ABSTRACT
The human immunodeficiency virus type 1 (HIV-1) accessory protein Nef is heavily targeted by CD8+ T lymphocytes (CTLs) during acute infection and therefore is included in many candidate vaccines. We investigated whether CTL targeting of Nef during acute infection contributes to immune control by disrupting the function of Nef. The sequence and function of Nef in parallel with CTL responses were assessed longitudinally from peak viremia until the viremia set point in a cohort of six subjects with acute infection. All but one individual had a single founder strain. Nef-specific CTL responses were detected in all subjects and declined in magnitude over time. These responses were associated with mutations, but none of the mutations were detected in important functional motifs. Nef-mediated downregulation of CD4 and major histocompatibility complex (MHC) class I molecules was better preserved in acute infection than in chronic infection. Finally, Nef-specific CTL responses were not associated with a reduction in viremia from its acute-phase peak. Our results indicate that CTLs targeting Nef epitopes outside critical functional domains have little effect on the pathogenic functions of Nef, rendering these responses ineffective in acute infection.
IMPORTANCE These data indicate that using the whole Nef protein as a vaccine immunogen likely allows immunodominance that leads to targeting of CTL responses that are rapidly escaped with little effect on Nef-mediated pathogenic functions. Pursuing vaccination approaches that can more precisely direct responses to vulnerable areas would maximize efficacy. Until vaccine-induced targeting can be optimized, other approaches, such as the use of Nef function inhibitors or the pursuit of immunotherapies such as T cell receptor gene therapy or adoptive transfer, may be more likely to result in successful control of viremia.
INTRODUCTION
During the first weeks of acute human immunodeficiency virus type 1 (HIV-1) infection, there is rapid adaptation in the context of the developing immune response. Early sequence diversification is coincident with the appearance of cytotoxic T lymphocytes (CTLs) (1–4). During this acute phase, a balance is achieved between CTL-mediated immune pressure and viral escape mutations, resulting in a stable viremia set point (2, 3, 5, 6). The accessory protein Nef is one of the proteins most heavily targeted by CTLs in early infection (5, 7–9), and because of its immunodominance, Nef is included as an immunogen in many vaccine preparations. Not surprisingly, a recent study of full-genome sequence evolution during the first 6 months of infection demonstrated that CTL escape occurred predominately in Nef (2), further demonstrating the effect of heavy selective pressure on Nef adaptation after acute infection.
Nef plays a key role in pathogenesis in vivo through a variety of functions (10). Among the best-understood functions of Nef are downregulation of major histocompatibility complex (MHC) class I (11, 12) and CD4 (13) molecules on the surfaces of infected cells. The downregulation of CD4 by Nef prevents the binding of CD4 to Env on nascent virions, thereby enhancing virion release (14). MHC class I downregulation has been shown to render infected cells less susceptible to HIV-specific CD8+ CTLs and thereby to confer resistance to CTL antiviral activity on infected cells (11, 15). Loss of some or all of these functions leads to attenuated infection in vivo (16).
Previous studies have shown that the function of Nef is shaped by CTL responses and differs according to the stage of disease; Nef-mediated CD4 and MHC class I downregulation is preserved in transmission (17) but is frequently diminished or absent by the end stage of AIDS (18). Primary isolates of Nef are highly variable in their sequences (19), and the quasispecies swarm can readily adapt to optimize its function. In a cohort of chronically infected subjects, it was observed that circulating quasispecies contained mixtures of two distinct populations of virus, one that fully downregulated MHC class I molecules and another with no ability to downregulate MHC class I molecules. Further, the proportion of functional quasispecies correlated with the breadth of the CTL response in these subjects, increasing in response to increased immune pressure, thus demonstrating the functional adaptation of Nef to the immune milieu of the host during chronic infection (20).
The role of Nef-specific CTLs in controlling viremia and shaping the functional adaptation of Nef in acute infection is not clear. Recent studies examining the relationship between CTL responses and viremia identified a correlation between Nef-specific CTLs and a lower viral set point in acute infection (21). However, results from vaccine studies in the simian immunodeficiency virus (SIV)-macaque model are contradictory: some data suggested an important contribution of Nef-specific CTLs to the control of viremia (22), while others showed that Nef-specific CTLs are easily escaped in acute infection and have little effect on viremia (23, 24).
Although Nef functions are largely preserved in transmission (17), there are few detailed data demonstrating how CTLs may shape the functional evolution of Nef during acute infection. During the rapid adaptation period from peak viremia to the viremic set point, Nef presumably evolves to achieve a balance between sequence mutation to escape Nef-directed CTL responses and the preservation of key functions that contribute to HIV-1 persistence and pathogenesis. We examined the interaction of CTLs with Nef during acute infection by a longitudinal study of Nef sequence evolution and function in parallel with Nef-specific gamma interferon (IFN-γ) ELISpot (enzyme-linked immunosorbent spot) assay-positive CD8+ CTL responses during the period from peak viremia until the establishment of a stable viremia set point.
MATERIALS AND METHODS
Subject selection and specimen collection.
Previously collected and deidentified plasma and peripheral blood mononuclear cells (PBMCs) were obtained from 7 subjects participating in the Acute Infection Early Disease Research Program (AIEDRP) and were certified exempt from IRB approval by the UCLA IRB. Six subjects were recruited during acute infection prior to seroconversion as determined by Western blotting, at Fiebig stages 2 to 4 (25). One subject (subject 07) was positive by Western blotting at enrollment, a finding consistent with recent, but not acute, infection. Serial samples were collected at regular intervals of approximately 2 weeks for the first 3 to 4 months after enrollment. Samples from 5 time points spanning the period from enrollment to the time of the viral set point for each subject were selected for study (see Fig. 1). The day of infection was determined on the basis of the likely exposure date given by the subject, if known, or was calculated as 7 days prior to the onset of symptoms if the exact exposure date was uncertain. Genomic DNA from PBMCs was used for intermediate-resolution HLA typing using a sequence-specific oligonucleotide (SSO) method as performed by the UCLA Immunogenetics clinical laboratory. Subjects were selected irrespective of their HLA type, and none had HLA-B*57, B*58. Additional plasma samples from 18 chronically infected subjects were obtained at a single time point according to a UCLA IRB-approved protocol.
FIG 1.
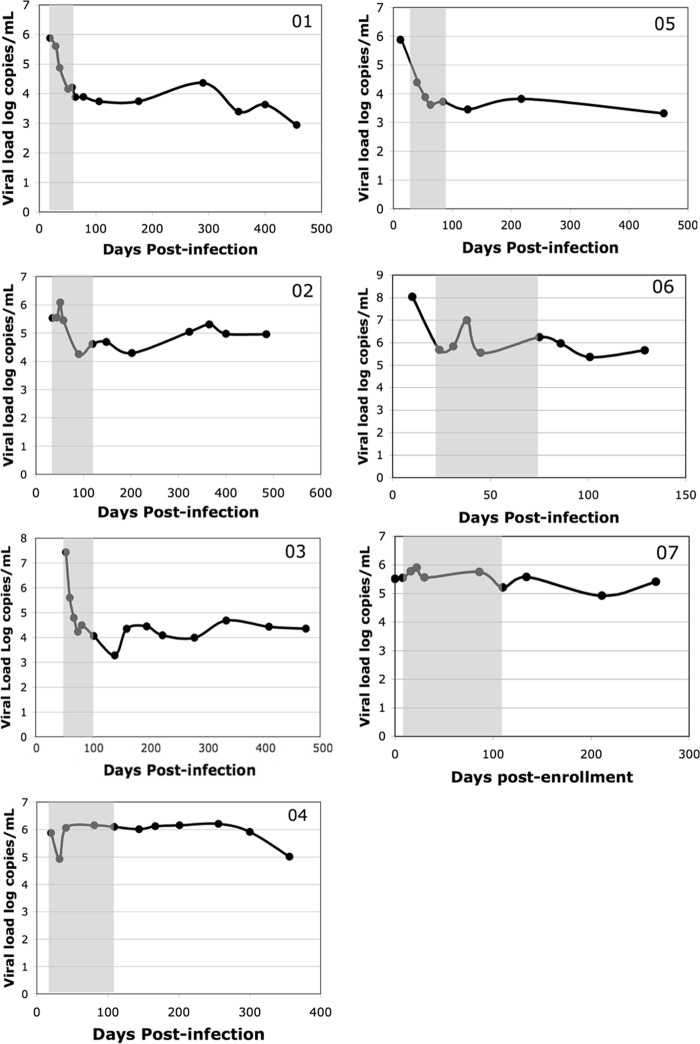
Decrease in the level of viremia after acute infection. Each panel shows the viral load (log copies of virus per milliliter of plasma) versus time in days postinfection. Subject identifiers are given in the upper right corners. The day of infection was assumed to be 7 to 10 days prior to the onset of symptoms of subjects presenting with acute retroviral syndrome or the day of the known exposure if reported by the subject. Note that subject 07 was in the early chronic phase of infection (positive by Western blotting at enrollment); therefore, for this subject, time is expressed as days postenrollment, not days postinfection. Each point corresponds to a study visit during which plasma and PBMCs were banked and clinical data were collected. The shaded area indicates the time points selected for study.
