Abstract
Bacteria must control the progression of their cell cycle in response to nutrient availability. This regulation can be mediated by guanosine tetra- or pentaphosphate [(p)ppGpp], which are synthesized by enzymes of the RelA/SpoT homologue (Rsh) family, particularly under starvation conditions. Here, we study the effects of (p)ppGpp on the cell cycle of Caulobacter crescentus, an oligotrophic bacterium with a dimorphic life cycle. C. crescentus divides asymmetrically, producing a motile swarmer cell that cannot replicate its chromosome and a sessile stalked cell that is replication competent. The swarmer cell rapidly differentiates into a stalked cell in appropriate conditions. An artificial increase in the levels of (p)ppGpp in nonstarved C. crescentus cells was achieved by expressing a truncated relA gene from Escherichia coli, encoding a constitutively active (p)ppGpp synthetase. By combining single-cell microscopy, flow cytometry approaches, and swarming assays, we show that an increase in the intracellular concentration of (p)ppGpp is sufficient to slow down the swarmer-to-stalked cell differentiation process and to delay the initiation of chromosome replication. We also present evidence that the intracellular levels of two master regulators of the cell cycle of C. crescentus, DnaA and CtrA, are modulated in response to (p)ppGpp accumulation, even in the absence of actual starvation. CtrA proteolysis and DnaA synthesis seem indirectly inhibited by (p)ppGpp accumulation. By extending the life span of the motile nonreproductive swarmer cell and thus promoting dispersal and foraging functions over multiplication under starvation conditions, (p)ppGpp may play a central role in the ecological adaptation of C. crescentus to nutritional stresses.
INTRODUCTION
Bacteria promote their survival in changing environments by continuously adjusting their growth and physiology in response to variations in nutrient availability (1). In particular, they need mechanisms to precisely control the progression of their cell cycle, ensuring that DNA replication, cell growth, and cell division remain coordinated.
The signaling molecules guanosine tetra- and pentaphosphate [(p)ppGpp] are instrumental in the so-called stringent response to limiting nutrients (2, 3). In most bacteria, (p)ppGpp accumulates as a consequence of a shortage in macronutrients and induces a massive switch in transcription by directly binding to and affecting the kinetic properties of the RNA polymerase (2–5). In addition, these small molecules regulate the concentration, stability, or activity of regulatory RNAs and key regulatory proteins, including at least two sigma factors (2, 6). The functional effects of the stringent response concur in reallocating the cellular resources from growth-oriented toward survival-oriented activities: the synthesis of DNA, stable RNAs, ribosomal proteins, and membrane components are usually inhibited, whereas factors essential for adaptation to nutrient limitation are activated. Enzymes of the Rsh family are the key regulators of the stringent response: most bacterial genomes encode at least one long bifunctional Rsh protein able to synthesize and hydrolyze (p)ppGpp (7). Enzymes found in copiotrophic bacteria, except SpoT in Escherichia coli, activate (p)ppGpp synthesis in response to ribosome stalling due to amino acid shortage (3). In the oligotrophic bacterium Caulobacter crescentus, the Rsh protein SpoT only synthesizes (p)ppGpp if it senses stalled ribosomes, together with carbon or nitrogen shortage, which is probably an adaptation to life in very diluted environments (8, 9).
The dimorphism of C. crescentus makes it an interesting model to study the impact of (p)ppGpp on the progression of the bacterial cell cycle. C. crescentus divides asymmetrically, giving a swarmer cell and a stalked cell (10). The swarmer cell is chemotactically competent and motile but is unable to replicate its chromosome (G1 phase) or to divide. In nutrient-replete conditions, the swarmer cell differentiates into a stalked cell after a short period of time. During this swarmer-to-stalked cell transition, the flagellum of the cell is ejected, pili are retracted, and a stalk grows at the pole of the cell previously occupied by the flagellum. The sessile stalked cell immediately initiates the replication of its chromosome (S phase) and starts preparing for cell division. The C. crescentus predivisional cell is asymmetrical, with multiple proteins preferentially localized at one of the two cell poles playing a central role in the regulation of the dimorphic cell cycle of C. crescentus (10, 11). The asymmetry in chromosome replication capacities is established before cell division, through the spatial regulation of the CtrA response regulator (12). CtrA binds to multiple sites on the chromosomal origin to inhibit the initiation of DNA replication by the conserved DnaA protein (13–15). A complex regulatory network controls the levels of active phosphorylated CtrA so that it only accumulates in the flagellated compartment of predivisional cells and in swarmer cells (10, 12). The proteolysis or the inactivation of CtrA during the swarmer-to-stalked cell transition, as well as the presence of active DnaA molecules, are needed for the G1-to-S phase transition (12–15).
The progression of the cell cycle of C. crescentus, and more specifically the duration of the period before the swarmer cell loses motility and initiates chromosome replication, is affected by nutrient availability. When C. crescentus swarmer cells are starved for carbon or nitrogen, the swarmer-to-stalked cell transition is delayed or blocked for a subset of the cells in the population (16–18). In addition, the G1-to-S phase transition is blocked in a large majority of the cells (16–19). In chemostat cultures exposed to nitrogen limitation, the swarmer-to-stalked cell transition is also significantly delayed (20). Whether these modulations of the cell cycle are dependent on (p)ppGpp or not was partially addressed using a (p)ppGpp-null mutant strain. C crescentus possesses a single dispensable Rsh enzyme, named SpoT (9, 16). Interestingly, C. crescentus swarmer cells lacking SpoT and exposed to carbon starvation initiate the replication of their chromosome, suggesting that the G1-to-S blockage upon starvation requires (p)ppGpp (16, 18). Further investigations demonstrated that DnaA was rapidly degraded in a SpoT-dependent manner, during these starvation experiments (16, 19). In addition, a recent study demonstrated that the basal levels of (p)ppGpp naturally present in nonstarved wild-type cells slightly slows down the swarmer-to-stalked cell transition (18).
Several questions about the links between (p)ppGpp and the regulation of the cell cycle of C. crescentus still remained unanswered. Indeed, the carbon and nitrogen starvation experiments previously used to trigger a stringent response in C. crescentus could not decouple direct effects of the (p)ppGpp regulatory network from indirect consequences of the lack of essential macronutrients. For example, wild-type and ΔspoT mutant cells exposed to nitrogen and carbon starvation were both impaired in DNA replication elongation (16, 19), which was expected since deoxyribonucleotide triphosphate pools must be severely depleted under these conditions. It is, however, possible that processes downstream of the initiation step of the DNA replication process are also specifically inhibited by (p)ppGpp, as is the case in Bacillus subtilis, for example (21). Moreover, mutant C. crescentus swarmer cells lacking SpoT and exposed to glucose starvation were often able to differentiate into stalked cells, but at a very low speed (18), possibly also because the metabolism of starved cells is profoundly slowed down. A method to test the specific regulatory effects that (p)ppGpp accumulation has on the progression of the cell cycle of C. crescentus in the absence of actual starvation would be instrumental in decoupling the stringent response from the dramatic effects that nutrient limitations have on cell growth.
Here, we report the construction of a C. crescentus strain that can be manipulated to accumulate high intracellular levels of (p)ppGpp in the absence of actual starvation. Using synchronized populations of cells, we observed that the accumulation of (p)ppGpp is sufficient to delay or slow down the swarmer-to-stalked cell transition and the initiation of chromosome replication. We also show that (p)ppGpp affects the intracellular levels of the DnaA and CtrA cell cycle regulators, even in nonstarved cells. Thus, our findings demonstrate that the (p)ppGpp alarmone is a very important component in the regulatory network that modulates the cell cycle of C. crescentus when faced with limiting nutrients.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
C. crescentus strains were grown in peptone-yeast extract (PYE) complex medium or M2G minimal medium at 28°C (22). Where indicated, 0.2% glucose (PYEG) or 0.2% glucose, together with 0.3% xylose (PYEGX), was added to the PYE medium. For (p)ppGpp detection, cells were grown in low-phosphate M5 medium (containing 0.2% glucose as a carbon source) supplemented with 1 mM glutamate (M5GG) (14). E. coli strains were grown in Luria broth medium at 37°C. The plasmids and strains used are listed in Table 1. The antibiotics used for C. crescentus liquid cultures included rifampin (15 μg/ml), gentamicin (1 μg/ml), kanamycin (5 μg/ml), and penicillin G (200 μg/ml). The antibiotics used for C. crescentus grown on PYE–1.5% agar plates included gentamicin (5 μg/ml) and kanamycin (25 μg/ml). The antibiotics used for E. coli liquid cultures/plates included gentamicin (15/20 μg/ml) and kanamycin (30/50 μg/ml). Plasmids were introduced into C. crescentus by electroporation. Bacteriophage ϕCR30 was used for generalized transduction into C. crescentus (22).
TABLE 1.
Strains and plasmids used in this study
| Plasmid or strain | Relevant characteristics, construction, or genotype | Source or reference |
|---|---|---|
| Plasmids | ||
| pXTCYC-4 | Nonreplicating plasmid for expression of genes under the control of the xylX promoter after integration at the xylX locus in the C. crescentus genome | 61 |
| pXTCYC-4-relA′-FLAG | relA′-FLAG under the control of the xylX promoter in pXTCYC-4 | This study |
| pXTCYC-4-relA′(E335Q)-FLAG | relA′(E335Q)-FLAG under the control of the xylX promoter in pXTCYC-4 | This study |
| pDnaN-RFP | dnaN-mcherry under the control of the native dnaN promoter once integrated into the chromosome | 47 |
| Strains | ||
| E. coli | ||
| TOP10 | Cloning strain | Invitrogen |
| MG1655 | Wild-type strain | 62 |
| C. crescentus | ||
| NA1000 | Synchronizable derivative of wild-type strain CB15 (CB15N) | 63 |
| JC388 | NA1000 dnaN::dnaN-RFP | 47 |
| TFLRa | NA1000 pleC::pleC-YFP divJ::divJ-RFP cpaE::cpaE-CFP | 42 |
| JC835 | NA1000 pXTCYC-4 integrated at the xylX chromosomal locus | This study |
| JC820 | NA1000 pXTCYC-4-relA′-FLAG integrated at the xylX chromosomal locus | This study |
| JC1198 | NA1000 pXTCYC-4-relA′(E335Q)-FLAG integrated at the xylX chromosomal locus | This study |
| JC845 | TFLR pXTCYC-4-relA′-FLAG integrated at the xylX chromosomal locus | This study |
| JC861 | JC388 pXTCYC-4-relA′-FLAG integrated at the xylX chromosomal locus | This study |
TLFR, triply fluorescently labeled reporter.
