Abstract
Neisseria adhesin A (NadA), involved in the adhesion and invasion of Neisseria meningitidis into host tissues, is one of the major components of Bexsero, a novel multicomponent vaccine licensed for protection against meningococcal serogroup B in Europe, Australia, and Canada. NadA has been identified in approximately 30% of clinical isolates and in a much lower proportion of carrier isolates. Three protein variants were originally identified in invasive meningococci and named NadA-1, NadA-2, and NadA-3, whereas most carrier isolates either lacked the gene or harbored a different variant, NadA-4. Further analysis of isolates belonging to the sequence type 213 (ST-213) clonal complex identified NadA-5, which was structurally similar to NadA-4, but more distantly related to NadA-1, -2, and -3. At the time of this writing, more than 89 distinct nadA allele sequences and 43 distinct peptides have been described. Here, we present a revised nomenclature system, taking into account the complete data set, which is compatible with previous classification schemes and is expandable. The main features of this new scheme include (i) the grouping of the previously named NadA-2 and NadA-3 variants into a single NadA-2/3 variant, (ii) the grouping of the previously assigned NadA-4 and NadA-5 variants into a single NadA-4/5 variant, (iii) the introduction of an additional variant (NadA-6), and (iv) the classification of the variants into two main groups, named groups I and II. To facilitate querying of the sequences and submission of new allele sequences, the nucleotide and amino acid sequences are available at http://pubmlst.org/neisseria/NadA/.
INTRODUCTION
Neisseria adhesin A (NadA) is a trimeric autotransporter protein, with a role in adhesion to and invasion of host tissues, which is present in a subset of meningococcal strains (1). Recent studies have shown that NadA is involved in interaction with and stimulation of immune cells during infection (2–4) and in binding to the human chaperone Hsp90 (5, 6). Its predicted molecular structure is very similar to the known virulence-associated factors Yersinia adhesin A (YadA) and UspA2 (7, 8), which suggests that NadA is a member of the OCA (oligomeric coiled-coil adhesin) family (9). NadA forms oligomers which are anchored via a transmembrane domain into the outer membrane and has been shown to be immunogenic in animal models (10). Further, children recovering from invasive meningococcal disease produce specific antibody responses against this antigen (11). NadA expression is regulated at the transcriptional level by the length variation of a tandem repeat region located in the gene promoter (12, 13), with expression regulated by NadR, a repressor molecule (14–16). Expression can be induced by the use of 4-hydroxyphenyl acetic acid, a small molecule secreted in human saliva, which inhibits NadR-dependent repression (14). Recent data indicate protein upregulation in vivo (15). In addition, MtrR, a transcriptional regulator, has a negative regulatory impact on NadA expression, especially when overexpressed (17).
Nucleotide sequence analyses of nadA have shown that the gene is present in approximately 30% of clinical isolates but in a much lower proportion (16%) of isolates obtained from asymptomatic carriage (10, 18–20). The nadA gene is present in almost all isolates belonging to the hyperinvasive sequence type 8 (ST-8), ST-11, ST-32, and ST-213 clonal complexes (cc) tested to date but is rarely present in ST-41/44 and ST-269 cc isolates. Three variants, named NadA-1, NadA-2, and NadA-3, were originally identified in pathogenic strains (10). nadA was initially grouped into three alleles based on the different sizes of the PCR patterns obtained with primers external to the nadA gene insertion site. Once discovered, the NadA-1, -2, and -3 proteins were inappropriately referred to as alleles because of the low diversity of the encoding genes, sharing 84 to 99% identity. In particular, NadA-2 and NadA-3 were very similar (the corresponding genes shared ≥97% identity). Each NadA variant was associated with meningococcal clonal complexes (21). In particular, NadA-1 was associated with 100% of ST-32 cc isolates tested. NadA-2 and NadA-3 were found in ST-8 and ST-11 ccs and in a few other meningococcal lineages (some ST-18 cc strains and some unassigned clonal complexes). NadA-3 was also present in ST-174 cc strains, mostly serogroup Y (22). NadA-1, -2, and -3 were shown to form high-molecular-weight oligomers on the meningococcal surface, to mediate adhesion and invasion into epithelial cells in vitro, to induce high titers of cross-reactive bactericidal antibodies, and to induce protection in the infant rat model (10).
