72.91%) out of 48 ESBL-producing bacteria. In addition, 29 (60.4%) out of 48 ESBL-producing isolates had OmpK36 and OmpK35, simultaneously. The existence of blaTEM, blaSHV and blaCTX-Mwas detected in 24 (50%), 30 (62.5%) and 28 (58.33%) ESBL-producing isolates, respectively and coexistence of resistance genes was also observed (Table 3). The prevalence of both oqxA and oqxB detected in K. pneumoniae was high: 50 (60.2%) and 50 (60.2%), respectively.
Table 1. Antimicrobial susceptibility testing results of 83 isolates of K. pneumoniae collected from Mofid Children and Taleghani Hospitals, Tehran, Iran.
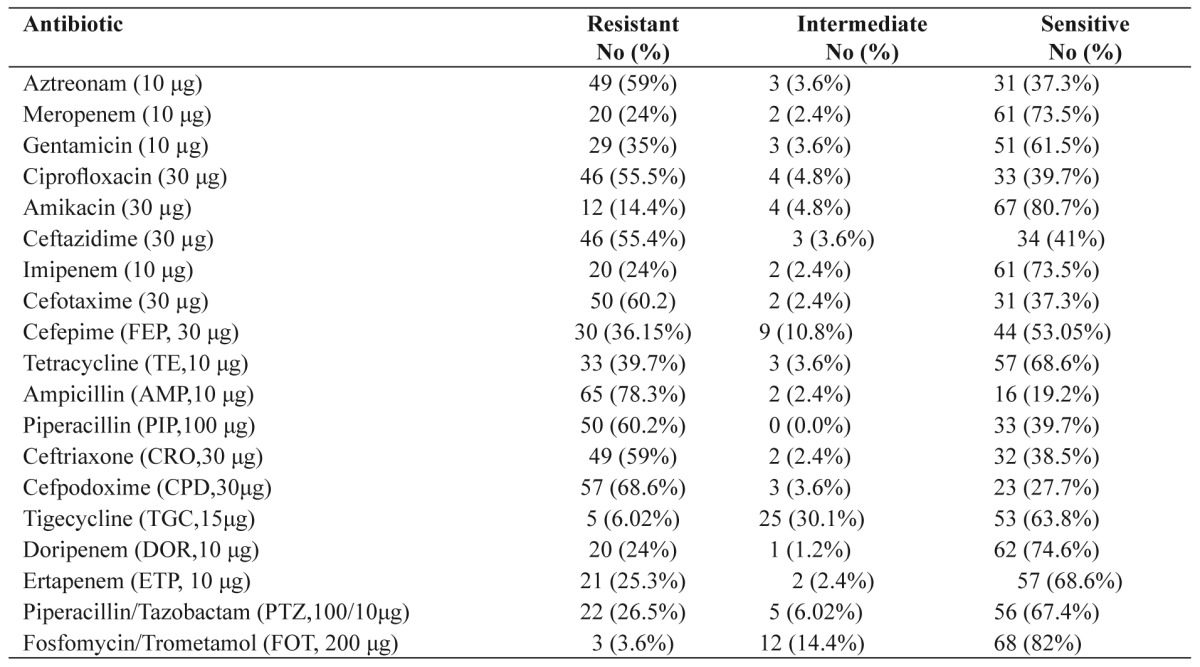
Table 2. Microbiological activities of various antimicrobial agents against 83 K. pneumoniae isolates.
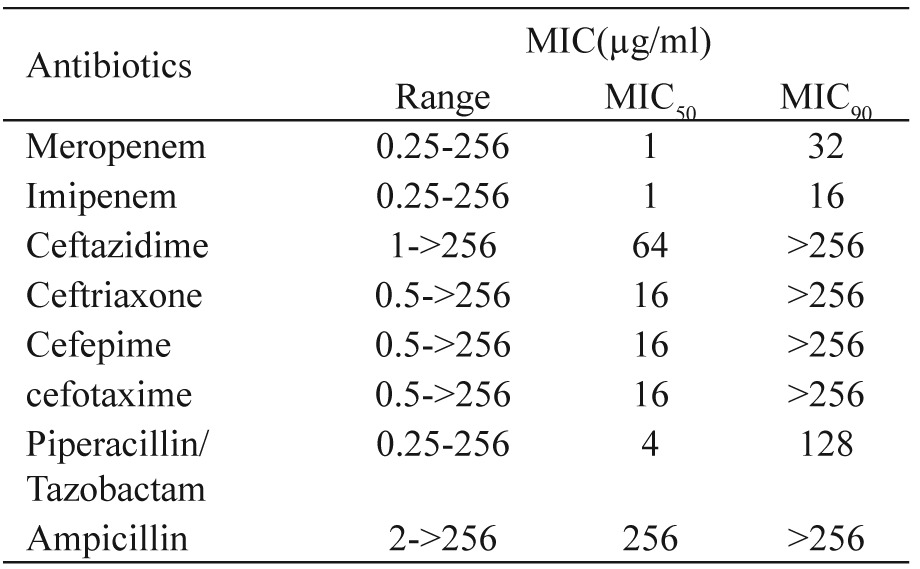
Table 3. Co-exitisting resistance genes in K. pneumoniae collected from Mofid Children and Taleghani Hospitals, Tehran, Iran.