Construction of plasmids and strains.
For the construction of the pXTCYC-4-relA′-FLAG and pXTCYC-4-relA′(E335Q)-FLAG plasmids, oligonucleotides 5′-GGAATTCCATATGGTTGCGGTAAGAAGTGCAC-3′ and 5′-CGACGCGTTTATTTATCATCATCATCTTTATAATCCATGCCCATCTGCAGCTGGTAGG-3′ were used to amplify the relA′ gene from the E. coli MG1655 chromosome by PCR. The second oligonucleotide adds a FLAG tag to the C terminus of the RelA′ and RelA′(E335Q) proteins. Two more oligonucleotides were used to create the mutated relA′(E335Q) coding sequence from a two-step PCR amplification procedure: 5′-GGAAAAACCGTTCAGATCCAAATC-3′ and 5′-GATTTGGATCTGAACGGTTTTTCC-3′. The relA′ and relA′(E335Q) PCR products were digested with NdeI and MluI and cloned into NdeI-MluI-digested pXTCYC-4, giving pXTCYC-4-relA′-FLAG or pXTCYC-4-relA′(E335Q)-FLAG.
For the construction of strains JC835, JC820, and JC1198, plasmids pXTCYC-4, pXTCYC-4-relA′-FLAG, and pXTCYC-4-relA′(E335Q)-FLAG, respectively, were integrated at the native xylX promoter (23) of strain NA1000 by a single integration event. It is noteworthy that the control strain JC835 grew more slowly (∼115-min generation time instead of ∼90 min) than a wild-type strain (NA1000) in PYEG, probably due to the required addition of gentamicin into the medium, to prevent the re-excision of the pXTCYC-4 vector by homologous recombination.
To construct strains JC845 and JC861, the integrated pXTCYC-4-relA′-FLAG plasmid from JC820 was transduced into the TFLR or JC388 strains, respectively.
Immunoblot analysis.
DnaA, CtrA, and FLAG-tagged RelA′ were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (24). Each sample on the same blot was normalized to the same optical density at 660 nm (OD660) to load similar amounts of cell extracts. The polyacrylamide concentrations were 10% for DnaA and 12% for CtrA and RelA′-FLAG. Proteins were electrotransferred to a polyvinylidene difluoride membrane (Millipore). Immunodetection was performed with polyclonal anti-DnaA and anti-CtrA antibodies, monoclonal anti-FLAG antibodies (Sigma-Aldrich), and horseradish peroxidase-conjugated anti-rabbit IgG antibodies (Sigma-Aldrich). All sera were diluted 1:10,000. A chemiluminescent reagent (Perkin-Elmer, Wellesley, MA) and Kodak (Rochester, NY) Bio-Max MR films were used for detection. Images were processed with Photoshop (Adobe, Mountain View, CA), and the relative band intensities were determined using ImageJ software version 1.43.
To compare the stability of DnaA and CtrA in strains JC835 and JC820, rifampin (10 μg/ml) was added to cultures to inhibit transcription and ultimately protein synthesis. Aliquots of cells were sampled at several time points after rifampin addition and immunoblotting was used to monitor DnaA and CtrA levels in these samples, as described above.
Detection of (p)ppGpp.
(p)ppGpp levels were estimated using a protocol adapted from a study by Lesley and Shapiro (16). Briefly, JC820 and JC835 strains were grown in M5GG medium to an OD660 of 0.25. Both cultures were then split into two, and 0.3% of xylose was added into one of the aliquots to induce the xylX promoter. After 1 h of induction, 3 MBq of H332PO4/ml with an activity of 222 TBq/mmol (Hartmann Analytic) was added to the four cultures, and the cells were incubated for 90 min at 28°C with agitation and fixed by the addition of 10% of 10 M formic acid. As a reference for cells experiencing a stringent response, a culture of NA1000 grown in M5GG to an OD660 of 0.25 was also split into two, and one of the aliquots was washed three times with M5 without glucose and glutamate. Both NA1000 cultures were labeled for 90 min and fixed as described before. All cell extracts were kept on ice for 30 min and then centrifuged for 5 min, and 6 × 2 μl of the supernatant [12 μl total] was spotted onto a polyethyleneimine (PEI) plate (Sigma-Aldrich). The PEI plate had been soaked in sterile distilled water overnight and dried at room temperature before spotting. The plate was developed in 1.5 M KH2PO4 (pH 3.4) in a saturated thin-layer chromatography (TLC) chamber for approximately 2 h and dried at room temperature. Nucleotides were detected via a phosphorimaging system (Tritium screen). pppGpp, ppGpp, and GTP were identified on the basis of their retardation factor (Rf) and by comparison with published autoradiographs (16, 25). Spots were quantified using ImageJ software version 1.43.
Culture synchronization.
Swarmer cells were isolated from mixed cultures through a modified version of the Percoll density centrifugation protocol (26). JC820 cells were grown to exponential phase in PYEG and then split and grown in PYEG or PYEGX for 4 h. Cells from each mixed culture (20 ml, OD660 ∼ 0.3) were pelleted, washed twice in 2 ml of ice-cold M2 salts (22), and resuspended in 0.9 ml of ice-cold M2 salts and 0.9 ml of ice-cold Percoll. The Percoll mixture was centrifuged at 11,000 rpm at 4°C for 20 min. Swarmer cells were isolated, washed three times with ice-cold M2 salts, and finally resuspended in PYEG or PYEGX medium. The swarmer cells were able to progress synchronously through the cell cycle.
Flow cytometry analysis.
Flow cytometry analysis was performed as previously described (27). A total of 20,000 cells from each biological sample was stained with Vybrant DyeCycle Orange (DNA stain [Invitrogen]) and analyzed. The data were collected using the FL-2 fluorescence to identify events and quantify DNA content. The data were analyzed and visualized with R (using the “prada” package developed by Florian Hahne, Wolfgang Huber, Markus Ruschhaupt, and Joern Toedling; Prada: data analysis for cell-based functional assays [R package version 1.24.0]). To count cells harboring 1N chromosomes in a mixed population not treated with rifampin, we used the following approach. First, we calculated the position of the 1N peak maximum as the median of the 20 highest FL-2 channels in the neighborhood of the 1N position. The number of cells with 1N chromosome was then approximated as twice the number of cells whose DNA content was smaller than the 1N peak maximum. When cultures were treated with rifampin before their analysis by flow cytometry, we considered that all cells whose DNA content was less than the average (midpoint) between the 1N peak and 2N peaks could be counted as cells with 1N chromosome(s). The addition of rifampin for 3 h in the culture medium of synchronized cells before the flow cytometry analysis blocked cell division and the initiation of chromosome replication but allowed ongoing rounds of replication to finish (28). The median forward scattering (FSC) of a population was used as an estimate of the cell size. To determine the FSC of cells containing 1N and 2N chromosomes, we calculated the median FSC of cells in the 20 channels centered around the 1N peak maximum, determined as described above, and in the 20 channels centered around the 2N peak maximum, whose position was estimated as the double of the fluorescence value of the 1N peak maximum, respectively.
Fluorescence microscopy.
Cells were immobilized on slides using a thin layer of medium plus 1% agarose. Phase-contrast (PH), Normarski differential interference contrast (DIC), and fluorescence microscopy images were taken with an AxioImager M1 microscope (Zeiss) with a cascade 1K EMCCD camera (Photometrics) controlled through Metamorph 7.5 (Universal Imaging, Downingtown, PA). Protein localization patterns were classified using overlay images between PH/DIC images and fluorescence images using Metamorph 7.5.
Swarming assay.
Strains were cultivated overnight at 28°C in PYEG. A total of 0.5 μl of each culture was stabbed into 0.3% PYEG agar plates supplemented with different concentrations of xylose. Plates were incubated at room temperature for 4 days in a humid container.
RESULTS AND DISCUSSION
Development of a heterologous system to induce (p)ppGpp accumulation in nonstarved C. crescentus cells.
We wanted to determine whether the accumulation of high levels of (p)ppGpp is sufficient to block or slow down the cell cycle of C. crescentus in nutrient-replete conditions. Although often used in copiotrophic bacteria, the methods based on amino acid analogues do not activate (p)ppGpp synthesis in C. crescentus (8, 9). Moreover, the native C. crescentus SpoT protein, containing both a (p)ppGpp synthase domain and a hydrolase domain, would be difficult to manipulate genetically into a constitutive (p)ppGpp synthetase because of the complex regulation of its activity (9, 16). Instead, we chose to develop a heterologous system, expressing in C. crescentus a truncated and constitutively active RelA protein from E. coli.