The NadA-4 variant was originally described in meningococci isolated from healthy people (19). NadA-4 consists of 323 amino acids and is shorter than the NadA-1, NadA-2, and NadA-3 variants, which are composed of 362, 398, and 405 amino acids, respectively. Although shorter in length, the overall secondary structure and domain organizations are conserved. NadA-4 is exposed on the bacterial surface, where it forms high-molecular-weight oligomers and binds to epithelial cells. Antibodies against the recombinant NadA-4 protein are bactericidal against homologous strains, whereas the activity against strains carrying the three other NadA variants is weak (19), suggesting that sequence variability is responsible for the absence of cross-reactivity with NadA-1, -2, and -3.
Further analysis of a number of strains belonging to a new emerging clonal complex, ST-213 cc, revealed the presence of an additional variant proposed as NadA-5, closely related to NadA-4 based on sequence similarities. The peculiarity of this additional variant is that, in the majority of isolates, the gene is apparently switched off at an internal poly(C) tract, unique to nadA-5 (18, 21, 23).
In the context of the licensure of Bexsero, identification of NadA variants remains important as NadA protein elicits bactericidal antibodies in humans and is one of the major components of this vaccine (24, 25). In order to evaluate/update the existing nomenclature scheme and to establish a publicly accessible nomenclature database, we examined all available nadA nucleotide sequences from a reference set of 363 bacteria isolated worldwide in the latter half of the twentieth century from samples obtained from both patients (the majority of isolates) and carriers (about 12% of isolates).
MATERIALS AND METHODS
Isolate culture, PCR, and nucleotide sequencing of nadA.
Two sets of data were used for nadA gene sequencing. The first set of data concerned sequences determined directly by PCR amplification and analysis of chromosomal DNA obtained from meningococcal isolates already described in previous publications (10, 18, 19, 21, 23, 25, 26). Meningococcal culture, chromosomal DNA preparation, PCR amplification, and nadA gene sequencing were performed as described. The second set of data derives from sequences received from different external laboratories worldwide that, when possible, were independently verified using the corresponding electropherograms.
Sequence analyses.
Sequence analyses were assembled and analyzed using Sequencher version 4.10.1 sequence analysis software (Gene Codes Corporation, Ann Arbor, MI, USA), BioEdit (27), Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0 (28), and Jalview (29). Phylogenetic trees were constructed using MEGA and SplitsTree version 4.11.3 (30).
Nucleotide sequence accession numbers.
New allele sequences were submitted to the PubMLST database (http://pubmlst.org/neisseria/NadA/) under accession numbers NEIS1969_1 to NEIS1969_127 (see Text S1 and S2 in the supplemental material).
RESULTS AND DISCUSSION
At the time of writing, 89 unique gene sequences and 43 different amino acid sequences had been identified. Two groups of sequence variant types, named groups I and II, were evident from the alignment of all unique nucleotide sequences and from the phylogenetic tree (Fig. 1).
FIG 1.
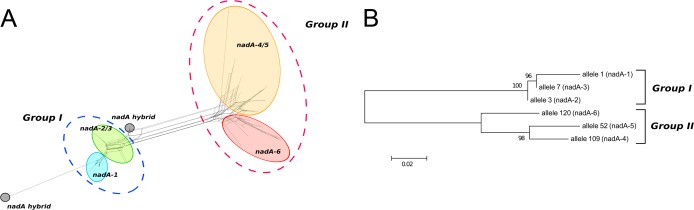
(A) SplitsTree representation showing the nadA gene variation. The genes encoding the different protein variants are circled and shown in different colors. The two main groups are indicated by traced circles. Two hybrid forms are also reported. (B) Neighbor-joining tree showing the distribution of different alleles corresponding to the six NadA protein variants.
Group I, nadA-1, -2, and -3.