Discussion
K. pneumoniae has become rapidly the most common ESBL producing bacteria, making its eradication difficult from high risk departments (burn units, ICUs and NICUs)12. The lowest rates of resistance in isolates were observed for fosfomycin 3 (3.6%), tigecycline 5 (6.02%), amikacin 12 (14.4%), ertapenem 21 (25.3%), doripenem 20 (24%), meropenem 20 (24%), imipenem 20 (24%) and piperacillin/tazobactam 22 (26.5%). The highest rates of resistance were observed for ampicillin 65 (78.3%), cefpodoxime 57 (68.6%), piperacillin 50 (60.2%), cefotaxime 50 (60.2%), aztreonam 49 (59%), ceftriaxone 49 (59%), ceftazidime 46 (55.4%) and ciprofloxacin 46 (55.5%). So, the best coverage against the study isolates was obtained with fosfomycin and tigecycline. Of the 83 K. pneumoniae, the prevalence of ESBL was 48 (57.5%). Forty-six ESBL-positive isolates were resistant to cefotaxime and ceftazidime, simultaneously. The high rate of ESBL prevalence in Iran and its widespread dissemination is causing concern. In our study, the existence of blaTEM, blaSHV and blaCTX-M was detected in 24 (50%), 30 (62.5%) and 28 (58.33%) ESBL-producing isolates, respectively. This is worrisome especially in Iran where the ESBL prevalence is very high. Efflux pump systems are an extremely important cause of multi-drug resistance13. The genes oqxA and oqxB are common in an operon and they encode OqxAB14. A surprisingly high prevalence, 50 (60.2%) of oqxAB was detected in K. pneumoniae isolates, significantly higher than previously reported for Denmark, Sweden (1.8%), and South Korea (0.4%) and fewer than China (75%)6,14. Plasmid-borne multidrug efflux pumps encoding both resistance to antimicrobials and disinfectants could cause serious problems as not only usage of antimicrobials but also of compounds used in everyday living could select for plasmids encoding resistance to important antimicrobials for human treatment6. ESBL-producing K. pneumoniae carrying plasmid-mediated OqxAB efflux pumps, can be a reservoir for the spread of these genes. Susceptibility to quinolones is reduced when this pump is highly expressed6. The efficacy of the RND-type multi-efflux pumps in Enterobacteriaceae is dependent on the presence of an outer membrane protein (OMP)5. The termination of translation by nonsense mutations, the disruption of the gene by insertion sequences, and the down-regulation of transcription by mutations occurring within the promoter may cause altered porin expression15. In our study, we assessed the presence of two major porins, OmpK35 and OmpK36. OmpK35 was detected in 30 (62.5%) of 48 ESBL-producing isolates while ompK36 was found in 35 (72.91%) out of 48 ESBL-producing bacteria. Ertapenem may be particularly affected by the concomitant loss of OmpK35 and OmpK36. Five ertapenem resistant strains did not express the OmpK35 protein but expressed OmpK36. Nine carbapenem resistant strains did not express either the OmpK35 or the OmpK36 porin. Loss of this porin may be one of the factors contributing to antibacterial resistance in ESBL-producing K. pneumoniae and may favour the selection of additional mechanisms of resistance. In conclusion, the prevalence of the OqxAB efflux pumps is high in ESBL-producing K. pneumoniae in Iran and represents a potential reservoir for its spread.
Conflict of Interest
The authors declare no competing financial interests.
Acknowledgements
The authors thank Dr. Mobayen (Islamic Azad University of Tabriz) for providing K. pneumoniae ATCC700603 strain.