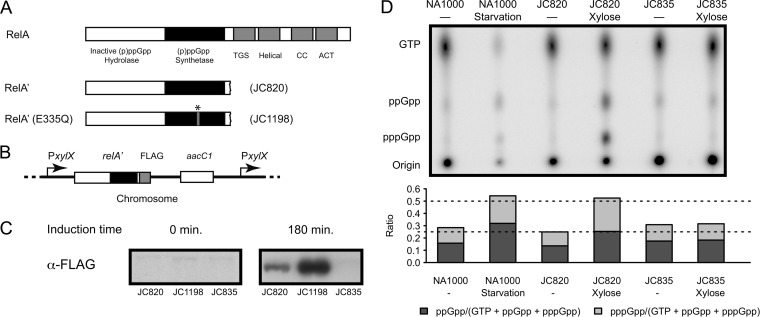
E. coli uses two different pathways to synthesize (p)ppGpp: the RelA-dependent and the SpoT-dependent pathways (3, 29). RelA is associated with the ribosome and it synthesizes (p)ppGpp from ATP and GTP (pppGpp) or GDP (ppGpp), in response to amino acid starvation when sensing uncharged tRNA in the ribosome (30). This enzyme contains a (p)ppGpp synthetase domain and a regulatory domain that inhibits the activity of the enzyme in replete conditions (Fig. 1A) (3, 7, 31). The expression of a truncated relA gene, which encodes a truncated RelA protein that lacks its C-terminal regulatory domain (thereafter called RelA′), induces an artificial accumulation of (p)ppGpp in E. coli and Legionella pneumophila cells, even in replete conditions (32–35). To test whether this system could also induce the accumulation of (p)ppGpp in C. crescentus cells, we engineered a strain (JC820) with an integrated relA′-FLAG construct under the control of the xylose-inducible xylX promoter at the native xylX chromosomal locus (Fig. 1B). This strain produced a FLAG-tagged RelA′ protein that was easily detectable by immunoblot experiments after 3 h (Fig. 1C) and at least 8 h (see Fig. S1 in the supplemental material) after the addition of xylose into the culture medium. To determine whether higher levels of (p)ppGpp accumulated in these cells than in wild-type cells, we used one-dimensional TLC. The (p)ppGpp/[GTP+(p)ppGpp] ratio was used as a relative measure of the intracellular concentration of (p)ppGpp (Fig. 1D). We observed that the (p)ppGpp/[GTP+(p)ppGpp] ratio of JC820 cells grown in xylose-containing medium for 2.5 h, was ∼2-fold higher than that of control JC835 cells (containing an integrated empty plasmid) grown in the same medium. Moreover, the (p)ppGpp/[GTP+(p)ppGpp] ratio of JC820 was similar to that of wild-type cells grown in carbon starvation conditions for 1.5 h. We concluded that the heterologous system that we developed was efficient to induce an artificial stringent response in nonstarved cells that was comparable to the natural stringent response that occurs during starvation in C. crescentus cells. We nevertheless noticed that the starved wild-type cells contained a slightly higher proportion of ppGpp than pppGpp, compared to the JC820 cells grown in xylose-containing medium. This difference in the pppGpp/ppGpp ratio may result from a difference in affinity for GTP and GDP between the C. crescentus SpoT and the RelA′ protein or from a difference in intracellular GTP concentration due to nutrient exhaustion in starvation conditions; it could also be influenced by the expression levels and activity of two putative pppGpp phosphohydrolases present in the annotated proteome of C. crescentus (3, 18, 36–38). Both pppGpp and ppGpp are able to elicit a stringent response in E. coli and B. subtilis, although their relative efficiencies differ: ppGpp seems to be more potent than pppGpp in E. coli (3), the reverse being true in B. subtilis for at least one aspect of the stringent response (21). The RelA′ system was subsequently used to evaluate the effect that (p)ppGpp has on the progression of the cell cycle of C. crescentus cells cultivated in rich medium.
FIG 1.
C. crescentus cells expressing RelA′-FLAG accumulate high levels of (p)ppGpp. (A) Schematics showing the organization of the different domains of the E. coli RelA protein and of the truncated RelA′ and RelA′(E335Q) proteins used in the present study. The TGS/CC/ACT domains form the regulatory region of the protein (3, 7). The star indicates the E335Q mutation. (B) Genetic constructs created to express RelA′-FLAG and RelA′(E335Q)-FLAG in C. crescentus. Plasmid pXTCYC-4-relA′-FLAG or pXTCYC-4-relA′(E335Q)-FLAG was integrated at the native xylX promoter in the C. crescentus chromosome, giving strains JC820 and JC1198, respectively. (C) RelA′-FLAG and RelA′(Q335A)-FLAG proteins are expressed in the JC820 and JC1198 strains, respectively, upon xylose addition in rich medium. JC820, JC1198 and JC835 (control strain) strains were cultivated in exponential phase in PYEG medium, and 0.3% xylose was added (PYEGX) at time zero. Cell extracts collected at time zero and 180 min after xylose addition were used to perform immunoblotting experiments using anti-FLAG antibodies. (D) Intracellular levels of (p)ppGpp in RelA′-FLAG-expressing cells, compared to starved wild-type cells. Strains NA1000 (wild-type strain), JC835 and JC820 were cultivated for 2.5 h in M5GG medium with or without 0.3% xylose. When indicated, NA1000 cells were starved in M5 medium for 90 min. The TLC autoradiograph image shown in the upper part of the figure was used to calculate the pppGpp/(GTP+ppGpp+pppGpp) and ppGpp/(GTP+ppGpp+pppGpp) ratios shown in the lower panel.
(p)ppGpp slows down the growth of C. crescentus.
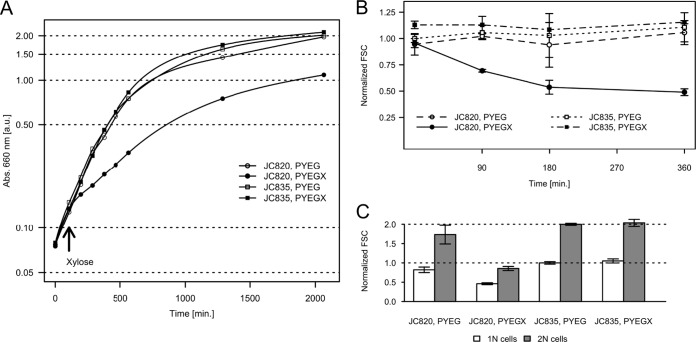
We observed the effect of the accumulation of (p)ppGpp on the growth rate of C. crescentus in exponential phase. The doubling time of strain JC820 grown in rich medium was ∼2-fold higher when it expressed RelA′-FLAG (xylose-containing medium) than when it did not (Fig. 2A). The doubling time of this RelA′-FLAG-expressing strain was also ∼2-fold higher (∼275 min) than that of the control strain JC835 (∼115 min) grown in the same medium (Fig. 2A). Similar results were obtained when these two strains were cultivated in minimal medium containing glucose and xylose (data not shown). To test whether cells remained viable when they accumulated excess (p)ppGpp, we compared the viability of JC820 and JC835 cells cultivated in PYEG supplemented with 0.3% xylose for 3 or 6 h using a Live/Dead staining procedure and fluorescence microscopy (39): both strains had similar viabilities with a very few occurrences of dead cells after 3 h (data not shown) or 6 h (see Fig. S2 in the supplemental material). We however observed that, when plated on solid PYEG medium, a JC820 culture formed ∼105 fewer visible CFU than a JC835 control culture after 3 h of growth in liquid PYEGX (Fig.S3), suggesting that (p)ppGpp-accumulating cells might have difficulties restarting growth on plates. Altogether, these observations suggest that the artificial induction of (p)ppGpp synthesis slows down C. crescentus growth and affects growth recovery on solid medium but that it does not affect cell viability in liquid medium. To confirm that (p)ppGpp accumulation, rather than the expression of the RelA′-FLAG protein, was responsible for the observed growth defect, we also engineered (Fig. 1A&B) and expressed (Fig. 1C) a relA′(E335Q)-FLAG construct encoding a mutant RelA′(E335Q)-FLAG protein in strain JC1198. The E335Q mutation in the (p)ppGpp synthetase domain of RelA is equivalent to the E319Q mutation in SpoT, which eliminates (p)ppGpp synthesis by SpoT(E319Q) in E. coli (40). We observed that the doubling time (see Fig. S4 in the supplemental material), the capacity to form colonies on solid medium (see Fig. S3 in the supplemental material), and the viability (see Fig. S2 in the supplemental material) of C. crescentus cells was not affected by the expression of the inactive RelA′(E335Q)-FLAG protein. This control confirms that the growth defect associated with RelA′-FLAG expression (Fig. 2A) is due to (p)ppGpp synthesis. We concluded that the artificial induction of (p)ppGpp synthesis in C. crescentus cells that are not starved slows down cell growth, as previously also observed for E. coli and L. pneumophila cells (32–35).
FIG 2.
The accumulation of (p)ppGpp slows down the growth of C. crescentus cells. (A) Growth of a population of cells upon accumulation of (p)ppGpp. Strains JC820 (expressing relA′-FLAG from the xylX promoter) and JC835 (control strain) were cultivated in exponential phase in PYEG. 0.3% xylose was added (PYEGX) at time 105 min in half of each culture. The absorbance at 660 nm was used as an estimate of cellular mass. (B) The size of cells accumulating (p)ppGpp decreases sharply. Strains JC820 and JC835 were cultivated in exponential phase in PYEG. Xylose at 0.3% was added (PXEGX) at time zero in half of each culture. The forward scattering (FSC) of 20,000 individual cells for each population was measured using a flow cytometer. The FSC is proportional to the size of the cell. Plotted values are the average median FSC value for each strain and condition over three biological replicates normalized by the average median FSC value of JC835 in PYEG at the time zero; error bars refer to the standard deviations. (C) The decrease in size equally affects cells containing one (1N) or two (2N) complete chromosomes. The median FSC value of cells clustering in the neighborhood of the 1N and 2N peak maximum was calculated for three biological replicates after 6 h in PYEG or PYEGX. The plotted values are averages over triplicates for each strain and condition normalized by the average of JC835, PYEG 1N cells; error bars indicate the standard deviations.
