Abstract
The mature T cell repertoire has the ability to orchestrate immunity to a wide range of potential pathogen challenges. This ability stems from thymic development producing individual T cell clonotypes that express T cell receptors (TCR) with unique patterns of antigen reactivity. The antigen specificity of TCRs is created from the combinatorial pairing of one of a set of germline encoded TCR variable (V) alpha and V beta gene segments with randomly created CDR3 sequences. How the amalgamation of germline encoded and randomly created TCR sequences results in antigen receptors with unique patterns of ligand specificity is not fully understood. Using cellular, biophysical and structural analyses, we show that CDR3α residues can modulate the geometry in which TCRs bind pMHC, governing whether and how germline encoded TCR Vα and Vβ residues interact with MHC. In addition, a CDR1α residue that is positioned distal to the TCR-pMHC binding interface is shown to contribute to the peptide specificity of T cells. These findings demonstrate that the specificity of individual T cell clonotypes arises not only from TCR residues which create direct contacts with the pMHC, but also from a collection of indirect effects which modulate how TCR residues are used to bind pMHC.
Introduction
The ability of a T cell repertoire to target the array of potential pathogen challenges stems from individual T cell clonotypes having unique peptide and host-MHC reactivity patterns (1). Thymocytes are equipped with TCRs during development within the thymus. The sequence of individual TCR clonotypes is created from the pairing of one of a limited set of Vα and Vβ gene segments with highly variable CDR3α and CDR3β sequences derived from V(D)J recombination (2). Following the expression of a complete TCR the process of T cell selection then determine the fate of developing thymocytes. Thymocytes expressing TCRs that have weak affinity for self-pMHC complexes, and thus are capable of recognizing ligands presented by host-MHC proteins, undergo positive selection and are exported as T cells to the mature repertoire (3–7). However, thymocytes are eliminated during development if they express overtly self-reactive TCRs or TCRs that are unable to bind self-peptide presented by host-MHC with even weak affinity (8, 9). Through the processes of thymic selection, a repertoire of mature T cells is created that express diverse TCR sequences, endowing the T cell repertoire with a vast breadth of antigen specificities (10, 11).
How the merging of germline encoded and randomly created sequences produces TCRs that recognize antigen presented on host-MHC molecules has not been fully defined. Prior to T cell selection, 5–20% of thymocytes expressing randomly generated TCRs react with cells presenting MHC molecules (12–14). These findings demonstrate that TCR-bearing pre-selection thymocytes are biased towards recognizing self-peptides presented by MHC ligands. Though this MHC-bias is undoubtedly helpful in creating a mature T cell repertoire that is reactive to cells presenting pMHC, the underlying mechanisms that drive pre-selection T cells to recognize self-pMHC, and how this repertoire is shaped into a foreign-antigen specific mature T cell repertoire remains controversial (15–18).
Through studying T cells isolated from mice with limited negative selection, we have provided evidence that T cells can have a range of pMHC cross-reactivity patterns (19–21). These and other T cell activation studies suggest that TCRs may have an intrinsic ability to bind pMHC, which is regulated by TCR V gene pairing or CDR3 sequences (12–14, 22–24). Structural studies have also been used to unravel how TCRs create specificity for pMHC complexes. Most TCRs bind MHC ligands within a semi-conserved diagonal orientation, which largely places the CDR3 loops atop the bound peptide and the germline encoded V gene CDR1 and CDR2 residues positioned over the MHC alpha helices (25). Examination of TCRs carrying similar TCR V genes engaged to similar MHC alleles have shown a more limited range of TCR-pMHC docking angles (26, 27). These structural observations have led to a hypothesis that particular germline encoded residues of TCR V genes have been evolutionarily selected to bind MHC in conserved ways and provide TCRs with a built-in specificity for MHC ligands (28, 29).
In contrast to the hypothesis that TCR V genes have evolved to specifically bind MHC in conserved ways, other experiments have suggested that T cell signaling may regulate the ligand specificity of TCRs (30–32). For example, a recent study of T cells that develop in mice devoid of MHC ligands argues that CD4 and CD8 T cell co-receptor signaling has a critical role in selecting T cells that can recognize MHC ligands, and in eliminating T cells that recognize non-MHC ligands (30). Although T cell signaling during positive selection can ensure that mature T cells express TCRs with specificity for ligands presented by host-MHC, these models do not explain why a high-frequency of pre-selection TCRs are preordained to recognize self-pMHC complexes.