Viral isolation and nef quasispecies amplification.
Viral RNA was isolated from 1 ml of plasma by using the UltraSens viral isolation kit (Qiagen) according to the manufacturer's protocol. Total nef quasispecies were amplified by two-step reverse transcriptase PCR (RT-PCR) using the SuperScript III RT kit (Invitrogen) and the high-fidelity polymerase Phusion (New England BioLabs) with previously published primers and cycle conditions (20, 26). Multiple PCRs were performed on each sample, and products were then pooled prior to cloning. Note that it has been shown that there is no inherent advantage of single-genome sequencing over the method described above in terms of measuring population diversity (27).
Cloning and reporter virus production.
Bulk nef amplification products were cloned into plasmid AA1305#18, an NL4-3-based proviral vector in which the reporter gene encoding murine CD24 (also known as heat-stable antigen [HSA]) has been inserted into vpr, and portions of vpu and env also known to contribute to CD4 downregulation have been deleted, as described previously (28). HEK293T cells were cotransfected with the AA1305 proviral constructs and a construct encoding vesicular stomatitis virus glycoprotein (VSV-G) to produce pseudotyped recombinant replication-defective reporter viruses. Cloning efficiencies of 85% or higher were confirmed by plating a portion of the cloning mixture and selecting 10 to 12 colonies to check for the proper restriction digest pattern. Preservation of the diversity of the quasispecies mixture after cloning and virus production was assessed by sequencing virus stocks and quantifying polymorphic positions by examination of electropherograms.
Sequencing and sequence analysis.
Multiple individual clones for each sample were isolated and sequenced. Sequences were aligned with the NL4-3 nef sequence and the Los Alamos National Laboratory (LANL) HIV Sequence Database clade B consensus nef sequence and were then manually edited by toggling the amino acid translation using the BioEdit program. All sequences were examined for G-to-A hypermutation using Hypermut, version 2.0, from the LANL HIV Sequence Database tools. Sequences were examined for evidence of recombination using SimPlot and Bootscan (29). Sequences with nonintact reading frames due to frameshift or nonsense mutations were excluded prior to the analysis for adaptive evolution. Phylogenetic trees were constructed with neighbor-joining (NJ) and maximum likelihood (ML) algorithms using PHYLIP, version 3.64 (30). The neighbor-joining tree was statistically evaluated with 1,000 bootstrap replicates. Sequence diversity within the quasispecies swarm and overall divergence from the HIV-1 clade B consensus (Consensus B) sequence were determined using the SENDBS program with the Hasegawa plus gamma model, and standard errors were estimated from 500 bootstrap replicates. All of the following analyses were performed using HyPhy. The MODELTEST program was used to determine that the best-fitting model for the data was HKY85. The ratio of global nonsynonymous to synonymous substitution rates (dN/dS ratio), along with its 95% confidence intervals (CI), was estimated after the maximum likelihood function for each subject and each time point was built and optimized. Individual amino acid positions with evidence of adaptive evolution were identified by three separate methods: single likelihood ancestor counting (SLAC), relative-effects likelihood (REL), and fixed-effects likelihood (FEL). A site was considered to be adapting under selective pressure if that site was identified by at least 2 of 3 methods with a significance level of at least 95%. Additionally, only those sites with dN/dS ratios significantly greater than 1 or less than 1 were considered positive.
ELISpot mapping.
Gamma interferon (IFN-γ) enzyme-linked immunospot (ELISpot) assays were performed with expanded CD8+ PBMCs as described previously (31). Nonspecific polyclonal expansion of CD8+ lymphocytes from PBMCs was performed using CD3-CD4-bispecific monoclonal antibodies as described previously (32). Briefly, the expanded CD8+ T lymphocytes were plated at 2 × 105/well and were exposed to a library of HIV-1 peptides (consecutive 15-mers overlapping by 11 amino acids) spanning all clade B consensus HIV-1 proteins, obtained from the NIH AIDS Research and Reference Reagent Repository (catalog numbers 5138, 5189, 6208, 6444, 6445, 6446, 6447, 8117, and 9480). Peptides were initially screened in 53 pools of 12 to 16 peptides each and were added to the wells at a final concentration of 5 μg/ml for each individual peptide, followed by deconvolution of positive pools. Based on the availability of cells, additional tests were performed using individual peptides representing common HLA-specific minimal HIV epitopes that had been reported previously. Individual IFN-γ-secreting cells (spot-forming cells [SFC]) were counted using an automated ELISpot counting system (Cellular Technology Ltd., Cleveland, OH). Assays with a negative-control background mean of fewer than 100 SFC/106 cells were considered valid. A positive response was defined as a mean higher than three times the mean of the negative controls or at least 60 SFC/106 cells, whichever was higher.
Measuring the downregulation of CD4 and MHC class I molecules.
As described previously (28), T1 cells were infected with VSV-G-pseudotyped reporter viruses carrying primary nef quasispecies or the “wild type” NL4-3, M20A Nef (specifically deficient in MHC class I downregulation) (33), LL→AA Nef (specifically deficient in CD4 downregulation) (34), or ΔNef control allele. Note that we have determined previously that measuring the function of the bulk quasispecies yields results comparable to those obtained by measuring multiple individual clones (28). On day 3 postinfection, 2 × 105 cells were stained with fluorescein isothiocyanate (FITC)-conjugated anti-murine CD24/HSA (BD Pharmingen), allophycocyanin (APC)-conjugated anti-human CD4 (BD Pharmingen), and phycoerythrin (PE)-conjugated anti-human HLA-A*02 (ProImmune). At least 5 × 104 live cells were counted using a FACScan flow cytometer, and data were analyzed using CellQuest software (Becton, Dickinson). Maximum levels of HLA A*02 and CD4 were determined using the ΔNef mutant. All infections and flow cytometry assays were performed in triplicate. Significant differences from NL4-3 Nef were determined using a two-tailed t test with unequal variance (35).
Statistical analysis.
Comparison of gain or loss of Nef function between acute and chronic infection was performed with a Fisher exact test. Significant differences in selective pressure were determined by comparison to the average dN/dS ratio of the entire data set; nonoverlapping 95% confidence intervals signify a significant difference. Regression analysis was performed for the detection of correlations between the following pairs of observations: (i) viremia level and CTL response (measured as the number of positive peptide pools and the number of SFC/million cells), (ii) Nef function and CTL response, (iii) Nef function and dN/dS ratio, (iv) Nef function and the total number of positively or negatively selected sites, (v) Nef function and diversity, (vi) CTL response and dN/dS ratio.
Nucleotide sequence accession numbers.
The nef sequences determined in this study have been deposited in GenBank under accession numbers KJ680562 to KJ680919.
RESULTS
Study subjects.
Plasma and PBMCs were obtained from 7 subjects participating in the Acute Infection Early Disease Research Program (AIEDRP) at the Los Angeles site, a cohort of subjects with signs or symptoms of an acute infection or evidence of recent HIV-1 infection (36) (Table 1). Six of the subjects were recruited during acute infection prior to positivity by Western blotting, corresponding to Fiebig stages 2 to 4 (25). One subject (subject 07) was Western blot positive at enrollment, a status consistent with recent, but not acute, infection. Serial plasma and PBMC samples were collected at regular intervals of approximately 2 weeks for the first 3 to 4 months after enrollment, followed by less frequent intervals. Samples from 5 time points spanning the time from enrollment to the establishment of steady-state viremia (the set point) for each subject were selected for study (Fig. 1), except for subject 07, who was determined to have been enrolled during early chronic infection and was included as a control.
TABLE 1.
Subject information
| Subject identifier | HLA typea | Fiebig stageb | Peak VLc | Viremia set pointd (VL) | Change in VL | CD4 count at set point |
|---|---|---|---|---|---|---|
| 01 | A*03,31 B*14,38 C*08,12 | 3–4 | 5.87 | 3.74 | −2.13 | 714 |
| 02 | A*01,32 B*08,64 C*07,08 | 3–4 | 5.53 | 4.61 | −0.92 | 517 |
| 03 | A*02,29 B*62,51 C*04,15 | 5 | 7.43 | 4.10 | −3.33 | 649 |
| 04 | A*01,02 B*08,18 C*07 | 3–4 | 5.87 | 6.10 | +0.23 | 441 |
| 05 | A*01,03 B*07,08 C*07 | 3–4 | 5.87 | 3.46 | −2.41 | 435 |
| 06 | A*01,68 B*08,40 C*02,07 | 2 | 8.04 | 6.25 | −1.79 | 280 |
| 07 | A*03,24 B*07,35 C*04,07 | 6 | NA | 5.58 | NA | 566 |
Intermediate-resolution typing of HLA class I A, B, and C loci, determined by reverse SSO hybridization.