In E. coli, a decrease in cell size was found to be associated with a reduced growth during the stringent response (34, 35, 41). We noticed that C. crescentus cells accumulating (p)ppGpp often appeared smaller under the microscope (see Fig. S2 in the supplemental material) and therefore chose to estimate more carefully the effect that (p)ppGpp has on the size of C. crescentus by measuring the forward scattering of individual cells by flow cytometry. Populations of JC820, JC835, and JC1198 cells grown in PYEGX were analyzed at several time points after xylose addition. We observed that the median size of the cells decreased significantly as (p)ppGpp accumulated: 3 h after the addition of the inducer into the medium, the median size of the JC820 cells was ∼2-fold smaller than that of the control JC835 (Fig. 2B) or JC1198 (see Fig. S4B in the supplemental material) cells grown in the same conditions. This decrease in cell size seemed homogeneous across the population: in rich medium containing xylose, the size of both JC820 cells with 1N chromosome (G1-phase swarmer cells) and JC820 cells with 2N chromosomes (late predivisional cells) decreased in the same proportion relative to the control JC835 population (Fig. 2C). This observation suggests that C. crescentus cells accumulating high (p)ppGpp levels tend to divide at a lower cell mass than wild-type cells.
(p)ppGpp delays or slows down the swarmer-to-stalked cell differentiation.
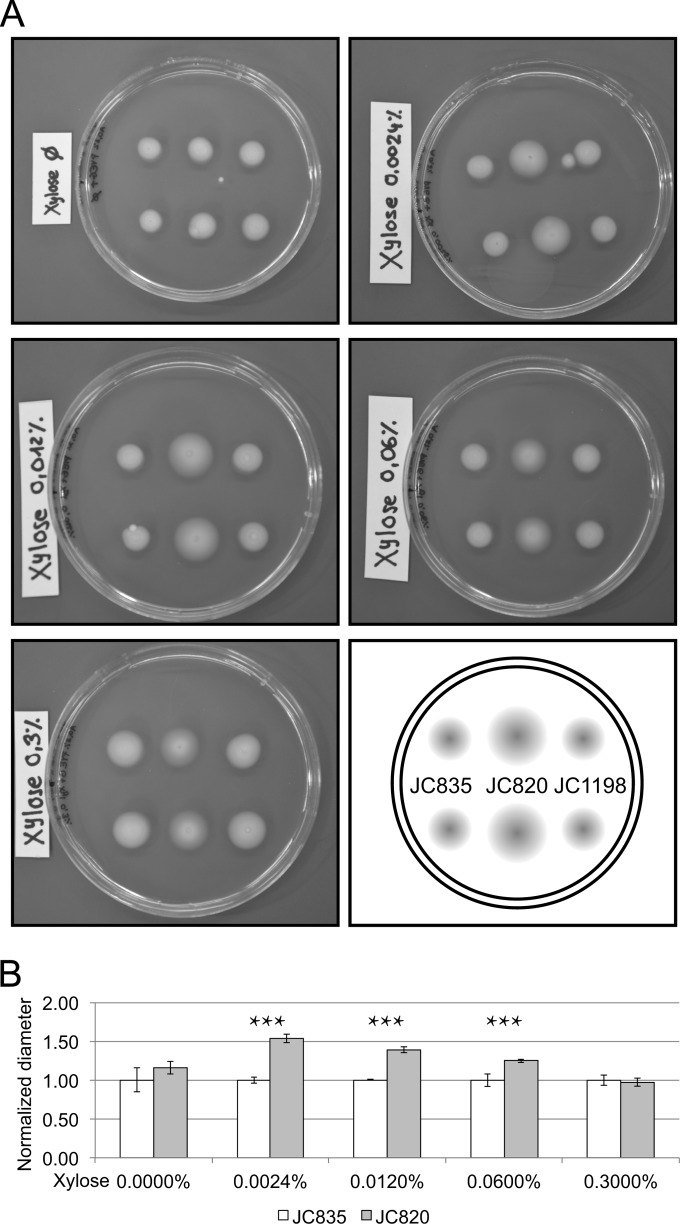
To determine whether a specific step of the cell cycle of C. crescentus was slowed down in response to (p)ppGpp accumulation, we observed the morphology and the motility of the JC820 cells using a light microscope after 4.5 h of growth in rich medium containing xylose (PYEGX) or not (PYEG). We found that the populations of cells accumulating (p)ppGpp in PYEGX contained a higher proportion of motile cells and a lower proportion of predivisional cells than control populations (see Fig. S2 in the supplemental material). We confirmed this conclusion by testing the motility of JC820, JC835, and JC1198 cells in low-percentage agar media containing different concentrations of the xylose inducer (Fig. 3). We observed that cells accumulating (p)ppGpp in the presence of 0.0024, 0.012, and 0.06% xylose formed significantly (Student t test, P < 0.05) larger halos than control cells injected into the same media, indicating that these populations comprised more motile cells (Fig. 3B). At the maximal concentration of xylose (0.3%), no significant difference in diameter between the halos formed by JC835 and JC820 strains was found; we conjecture that this is due to the fact that the increase in motility is compensated by a severe reduction in growth rate, as is suggested by the increased transparency of the halo formed by JC820 cells (Fig. 3A). Altogether, these results suggested that the swarmer-to-stalked cell transition may be slowed down in response to (p)ppGpp accumulation in C. crescentus cells.
FIG 3.
Swarming motility of cells expressing different levels of RelA′-FLAG compared to control cells. Strains JC835 (control strain), JC820 (expressing RelA′-FLAG) and JC1198 [expressing RelA′(E335Q)-FLAG] cultivated into PYEG were stabbed into 0.3% PYEG agar containing 0, 0.0024, 0.012, 0.06, or 0.3% xylose and incubated for at room temperature for 4 days. (A) Images of plates with two swarming halos per strain. (B) Relative quantification of the diameters of swarming halos from strains JC835 and JC820 using images shown in panel A. Error bars indicate the standard deviations; stars indicate significance (Student t test, P < 0.05).
One cannot simply look for the presence of a flagellum or a stalk at the cell pole to determine whether the swarmer-to-stalked cell transition was delayed when cells accumulated (p)ppGpp, because the flagellum is very fragile and the newborn stalk is very small in rich medium. Instead, we chose to look at the subcellular localization of three dynamic proteins as a function of the cell cycle. First, we transduced the relA′-FLAG construct into a strain encoding three fluorescent markers of cell differentiation in C. crescentus, generating strain JC845. These three molecular markers are the CpaE-CFP, PleC-YFP, and DivJ-RFP proteins, all functional and synthesized instead of the native proteins (42). CpaE activates the assembly of pili at flagellated cell poles, while PleC and DivJ are localized histidine kinases regulating cell fate asymmetry in C. crescentus predivisional cells (43, 44). CpaE-CFP and PleC-YFP normally localize as fluorescent foci at the flagellated pole of swarmer cells and then delocalize to the cytoplasm during the swarmer-to-stalked cell transition (Fig. 4A) (44–46). DivJ-RFP is diffuse in the cytoplasm of swarmer cells until it forms a fluorescent focus at the future stalked pole of the cell during the swarmer-to-stalked cell transition (Fig. 4A) (44, 46). Swarmer cells from strain JC845 grown in rich medium containing xylose or not were isolated and then analyzed by fluorescence microscopy for 90 min. Every 30 min, we analyzed the cells by fluorescence microscopy (examples of images are shown in Fig. S5 in the supplemental material) to evaluate the proportion of cells (Fig. 4) showing a PleC-YFP focus or a CpaE-CFP focus, but no DivJ-RFP focus, to estimate the proportion of swarmer cells in the population. Similarly, the proportion of cells showing a DivJ-RFP focus was evaluated (see Fig. S5 in the supplemental material and Fig. 4), as an estimate of the proportion of stalked cells in the population. Using this method, we observed that the decrease in the proportion of swarmer cells and, conversely, the increase in the proportion of stalked cells, were significantly slowed down in JC845 populations cultivated in rich medium containing xylose compared to populations cultivated in rich medium lacking xylose (Fig. 4B and C). For example, 90 min after the synchronization procedure, the proportion of cells showing a DivJ-RFP focus was still <50% in the population of cells accumulating (p)ppGpp, whereas it was >90% in the control population. We also noticed an increase in the proportion of cells with no fluorescent focus in cells accumulating (p)ppGpp, especially at the 60 min time point after the synchronization procedure (Fig. 4C). These cells probably correspond to cells imaged during the swarmer-to-stalked cell transition (no more PleC and CpaE foci, no DivJ focus yet). It is worth noting that, at late time points, the population of cells accumulating (p)ppGpp showed more heterogeneity than the control population, containing a mixture of delayed swarmer cells, swarmer cells transitioning into stalked cells, and stalked cells. This was expected given the longer duration of the swarmer stage; swarmer cells isolated during the synchronization procedure may then be at different stages in the continuum of their slow differentiation process. All of these results were consistent with the previously observed enrichment in motile cells in populations producing RelA′-FLAG (Fig. 3), we concluded that the completion of the swarmer-to-stalked cell transition is significantly delayed in cells that accumulate (p)ppGpp. This result suggests that high intracellular (p)ppGpp levels are a signal sufficient to delay or slow down the swarmer-to-stalked cell transition, independently of other (p)ppGpp-independent regulatory pathways that may be activated in response to starvation. Thus, the swarmer-to-stalked cell transition delay that was observed under starvation conditions (16, 18, 19) was very likely due to (p)ppGpp accumulation during the stringent response and not just to a lack of nutrients.
FIG 4.
