Abstract
Human olfactory cells obtained by rapid nasal biopsy have been suggested to be a good surrogate system to address brain disease-associated molecular changes. Nonetheless, whether use of this experimental strategy is justified remains unclear. Here we compared expression profiles of olfactory cells systematically with those from the brain tissues and other cells. Principal component analysis indicated that the expression profiles of olfactory cells are very different from those of blood cells, but are closer to those of stem cells, in particular mesenchymal stem cells, that can be differentiated into the cells of the central nervous system.
Keywords: Olfactory cells, lymphoblasts, stem cells, gene expression profile, schizophrenia, bipolar disorder, mental disorder, neurological disorder
Comparison of expression and epigenetic profiles from biospecimens between patients and controls is currently one of the major strategies to elucidate disease mechanisms and to identify biomarkers (Cooper-Knock et al., 2012). A major barrier in applying this strategy for brain disorders is the difficulty in accessing cells and tissues relevant to the disease pathophysiology (Dolmetsch and Geschwind, 2011). Olfactory tissue obtained by nasal biopsy has been suggested as a surrogate biospecimen for the brain (Borgmann-Winter et al., 2009; Cascella et al., 2007; Feron et al., 1995; Feron et al., 1998; Girard et al., 2011; Gomez et al., 2000; Hahn et al., 2005; Mackay-Sim et al., 2008; Murrell et al., 2005; Sawa and Cascella, 2009; Talamo et al., 1989; Trojanowski et al., 1991). Nonetheless, the validity and limitations of using olfactory tissues have not been systematically addressed. In this study, we aim to address this question by comparing expression profiles among olfactory cells, blood cells, stem cells, and brain tissues.
Lymphoblasts were generated according to a published protocol (Bader et al., 2012; Penno, 1993). Dissociated olfactory cells were purified from the olfactory epithelium, which was obtained via rapid nasal biopsy under local anesthesia, as previously described (Kano et al., 2013; Tajinda et al., 2010). Induced pluripotent stem cells and the neurospheres originated from stem cells were generated according to a published protocol (Imaizumi et al., 2012). The experimental details of cells in this study are summarized in Supplementary Table. This study was approved by Institutional Review Boards at Johns Hopkins University (NA00037204) and Keio University (20-16-18).
We conducted expression profile analysis using Affymetrix U133 Plus 2.0 GeneChip® with standard protocols (Affimetrix), as published (Mao et al., 2005). These transcripts corresponded to 22,500 human transcript sets. Raw data from U133A Gene-Chips were normalized by using robust multi-array average (RMA) normalization (R version2.1.0) from BioConductor, or GC-content correction RMA (GC-RMA) using Partek® Genome Suite version 6.0 software. The results using either GC-RMA or RMA were very similar (data not shown), and we used GC-RMA because of its ability to estimate probe affinity to non-specific binding. In addition to the in-house expression data with biospecimens from healthy subjects, the expression profiles of adult and fetal brain tissues, lymphocytes, lymphoblasts, induced pluripotent stem cells, embryonic stem cells, and mesenchymal stem cells in healthy controls were imported from Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/). We also compared the profiles of lymphoblasts and induced pluripotent stem cells between in-house data and public databases, and confirmed the compatibility between these two. In the overall analysis, we integrated all the data together.
Exploratory analyses by use of principal component analysis (PCA) was performed with Partek® software (http://www.partek.com). For PCA, we used the covariance dispersion matrix option. PCA allows visualization of highly dimensional data along principal component axes. These axes reflect the degree of variance in the data allowing the identification of groups of data having possible biological relevance (Kondo et al., 2013; Mao et al., 2005). To test which biological functions contribute these distributions, we applied Gene Ontology Consortium functional analysis (www.geneontology.org) on the TIBCO Spotfire DecisionSite analytic platform, according to the established protocol (Tavazoie et al., 1999).