Determined according to the method of Fiebig et al. (25) at the time of enrollment and at the first study visit.
VL, viral load (expressed as log copies of virus per milliliter of plasma); NA, not applicable.
Defined as the first point at which a subject reached a stable level of viremia in the absence of therapy (refer to Fig. 1).
Evolution of nef quasispecies in acute infection.
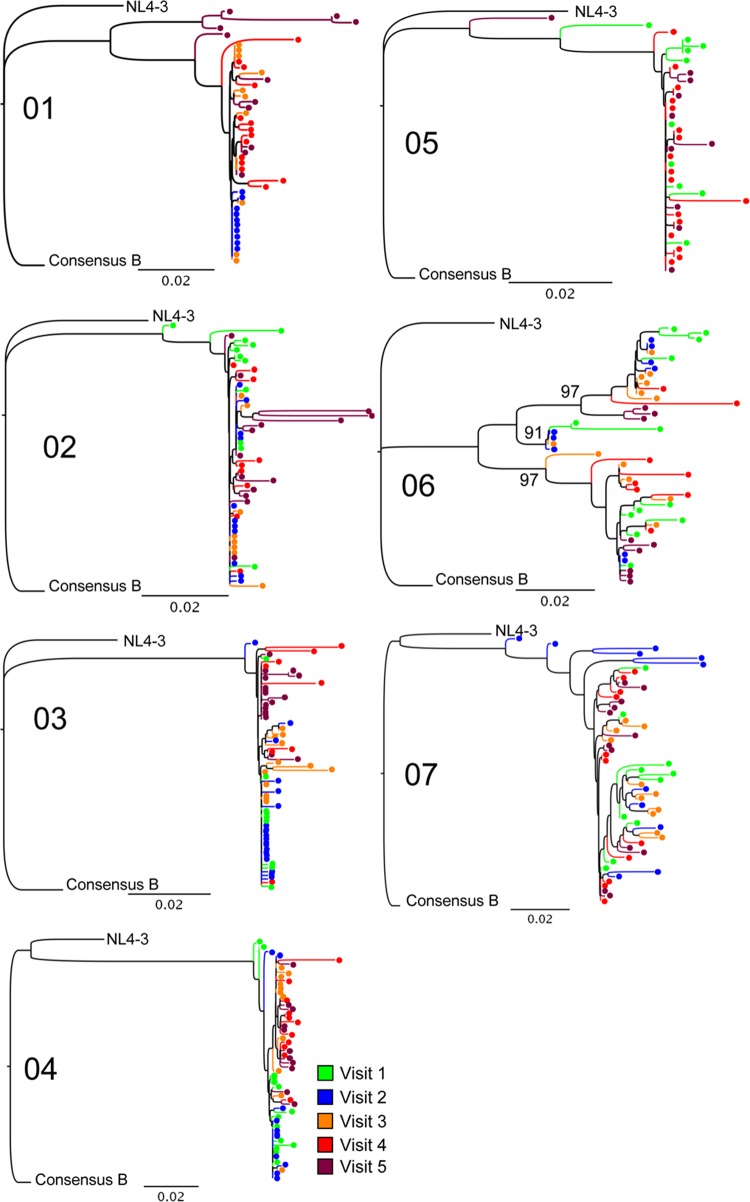
Multiple individual nef alleles were sequenced from plasma collected at each time point (mean, 11 [range, 8 to 21] for each time point for each subject). Neighbor-joining phylogenetic trees demonstrated that each subject was infected with clade B virus, and all sequences derived from each individual formed independent clusters supported by 100% of bootstrap replicates (Fig. 2). Five of six acutely infected individuals had trees suggesting a single founder strain, and subject 06 had evidence of multiple transmitted variants with 3 statistically significant clusters supported by high bootstrap values (91%, 97%, and 97%). Subject 07 was in the early chronic phase of infection; the data for this individual were used for purposes of comparison.
FIG 2.
Phylogenetic trees demonstrate transmitted virus and early diversification. Multiple full-length nef isolates were sequenced at 3 to 5 time points, about 2 weeks apart (Fig. 1). Isolates are color-coded by study visit number as indicated in the key. Sequences were aligned, edited, and translated in comparison to both the NL4-3 and the clade B consensus (“Consensus B”) sequence. Neighbor-joining trees were constructed and evaluated with 1,000 bootstrap replicates. Sequences from each subject formed independent clusters with 100% bootstrap support. As noted, the sequences for subject 07, who was in the early chronic phase of infection, served as a control for comparison. Of the acutely infected subjects, only subject 06 had evidence of multiple transmitted variants, with 3 statistically significant clusters supported by high bootstrap values (91%, 97%, and 97%), as indicated on the tree. Bars, 0.02 genetic distance.
The diversity of the quasispecies and their divergence from the Consensus B sequence were measured at each time point for the acutely infected subjects (Fig. 3). There was a nadir in quasispecies diversity for each subject between days 20 and 40 postinfection, in agreement with other reports (37). This dip in diversity corresponded to the presence of a largely monotypic quasispecies population, indicating, at least in some cases, another genetic bottleneck occurring after the time of transmission (Fig. 3A). There was an increase in diversity over time after the initial bottleneck in each subject. There was no overall pattern of convergence or divergence in relationship to the Consensus B sequence over this time, although subject 01 did show a trend toward convergence (Fig. 3B).
FIG 3.
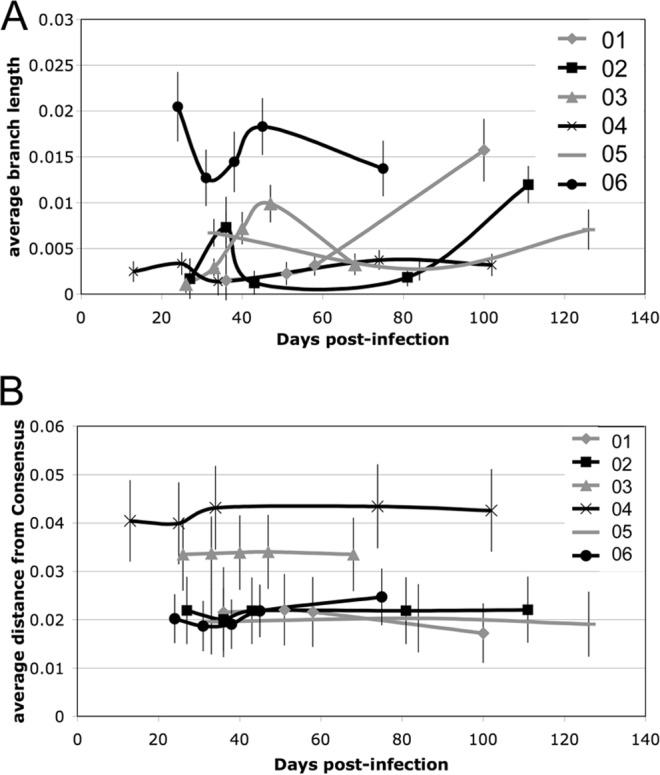
Sequence diversity and divergence from consensus over time after acute infection. (A) The amount of diversity, expressed as the average branch length for all sequences isolated at each time point, is plotted relative to the number of days postinfection. (B) The average divergence from the clade B consensus for all sequences at each time point is plotted relative to the number of days postinfection. Both diversity and divergence estimates were evaluated with 500 bootstrap replicates to give standard errors of the means (indicated by error bars).
Nef is heavily targeted by CTLs in acute infection.
In order to correlate changes in viral sequences with cellular immune responses, HIV-specific CTL targeting of the whole virus proteome was measured by IFN-γ ELISpot assays at the same time points (except for subject 06, who was tested only with HLA-matched minimal-epitope peptides and Gag and Pol Consensus B peptide pools due to limitations in the availability of PBMCs). In agreement with previous studies reporting the immunodominance of Nef in acute infection, Nef-specific CTL responses were detected in all subjects (Fig. 4). There was no overall pattern in the kinetics of Nef-specific CTL targeting. Subject 04 had a significant Nef-specific response as early as <20 days postinfection, while the first response detected in subject 06 developed between days 31 and 38 (Fig. 4B). Nef-specific responses accounted for as much as 58% of the total magnitude of detected CTL responses against HIV-1 (average, 16%; range, 0 to 58% for individual time points [Fig. 4C]).
FIG 4.