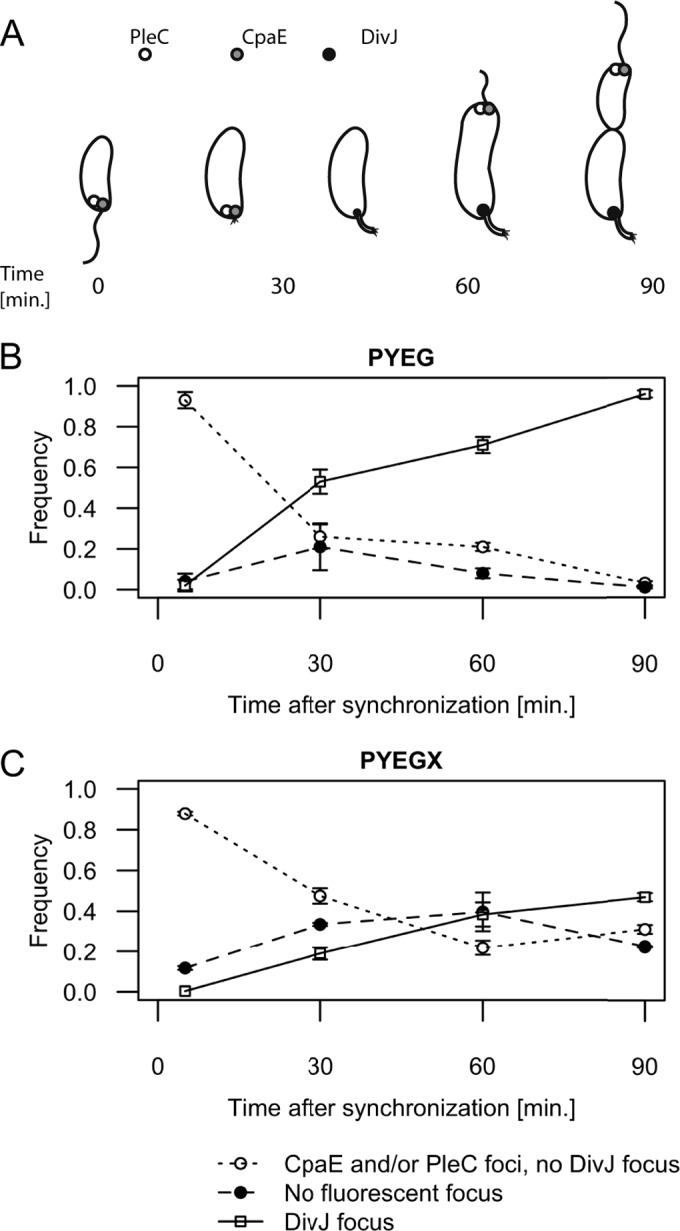
The accumulation of (p)ppGpp delays the swarmer-to-stalked cell transition. (A) Schematic showing the subcellular localization of CpaE, PleC, and DivJ as a function of the cell cycle when C. crescentus is grown in PYE medium. (B) Quantification of fluorescence microscopy experiments using isolated swarmer cells from strain JC845 (NA1000 pleC::pleC-YFP divJ::divJ-RFP cpaE::cpaE-CFP pXTCYC-4-relA′-FLAG) grown in PYEG. (C) Quantification of fluorescence microscopy experiments using isolated swarmer cells from strain JC845 grown in PYEGX after the synchronization procedure. The graphs shown in panels B and C correspond to the frequency of cells with detectable PleC-YFP or CpaE-CFP foci but no detectable DivJ focus (○), with visible DivJ-RFP foci (□) or with no visible focus (●). The time corresponds to the time after resuspension of the freshly isolated swarmer cells into PYEG or PYEGX. Typical images acquired at 0 and 60 min and used for this quantification are shown in Fig. S5 in the supplemental material. Plotted values are the averages of observations in three microscope fields; error bars indicate the standard deviations.
(p)ppGpp delays the initiation of chromosome replication.
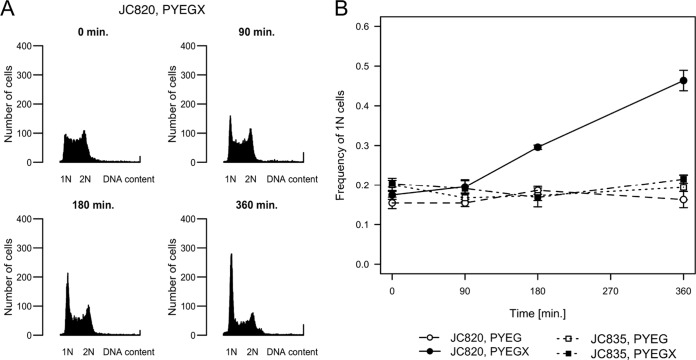
In wild-type C. crescentus cells grown in replete conditions, the swarmer-to-stalked cell transition is coincident with the initiation of chromosome replication. Thus, we decided to test whether the initiation of DNA replication was also delayed in cells accumulating (p)ppGpp. Using flow cytometry, we measured the DNA content of JC820 cells grown in rich medium. Upon induction of RelA′-FLAG synthesis, the proportion of cells containing only one complete chromosome increased over time (Fig. 5), reaching nearly 50% of the population after exposure to xylose for 6 h (Fig. 5B). Compared to populations in which (p)ppGpp synthesis was not induced, the frequency of nonreplicating G1 cells was ∼3-fold higher. Similar comparisons were made with JC835 and JC1198 populations of control cells that do not accumulate excess (p)ppGpp (Fig. 5B and see Fig. S6 in the supplemental material). These results suggested that the G1-to-S phase transition, which normally occurs at approximately the same time of the cell cycle as the swarmer-to-stalked cell transition (Fig. 6A), may also be severely delayed upon accumulation of (p)ppGpp, leading to an accumulation of G1 cells over time.
FIG 5.
The accumulation of (p)ppGpp increases the proportion of cells with one chromosome. (A) Histograms of the DNA content of cells accumulating (p)ppGpp. Representative profiles were obtained by flow cytometry analysis of unsynchronized cells from strain JC820 expressing RelA′-FLAG upon xylose addition. Cells were grown to exponential phase in PYEG before 0.3% xylose (PYEGX) was added at time zero. Cells were fixed at the indicated times and stained prior to flow cytometry analysis. The horizontal axis indicates the fluorescence intensity of individual cells and the number (N) of complete chromosomes. The vertical axis indicates the number of cells. (B) The proportion of cells with one chromosome increases when (p)ppGpp accumulates. Quantification of the results of flow cytometry experiments using strain JC820 and the control strain JC835. Cells were grown to exponential phase in PYEG before 0.3% xylose (PYEGX) was added to half of the culture at time zero. The y axis corresponds to the average proportion of cells containing 1N chromosome per cell at the indicated times after xylose addition. Plotted values are the averages of three independent populations; the standard deviations are also indicated.
FIG 6.
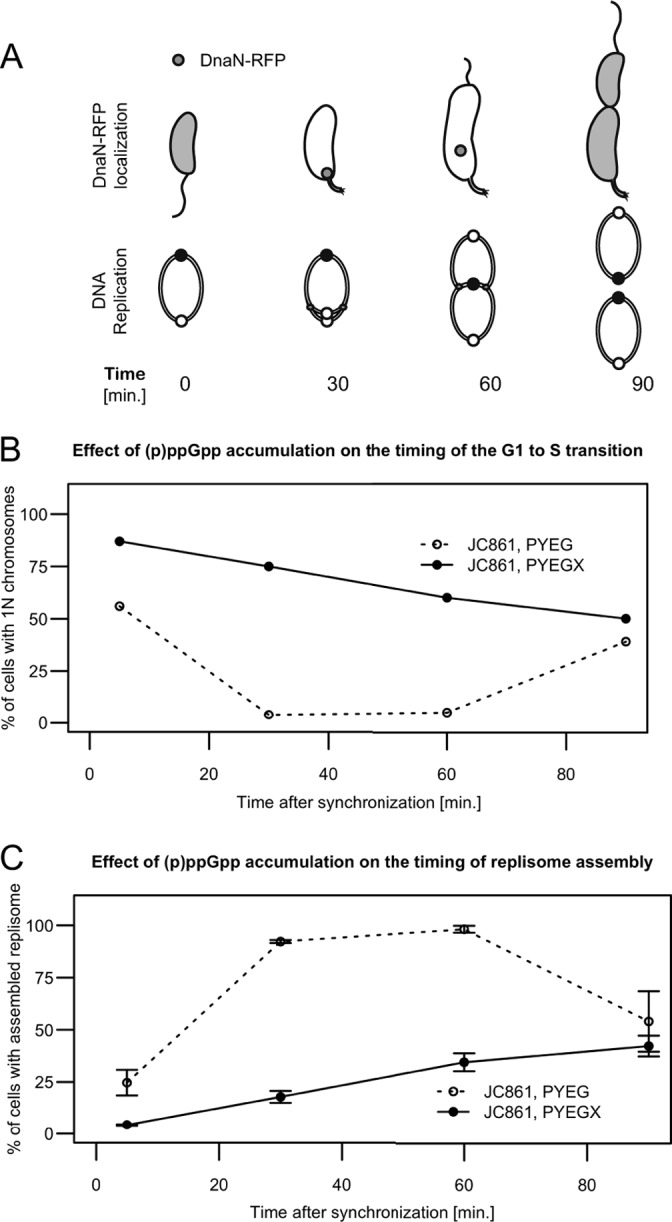
The accumulation of (p)ppGpp delays the initiation of chromosome replication. (A) Schematics showing the status of chromosome replication as a function of time when swarmer cells are grown in rich medium. The upper schematic shows that DnaN-RFP localizes as a moving focus only when DNA replication is ongoing. The lower schematic shows the replicating circular chromosome of C. crescentus as a function of the cell cycle. The small white and black circles on the lower schematic indicate the chromosomal origin and terminus, respectively. (B) (p)ppGpp accumulation delays the initiation of chromosome replication in swarmer cells. Quantification of flow cytometry experiments using isolated swarmer cells from strain JC861 (NA1000 dnaN::dnaN-RFP pXTCYC-4-relA′-FLAG) grown in PYEG or PYEGX. Cells were sampled at the indicated times, treated with rifampin, and then fixed and stained prior to flow cytometry analysis. The graph shows the percentage of cells with 1N chromosome, corresponding to cells that had not initiated chromosome replication at the indicated times of the cell cycle. (C) (p)ppGpp accumulation delays the assembly of the replisome in swarmer cells. Quantification of fluorescence microscopy experiments using isolated swarmer cells from strain JC861 grown in PYEG or PYEGX. Typical images acquired at different times under each condition and used for this quantification are shown in Fig. S8 in the supplemental material. The graph shows the percentage of cells with visible DnaN-RFP fluorescent foci, corresponding to cells that had initiated DNA replication at the indicated times of the cell cycle. The plotted values are the average percentages over three microscope fields; error bars indicate the standard deviations.