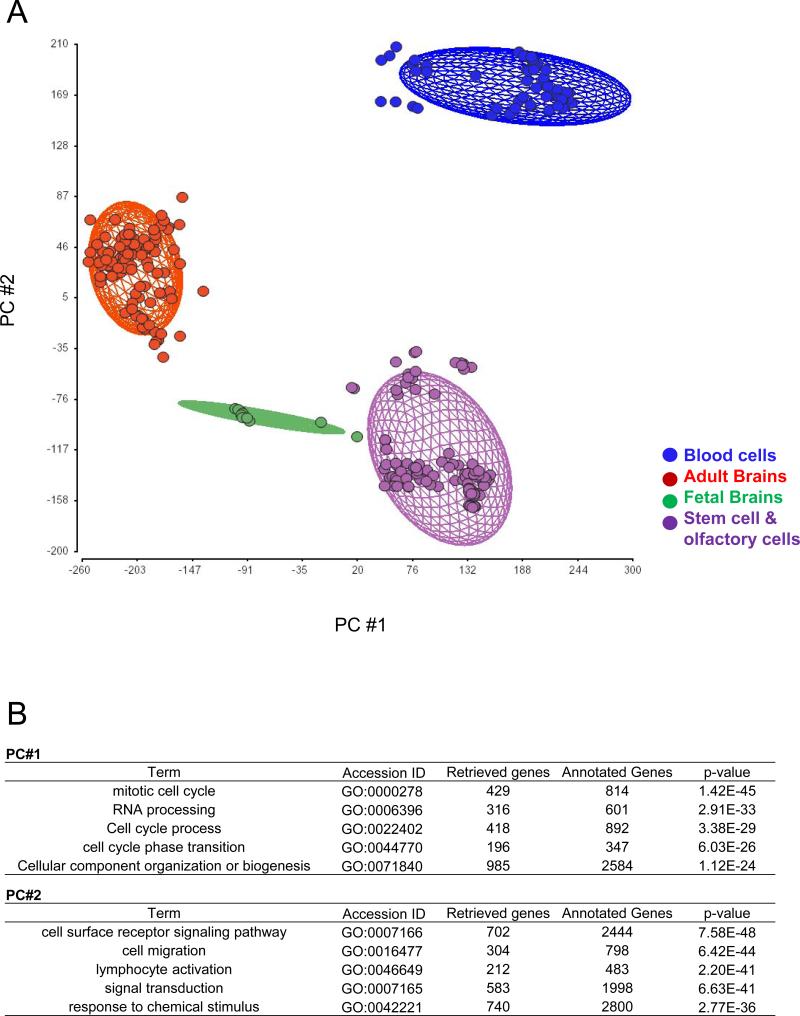
First, we examined the general landscape of expression profiles among entire tissues and cells in this study (Fig. 1A). We obtained similar results using hierarchical clustering (data not shown). According to the PCA, the samples were separated into four groups; adult brain tissue group (red), fetal brain tissue group (green), blood cell group (blue) consisting of lymphoblasts and lymphocyte cells, and stem cell group (purple) that comprise embryonic and induced pluripotent stem cells, neurosphere and neural progenitor cells derived from these stem cells, and mesenchymal stem cells. Interestingly, olfactory cells belong to the last stem cell group. Compared to the blood cells, the expression profiles of the stem cell group were similar to those of the adult and fetal brain groups. The Gene Ontology biological processes contributing to these differential distributions are shown in Fig. 1B. We also performed Analysis of Covariance (ANCOVA) (data not shown). We acknowledge that milder effects of age (fetal versus adult) and scan date, as is typical of microarray studies, existed in addition to the major effects by cell type (Supplementary Fig.).
Fig. 1.
A) Sample distribution among various tissues obtained by principal component analysis (PCA) in Partek Genomics Suite using a data set of gene expression from 279 experiments. The ellipsoids represent two standard deviations beyond the mean of the centroid of each tissue group. Data points correspond to samples (red, adult brains; green, fetal brains; blue, blood cells; purple, stem cells and olfactory cells). B) Representative biological processes that contribute to the distribution of PC#1 and PC#2 (p <1.0E-25) through gene ontology assessment are depicted.