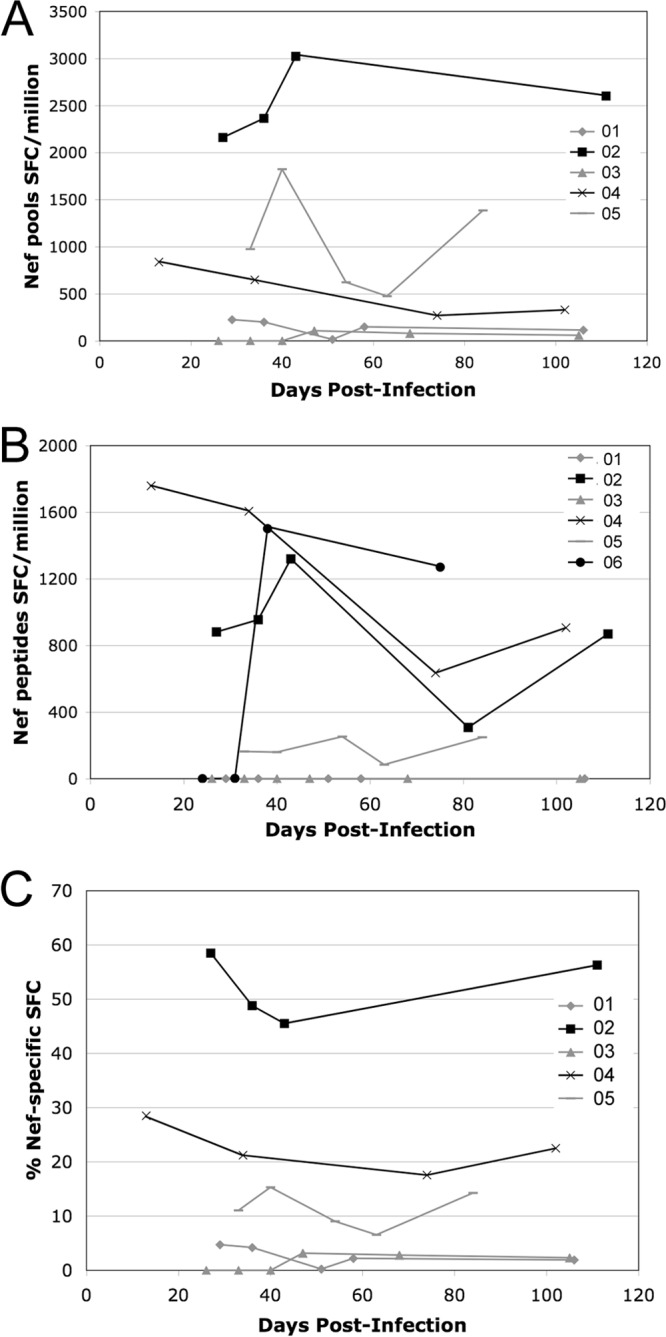
Nef-specific CTL responses over time after acute infection. Nef-specific CTL responses were measured by IFN-γ ELISpot assays using polyclonally expanded CD8+ cells from each study visit. (A and B) Cells were tested either with a library of overlapping 15-mers corresponding to the clade B consensus Nef sequence in pools of 12 to 16 peptides each (A) or with individual peptides representing HLA-matched reported minimal epitopes corresponding to each subject's HLA type (B). Responses are reported as the number of spot-forming cells (SFC) per million CD8+ cells and are plotted over time, expressed as days postinfection. Due to limited cell numbers, subject 06 was tested only with the HLA-matched minimal-epitope peptides and not with the Consensus B peptide pools. (C) The percentage of total SFCs attributable to Nef-specific responses is plotted for subjects tested with the full library of 15-mer pools.
Nef-specific CTL responses did not result in mutations in important functional residues or motifs.
Nef sequences were examined for amino acid changes that may have resulted from CTL-mediated immune selective pressure (Table 2). A range of 0 to 6 epitopes (mean, 2) was targeted per subject, as determined by the number of HLA-matched minimal-epitope peptides eliciting a positive ELISpot response. Amino acid changes were detected within 50% (6 of 12) of targeted epitopes (Fig. 5). As an additional means of detecting possible CTL-driven mutations that may have been missed by ELISpot mapping with Consensus B peptides, sequences were examined for sites under significant positive selective pressure. The dN/dS ratio, a measure of adaptive evolution, was estimated for each subject at each amino acid site. Four sites from 3 subjects were identified as undergoing significant positive selective pressure, defined by a dN/dS ratio of >1. Of these 4 positively selected sites, 3 lie within previously reported epitopes that are restricted by an HLA allele present in the subject's haplotype (Fig. 5). All putative escape mutations fell outside known domains associated with overall Nef functionality (the G2 myristylation site and the D123 dimerization site), the specific functions of CD4 and MHC class I downregulation (RxR17–19, M20, E62–65, PxxP72–81, L164–165), or the effects of Nef on cellular trafficking and signaling (L174–175).
TABLE 2.
Summary of amino acid changes in Nef during acute infectiona
| Subject | Amino acid changeb | Reversion to consensus |
|---|---|---|
| 01 | D38X | No |
| H40Y | No | |
| T133I | Yes | |
| R190K | Yes | |
| H192P/R | No | |
| M194V | No | |
| 02 | Q22P | No |
| D28E | No | |
| R184K | Yes | |
| 03 | R22P | No |
| H199Y | No | |
| 04 | P14L | No |
| E94Q | No | |
| I114 M | No | |
| H115Y | Yes | |
| H192R | No | |
| V194 M | Yes | |
| 05 | K105E/R | No |
| 06 | E93K | No |
| H102Y | Yes | |
| Q104P | No | |
| D108E | No |
Defined as a fixed substitution or dominant polymorphism (>50% of clones) appearing between the first and last time points.
Numbering according to the HXB2 reference sequence. X, any amino acid.
FIG 5.
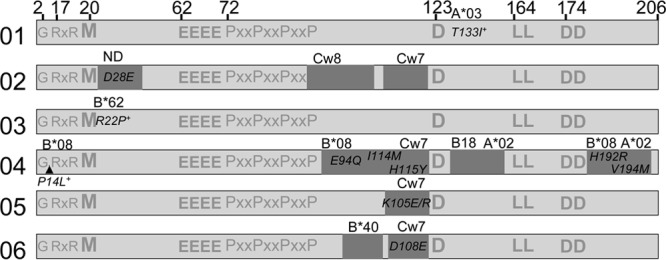
Map of Nef-specific CTL responses and associated amino acid changes relative to important functional sites. For each acutely infected subject, a line diagram of Nef shows the sites targeted by CTLs (dark shading) relative to known residues and domains important for Nef function. CTL responses were mapped by IFN-γ ELISpot assays using HLA-matched minimal-epitope peptides as shown in Fig. 4B. The amino acid positions of important functional sites are listed at the top. See Results for details of the functions associated with these positions. Within the shaded boxes indicating targeted epitopes are corresponding fixed amino acid substitutions or polymorphisms that appeared between the first and last study visits. Additional amino acid sites under positive selective pressure but not detected by ELISpot assays are indicated by plus signs. The HLA restriction of the epitope is indicated above each shaded box.
A peak in positive selective pressure on nef was followed by purifying selection by the time of the viral set point.
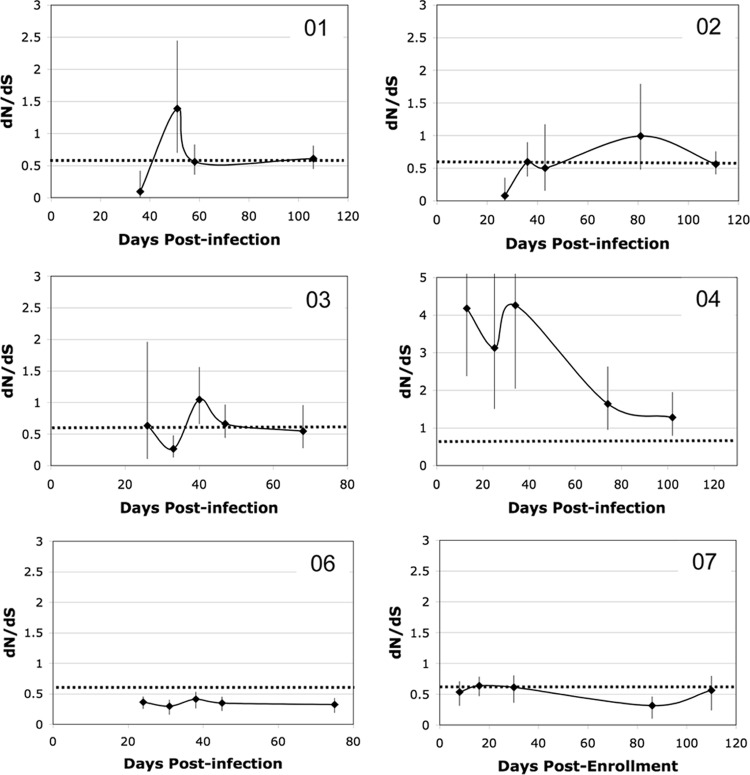
Although Nef-specific CTLs did not target functionally important sites within Nef, they did exert detectable selective pressure resulting in adaptive evolution of the quasispecies. The global dN/dS ratio was calculated for each subject at each time to determine the average selective pressure on the whole protein over time (Fig. 6). By use of a dN/dS ratio of 0.6 as a baseline point of reference that was previously defined for nef during chronic infection (26), several different patterns of adaptive evolution during acute infection were apparent. Three of 6 acutely infected subjects (subjects 01, 02, and 03) showed a pattern of intense purifying selection followed by positive selection before stabilizing at a dN/dS ratio of 0.6 in early chronic infection. This pattern is consistent with the observed genetic bottleneck coincident with very strong purifying selection followed by a period of intense adaptation to the new host as the virus explores many different escape pathways before reaching a steady state. The Nef of subject 04 was under significant positive selective pressure (dN/dS ratio, around 4) from the earliest time points, with no initial period of intense purifying selection detected, and then dropped toward the baseline. Finally, the Nef of subject 06 was under more intense purifying selection (dN/dS ratio, <0.6) at every time point, whereas the chronically infected subject 07 had a stable dN/dS ratio around 0.6 at each time point tested. Changes in the dN/dS ratio over time did not clearly correlate with changes in the magnitude of the Nef-specific CTL response (compare Fig. 4 and 6), but the dN/dS ratios for epitope regions were higher than those for nonepitope regions in all but one subject (see Fig. S1 in the supplemental material). There was a trend for an inverse correlation between the dN/dS ratio and diversity (P, 0.075), suggesting that greater selective pressure (a higher dN/dS ratio) drove lower sequence diversity.