To confirm this hypothesis, we estimated the time needed for swarmer cells to initiate DNA replication in the presence or absence of (p)ppGpp accumulation. We used strain JC861, which expresses a functional DnaN-RFP fusion protein instead of the native DnaN protein (47), together with the relA′-FLAG construct controlled by the xylX promoter. We isolated swarmer cells from a JC861 mixed population previously cultivated for 3 h in PYEG or PYEGX, and resuspended them in the same media. We took aliquots from these populations at different times after the synchronization procedure and treated them with rifampin for three more hours. The DNA content of the cells was then analyzed by flow cytometry (representative images are shown in Fig. S7 in the supplemental material). Rifampin blocks the initiation of DNA replication and cell division in C. crescentus but allows the completion of ongoing rounds of replication (28). We estimated the proportion of cells that had not initiated DNA replication, corresponding to cells containing only one chromosome, at each time point. We found that the average half-life of a G1 cell, defined as the time when half of the cells in a population have initiated DNA replication, was less than 10 min in a population grown in rich medium lacking xylose (Fig. 6B). When these same cells were grown in rich medium containing xylose to induce (p)ppGpp accumulation, this period lasted for about 90 min. This result indicated that the G1-to-S transition is strongly delayed by the accumulation of (p)ppGpp, even when the cells are grown in replete conditions. To confirm this result, we observed the same cells by fluorescence microscopy (representative images are shown in Fig. S8 in the supplemental material). The DnaN protein is the β-clamp of the DNA polymerase, and fluorescently tagged DnaN proteins form a focus at the stalked pole of the cell once the replisome complex is assembled onto the DNA during the initiation of chromosome replication (47) (Fig. 6A). Thus, the localization of DnaN-RFP indicates that DNA replication is ongoing. We observed that >95% of the JC861 cells cultivated in rich medium lacking xylose contained fluorescent DnaN-RFP foci 1 h after synchronization (Fig. 6C). In contrast, <40% of the cells grown in rich medium containing xylose showed fluorescent foci at the same time point. To show that most swarmer cells accumulating excess (p)ppGpp could initiate and complete chromosome replication if given enough time, we treated JC861 swarmer populations with penicillin and analyzed the DNA content of individual cells by flow cytometry over time (see Fig. S9 in the supplemental material). Penicillin blocks cell division (48) but did not affect the completion of the first round of DNA replication (see Fig. S9 in the supplemental material). Consistent with previous results (Fig. 6B), we observed that only a minority (34%) of the (p)ppGpp accumulating cells had finished the replication of their chromosomes after 120 min (see Fig. S9 in the supplemental material), confirming that DNA replication is delayed in most cells. However, after a longer time period of 285 min, the vast majority of the cells (83%) had finally finished the replication of their chromosome (see Fig. S9 in the supplemental material). Altogether, these experiments demonstrate that swarmer cells accumulating high levels of (p)ppGpp in rich medium wait longer before they initiate the replication of their chromosome compared to cells with normal levels of (p)ppGpp but that most cells will finally do so. Our findings also suggest that (p)ppGpp accumulation is sufficient to delay the initiation of chromosome replication during the stringent response, independently of other (p)ppGpp-independent regulatory pathways that may be activated in response to starvation.
The DNA primases from E. coli and B. subtilis are inhibited by (p)ppGpp (21, 49–51). This leads to an inhibition of both the initiation and the elongation of DNA replication in B. subtilis cells during the stringent response. Previous flow cytometry assays performed on starved wild-type and spoT mutant C. crescentus cells could not address whether the elongation of DNA replication was also specifically affected by (p)ppGpp, since starved C. crescentus cells lacked nutrients to complete DNA replication (16). We found that nonstarved cells accumulating (p)ppGpp and treated with rifampin had no difficulties to complete chromosome replication within 3 h, since we observed that these cells contained either one or two complete chromosomes, but no incompletely replicated chromosomes by flow cytometry (data not shown). We conclude that it is only the initiation, and not the elongation, of chromosome replication that is strongly inhibited by (p)ppGpp in C. crescentus.
(p)ppGpp inhibits DnaA accumulation and CtrA degradation.
DnaA mediates the initiation of chromosome replication in most bacterial species (52), including C. crescentus (15). In E. coli, it was shown that the rates of dnaA transcription are inversely correlated with (p)ppGpp intracellular levels, suggesting that the delay in DNA replication may be due to a shortage in DnaA (53, 54). In C. crescentus, DnaA is actively proteolyzed in response to starvation (19), and it was shown that (p)ppGpp is required for this process (16). In contrast, basal (p)ppGpp levels in nonstarved cells did not affect DnaA levels (18). To test whether high levels of (p)ppGpp are sufficient to inhibit the accumulation of DnaA, we measured the levels of DnaA by immunoblots using cell extracts from strain JC820 grown in rich medium containing xylose to induce an artificial accumulation of (p)ppGpp for up to two generations (8 h). We observed that the levels of DnaA were ∼5-fold lower in cells accumulating (p)ppGpp for 8 h (grown with the xylose inducer) than in cells with basal levels of (p)ppGpp (grown without xylose) (Fig. 7A). Thus, the accumulation of (p)ppGpp in nonstarved cells is sufficient to inhibit the accumulation of DnaA. Given that the degradation of DnaA is accelerated during starvation, we reasoned that the proteolysis of DnaA might be stimulated in response to (p)ppGpp accumulation. To determine whether (p)ppGpp affects DnaA proteolysis in nonstarved cells, we compared the stability of DnaA in strains JC820 and JC835. Cells were cultivated in xylose-containing media for 2.5 h before the addition of rifampin to block dnaA transcription and ultimately DnaA synthesis; DnaA levels were then regularly monitored for 90 min by immunoblotting using cell extracts (Fig. 7B). The relative quantifications of these blots did not show an obvious difference in DnaA turnover rates between the two strains (Fig. 7E), suggesting that the depletion of DnaA in response to (p)ppGpp accumulation (Fig. 7A) might be linked to a probably indirect inhibition of DnaA synthesis rather than to the increased degradation of the protein. The shortage in DnaA that we observed might partly explain why (p)ppGpp-accumulating cells stay in G1 phase for so long. Besides acting as the initiator of DNA replication, DnaA is also an important transcriptional regulator in C. crescentus (27, 47, 55, 56): it activates the transcription of minimum 14 genes encoding proteins involved in various functions such as DNA replication and cell division. DnaA is nevertheless not needed for the swarmer-to-stalked cell transition, since DnaA-depleted swarmer cells still differentiate into stalked cells (15). Thus, it is unlikely that the inhibition of DnaA accumulation accounts for the swarmer-to-stalked cell transition delay that we observed in (p)ppGpp-accumulating cells grown in rich medium.
FIG 7.
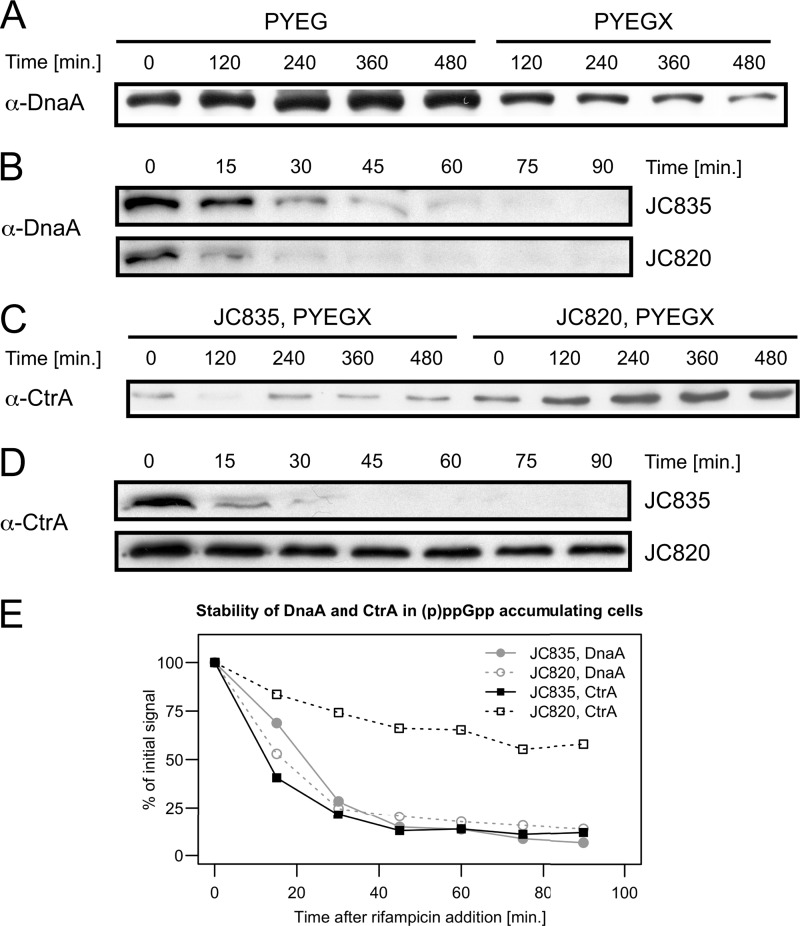
Effects of the accumulation of (p)ppGpp on the intracellular levels and the proteolysis of the DnaA and CtrA master regulators of the C. crescentus cell cycle. (A) The levels of DnaA are decreased upon (p)ppGpp accumulation. Strain JC820 (NA1000 pXTCYC-4-relA′-FLAG) was grown to exponential phase in PYEG and 0.3% xylose was added (PYEGX) to half of the culture at time zero. Cells were collected at the indicated times for immunoblot analysis using antibodies raised against DnaA. (B) The proteolysis of DnaA is not conspicuously faster upon (p)ppGpp accumulation. Strains JC835 (NA1000 pXTCYC-4) and JC820 were grown to exponential phase in PYEG and 0.3% xylose was added to the medium 150 min before the addition of rifampin (10 μg/ml). The addition of rifampin blocked further dnaA transcription and DnaA synthesis. DnaA decay in strains JC820 and JC835 were compared by immunoblot analysis using antibodies raised against DnaA; samples were collected 0, 15, 30, 45, 60, 75, and 90 min after rifampin addition. (C) The levels of CtrA are increased upon (p)ppGpp accumulation. Strain JC820 and JC835 (NA1000 pXTCYC-4) were grown to exponential phase in PYEG and 0.3% xylose was added (PYEGX) to the cultures at time zero. Cells were collected at the indicated times for immunoblot analysis using antibodies raised against CtrA. (D) CtrA proteolysis is inhibited upon (p)ppGpp accumulation. Strains JC835 and JC820 were grown as indicated in panel B, and the stability of CtrA was estimated by immunoblot analysis with antibodies raised against CtrA as in panel B. (E) DnaA and CtrA levels as a function of time after rifampin addition were estimated from the immunoblots shown in panels B and D.