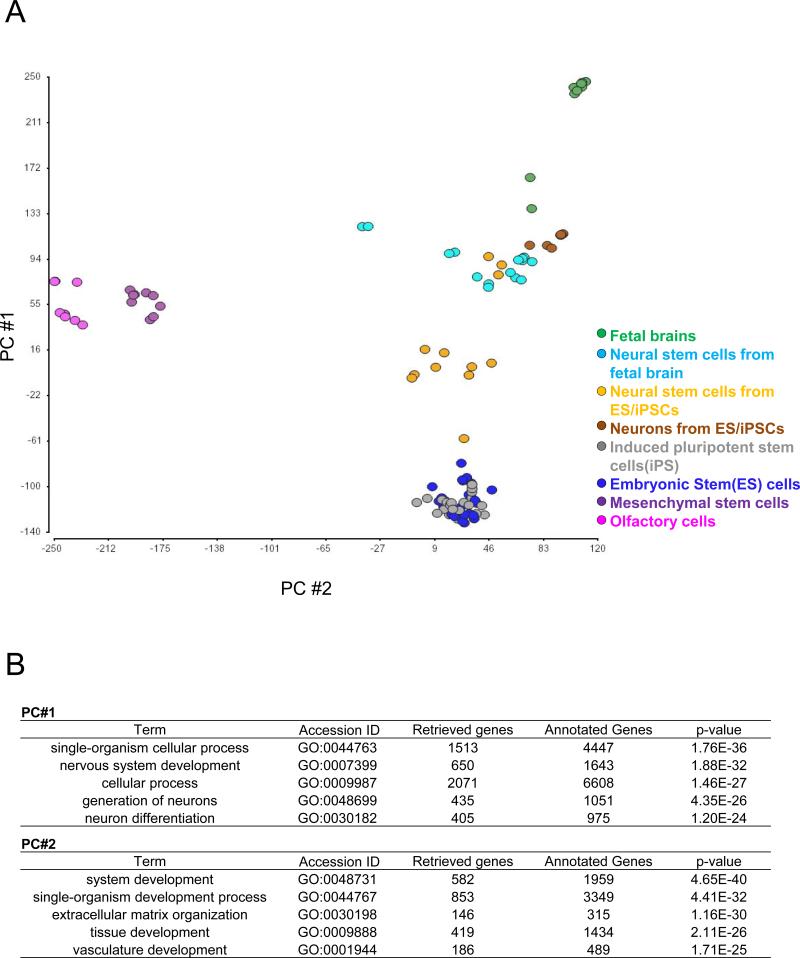
Next, we further characterized the expression profiles inside the stem cell group (depicted in purple in Fig. 1.) (Fig. 2). We observed that expression profiles of the embryonic and induced pluripotent stem cells-derived neurospheres are close to those of the fetal brain-derived neurospheres. These neurospheres were plotted between the original stem cells and the fetal brain tissues in the PCA, reflecting their developmental trajectory. In contrast, the expression profiles of olfactory cells and mesenchymal stem cells were similar with each other, but different from those of embryonic and induced pluripotent stem cells. The processes contributing to these differential distributions are shown in Fig. 2B.
Fig. 2.
A) Results of principal component analysis (PCA) from stem cell group samples. Data points correspond to samples (green, fetal brains; light blue, neural stem cells from fetal brains; orange, neural stem cells from embryonic and induced pluripotent stem cells; blue, embryonic stem cells; gray, induced pluripotent stem cells; purple, mesenchymal stem cells; and pink, olfactory cells).
B) Representative biological processes that contribute to the distribution of PC#1 and PC#2 (p <1.0E-20) through gene ontology assessment are depicted.