The majority of nucleotide sequences (56%) clustered in group I and shared 84 to 99% homology. The encoded proteins NadA-1, NadA-2, and NadA-3 showed high similarity in terms of both size and sequence (>90% homology). In previous studies, protein variants corresponding to NadA-1, -2, and -3 were shown to induce antibodies in mice with cross-bactericidal activity. Bactericidal activity on strains carrying different variants was not influenced by the sequence diversity but rather by the antigen expression level (10, 25). Several genes encoding NadA-1, -2, and -3 with particular features were found, as described below. More than 40 isolates, all isolated in Australia and belonging to the ST-32 cc, showed a single-base deletion (C at position 984 with respect to nadA allele 1) without a downstream insertion, generating a putative atypical NadA-1, with a longer anchor domain (65 amino acids) and an altered distal portion (starting from position 329 of the amino acid sequence), including a different stop codon (TGA instead of TAA). Moreover, genes with frameshift point mutations introducing premature stop codons, probably encoding truncated NadA-1, -2, and -3 proteins, were identified. In addition, genes encoding putative NadA-1, -2, and -3 proteins with in-frame/not-in-frame deletions of the coiled-coil domain were also found. In several cases the gene was interrupted by the insertion of the genetic element IS1301 in the coding sequence of NadA-2 and NadA-3. IS1301 insertion always occurred at the consensus recognition sequence 5′-ACTAG-3′. This recognition sequence was actually present twice in the nadA gene, in positions 474 and 583 of the nadA-3 coordinates. In all but one sequence, IS1301 insertion occurred at position 474 of the nucleotide sequence. The insertion was found in both orientations. A unique protein variant, intermediate between NadA-2 and -3, was found in strains obtained from cases related to the epidemic occurring among pilgrims to the Hajj, Saudi Arabia, in 2000 to 2001 (31–33). The analyzed isolates (W135:2a:P1.5,2:cc11) were recovered from several countries to which the epidemic strain spread via returning pilgrims and were epidemiologically related to the original Hajj strain, though showing, in some cases, different profiles by pulsed-field gel electrophoresis (M. K. Taha, personal communications).
Group II, nadA-4, -5, and -6.
The rest of the nucleotide sequences were assigned to group II and shared 75 to 99% homology. The encoded proteins NadA-4, NadA-5, and NadA-6 showed high similarity in terms of both size and sequence (more than 75% homology), whereas they shared only 46 to 50% identity with NadA-1, -2, and -3 (Fig. 2). NadA-4, originally described in carrier isolates, was also found in invasive isolates that, in terms of their allelic profiles, were relatively distant from the hyperinvasive clonal complexes already established in the public multilocus sequence typing (MLST) database, available at http://pubmlst.org/neisseria (34). We cannot exclude the possibility that these were meningococci of low pathogenic potential that had caused invasive disease.
FIG 2.
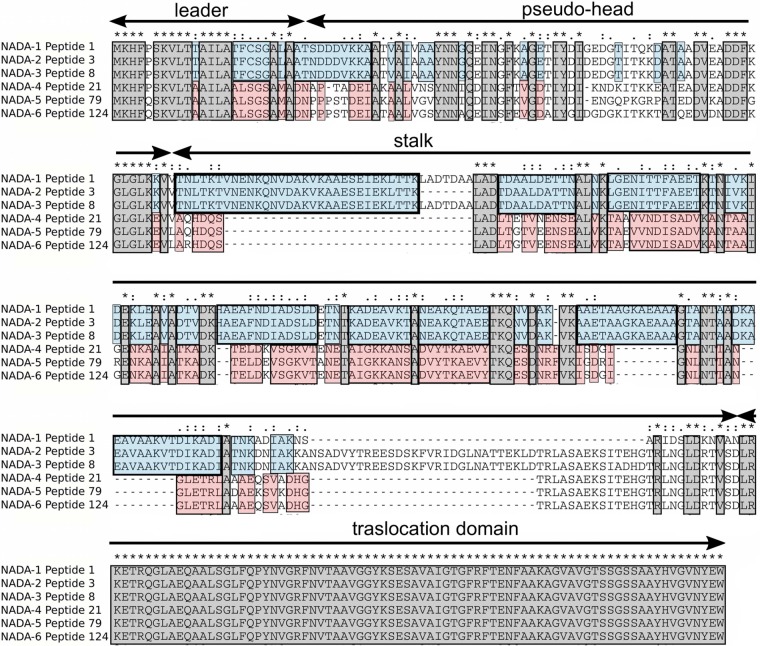
Sequence alignment of the six protein variants (the corresponding alleles are indicated in Fig. 1B). The alignment highlights the high level of identity between NadA-1, NadA-2, and NadA-3, and NadA-4, NadA-5, and NadA-6. The blue boxes indicate the amino acid residues specific for group I, and the red boxes indicate the amino acid residues specific for group II. The amino acid residues common to all NadA variants are shown in gray. The leader, the extracellular passenger domain consisting of a pseudohead plus a stalk region with high coiled-coil propensity, and the C-terminal membrane-anchoring region or translocation domain are indicated by arrows.