References
- 1.Veleba M, Higgins PG, Gonzalez G, Seifert H, Schneiders T. Characterization of RarA, a novel AraC family multidrug resistance regulator in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2012;56:4450–4458. doi: 10.1128/AAC.00456-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manoharan A, Premalatha K, Chatterjee S, Mathai D. SARI Study Group. Correlation of TEM, SHV and CTX-M extended-spectrum beta lactamases among Enterobacteriaceae with their in vitro antimicrobial susceptibility. Indian J Med Microbiol. 2011;29:161–164. doi: 10.4103/0255-0857.81799. [DOI] [PubMed] [Google Scholar]
- 3.Chen X, Zhang W, Pan W, Yin J, Pan Z, Gao S, et al. Prevalence of qnr, aac(6’)-Ib-cr, qepA, and oqxAB in Escherichia coli isolates from humans, animals, and the environment. Antimicrob Agents Chemother. 2012;56:3423–3427. doi: 10.1128/AAC.06191-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruiz E, Sáenz Y, Zarazaga M, Rocha-Gracia R, Martínez-Martínez L, Arlet G, et al. qnr, aac(6’)-Ib-cr and qepA genes in Escherichia coli and Klebsiella spp.: genetic environments and plasmid and chromosomal location. J Antimicrob Chemother. 2012;67:886–897. doi: 10.1093/jac/dkr548. [DOI] [PubMed] [Google Scholar]
- 5.Hansen LH, Jensen LB, Sørensen HI, Sørensen SJ. Substrate specificity of the OqxAB multidrug resistance pump in Escherichia coli and selected enteric bacteria. J Antimicrob Chemother. 2007;60:145–147. doi: 10.1093/jac/dkm167. [DOI] [PubMed] [Google Scholar]
- 6.Rodríguez-Martínez JM, Díaz de Alba P, Briales A, Machuca J, Lossa M, Fernández-Cuenca F, et al. Contribution of OqxAB efflux pumps to quinolone resistance in extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae. J Antimicrob Chemother. 2013;68:68–73. doi: 10.1093/jac/dks377. [DOI] [PubMed] [Google Scholar]
- 7.Tsai YK, Fung CP, Lin JC, Chen JH, Chang FY, Chen TL, et al. Klebsiella pneumoniae outer membrane porins OmpK35 and OmpK36 play roles in both antimicrobial resistance and virulence. Antimicrob Agents Chemother. 2011;55:1485–1493. doi: 10.1128/AAC.01275-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute.Performance standards for antimicrobial susceptibility testing; Twenty-second informational supplement. Document M100-S22 Wayne, PA, CLSI, 2012; 32(3) availiable at: http://antimicrobianos.com.ar/ATB/wp-content/uploads/2012/11/M100S22E.pdf, accessed on January 2012. [Google Scholar]
- 9.Ma L, Chang FY, Fung CP, ChenTL JH, Lin JC, Lu PL, et al. Variety of TEM-, SHV-, and CTX-M-type beta-lactamases present in recent clinical isolates of Escherichia coli, Klebsiella pneumoniae, and Enterobacter cloacae from Taiwan. Microb Drug Resist. 2005;11:31–39. doi: 10.1089/mdr.2005.11.31. [DOI] [PubMed] [Google Scholar]
- 10.Landman D, Bratu S, Quale J. Contribution of OmpK36 to carbapenem susceptibility in KPC-producing Klebsiella pneumoniae. J Med Microbiol. 2009;58:1303–1308. doi: 10.1099/jmm.0.012575-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu BT, Liao XP, Yang SS, Wang XM, Li LL, Sun J, et al. Detection of mutations in the gyrA and parC genes in Escherichia coli isolates carrying plasmid-mediated quinolone resistance genes from diseased food-producing animals. J Med Microbiol. 2012;61:1591–1599. doi: 10.1099/jmm.0.043307-0. [DOI] [PubMed] [Google Scholar]
- 12.Eftekhar F, Rastegar M, Golalipoor M, Mansoursamaei N. Detection of Extended Spectrum B-Lactamases in Urinary Isolates of Klebsiella pneumoniae in Relation to Bla, Bla and Bla Gene Carriage. Iran J Public Health. 2012;41:127–132. [PMC free article] [PubMed] [Google Scholar]
- 13.Meletis G, Exindari M, Vavatsi N, Sofianou D, Diza E. Mechanisms responsible for the emergence of carbapenem resistance in Pseudomonas aeruginosa. Hippokratia. 2012;16:303–307. [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao J, Chen Z, Chen S, Deng Y, Liu Y, Tian W, et al. Prevalence and dissemination of oqxAB in Escherichia coli isolates from animals, farmworkers, and the environment. Antimicrob Agents Chemother. 2010;54:4219–4224. doi: 10.1128/AAC.00139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García-Fernández A, Miriagou V, Papagiannitsis CC, Giordano A, Venditti M, Mancini C, et al. An ertapenem-resistant extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae clone carries a novel OmpK36 porin variant. Antimicrob Agents Chemother. 2010;54:4178–4184. doi: 10.1128/AAC.01301-09. [DOI] [PMC free article] [PubMed] [Google Scholar]