FIG 6.
Change in selective pressure on Nef over time after acute infection. The amount of adaptive evolution was measured by calculating a maximum likelihood estimate of the dN/dS ratio with the 95% CI for all sequences at each time point. Values are plotted against time, expressed as days postinfection (or as days postenrollment for chronically infected subject 07). Data for all subjects are shown on the same scale except for subject 04, who had much higher dN/dS values than the others. The dotted lines mark a baseline dN/dS ratio of 0.6, the average value reported for Nef from chronically infected subjects. The sequence data obtained for subject 05 were not adequate for determination of the pattern of adaptive evolution in this individual, but at the viral set point, the dN/dS ratio had stabilized near baseline at 0.53.
Nef-mediated CD4 and MHC class I downregulation functions are well preserved in acute infection as opposed to chronic infection.
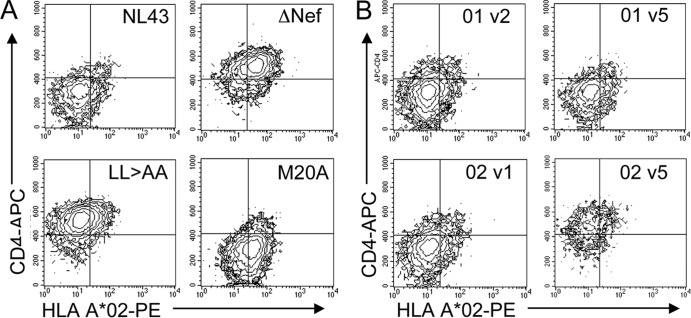
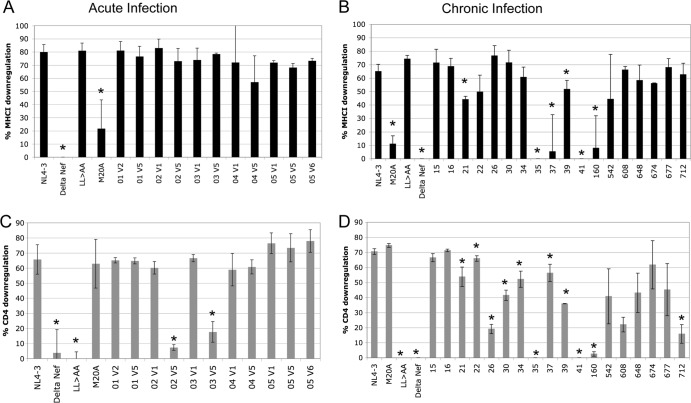
Next, levels of Nef-mediated downregulation of CD4 and MHC class I molecules were measured at the first and last time points in order to determine whether the sequence evolution observed in acute infection resulted in significant functional impairment. These functions were selected given their well-documented roles in HIV-1 pathogenesis. Levels of downregulation of CD4 and the MHC class I molecule HLA A*02 by Nef were measured on infected cells and were compared to those with the NL4-3 Nef (fully functional), ΔNef (deficient in both functions), LL→AA Nef (deficient in CD4 downregulation), and M20A Nef (deficient in MHC class I downregulation) control alleles (Fig. 7). In general, the function of Nef from acutely infected subjects at the first and last time points fell into two different patterns: fully functional at both time points (Fig. 7B, subject 01) or fully functional at the first time point and then deficient in CD4 downregulation at the last time point, when the viremia set point was achieved (Fig. 7B, subject 02). In comparison to the function of nef quasispecies for a cohort of chronically infected subjects, all acutely infected subjects had fully preserved MHC class I downregulation at both the first and the last time point (Fig. 8A), while 2 of 6 (subjects 03 and 02) had lost Nef-mediated CD4 downregulation by the last time point (Fig. 8C). Although there were no mutations at sites known to affect CD4 downregulation in the sequences from these time points, there were substitutions and polymorphisms at other sites that, alone or in combination, may account for the difference in function (Table 2; see also Fig. S2 in the supplemental material). Comparison to chronically infected subjects (Fig. 8B and D) using a Fisher exact test showed overall statistically significant preservation of downregulation of both CD4 and MHC class I molecules by Nef in acute infection (P, 0.0275 and 0.0250, respectively).
FIG 7.
Downregulation of CD4 and MHC class I molecules by Nef. The CEM T1 permissive cell line was infected with VSV-G-pseudotyped recombinant reporter viruses, and levels of CD4 and HLA-A*02 were measured on all reporter-positive cells 3 days later. (A) Levels of Nef-mediated downregulation were compared to expression by a panel of control viruses, which included the NL4-3 Nef (“wild type”), ΔNef, Nef LL→AA (deficient only in CD4 downregulation), and Nef M20A (deficient only in MHC class I downregulation) viruses. (B) nef quasispecies from acutely infected subjects from the first and last study visits (designated “v1” for visit 1, etc.) were cloned and tested in bulk. Representative dot plots for subjects 01 and 02 are shown.
FIG 8.
Comparison of CD4 and MHC class I downregulation by acutely and chronically infected individuals. (A and B) MHC class I downregulation by nef isolates from subjects with acute and chronic infections, respectively. (C and D) CD4 downregulation by acute- and chronic-phase isolates, respectively. The percentage of downregulation was calculated relative to maximum expression levels of CD4 or MHC class I molecules, determined by using the ΔNef mutant. Data are means and standard deviations of results from at least 3 replicate infections. For acutely infected subjects, samples from the first and last study visit were tested (indicated by “V1” for visit 1, etc.). Asterisks indicate significant differences (P, <0.05) in function from NL4-3 Nef.
Total CTL responses correlated with reductions in viremia levels, but Nef-specific CTL responses correlated with higher viremia levels.
Finally, the relationships between CTL response, viremia, and Nef function were examined. Despite the small sample size, there was a significant inverse correlation between the total number of recognized peptide pools and the magnitude of the drop in the viremia level from peak viremia to the set point (Fig. 9A). Overall, these correlations are consistent with previous reports (3) that CTL responses are a major driver of the reduction in the viremia level after acute infection.
FIG 9.