In all gene sequences encoding NadA-5, an internal poly(C) tract in the coding region was found in addition to the variable number of tetranucleotide tandem repeats present in the promoter region of the gene. The overall number of C residues differed among isolates, resulting in frameshift mutations in the coding sequence that may lead to a truncated protein. However, expression of the whole NadA-5 protein was demonstrated in all of the isolates, including isolates with an apparent frameshift due to the existence in the same cell culture of colonies with a different number of Cs (unpublished data). This suggested that these cultures consist of a heterogeneous population, containing both nadA-5-on and nadA-5-off genes. The amount of the protein expressed was a balance between the nadA-5-on- and nadA-5-off-harboring cells. It has been shown that graduated reductions in the surface expression of outer membrane proteins mediated by phase variation enable meningococci to escape killing in vitro by bactericidal antibodies (35). Three nadA sequences were found in frame at the internal poly(C) tract, suggesting the presence of isolates in which the majority of colonies contained nadA-5-on genes. In these isolates, the amount of protein expressed was higher with respect to isolates with the majority of nadA-5-off genes (unpublished data).
NadA-6 was a new variant, which was characterized by sequence homology with NadA-4 and -5, originally described in one isolate belonging to the ST-11 cc (23). This sequence has subsequently been found in a large group of isolates of the B:2a:P1.5 phenotype, mostly isolated in Spain from 2002 to 2008, and belonging to the ST-11 cc. These may have arisen by horizontal transfer of the capsular operon (36). In the majority of isolates analyzed, the nadA gene was inactivated by frameshift mutations. The effect of this frameshift mutation was the absence of NadA in the strain whole-protein sample, as previously assessed by Western blot analysis (23). However, in several cases the gene was in frame and probably encoded the whole NadA-6 protein. In addition, two sequences with an internal poly(G) tract in the coding region in addition to the variable number of tetranucleotide tandem repeats present at the promoter level encoded a protein variant homologous to NadA-6. The overall number of G residues differed in the two strains. This suggested possible phase variability and perhaps the presence of a heterogeneous population such as already supposed in the case of NadA-5.
Sequences corresponding to chimeric NadA-4, -5, and -6 forms were also found, such as hybrid forms between the two main groups. Moreover, a longer in-frame nadA sequence was described in one isolate of Neisseria cinerea, a species that usually lacks the nadA gene (37). The amino acid sequence showed higher homology at the C-terminal portion with NadA-4, -5, and -6 variants.
Despite sequence diversity, the same overall trimeric structure, characteristic of the OCA family, was predicted for all NadA variants, an extracellular passenger domain consisting of a pseudohead plus a stalk region with high coiled-coil propensity, and a C-terminal membrane-anchoring region or translocation domain (Fig. 2). In previous studies it has been demonstrated that NadA-4 was present on the bacterial surface as heat-stable high-molecular-weight oligomers (19). Recombinant NadA-4 was used in an epithelial cell binding assay to verify that this protein has the same binding activity as group I variants. Moreover, it was demonstrated that antibodies against a recombinant NadA-4 protein are bactericidal against homologous strains (19). Even if the functional features of NadA variants belonging to group I were maintained, NadA-4 was poorly immunologically cross-reactive with NadA-1, -2, and -3 (19). Concerning NadA-5 and NadA-6, more data are needed to confirm protein expression and functional/immunological activity. However, their relatively high levels of sequence identity with NadA-4 suggest that functional and immunological properties should be conserved.
Revised nomenclature scheme.
On the basis of all of the sequence data available, we propose a new nomenclature scheme largely compatible with the previous scheme. Due to the very high level of homology and the presence of peptides with an intermediate sequence between those of NadA-2 and -3, such as in Hajj-related isolates, we group sequences in one unique variant, NadA-2/3. The same applies for NadA-4 and NadA-5, which we propose to group in the variant NadA-4/5. A comparison between previous classification (21, 38) and the new nomenclature scheme is provided (see Table S1 in the supplemental material).