The main finding of the present study is the demonstration that expression profiles of olfactory cells are similar to those of stem cells, differentiated neurons from these cells, and the brain tissue, which is in contrast to those from peripheral lymphoblasts. Interestingly, a closer look at expression profiles indicated more similarity of olfactory cells to mesenchymal stem cells than to embryonic and induced pluripotent stem cells. These observations are consistent with a recent report that miR382 expression is decreased in olfactory cells from patients with schizophrenia compared with those from normal controls (Mor et al., 2013), which is consistent with a decrease in the microRNA in the patient brains (Wang et al., 2011). Even more importantly, the expression of miR382 was below detectable levels in lymphoblasts (Mor et al., 2013). These results highlight an advantage of using olfactory cells to study brain-relevant and disease-associated molecular changes. In contrast to autopsied brains, olfactory cells can be used for functional assays: for example, it may be very important to test whether and how disease-associated target molecules are dynamically changed upon environmental stress.
The data in the present study show that the molecular profiles of olfactory cells are similar to those of mesenchymal stem cells. It is well known that mesenchymal stem cells can be good resources for generating mature neurons and glia (Neirinckx et al., 2013; Scuteri et al., 2011). As far as we are aware, no group has demonstrated the generation of mature neurons with action potential and mature cells of other glial lineage from human olfactory cells. However, the molecular profiles shown here can support future endeavors to generate these mature cells from olfactory cells obtained by nasal biopsy. Currently, induced pluripotent stem cells are considered to provide a powerful strategy to obtain and characterize central nervous system-relevant cells in vitro (Grskovic et al., 2011; Ito et al., 2012). However, this experimental strategy is still very costly, time-consuming, and laborious. Furthermore, even if a methodology that excludes the integration of exogenous factors has been established, artifacts elicited by robust reprogramming cannot be fully eliminated. Thus, olfactory cells may provide an alternative opportunity to obtain central nervous system-relevant cells without introducing exogenous factors and reprogramming to the stem cell stage.
Recent studies using olfactory cells and tissues have been reported to address molecular changes associated with mental illnesses, such as schizophrenia (Fan et al., 2013; Kano et al., 2013; Mor et al., 2013; Toritsuka et al., in press). The next question may be whether and how olfactory cells and nasal biopsy can be used more easily in clinical settings, including rapid diagnostic procedures, rather than remaining as methods for exploring biomarkers. A recent brushing technique to obtain olfactory tissues may expand this possibility (Benitez-King et al., 2011; Levy et al., 2004; Ono et al., 2012). Given the high potential of olfactory tissues to detect brain-associated molecular changes, it will be important to develop means for safer and easier acquisition.
Supplementary Material
Highlights.
Expression patterns of olfactory cells are similar to developing brain cells.
Blood cells are different from brain cells in regard to expression patterns.
Olfactory cells can be used a peripherally accessible surrogate system.
Acknowledgments
We thank Dr. Pamela Talalay for critical reading of this manuscript. We appreciate Yukiko Y. Lema for organizing the figures and manuscript. This work was supported by USPHS grants MH-084018 (A.S.), MH-094268 Silvo O. Conte center (A.S.), MH-069853 (A.S.), MH-085226 (A.S.), MH-088753 (A.S.), MH-092443 (A.S.), MH-96208 (K.I.) and K99MH-093458 (S-I.K.), grants from Stanley (A.S.), RUSK (A.S.), S-R foundations (A.S.), NARSAD (A.S., S-I.K., and K.I.), the Japanese Ministry of Education, Culture, Sports, Science and Technology (H.O.) and Maryland Stem Cell Research Fund (A.S., Y.H., and K.I.). S-I.K. is also supported by the Hammerschlag family (S-I.K)
Abbreviations
- PCA
principal component analysis
- RMA
robust multi-array average
- GC-RMA
GC-content correction robust multi-array average
- ANCOVA
analysis of covariance
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Figure Legend
Sources of variation in gene expression data from ANCOVA. We performed two-way ANCOVA and observed a mean F ratio with a major contribution from tissue/cell type, with less influence of scan date and age.