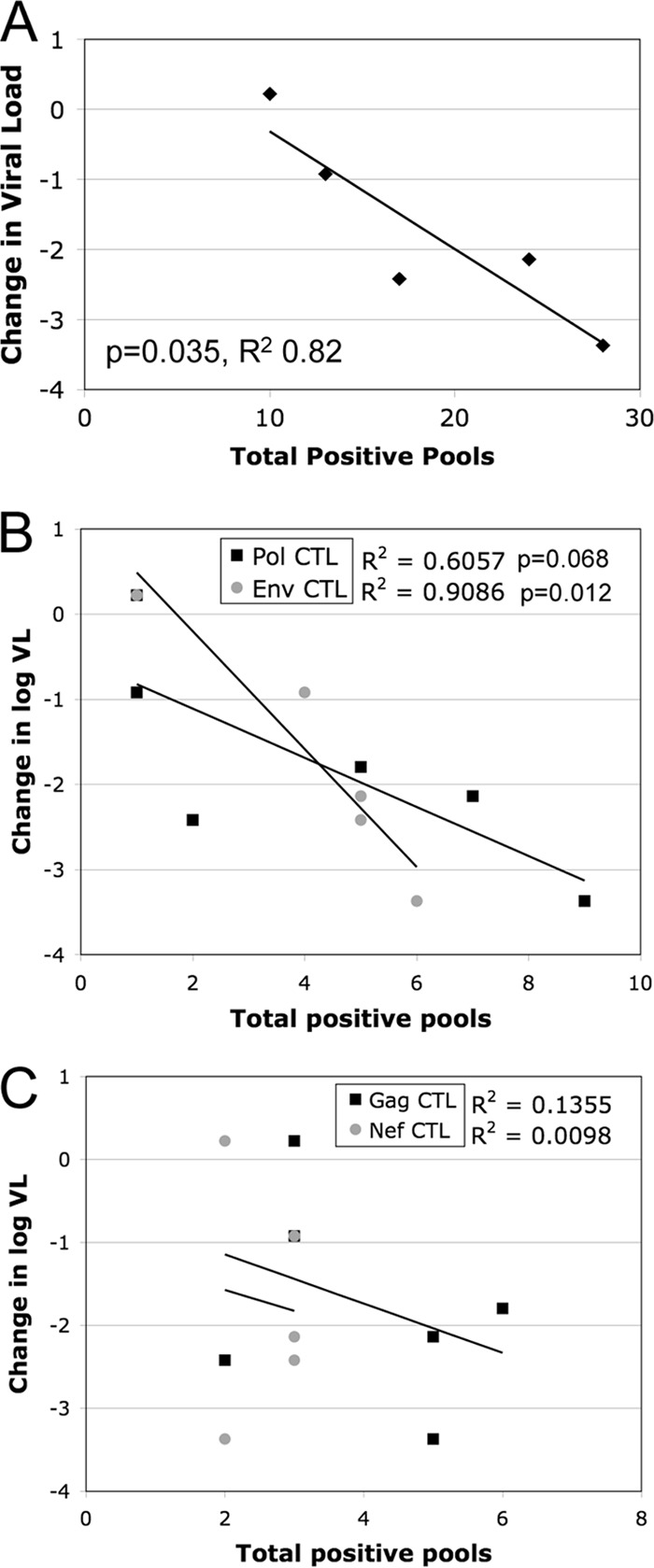
Correlations between CTL responses and viremia levels after acute infection. (A) Using regression analysis, a statistically significant inverse relationship was observed between the change in the log viral load (VL) between peak and set point viremia and the total number of IFN-γ ELISpot assay-positive peptide pools. (B and C) Analysis of the relationship between the magnitude of protein-specific responses and the decline in viremia revealed significant inverse relationships with Env- and Pol-specific subsets (P, 0.012 and 0.068, respectively) (B) but no relationship with Nef- and Gag-specific responses (P, 0.87 and 0.47, respectively) (C). For each correlation, significant P values and R2 are shown.
The effects of specific subsets of CTL responses on viremia were then examined. Significant inverse correlations were observed between the decrease in the viremia level from peak to set point and both Pol-specific and Env-specific CTL responses (P, 0.068 and 0.012, respectively) (Fig. 9B). However, there was no significant correlation between the change in the viremia level and Nef-specific or Gag-specific CTL responses (P, 0.87 and 0.47, respectively) (Fig. 9C).
Finally, the relationship between CTL response and Nef function was examined. In agreement with the prior demonstration of a positive correlation between CTL breadth and the preservation of MHC class I downregulation in a chronic infection cohort, in this acutely infected cohort there was a trend (P, 0.090; R2, 0.670) for a positive association between CTL breadth and the degree of MHC class I downregulation (see Fig. S3 in the supplemental material).
DISCUSSION
We show that during acute infection, the CTL response heavily targets Nef in individuals without protective MHC class I haplotypes and exerts significant selective pressure yet has little effect on Nef functions, particularly MHC class I downregulation. In agreement with the findings of prior studies of acutely infected persons (5–7, 9), Nef is targeted far out of proportion to its size, accounting for approximately 8% of the viral proteome yet yielding more than 50% of the entire HIV-1-specific CTL response in some persons. Given the demonstration of significant selective pressure leading to sequence evolution, the lack of association of the Nef-specific CTL response with the drop in the viremia level in this acute-infection cohort is likely due to the pattern of epitope targeting and the capacity of Nef to escape without loss of function.
It is noteworthy that many other factors in addition to CTL selective pressure influence the overall adaptive evolution of Nef and contribute to shifts in the dN/dS ratio (37). The fact that the dN/dS ratio does not correlate clearly with CTL responses likely indicates that other factors, such as target cell availability and host restriction factors, also exert selective pressure on the virus during acute infection. The intense purifying selection seen at the earliest time points, coincident with the genetic bottleneck prior to the peak of the CTL response, reflects such other factors.
Studies of the utility of Nef-specific CTLs in controlling viremia in natural infection and in vaccination have come to mixed conclusions. Our finding that Nef-specific CTLs have no effect on the viremia set point after acute infection is similar to recent data from Turk et al. (38), but it contradicts another recent study by Riou et al., in which a correlation between polyfunctional Nef-specific CTLs and a lower viremia set point was observed (21). Similarly, there are contradictory results from recent SIV-macaque vaccine studies, some of which showed an important contribution of Nef-specific CTLs to the control of viremia (22) while others showed that Nef-specific CTLs failed to control viremia and are easily escaped from in acute infection (23, 24). Price et al. also provide evidence of rapid escape from Nef-specific CTLs in acute HIV-1 infection (39). Finally, the infusion of a highly expanded autologous Nef-specific CTL clone for adoptive immunotherapy unfortunately resulted in epitope escape, increased viremia, and disease progression (40). Although the generality of our results is tempered by our small sample size, there is ample evidence to show that CTL targeting of Nef can be highly effective or, alternatively, easily escaped.
A likely explanation for the differences observed in the efficacy of Nef-specific CTLs is variability in targeting associated with the MHC class I type. Of note, our cohort included no individuals with haplotypes associated with superior immune control of HIV-1 infection. Evidence suggests that some protective MHC class I types are associated with effective CTL targeting of Nef. In the HVTN 502 (STEP) trial, one of the six vaccine-induced epitopes that predicted efficacy in reducing the viremia set point was the HLA-B*57-restricted Nef epitope HW9 (Nef amino acids 116 to 124) (41), and separately, Navis et al. also showed that CTL targeting of this epitope is associated with long-term nonprogression (42). On the other hand, responses targeting A*03-restricted QK10 (Nef amino acids 73 to 82) and B*08-restricted FL8 (Nef amino acids 90 to 97) resulted in escape and disease progression (39, 40). However, Nef-specific CTL responses with other MHC class I restrictions have also been associated with control of viremia, such as targeting of the A*02-restricted LV10 epitope (Nef amino acids 137 to 146) in the STEP vaccine trial (41). Notably, the epitopes associated with control of viremia (HW9 and LV10) cover critical amino acid residues, D123 and G140, essential for multiple Nef functions (26, 43); mutation at position 140 in Nef results in a significant decrease in the replication capacity in primary CD4+ T lymphocytes (our unpublished data). Ueno and colleagues demonstrated the balance between escape mutations and functional conservation by showing that mutations in the functionally important proline-rich region leading to CTL escape resulted in the impairment of multiple functions (44, 45). Thus, the different results of in vivo correlations of viremia with Nef-specific CTLs can be reconciled by MHC class I-determined targeting in relation to key functional domains of Nef.
Infectivity and T cell activation are other key pathogenic functions of Nef (46, 47) that were not assessed in this study, and it is possible that CTLs did affect these or other Nef activities. The specific domains and amino acid residues associated with these functions are not as well defined as those associated with MHC class I and CD4 downregulation. The lack of association between the Nef-specific CTL responses and control of viremia in our cohort suggested that CTL targeting did not result in significant effects on the role of Nef in viral fitness.
The significant relationship observed between Env-specific CTLs and reductions in viremia from peak to set point in this cohort has not been seen in other studies. The small sample size and the lack of corroborating reports make it difficult to determine whether this finding has broad significance. Examination of the details of the Env-specific responses in this cohort revealed some unusual characteristics that suggest that the association is specific to this small cohort. Eight of the 23 responses that were mapped with individual peptides targeted gp120 (35%), while 15 targeted gp41 (65%). Seventeen responses (74%) targeted regions with known functional or structural significance (signal peptide, leucine zipper, fusion peptide, cysteine bridge, transmembrane domain), and only one targeted a variable loop. There was a predominance of HLA-C-restricted epitopes targeted (8 of 23 [35%]). Specifically, 3 of 6 subjects targeted the HLA-C*07-restricted gp41 epitope FT20 (amino acids 519 to 538), which targets the fusion peptide. Additionally 4 of 6 subjects targeted the HLA-C*07-restricted gp41 epitope YC13 (amino acids 586 to 598), which targets a leucine zipper motif. A fifth subject targeted the same leucine zipper motif with an HLA-B*14-restricted response. Two subjects targeted the signal peptide cleavage sequence. In contrast to the Nef-specific responses in this cohort, the Env-specific responses appeared to target more effectively critical functional and structural regions that may be less able to mutate so as to escape CTL selective pressure.