We further propose the introduction of the NadA-6 variant, even though at present a limited number of encoding genes with in-frame sequences have been described in the Neisseria meningitidis isolates so far sampled. In summary, the protein variants proposed are NadA-1, NadA-2/3, NadA-4/5, and NadA-6 (Fig. 1). The nomenclature scheme would be the prefix “NadA” with a dash and then the variant number, followed by a dot and the peptide subvariant; e.g., NadA-1.1 refers to variant 1, peptide 1. Finally, we propose the classification of all nucleotide and amino acid sequences into group I and group II.
Sequences obtained as part of this study (see Materials and Methods; see also Text S1 and S2 in the supplemental material) are available at the web-accessible PubMLST database (http://pubmlst.org/neisseria/NadA/) to facilitate querying of sequences and submission of new allele sequences. Unique nucleotide sequences have been assigned an arbitrary numerical identifier (id) in order of submission.
For consistency with every other locus within the PubMLST database, classification includes the complete coding region, with both the leader peptide and the stop codon. This is also done for the corresponding peptide for which a sequence is assigned only if the reading frame is maintained. In case of insertion sequences or frameshift mutations leading to truncated or apparently truncated proteins, the corresponding peptide sequence identifier is not assigned. The variant is, however, assigned, and specific comments/flags are reported (e.g., in the case of nadA allele id 16, which is interrupted by the IS1301, the peptide id is not assigned, whereas the variant assigned is NadA-2/3).
In the database, the fields available for each sequence are locus (nadA or Neis1969 or NMC1969), allele id, sequence (including the nucleotide and amino acid sequences), peptide id (assigned only if the coding region is in frame), length, and status (forward and reverse sequence trace files or electropherograms available and checked). Detailed features such as the presence of IS1301 and the presence of frame-shift mutations, premature stop codons, in frame/not in frame deletions, etc., are also reported as comments and/or flags. This also allows distinguishing and classification of sequences for which the coding region is not in frame. Additional available allele information is date entered, date stamp, sender (the submitter of the data), and database curator. Moreover, Table S1 in the supplemental material, which reconciles new peptide ids with those already published (21, 38), is also available in the PubMLST database.
In order for the submitter to identify nadA alleles and peptides, both single- and batch-sequence queries can be performed by pasting in either DNA or peptide sequences in the FASTA format. Query sequences will be checked first for an exact match against the chosen (or all) loci. The nearest partial matches will be identified if an exact match is not found. In order to search by sequence attributes, specific search criteria can be selected or, alternatively, to return all records, the respective field may be left blank prior to submission. As an example, a screenshot of the site is provided (see Fig. S1 in the supplemental material).
Newly submitted sequences will be available through the website. This is a curated database and sequence trace files (electropherograms) are appreciated where appropriate in order to enhance confidence in the assignment of new variants. Submissions based on high-quality high-coverage genome sequences are also now acceptable. The submitter of the data is also invited to provide as much information as possible about the isolate the sequence comes from, if available, and also, where appropriate, GenBank or PubMed accession numbers (not mandatory).
The web-accessible database enables alternative NadA nomenclature to be cross-referenced and harmonized, mapping the allele and peptide sequence identifiers to each other and to any alternative names.
The knowledge of the extent and structuring of vaccine component diversity is essential, not only for vaccine formulation (to ensure maximum vaccine coverage), but also for pre-/postlicensure monitoring. Although the use of functional assays is important, nucleotide and peptide sequence diversity is a relevant guide for these processes. Moreover, a standardized nomenclature and the availability of a public web-accessible database are essential for enabling comparisons among different studies.
Supplementary Material
Footnotes
Published ahead of print 7 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/CVI.00825-13.