CtrA is a response regulator controlling essential aspects of the cell cycle of C. crescentus and of other Alphaproteobacteria (57). When abundant and phosphorylated in swarmer cells and in the swarmer compartment of predivisional cells, it inhibits the initiation of chromosome replication by directly binding to the origin of replication (58). It acts at the same time as a transcription factor directly controlling the expression of about 95 genes in C. crescentus (13, 14, 59). The proteolysis of CtrA during the swarmer-to-stalked cell transition is dependent on the dephosphorylation of the CckA histidine kinase, which is itself dependent on the phosphorylation of the DivK response regulator (10, 60). Interestingly, the phosphorylation of DivK during the swarmer-to-stalked cell transition requires the delocalization of PleC from the flagellated cell pole and the localization of DivJ at the future stalked pole. Thus, our prediction was that CtrA may be stabilized in response to (p)ppGpp accumulation, since we observed that PleC, but not DivJ remained at the flagellated cell pole for longer when the expression of RelA′-FLAG was activated (Fig. 4). To test this prediction, we first measured CtrA levels by immunoblots using cell extracts from strain JC820 grown in rich medium containing xylose to induce an artificial accumulation of (p)ppGpp. We observed that the levels of CtrA were ∼4-fold higher in JC820 cells grown for 8 h in rich medium containing xylose than in control cells that did not accumulate high (p)ppGpp levels (Fig. 7B). This result suggested that the accumulation of (p)ppGpp might be sufficient to slow down the proteolysis of CtrA in replete conditions. To confirm this prediction, we compared the stability of CtrA in JC820 and JC835 populations, as previously done for DnaA. We found that CtrA was degraded at a much lower rate in JC820 cells than JC835 cells (Fig. 7D and E), showing that (p)ppGpp inhibits the proteolysis of CtrA in nonstarved C. crescentus cells. A recent study based on a proteomic analysis indicated that the SigT ECF sigma factor might activate the proteolysis of CtrA in response to carbon starvation (17). Our results suggest that the degradation of CtrA during starvation is not a consequence of (p)ppGpp accumulation, but rather a consequence of a more general stress response that may involve SigT, since CtrA is stabilized rather than destabilized by (p)ppGpp in nonstarved cells. The stabilization of CtrA in nonstarved cells accumulating excess (p)ppGpp may strengthen the G1-to-S delay caused by the (p)ppGpp-mediated inhibition of DnaA synthesis.
Conclusion.
In the present study, we report the development of an inducible system that proved efficient to characterize the effects that (p)ppGpp has on the progression of the C. crescentus cell cycle and decouple them from the physiological consequences of starvation. Upon expression of RelA′-FLAG in this work, like during carbon or nitrogen starvation (9, 16–18, 20), (p)ppGpp accumulation leads to a frequent increase in the life span of the swarmer cell and to a general delay in the initiation of DNA replication. We observed that (p)ppGpp affects the levels (DnaA, CtrA) or the localization (PleC, DivJ) of several important components of the regulatory network controlling the progression of the cell cycle, indicating that (p)ppGpp might be a central player in the network regulating the C. crescentus cell cycle as previously suggested by Boutte et al. (18). In C. crescentus, as in E. coli and other bacterial models, the (p)ppGpp signaling pathway does not uniformly affect the progression of the cell cycle; it preferentially delays it at steps when chromosomes are complete, at the swarmer/G1 phase (1N) in C. crescentus. For a bacterial cell, facing severe starvation in the middle of the DNA replication or the cell division processes might be difficult to handle. The swarmer/G1 phase might therefore involve a checkpoint facilitating coordinated arrest of the cell cycle and restart when conditions improve (23). Moreover, our data are consistent with the hypothesis that variations in the concentration of intracellular (p)ppGpp may modulate the relative duration of the swarmer stage in C. crescentus in a continuous way (18). Indeed, in our experiments, the intracellular accumulation of (p)ppGpp under replete conditions does not appear to completely block the progression of the cell cycle; it merely slows it down. This suggests that there may be a correlation between the levels of (p)ppGpp and the duration of the swarmer/G1 phase, rather than a threshold effect leading to cell cycle arrest. In this perspective, the (p)ppGpp molecule would not only be the mediator of an emergency stress response slowing down cell cycle progression, but also the basis of a fine-tunable regulatory system whereby the relative proportion of foraging swarmer cells and reproductive stalked cells in a C. crescentus population would be continuously adjusted in response to nutrient availability. Although the exact mechanisms slowing down the swarmer-to-stalked cell transition and delaying the initiation of DNA replication are still to be discovered, the RelA′-FLAG system that we developed in C. crescentus will be an interesting tool for future studies on cell cycle regulation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Carmen Fernández-Fernández and Katharina Eich for helpful discussions. We also thank Lucy Shapiro and Beat Christen for the TFLR C. crescentus strain. We are grateful to Noémie Matthey for her technical contribution to this work.
This study was supported by the Swiss National Science Foundation (SNSF) fellowships 3100A0_122541 and 31003A_140758 and SNSF R'Equip grant 316030_121391.
Footnotes
Published ahead of print 2 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01575-14.
REFERENCES
- 1.Wang JD, Levin PA. 2009. Metabolism, cell growth, and the bacterial cell cycle. Nat. Rev. Microbiol. 7:822–827. 10.1038/nrmicro2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dalebroux ZD, Swanson MS. 2012. ppGpp: magic beyond RNA polymerase. Nat. Rev. Microbiol. 10:203–212. 10.1038/nrmicro2720 [DOI] [PubMed] [Google Scholar]
- 3.Potrykus K, Cashel M. 2008. (p)ppGpp: still magical? Annu. Rev. Microbiol. 62:35–51. 10.1146/annurev.micro.62.081307.162903 [DOI] [PubMed] [Google Scholar]
- 4.Jishage M, Kvint K, Shingler V, Nystrom T. 2002. Regulation of sigma factor competition by the alarmone ppGpp. Genes Dev. 16:1260–1270. 10.1101/gad.227902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gummesson B, Magnusson LU, Lovmar M, Kvint K, Persson O, Ballesteros M, Farewell A, Nystrom T. 2009. Increased RNA polymerase availability directs resources toward growth at the expense of maintenance. EMBO J. 28:2209–2219. 10.1038/emboj.2009.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edwards AN, Patterson-Fortin LM, Vakulskas CA, Mercante JW, Potrykus K, Vinella D, Camacho MI, Fields JA, Thompson SA, Georgellis D, Cashel M, Babitzke P, Romeo T. 2011. Circuitry linking the Csr and stringent response global regulatory systems. Mol. Microbiol. 80:1561–1580. 10.1111/j.1365-2958.2011.07663.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Atkinson GC, Tenson T, Hauryliuk V. 2011. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One 6:e23479. 10.1371/journal.pone.0023479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiaverotti TA, Parker G, Gallant J, Agabian N. 1981. Conditions that trigger guanosine tetraphosphate accumulation in Caulobacter crescentus. J. Bacteriol. 145:1463–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boutte CC, Crosson S. 2011. The complex logic of stringent response regulation in Caulobacter crescentus: starvation signaling in an oligotrophic environment. Mol. Microbiol. 80:695–714. 10.1111/j.1365-2958.2011.07602.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curtis PD, Brun YV. 2010. Getting in the loop: regulation of development in Caulobacter crescentus. Microbiol. Mol. Biol. Rev. 74:13–41. 10.1128/MMBR.00040-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collier J, Shapiro L. 2007. Spatial complexity and control of a bacterial cell cycle. Curr. Opin. Biotechnol. 18:333–340. 10.1016/j.copbio.2007.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collier J. 2012. Regulation of chromosomal replication in Caulobacter crescentus. Plasmid 67:76–87. 10.1016/j.plasmid.2011.12.007 [DOI] [PubMed] [Google Scholar]
- 13.Quon KC, Marczynski GT, Shapiro L. 1996. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84:83–93. 10.1016/S0092-8674(00)80995-2 [DOI] [PubMed] [Google Scholar]
- 14.Domian IJ, Quon KC, Shapiro L. 1997. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell 90:415–424. 10.1016/S0092-8674(00)80502-4 [DOI] [PubMed] [Google Scholar]
- 15.Gorbatyuk B, Marczynski GT. 2001. Physiological consequences of blocked Caulobacter crescentus dnaA expression, an essential DNA replication gene. Mol. Microbiol. 40:485–497. 10.1046/j.1365-2958.2001.02404.x [DOI] [PubMed] [Google Scholar]
- 16.Lesley JA, Shapiro L. 2008. SpoT regulates DnaA stability and initiation of DNA replication in carbon-starved Caulobacter crescentus. J. Bacteriol. 190:6867–6880. 10.1128/JB.00700-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Britos L, Abeliuk E, Taverner T, Lipton M, McAdams H, Shapiro L. 2011. Regulatory response to carbon starvation in Caulobacter crescentus. PLoS One 6:e18179. 10.1371/journal.pone.0018179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boutte CC, Henry JT, Crosson S. 2011. ppGpp and polyphosphate modulate cell cycle progression in Caulobacter crescentus. J. Bacteriol. 194:28–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorbatyuk B, Marczynski GT. 2005. Regulated degradation of chromosome replication proteins DnaA and CtrA in Caulobacter crescentus. Mol. Microbiol. 55:1233–1245. 10.1111/j.1365-2958.2004.04459.x [DOI] [PubMed] [Google Scholar]