Given the data generated by studies of acutely infected individuals and of human and nonhuman primate vaccines, it seems clear that Nef-specific CTL responses do have the potential to significantly curtail and control viral replication if appropriately directed against important functional domains that potentiate viral persistence in vivo, such those for MHC class I downregulation. It is not clear how to achieve this goal with a vaccine, but there is growing evidence that vaccines should preferentially target highly conserved regions, which are likely to be important functional domains (5, 48, 49). The use of the whole Nef protein as an immunogen likely results in immunodominance patterns in the majority of persons that hit nonessential, mutation-tolerant sites. A combination of better information on critical functional domains appropriate for targeting (26) and better technology to generate specifically targeted responses across diverse human populations is needed to overcome this hurdle to an effective HIV-1 vaccine. Meanwhile, alternative approaches to blocking Nef, such as small-molecule inhibitors, or immunotherapies such as adoptive transfer or T cell receptor gene therapy may be useful.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the following grants: NIH/NIAID AI083083 (to M.J.L.), AI051970 and AI043203 (to O.O.Y.), and ULITR000124 (to E.S.D.).
We thank Diane Reed and Kiara Munir for assistance with sequence collection. We also thank our study participants, who generously donated time and blood for these studies.
Footnotes
Published ahead of print 30 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00482-14.
REFERENCES
- 1.Salazar-Gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, Keele BF, Derdeyn CA, Farmer P, Hunter E, Allen S, Manigart O, Mulenga J, Anderson JA, Swanstrom R, Haynes BF, Athreya GS, Korber BT, Sharp PM, Shaw GM, Hahn BH. 2008. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J. Virol. 82:3952–3970. 10.1128/JVI.02660-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abrahams MR, Treurnicht FK, Ngandu NK, Goodier SA, Marais JC, Bredell H, Thebus R, de Assis Rosa D, Mlisana K, Seoighe C, Karim SA, Gray CM, Williamson C. 2013. Rapid, complex adaptation of transmitted HIV-1 full-length genomes in subtype C-infected individuals with differing disease progression. AIDS 27:507–518. 10.1097/QAD.0b013e32835cab64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goonetilleke N, Liu MK, Salazar-Gonzalez JF, Ferrari G, Giorgi E, Ganusov VV, Keele BF, Learn GH, Turnbull EL, Salazar MG, Weinhold KJ, Moore S, Letvin N, Haynes BF, Cohen MS, Hraber P, Bhattacharya T, Borrow P, Perelson AS, Hahn BH, Shaw GM, Korber BT, McMichael AJ. 2009. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J. Exp. Med. 206:1253–1272. 10.1084/jem.20090365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kearney M, Maldarelli F, Shao W, Margolick JB, Daar ES, Mellors JW, Rao V, Coffin JM, Palmer S. 2009. Human immunodeficiency virus type 1 population genetics and adaptation in newly infected individuals. J. Virol. 83:2715–2727. 10.1128/JVI.01960-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mlotshwa M, Riou C, Chopera D, de Assis Rosa D, Ntale R, Treunicht F, Woodman Z, Werner L, van Loggerenberg F, Mlisana K, Abdool Karim S, Williamson C, Gray CM. 2010. Fluidity of HIV-1-specific T-cell responses during acute and early subtype C HIV-1 infection and associations with early disease progression. J. Virol. 84:12018–12029. 10.1128/JVI.01472-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Streeck H, Jolin JS, Qi Y, Yassine-Diab B, Johnson RC, Kwon DS, Addo MM, Brumme C, Routy JP, Little S, Jessen HK, Kelleher AD, Hecht FM, Sekaly RP, Rosenberg ES, Walker BD, Carrington M, Altfeld M. 2009. Human immunodeficiency virus type 1-specific CD8+ T-cell responses during primary infection are major determinants of the viral set point and loss of CD4+ T cells. J. Virol. 83:7641–7648. 10.1128/JVI.00182-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malhotra U, Li F, Nolin J, Allison M, Zhao H, Mullins JI, Self S, McElrath MJ. 2007. Enhanced detection of human immunodeficiency virus type 1 (HIV-1) Nef-specific T cells recognizing multiple variants in early HIV-1 infection. J. Virol. 81:5225–5237. 10.1128/JVI.02564-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Addo MM, Yu XG, Rathod A, Cohen D, Eldridge RL, Strick D, Johnston MN, Corcoran C, Wurcel AG, Fitzpatrick CA, Feeney ME, Rodriguez WR, Basgoz N, Draenert R, Stone DR, Brander C, Goulder PJ, Rosenberg ES, Altfeld M, Walker BD. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 77:2081–2092. 10.1128/JVI.77.3.2081-2092.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Betts MR, Ambrozak DR, Douek DC, Bonhoeffer S, Brenchley JM, Casazza JP, Koup RA, Picker LJ. 2001. Analysis of total human immunodeficiency virus (HIV)-specific CD4+ and CD8+ T-cell responses: relationship to viral load in untreated HIV infection. J. Virol. 75:11983–11991. 10.1128/JVI.75.24.11983-11991.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirchhoff F. 2010. Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe 8:55–67. 10.1016/j.chom.2010.06.004 [DOI] [PubMed] [Google Scholar]
- 11.Collins KL, Chen BK, Kalams SA, Walker BD, Baltimore D. 1998. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature 391:397–401. 10.1038/34929 [DOI] [PubMed] [Google Scholar]
- 12.Schwartz O, Marechal V, Le Gall S, Lemonnier F, Heard JM. 1996. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat. Med. 2:338–342. 10.1038/nm0396-338 [DOI] [PubMed] [Google Scholar]
- 13.Garcia JV, Miller AD. 1991. Serine phosphorylation-independent downregulation of cell-surface CD4 by nef. Nature 350:508–511. 10.1038/350508a0 [DOI] [PubMed] [Google Scholar]
- 14.Lama J, Mangasarian A, Trono D. 1999. Cell-surface expression of CD4 reduces HIV-1 infectivity by blocking Env incorporation in a Nef- and Vpu-inhibitable manner. Curr. Biol. 9:622–631. 10.1016/S0960-9822(99)80284-X [DOI] [PubMed] [Google Scholar]
- 15.Yang OO, Nguyen PT, Kalams SA, Dorfman T, Gottlinger HG, Stewart S, Chen IS, Threlkeld S, Walker BD. 2002. Nef-mediated resistance of human immunodeficiency virus type 1 to antiviral cytotoxic T lymphocytes. J. Virol. 76:1626–1631. 10.1128/JVI.76.4.1626-1631.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geffin R, Wolf D, Muller R, Hill MD, Stellwag E, Freitag M, Sass G, Scott GB, Baur AS. 2000. Functional and structural defects in HIV type 1 nef genes derived from pediatric long-term survivors. AIDS Res. Hum. Retroviruses 16:1855–1868. 10.1089/08892220050195810 [DOI] [PubMed] [Google Scholar]
- 17.Noviello CM, Pond SL, Lewis MJ, Richman DD, Pillai SK, Yang OO, Little SJ, Smith DM, Guatelli JC. 2007. Maintenance of Nef-mediated modulation of major histocompatibility complex class I and CD4 after sexual transmission of human immunodeficiency virus type 1. J. Virol. 81:4776–4786. 10.1128/JVI.01793-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carl S, Greenough TC, Krumbiegel M, Greenberg M, Skowronski J, Sullivan JL, Kirchhoff F. 2001. Modulation of different human immunodeficiency virus type 1 Nef functions during progression to AIDS. J. Virol. 75:3657–3665. 10.1128/JVI.75.8.3657-3665.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mwimanzi P, Markle TJ, Ueno T, Brockman MA. 2012. Human leukocyte antigen (HLA) class I down-regulation by human immunodeficiency virus type 1 negative factor (HIV-1 Nef): what might we learn from natural sequence variants? Viruses 4:1711–1730. 10.3390/v4091711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis MJ, Balamurugan A, Ohno A, Kilpatrick S, Ng HL, Yang OO. 2008. Functional adaptation of Nef to the immune milieu of HIV-1 infection in vivo. J. Immunol. 180:4075–4081. 10.4049/jimmunol.180.6.4075 [DOI] [PubMed] [Google Scholar]