REFERENCES
- 1.Capecchi B, Adu-Bobie J, Di Marcello F, Ciucchi L, Masignani V, Taddei A, Rappuoli R, Pizza M, Arico B. 2005. Neisseria meningitidis NadA is a new invasin which promotes bacterial adhesion to and penetration into human epithelial cells. Mol. Microbiol. 55:687–698. 10.1111/j.1365-2958.2004.04423.x [DOI] [PubMed] [Google Scholar]
- 2.Franzoso S, Mazzon C, Sztukowska M, Cecchini P, Kasic T, Capecchi B, Tavano R, Papini E. 2008. Human monocytes/macrophages are a target of Neisseria meningitidis adhesin A (NadA). J. Leukoc. Biol. 83:1100–1110. 10.1189/jlb.1207810 [DOI] [PubMed] [Google Scholar]
- 3.Tavano R, Franzoso S, Cecchini P, Cartocci E, Oriente F, Arico B, Papini E. 2009. The membrane expression of Neisseria meningitidis adhesin A (NadA) increases the proimmune effects of MenB OMVs on human macrophages, compared with NadA-OMVs, without further stimulating their proinflammatory activity on circulating monocytes. J. Leukoc. Biol. 86:143–153. 10.1189/jlb.0109030 [DOI] [PubMed] [Google Scholar]
- 4.Mazzon C, Baldani-Guerra B, Cecchini P, Kasic T, Viola A, de Bernard M, Arico B, Gerosa F, Papini E. 2007. IFN-gamma and R-848 dependent activation of human monocyte-derived dendritic cells by Neisseria meningitidis adhesin A. J. Immunol. 179:3904–3916. 10.4049/jimmunol.179.6.3904 [DOI] [PubMed] [Google Scholar]
- 5.Cecchini P, Tavano R, Polverino de Laureto P, Franzoso S, Mazzon C, Montanari P, Papini E. 2011. The soluble recombinant Neisseria meningitidis adhesin NadA(Δ351-405) stimulates human monocytes by binding to extracellular Hsp90. PLoS One 6:e25089. 10.1371/journal.pone.0025089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montanari P, Bozza G, Capecchi B, Caproni E, Barrile R, Norais N, Capitani M, Sallese M, Cecchini P, Ciucchi L, Gao Z, Rappuoli R, Pizza M, Arico B, Merola M. 2012. Human heat shock protein (Hsp) 90 interferes with Neisseria meningitidis adhesin A (NadA)-mediated adhesion and invasion. Cell. Microbiol. 14:368–385. 10.1111/j.1462-5822.2011.01722.x [DOI] [PubMed] [Google Scholar]
- 7.Cornelis GR, Boland A, Boyd AP, Geuijen C, Iriarte M, Neyt C, Sory MP, Stainier I. 1998. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62:1315–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen D, Barniak V, VanDerMeid KR, McMichael JC. 1999. The levels and bactericidal capacity of antibodies directed against the UspA1 and UspA2 outer membrane proteins of Moraxella (Branhamella) catarrhalis in adults and children. Infect. Immun. 67:1310–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roggenkamp A, Ackermann N, Jacobi CA, Truelzsch K, Hoffmann H, Heesemann J. 2003. Molecular analysis of transport and oligomerization of the Yersinia enterocolitica adhesin YadA. J. Bacteriol. 185:3735–3744. 10.1128/JB.185.13.3735-3744.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Comanducci M, Bambini S, Brunelli B, Adu-Bobie J, Arico B, Capecchi B, Giuliani MM, Masignani V, Santini L, Savino S, Granoff DM, Caugant DA, Pizza M, Rappuoli R, Mora M. 2002. NadA, a novel vaccine candidate of Neisseria meningitidis. J. Exp. Med. 195:1445–1454. 10.1084/jem.20020407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Litt DJ, Savino S, Beddek A, Comanducci M, Sandiford C, Stevens J, Levin M, Ison C, Pizza M, Rappuoli R, Kroll JS. 2004. Putative vaccine antigens from Neisseria meningitidis recognized by serum antibodies of young children convalescing after meningococcal disease. J. Infect. Dis. 190:1488–1497. 10.1086/424464 [DOI] [PubMed] [Google Scholar]
- 12.Martin P, van de Ven T, Mouchel N, Jeffries AC, Hood DW, Moxon ER. 2003. Experimentally revised repertoire of putative contingency loci in Neisseria meningitidis strain MC58: evidence for a novel mechanism of phase variation. Mol. Microbiol. 50:245–257. 10.1046/j.1365-2958.2003.03678.x [DOI] [PubMed] [Google Scholar]