- 20.England JC, Perchuk BS, Laub MT, Gober JW. 2010. Global regulation of gene expression and cell differentiation in Caulobacter crescentus in response to nutrient availability. J. Bacteriol. 192:819–833. 10.1128/JB.01240-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang JD, Sanders GM, Grossman AD. 2007. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell 128:865–875. 10.1016/j.cell.2006.12.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ely B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol. 204:372–384. 10.1016/0076-6879(91)04019-K [DOI] [PubMed] [Google Scholar]
- 23.Meisenzahl AC, Shapiro L, Jenal U. 1997. Isolation and characterization of a xylose-dependent promoter from Caulobacter crescentus. J. Bacteriol. 179:592–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 25.Cashel M, Gallant J. 1969. Two compounds implicated in the function of the RC gene of Escherichia coli. Nature 221:838–841. 10.1038/221838a0 [DOI] [PubMed] [Google Scholar]
- 26.Tsai JW, Alley MR. 2001. Proteolysis of the Caulobacter McpA chemoreceptor is cell cycle regulated by a ClpX-dependent pathway. J. Bacteriol. 183:5001–5007. 10.1128/JB.183.17.5001-5007.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernandez-Fernandez C, Gonzalez D, Collier J. 2011. Regulation of the activity of the dual-function DnaA protein in Caulobacter crescentus. PLoS One 6:e26028. 10.1371/journal.pone.0026028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newton A. 1972. Role of transcription in the temporal control of development in Caulobacter crescentus (stalk-rifampin-RNA synthesis-DNA synthesis-motility). Proc. Natl. Acad. Sci. U. S. A. 69:447–451. 10.1073/pnas.69.2.447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magnusson LU, Farewell A, Nystrom T. 2005. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 13:236–242. 10.1016/j.tim.2005.03.008 [DOI] [PubMed] [Google Scholar]
- 30.Haseltine WA, Block R. 1973. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc. Natl. Acad. Sci. U. S. A. 70:1564–1568. 10.1073/pnas.70.5.1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gropp M, Strausz Y, Gross M, Glaser G. 2001. Regulation of Escherichia coli RelA requires oligomerization of the C-terminal domain. J. Bacteriol. 183:570–579. 10.1128/JB.183.2.570-579.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hammer BK, Swanson MS. 1999. Coordination of Legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol. Microbiol. 33:721–731. 10.1046/j.1365-2958.1999.01519.x [DOI] [PubMed] [Google Scholar]
- 33.Schreiber G, Metzger S, Aizenman E, Roza S, Cashel M, Glaser G. 1991. Overexpression of the relA gene in Escherichia coli. J. Biol. Chem. 266:3760–3767 [PubMed] [Google Scholar]
- 34.Schreiber G, Ron EZ, Glaser G. 1995. ppGpp-mediated regulation of DNA replication and cell division in Escherichia coli. Curr. Microbiol. 30:27–32. 10.1007/BF00294520 [DOI] [PubMed] [Google Scholar]
- 35.Ferullo DJ, Lovett ST. 2008. The stringent response and cell cycle arrest in Escherichia coli. PLoS Genet. 4:e1000300. 10.1371/journal.pgen.1000300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keasling JD, Bertsch L, Kornberg A. 1993. Guanosine pentaphosphate phosphohydrolase of Escherichia coli is a long-chain exopolyphosphatase. Proc. Natl. Acad. Sci. U. S. A. 90:7029–7033. 10.1073/pnas.90.15.7029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuroda A, Murphy H, Cashel M, Kornberg A. 1997. Guanosine tetra- and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J. Biol. Chem. 272:21240–21243. 10.1074/jbc.272.34.21240 [DOI] [PubMed] [Google Scholar]
- 38.Ruffing AM, Chen RR. 2012. Transcriptome profiling of a curdlan-producing Agrobacterium reveals conserved regulatory mechanisms of exopolysaccharide biosynthesis. Microb. Cell Fact. 11:17. 10.1186/1475-2859-11-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gonzalez D, Collier J. 2013. DNA methylation by CcrM activates the transcription of two genes required for the division of Caulobacter crescentus. Mol. Microbiol. 88:203–218. 10.1111/mmi.12180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harinarayanan R, Murphy H, Cashel M. 2008. Synthetic growth phenotypes of Escherichia coli lacking ppGpp and transketolase A (tktA) are due to ppGpp-mediated transcriptional regulation of tktB. Mol. Microbiol. 69:882–894. 10.1111/j.1365-2958.2008.06317.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bergmiller T, Pena-Miller R, Boehm A, Ackermann M. 2011. Single-cell time-lapse analysis of depletion of the universally conserved essential protein YgjD. BMC Microbiol. 11:118. 10.1186/1471-2180-11-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Christen B, Fero MJ, Hillson NJ, Bowman G, Hong SH, Shapiro L, McAdams HH. 2010. High-throughput identification of protein localization dependency networks. Proc. Natl. Acad. Sci. U. S. A. 107:4681–4686. 10.1073/pnas.1000846107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skerker JM, Shapiro L. 2000. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J. 19:3223–3234. 10.1093/emboj/19.13.3223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wheeler RT, Shapiro L. 1999. Differential localization of two histidine kinases controlling bacterial cell differentiation. Mol. Cell 4:683–694. 10.1016/S1097-2765(00)80379-2 [DOI] [PubMed] [Google Scholar]
- 45.Viollier PH, Sternheim N, Shapiro L. 2002. Identification of a localization factor for the polar positioning of bacterial structural and regulatory proteins. Proc. Natl. Acad. Sci. U. S. A. 99:13831–13836. 10.1073/pnas.182411999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matroule JY, Lam H, Burnette DT, Jacobs-Wagner C. 2004. Cytokinesis monitoring during development; rapid pole-to-pole shuttling of a signaling protein by localized kinase and phosphatase in Caulobacter. Cell 118:579–590. 10.1016/j.cell.2004.08.019 [DOI] [PubMed] [Google Scholar]
- 47.Collier J, Shapiro L. 2009. Feedback control of DnaA-mediated replication initiation by replisome-associated HdaA protein in Caulobacter. J. Bacteriol. 191:5706–5716. 10.1128/JB.00525-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Terrana B, Newton A. 1976. Requirement of a cell division step for stalk formation in Caulobacter crescentus. J. Bacteriol. 128:456–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Levine A, Vannier F, Dehbi M, Henckes G, Seror SJ. 1991. The stringent response blocks DNA replication outside the ori region in Bacillus subtilis and at the origin in Escherichia coli. J. Mol. Biol. 219:605–613. 10.1016/0022-2836(91)90657-R [DOI] [PubMed] [Google Scholar]
- 50.Gourse RL, Keck JL. 2007. Magic spots cast a spell on DNA primase. Cell 128:823–824. 10.1016/j.cell.2007.02.020 [DOI] [PubMed] [Google Scholar]
- 51.Maciag M, Kochanowska M, Lyzen R, Wegrzyn G, Szalewska-Palasz A. 2010. ppGpp inhibits the activity of Escherichia coli DnaG primase. Plasmid 63:61–67. 10.1016/j.plasmid.2009.11.002 [DOI] [PubMed] [Google Scholar]
- 52.Katayama T, Ozaki S, Keyamura K, Fujimitsu K. 2010. Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat. Rev. Microbiol. 8:163–170. 10.1038/nrmicro2314 [DOI] [PubMed] [Google Scholar]
- 53.Chiaramello AE, Zyskind JW. 1990. Coupling of DNA replication to growth rate in Escherichia coli: a possible role for guanosine tetraphosphate. J. Bacteriol. 172:2013–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zyskind JW, Smith DW. 1992. DNA replication, the bacterial cell cycle, and cell growth. Cell 69:5–8. 10.1016/0092-8674(92)90112-P [DOI] [PubMed] [Google Scholar]
- 55.Hottes AK, Shapiro L, McAdams HH. 2005. DnaA coordinates replication initiation and cell cycle transcription in Caulobacter crescentus. Mol. Microbiol. 58:1340–1353. 10.1111/j.1365-2958.2005.04912.x [DOI] [PubMed] [Google Scholar]
- 56.Collier J, Murray SR, Shapiro L. 2006. DnaA couples DNA replication and the expression of two cell cycle master regulators. EMBO J. 25:346–356. 10.1038/sj.emboj.7600927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brilli M, Fondi M, Fani R, Mengoni A, Ferri L, Bazzicalupo M, Biondi EG. 2010. The diversity and evolution of cell cycle regulation in alpha-proteobacteria: a comparative genomic analysis. BMC Syst. Biol. 4:52. 10.1186/1752-0509-4-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quon KC, Yang B, Domian IJ, Shapiro L, Marczynski GT. 1998. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc. Natl. Acad. Sci. U. S. A. 95:120–125. 10.1073/pnas.95.1.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Laub MT, Chen SL, Shapiro L, McAdams HH. 2002. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc. Natl. Acad. Sci. U. S. A. 99:4632–4637. 10.1073/pnas.062065699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsokos CG, Perchuk BS, Laub MT. 2011. A dynamic complex of signaling proteins uses polar localization to regulate cell-fate asymmetry in Caulobacter crescentus. Dev. Cell 20:329–341. 10.1016/j.devcel.2011.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thanbichler M, Iniesta AA, Shapiro L. 2007. A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res. 35:e137. 10.1093/nar/gkm818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guyer MS, Reed RR, Steitz JA, Low KB. 1981. Identification of a sex-factor-affinity site in Escherichia coli as gamma delta. Cold Spring Harbor Symp. Quant. Biol. 45(Pt 1):135–140 [DOI] [PubMed] [Google Scholar]
- 63.Evinger M, Agabian N. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 132:294–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.