Reference
- Bader V, Tomppo L, Trossbach SV, Bradshaw NJ, Prikulis I, Leliveld SR, Lin CY, Ishizuka K, Sawa A, Ramos A, Rosa I, Garcia A, Requena JR, Hipolito M, Rai N, Nwulia E, Henning U, Ferrea S, Luckhaus C, Ekelund J, Veijola J, Jarvelin MR, Hennah W, Korth C. Proteomic, genomic and translational approaches identify CRMP1 for a role in schizophrenia and its underlying traits. Hum Mol Genet. 2012;21:4406–4418. doi: 10.1093/hmg/dds273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez-King G, Riquelme A, Ortiz-Lopez L, Berlanga C, Rodriguez-Verdugo MS, Romo F, Calixto E, Solis-Chagoyan H, Jimenez M, Montano LM, Ramirez-Rodriguez G, Morales-Mulia S, Dominguez-Alonso A. A non-invasive method to isolate the neuronal linage from the nasal epithelium from schizophrenic and bipolar diseases. J Neurosci Methods. 2011;201:35–45. doi: 10.1016/j.jneumeth.2011.07.009. [DOI] [PubMed] [Google Scholar]
- Borgmann-Winter KE, Rawson NE, Wang HY, Wang H, Macdonald ML, Ozdener MH, Yee KK, Gomez G, Xu J, Bryant B, Adamek G, Mirza N, Pribitkin E, Hahn CG. Human olfactory epithelial cells generated in vitro express diverse neuronal characteristics. Neuroscience. 2009;158:642–653. doi: 10.1016/j.neuroscience.2008.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascella NG, Takaki M, Lin S, Sawa A. Neurodevelopmental involvement in schizophrenia: the olfactory epithelium as an alternative model for research. J Neurochem. 2007;102:587–594. doi: 10.1111/j.1471-4159.2007.04628.x. [DOI] [PubMed] [Google Scholar]
- Cooper-Knock J, Kirby J, Ferraiuolo L, Heath PR, Rattray M, Shaw PJ. Gene expression profiling in human neurodegenerative disease. Nat Rev Neurol. 2012;8:518–530. doi: 10.1038/nrneurol.2012.156. [DOI] [PubMed] [Google Scholar]
- Dolmetsch R, Geschwind DH. The human brain in a dish: the promise of iPSC-derived neurons. Cell. 2011;145:831–834. doi: 10.1016/j.cell.2011.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Abrahamsen G, Mills R, Calderon CC, Tee JY, Leyton L, Murrell W, Cooper-White J, McGrath JJ, Mackay-Sim A. Focal Adhesion Dynamics Are Altered in Schizophrenia. Biol Psychiatry. 2013 doi: 10.1016/j.biopsych.2013.01.020. [DOI] [PubMed] [Google Scholar]
- Feron F, Lizard G, Sicard G. Isolation of mature olfactory neurones using retrograde labelling and flow cytometry. J Neurosci Methods. 1995;57:9–14. doi: 10.1016/0165-0270(94)00105-p. [DOI] [PubMed] [Google Scholar]
- Feron F, Perry C, McGrath JJ, Mackay-Sim A. New techniques for biopsy and culture of human olfactory epithelial neurons. Arch Otolaryngol Head Neck Surg. 1998;124:861–866. doi: 10.1001/archotol.124.8.861. [DOI] [PubMed] [Google Scholar]
- Girard SD, Deveze A, Nivet E, Gepner B, Roman FS, Feron F. Isolating nasal olfactory stem cells from rodents or humans. J Vis Exp. 2011 doi: 10.3791/2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez G, Rawson NE, Hahn CG, Michaels R, Restrepo D. Characteristics of odorant elicited calcium changes in cultured human olfactory neurons. J Neurosci Res. 2000;62:737–749. doi: 10.1002/1097-4547(20001201)62:5<737::AID-JNR14>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Grskovic M, Javaherian A, Strulovici B, Daley GQ. Induced pluripotent stem cells--opportunities for disease modelling and drug discovery. Nat Rev Drug Discov. 2011;10:915–929. doi: 10.1038/nrd3577. [DOI] [PubMed] [Google Scholar]