- 13.Martin P, Makepeace K, Hill SA, Hood DW, Moxon ER. 2005. Microsatellite instability regulates transcription factor binding and gene expression. Proc. Natl. Acad. Sci. U. S. A. 102:3800–3804. 10.1073/pnas.0406805102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Metruccio MM, Pigozzi E, Roncarati D, Berlanda Scorza F, Norais N, Hill SA, Scarlato V, Delany I. 2009. A novel phase variation mechanism in the meningococcus driven by a ligand-responsive repressor and differential spacing of distal promoter elements. PLoS Pathog. 5:e1000710. 10.1371/journal.ppat.1000710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fagnocchi L, Biolchi A, Ferlicca F, Boccadifuoco G, Brunelli B, Brier S, Norais N, Chiarot E, Bensi G, Kroll JS, Pizza M, Donnelly J, Giuliani MM, Delany I. 2013. Transcriptional regulation of the nadA gene in Neisseria meningitidis impacts the prediction of coverage of a multicomponent meningococcal serogroup B vaccine. Infect. Immun. 81:560–569. 10.1128/IAI.01085-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fagnocchi L, Pigozzi E, Scarlato V, Delany I. 2012. In the NadR regulon, adhesins and diverse meningococcal functions are regulated in response to signals in human saliva. J. Bacteriol. 194:460–474. 10.1128/JB.06161-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cloward JM, Shafer WM. 2013. MtrR control of a transcriptional regulatory pathway in Neisseria meningitidis that influences expression of a gene (nadA) encoding a vaccine candidate. PLoS One 8: e56097. 10.1371/journal.pone.0056097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lucidarme J, Comanducci M, Findlow J, Gray SJ, Kaczmarski EB, Guiver M, Kugelberg E, Vallely PJ, Oster P, Pizza M, Bambini S, Muzzi A, Tang CM, Borrow R. 2009. Characterization of fHbp, nhba (gna2132), nadA, porA, sequence type (ST), and genomic presence of IS1301 in group B meningococcal ST269 clonal complex isolates from England and Wales. J. Clin. Microbiol. 47:3577–3585. 10.1128/JCM.00936-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Comanducci M, Bambini S, Caugant DA, Mora M, Brunelli B, Capecchi B, Ciucchi L, Rappuoli R, Pizza M. 2004. NadA diversity and carriage in Neisseria meningitidis. Infect. Immun. 72:4217–4223. 10.1128/IAI.72.7.4217-4223.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobsson S, Hedberg ST, Molling P, Unemo M, Comanducci M, Rappuoli R, Olcen P. 2009. Prevalence and sequence variations of the genes encoding the five antigens included in the novel 5CVMB vaccine covering group B meningococcal disease. Vaccine 27:1579–1584. 10.1016/j.vaccine.2008.12.052 [DOI] [PubMed] [Google Scholar]
- 21.Bambini S, Muzzi A, Olcen P, Rappuoli R, Pizza M, Comanducci M. 2009. Distribution and genetic variability of three vaccine components in a panel of strains representative of the diversity of serogroup B meningococcus. Vaccine 27:2794–2803. 10.1016/j.vaccine.2009.02.098 [DOI] [PubMed] [Google Scholar]
- 22.Ladhani SN, Lucidarme J, Newbold LS, Gray SJ, Carr AD, Findlow J, Ramsay ME, Kaczmarski EB, Borrow R. 2012. Invasive meningococcal capsular group Y disease, England and Wales, 2007–2009. Emerg. Infect. Dis. 18:63–70. 10.3201/eid1801.110901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lucidarme J, Comanducci M, Findlow J, Gray SJ, Kaczmarski EB, Guiver M, Vallely PJ, Oster P, Pizza M, Bambini S, Muzzi A, Borrow R. 2010. Characterization of fHbp, nhba (gna2132), nadA, porA, and sequence type in group B meningococcal case isolates collected in England and Wales during January 2008 and potential coverage of an investigational group B meningococcal vaccine. Clin. Vaccine Immunol. 17:919–929. 10.1128/CVI.00027-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pizza M, Scarlato V, Masignani V, Giuliani MM, Arico B, Comanducci M, Jennings GT, Baldi L, Bartolini E, Capecchi B, Galeotti CL, Luzzi E, Manetti R, Marchetti E, Mora M, Nuti S, Ratti G, Santini L, Savino S, Scarselli M, Storni E, Zuo P, Broeker M, Hundt E, Knapp B, Blair E, Mason T, Tettelin H, Hood DW, Jeffries AC, Saunders NJ, Granoff DM, Venter JC, Moxon ER, Grandi G, Rappuoli R. 2000. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 287:1816–1820. 10.1126/science.287.5459.1816 [DOI] [PubMed] [Google Scholar]