- 21.Riou C, Burgers WA, Mlisana K, Koup RA, Roederer M, Abdool Karim SS, Williamson C, Gray CM. 2014. Differential impact of magnitude, polyfunctional capacity, and specificity of HIV-specific CD8+ T cell responses on HIV set point. J. Virol. 88:1819–1824. 10.1128/JVI.02968-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahashi N, Nomura T, Takahara Y, Yamamoto H, Shiino T, Takeda A, Inoue M, Iida A, Hara H, Shu T, Hasegawa M, Sakawaki H, Miura T, Igarashi T, Koyanagi Y, Naruse TK, Kimura A, Matano T. 2013. A novel protective MHC-I haplotype not associated with dominant Gag-specific CD8+ T-cell responses in SIVmac239 infection of Burmese rhesus macaques. PLoS One 8:e54300. 10.1371/journal.pone.0054300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwamoto N, Takahashi N, Seki S, Nomura T, Yamamoto H, Inoue M, Shu T, Naruse TK, Kimura A, Matano T. 2014. Control of simian immunodeficiency virus replication by vaccine-induced Gag- and Vif-specific CD8+ T cells. J. Virol. 88:425–433. 10.1128/JVI.02634-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mudd PA, Ericsen AJ, Burwitz BJ, Wilson NA, O'Connor DH, Hughes AL, Watkins DI. 2012. Escape from CD8+ T cell responses in Mamu-B*00801+ macaques differentiates progressors from elite controllers. J. Immunol. 188:3364–3370. 10.4049/jimmunol.1102470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fiebig EW, Wright DJ, Rawal BD, Garrett PE, Schumacher RT, Peddada L, Heldebrant C, Smith R, Conrad A, Kleinman SH, Busch MP. 2003. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS 17:1871–1879. 10.1097/00002030-200309050-00005 [DOI] [PubMed] [Google Scholar]
- 26.Lewis MJ, Lee P, Ng HL, Yang OO. 2012. Immune selection in vitro reveals human immunodeficiency virus type 1 Nef sequence motifs important for its immune evasion function in vivo. J. Virol. 86:7126–7135. 10.1128/JVI.00878-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jordan MR, Kearney M, Palmer S, Shao W, Maldarelli F, Coakley EP, Chappey C, Wanke C, Coffin JM. 2010. Comparison of standard PCR/cloning to single genome sequencing for analysis of HIV-1 populations. J. Virol. Methods 168:114–120. 10.1016/j.jviromet.2010.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ali A, Realegeno S, Yang OO, Lewis MJ. 2009. Simultaneous assessment of CD4 and MHC-I downregulation by Nef primary isolates in the context of infection. J. Virol. Methods 161:297–304. 10.1016/j.jviromet.2009.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Felsenstein J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164–166 [Google Scholar]
- 31.Shacklett BL, Yang O, Hausner MA, Elliott J, Hultin L, Price C, Fuerst M, Matud J, Hultin P, Cox C, Ibarrondo J, Wong JT, Nixon DF, Anton PA, Jamieson BD. 2003. Optimization of methods to assess human mucosal T-cell responses to HIV infection. J. Immunol. Methods 279:17–31. 10.1016/S0022-1759(03)00255-2 [DOI] [PubMed] [Google Scholar]
- 32.Wong JT, Colvin RB. 1991. Selective reduction and proliferation of the CD4+ and CD8+ T cell subsets with bispecific monoclonal antibodies: evidence for inter-T cell-mediated cytolysis. Clin. Immunol. Immunopathol. 58:236–250. 10.1016/0090-1229(91)90139-2 [DOI] [PubMed] [Google Scholar]
- 33.Akari H, Arold S, Fukumori T, Okazaki T, Strebel K, Adachi A. 2000. Nef-induced major histocompatibility complex class I down-regulation is functionally dissociated from its virion incorporation, enhancement of viral infectivity, and CD4 down-regulation. J. Virol. 74:2907–2912. 10.1128/JVI.74.6.2907-2912.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Craig HM, Pandori MW, Guatelli JC. 1998. Interaction of HIV-1 Nef with the cellular dileucine-based sorting pathway is required for CD4 down-regulation and optimal viral infectivity. Proc. Natl. Acad. Sci. U. S. A. 95:11229–11234. 10.1073/pnas.95.19.11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ali A, Jamieson BD, Yang OO. 2003. Half-genome human immunodeficiency virus type 1 constructs for rapid production of reporter viruses. J. Virol. Methods 110:137–142. 10.1016/S0166-0934(03)00110-1 [DOI] [PubMed] [Google Scholar]
- 36.Little SJ, Holte S, Routy JP, Daar ES, Markowitz M, Collier AC, Koup RA, Mellors JW, Connick E, Conway B, Kilby M, Wang L, Whitcomb JM, Hellmann NS, Richman DD. 2002. Antiretroviral-drug resistance among patients recently infected with HIV. N. Engl. J. Med. 347:385–394. 10.1056/NEJMoa013552 [DOI] [PubMed] [Google Scholar]
- 37.Herbeck JT, Rolland M, Liu Y, McLaughlin S, McNevin J, Zhao H, Wong K, Stoddard JN, Raugi D, Sorensen S, Genowati I, Birditt B, McKay A, Diem K, Maust BS, Deng W, Collier AC, Stekler JD, McElrath MJ, Mullins JI. 2011. Demographic processes affect HIV-1 evolution in primary infection before the onset of selective processes. J. Virol. 85:7523–7534. 10.1128/JVI.02697-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turk G, Ghiglione Y, Falivene J, Socias ME, Laufer N, Coloccini RS, Rodriguez AM, Ruiz MJ, Pando MA, Giavedoni LD, Cahn P, Sued O, Salomon H, Gherardi MM. 2013. Early Gag immunodominance of the HIV-specific T-cell response during acute/early infection is associated with higher CD8+ T-cell antiviral activity and correlates with preservation of the CD4+ T-cell compartment. J. Virol. 87:7445–7462. 10.1128/JVI.00865-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price DA, Goulder PJ, Klenerman P, Sewell AK, Easterbrook PJ, Troop M, Bangham CR, Phillips RE. 1997. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc. Natl. Acad. Sci. U. S. A. 94:1890–1895. 10.1073/pnas.94.5.1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koenig S, Conley AJ, Brewah YA, Jones GM, Leath S, Boots LJ, Davey V, Pantaleo G, Demarest JF, Carter C, Wannebo C, Yannelli JR, Rosenberg SA, Lane HC. 1995. Transfer of HIV-1-specific cytotoxic T lymphocytes to an AIDS patient leads to selection for mutant HIV variants and subsequent disease progression. Nat. Med. 1:330–336. 10.1038/nm0495-330 [DOI] [PubMed] [Google Scholar]
- 41.Brumme ZL, John M, Carlson JM, Brumme CJ, Chan D, Brockman MA, Swenson LC, Tao I, Szeto S, Rosato P, Sela J, Kadie CM, Frahm N, Brander C, Haas DW, Riddler SA, Haubrich R, Walker BD, Harrigan PR, Heckerman D, Mallal S. 2009. HLA-associated immune escape pathways in HIV-1 subtype B Gag, Pol and Nef proteins. PLoS One 4:e6687. 10.1371/journal.pone.0006687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Navis M, Schellens IM, van Swieten P, Borghans JA, Miedema F, Kootstra NA, van Baarle D, Schuitemaker H. 2008. A nonprogressive clinical course in HIV-infected individuals expressing human leukocyte antigen B57/5801 is associated with preserved CD8+ T lymphocyte responsiveness to the HW9 epitope in Nef. J. Infect. Dis. 197:871–879. 10.1086/528695 [DOI] [PubMed] [Google Scholar]
- 43.Liu LX, Heveker N, Fackler OT, Arold S, Le Gall S, Janvier K, Peterlin BM, Dumas C, Schwartz O, Benichou S, Benarous R. 2000. Mutation of a conserved residue (D123) required for oligomerization of human immunodeficiency virus type 1 Nef protein abolishes interaction with human thioesterase and results in impairment of Nef biological functions. J. Virol. 74:5310–5319. 10.1128/JVI.74.11.5310-5319.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ueno T, Motozono C, Dohki S, Mwimanzi P, Rauch S, Fackler OT, Oka S, Takiguchi M. 2008. CTL-mediated selective pressure influences dynamic evolution and pathogenic functions of HIV-1 Nef. J. Immunol. 180:1107–1116. 10.4049/jimmunol.180.2.1107 [DOI] [PubMed] [Google Scholar]
- 45.Mwimanzi P, Hasan Z, Tokunaga M, Gatanaga H, Oka S, Ueno T. 2010. Naturally arising HIV-1 Nef variants conferring escape from cytotoxic T lymphocytes influence viral entry co-receptor expression and susceptibility to superinfection. Biochem. Biophys. Res. Commun. 403:422–427. 10.1016/j.bbrc.2010.11.047 [DOI] [PubMed] [Google Scholar]
- 47.Münch J, Rajan D, Schindler M, Specht A, Rucker E, Novembre FJ, Nerrienet E, Muller-Trutwin MC, Peeters M, Hahn BH, Kirchhoff F. 2007. Nef-mediated enhancement of virion infectivity and stimulation of viral replication are fundamental properties of primate lentiviruses. J. Virol. 81:13852–13864. 10.1128/JVI.00904-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang OO, Daar ES, Ng HL, Shih R, Jamieson BD. 2011. Increasing CTL targeting of conserved sequences during early HIV-1 infection is correlated to decreasing viremia. AIDS Res. Hum. Retroviruses 27:391–398. 10.1089/aid.2010.0183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kulkarni V, Valentin A, Rosati M, Alicea C, Singh AK, Jalah R, Broderick KE, Sardesai NY, Le Gall S, Mothe B, Brander C, Rolland M, Mullins JI, Pavlakis GN, Felber BK. 2014. Altered response hierarchy and increased T-cell breadth upon HIV-1 conserved element DNA vaccination in macaques. PLoS One 9:e86254. 10.1371/journal.pone.0086254 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.