- Hahn CG, Gomez G, Restrepo D, Friedman E, Josiassen R, Pribitkin EA, Lowry LD, Gallop RJ, Rawson NE. Aberrant intracellular calcium signaling in olfactory neurons from patients with bipolar disorder. Am J Psychiatry. 2005;162:616–618. doi: 10.1176/appi.ajp.162.3.616. [DOI] [PubMed] [Google Scholar]
- Imaizumi Y, Okada Y, Akamatsu W, Koike M, Kuzumaki N, Hayakawa H, Nihira T, Kobayashi T, Ohyama M, Sato S, Takanashi M, Funayama M, Hirayama A, Soga T, Hishiki T, Suematsu M, Yagi T, Ito D, Kosakai A, Hayashi K, Shouji M, Nakanishi A, Suzuki N, Mizuno Y, Mizushima N, Amagai M, Uchiyama Y, Mochizuki H, Hattori N, Okano H. Mitochondrial dysfunction associated with increased oxidative stress and alpha-synuclein accumulation in PARK2 iPSC-derived neurons and postmortem brain tissue. Mol Brain. 2012;5:35. doi: 10.1186/1756-6606-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito D, Okano H, Suzuki N. Accelerating progress in induced pluripotent stem cell research for neurological diseases. Ann Neurol. 2012;72:167–174. doi: 10.1002/ana.23596. [DOI] [PubMed] [Google Scholar]
- Kano S, Colantuoni C, Han F, Zhou Z, Yuan Q, Wilson A, Takayanagi Y, Lee Y, Rapoport J, Eaton W, Cascella N, Ji H, Goldman D, Sawa A. Genome-wide profiling of multiple histone methylations in olfactory cells: further implications for cellular susceptibility to oxidative stress in schizophrenia. Mol Psychiatry. 2013;18:740–742. doi: 10.1038/mp.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo MA, Tajinda K, Colantuoni C, Hiyama H, Seshadri S, Huang B, Pou S, Furukori K, Hookway C, Jaaro-Peled H, Kano SI, Matsuoka N, Harada K, Ni K, Pevsner J, Sawa A. Unique pharmacological actions of atypical neuroleptic quetiapine: possible role in cell cycle/fate control. Transl Psychiatry. 2013;3:e243. doi: 10.1038/tp.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy YS, Stroomza M, Melamed E, Offen D. Embryonic and adult stem cells as a source for cell therapy in Parkinson's disease. J Mol Neurosci. 2004;24:353–386. doi: 10.1385/JMN:24:3:353. [DOI] [PubMed] [Google Scholar]
- Mackay-Sim A, Feron F, Cochrane J, Bassingthwaighte L, Bayliss C, Davies W, Fronek P, Gray C, Kerr G, Licina P, Nowitzke A, Perry C, Silburn PA, Urquhart S, Geraghty T. Autologous olfactory ensheathing cell transplantation in human paraplegia: a 3-year clinical trial. Brain. 2008;131:2376–2386. doi: 10.1093/brain/awn173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao R, Wang X, Spitznagel EL, Jr., Frelin LP, Ting JC, Ding H, Kim JW, Ruczinski I, Downey TJ, Pevsner J. Primary and secondary transcriptional effects in the developing human Down syndrome brain and heart. Genome Biol. 2005;6:R107. doi: 10.1186/gb-2005-6-13-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor E, Kano S, Colantuoni C, Sawa A, Navon R, Shomron N. MicroRNA-382 expression is elevated in the olfactory neuroepithelium of schizophrenia patients. Neurobiol Dis. 2013;55:1–10. doi: 10.1016/j.nbd.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell W, Feron F, Wetzig A, Cameron N, Splatt K, Bellette B, Bianco J, Perry C, Lee G, Mackay-Sim A. Multipotent stem cells from adult olfactory mucosa. Dev Dyn. 2005;233:496–515. doi: 10.1002/dvdy.20360. [DOI] [PubMed] [Google Scholar]