- 25.Giuliani MM, Adu-Bobie J, Comanducci M, Arico B, Savino S, Santini L, Brunelli B, Bambini S, Biolchi A, Capecchi B, Cartocci E, Ciucchi L, Di Marcello F, Ferlicca F, Galli B, Luzzi E, Masignani V, Serruto D, Veggi D, Contorni M, Morandi M, Bartalesi A, Cinotti V, Mannucci D, Titta F, Ovidi E, Welsch JA, Granoff D, Rappuoli R, Pizza M. 2006. A universal vaccine for serogroup B meningococcus. Proc. Natl. Acad. Sci. U. S. A. 103:10834–10839. 10.1073/pnas.0603940103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bambini S, Piet J, Muzzi A, Keijzers W, Comandi S, De Tora L, Pizza M, Rappuoli R, van de Beek D, van der Ende A, Comanducci M. 2013. An analysis of the sequence variability of meningococcal fHbp, NadA and NHBA over a 50-year period in the Netherlands. PLoS One 8:e65043. 10.1371/journal.pone.0065043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 28.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular biology and evolution. 24:1596–1599. 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- 29.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254–267. 10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- 31.Taha MK, Achtman M, Alonso JM, Greenwood B, Ramsay M, Fox A, Gray S, Kaczmarski E. 2000. Serogroup W135 meningococcal disease in Hajj pilgrims. Lancet 356:2159. 10.1016/S0140-6736(00)03502-9 [DOI] [PubMed] [Google Scholar]
- 32.Mayer LW, Reeves MW, Al-Hamdan N, Sacchi CT, Taha MK, Ajello GW, Schmink SE, Noble CA, Tondella ML, Whitney AM, Al-Mazrou Y, Al-Jefri M, Mishkhis A, Sabban S, Caugant DA, Lingappa J, Rosenstein NE, Popovic T. 2002. Outbreak of W135 meningococcal disease in 2000: not emergence of a new W135 strain but clonal expansion within the electophoretic type-37 complex. J. Infect. Dis. 185:1596–1605. 10.1086/340414 [DOI] [PubMed] [Google Scholar]
- 33.Taha MK, Giorgini D, Ducos-Galand M, Alonso JM. 2004. Continuing diversification of Neisseria meningitidis W135 as a primary cause of meningococcal disease after emergence of the serogroup in 2000. J. Clin. Microbiol. 42:4158–4163. 10.1128/JCM.42.9.4158-4163.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. 10.1186/1471-2105-11-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tauseef I, Ali YM, Bayliss CD. 2013. Phase variation of PorA, a major outer membrane protein, mediates escape of bactericidal antibodies by Neisseria meningitidis. Infect. Immun. 81:1374–1380. 10.1128/IAI.01358-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castilla J, Vazquez JA, Salcedo C, Garcia Cenoz M, Garcia Irure JJ, Torroba L, Beristain X, Abad R, Barricarte A. 2009. B:2a:p1.5 meningococcal strains likely arisen from capsular switching event still spreading in Spain. J. Clin. Microbiol. 47:463–465. 10.1128/JCM.01495-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muzzi A, Mora M, Pizza M, Rappuoli R, Donati C. 2013. Conservation of meningococcal antigens in the genus Neisseria. mBio 4:e00163–13. 10.1128/mBio.00163-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Findlow J, Borrow R, Snape MD, Dawson T, Holland A, John TM, Evans A, Telford KL, Ypma E, Toneatto D, Oster P, Miller E, Pollard AJ. 2010. Multicenter, open-label, randomized phase II controlled trial of an investigational recombinant meningococcal serogroup B vaccine with and without outer membrane vesicles, administered in infancy. Clin. Infect. Dis. 51:1127–1137. 10.1086/656741 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.