- Neirinckx V, Coste C, Rogister B, Wislet-Gendebien S. Concise review: adult mesenchymal stem cells, adult neural crest stem cells, and therapy of neurological pathologies: a state of play. Stem Cells Transl Med. 2013;2:284–296. doi: 10.5966/sctm.2012-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M, Hamada Y, Horiuchi Y, Matsuo-Takasaki M, Imoto Y, Satomi K, Arinami T, Hasegawa M, Fujioka T, Nakamura Y, Noguchi E. Generation of induced pluripotent stem cells from human nasal epithelial cells using a Sendai virus vector. PLoS One. 2012;7:e42855. doi: 10.1371/journal.pone.0042855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penno MB, Pedrotti-Krueger M, Ray T. Cryopreservation of whole blood and isolated lymphocytes for B-call immortalization. J Tiss Culture Meth. 1993;15:43–48. [Google Scholar]
- Sawa A, Cascella NG. Peripheral olfactory system for clinical and basic psychiatry: a promising entry point to the mystery of brain mechanism and biomarker identification in schizophrenia. Am J Psychiatry. 2009;166:137–139. doi: 10.1176/appi.ajp.2008.08111702. [DOI] [PubMed] [Google Scholar]
- Scuteri A, Miloso M, Foudah D, Orciani M, Cavaletti G, Tredici G. Mesenchymal stem cells neuronal differentiation ability: a real perspective for nervous system repair? Curr Stem Cell Res Ther. 2011;6:82–92. doi: 10.2174/157488811795495486. [DOI] [PubMed] [Google Scholar]
- Tajinda K, Ishizuka K, Colantuoni C, Morita M, Winicki J, Le C, Lin S, Schretlen D, Sawa A, Cascella NG. Neuronal biomarkers from patients with mental illnesses: a novel method through nasal biopsy combined with laser-captured microdissection. Mol Psychiatry. 2010;15:231–232. doi: 10.1038/mp.2009.73. [DOI] [PubMed] [Google Scholar]
- Talamo BR, Rudel R, Kosik KS, Lee VM, Neff S, Adelman L, Kauer JS. Pathological changes in olfactory neurons in patients with Alzheimer's disease. Nature. 1989;337:736–739. doi: 10.1038/337736a0. [DOI] [PubMed] [Google Scholar]
- Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, Church GM. Systematic determination of genetic network architecture. Nat Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- Toritsuka M, Kimoto, Landek-Salgado AM, Yoshida A, Yamamoto N, Horiuchi Y, Hiyama H, Tajinda K, Kenie N, Illingworthf E, Iwamoto T, Kishimoto T, Sawa A, Tanigaki K. Deficits in microRNA-mediated control of Cxcr4/Cxcl12 signaling underlie developmental abnormalities in a 22q11-deletion syndrome mouse model. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1312661110. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojanowski JQ, Newman PD, Hill WD, Lee VM. Human olfactory epithelium in normal aging, Alzheimer's disease, and other neurodegenerative disorders. J Comp Neurol. 1991;310:365–376. doi: 10.1002/cne.903100307. [DOI] [PubMed] [Google Scholar]
- Wang WX, Huang Q, Hu Y, Stromberg AJ, Nelson PT. Patterns of microRNA expression in normal and early Alzheimer's disease human temporal cortex: white matter versus gray matter. Acta Neuropathol. 2011;121:193–205. doi: 10.1007/s00401-010-0756-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.