Several lines of evidence suggest that CDR3 sequences may strongly contribute to regulating how TCRs engage pMHC ligands. The CDR3 residues often contribute a large portion of the TCR binding site, with some TCRs engaging pMHC ligands with minimal contribution of CDR1 and CDR2 residues (18, 33, 34). Furthermore, MHC proteins are the most polymorphic genes in humans (35), and differential TCR V gene pairing creates TCRs that express different amino acid residues within CDR1 and CDR2 loops at common sites of contact with the MHC (2, 28). Perhaps as a way to accommodate this variability, a strict bifurcation of CDR1- and CDR2-MHC and CDR3-peptide residue contacts does not occur, with CDR3 residues contributing to MHC as well as peptide contacts and the V-gene encoded CDR1 and CDR2 contributing to both peptide and MHC contacts (18). Thus CDR3 sequences may play a primary role in creating both peptide and MHC specificity.
Studies exploring how TCRs generate specificity for pMHC ligands primarily have focused on identifying how direct contacts between the TCR and the pMHC contribute to the binding reaction. Here, we investigate a variety of indirect effects, whereby residues distal to the binding interface can have profound affects on MHC-TCR binding specificity. We demonstrate that CDR3α residues can affect the overall geometry in which a TCR engages a pMHC ligand, thereby indirectly affecting how different TCR Vα and TCR Vβ residues interact with pMHC residues. We further demonstrate that a CDR1 residue located outside of the TCR-pMHC binding interface can modulate the peptide specificity of T cells. Thus, in addition to providing direct pMHC contacts, TCR V gene and CDR3 residues create indirect effects that modulate TCR-peptide and TCR-MHC interactions.
Material and Methods
Mice and peptides
C57BL/6 mice were purchased from The Jackson Laboratory (Bar Harbor, ME). H-2Ab1−/− (MHC class II-deficient) mice were purchased from Taconic (Germantown, NY). Mice expressing the YAe62 TCRβ chain (21) were maintained in a pathogen-free environment in accordance with institutional guidelines in the Animal Care Facility at the University of Massachusetts Medical School. The 3K peptide is FEAQKAKANKAVD, numbered P-2 to P11; FMRKA is FEAFMARAKAAVD; QKKLK is FEAQKAKALKAVD; QTKRG is FEAQTAKARGAVD; RCKST is FEARCAKASTAVD; SKKRP is FEASKAKARPAVD; YTRRT is FEAYTARARTAVD.
Viral infection, generation of T cell hybridomas and transfectants, and TCR cloning
Mice were infected with 5 × 106 pfu Vac:IAb-3K. Responding CD4 T cells were isolated by positive selection using antibody affinity columns (Cederlane) and converted into hybridomas as previously described (36). 13.B1, 13.D5, 14.A6, 14.C6 and T cell hybridomas were constructed from MHCwt YAe TCRβ Tg mice (21). To clone TCR chains, RNA was isolated from T cell hybridomas, converted to cDNA, and PCR screened for rearranged TCRα chains using a set of degenerated PCR primers corresponding to all mouse Vα gene families. Identified rearranged TCR Vα genes were PCR cloned from the IAb-3K-specific hybridomas and fused to the TCR Cα region, using the following PCR primers:
Vα2 – GGGGGCTCGAGAGGAATGGACACGATCCTGACAGCA
Cα – CTGGTACACAGCAGGTTCCGGATTCTGGATGT
The J809.B5, J809.G3, J809.H1, 2W1S 20.4, YAe62 and 75–55 hybridomas and cloned TCRs were previously published (19, 21). To generate T cell hybridoma transfectants, TCRα or TCRβ chains were cloned into MSCV based vectors with an IRES element containing resistance genes for puromycin or neomycin, respectively. Retroviral vectors were cotransfected with the pCLEco accessory plasmid into the Phoenix packaging line using Lipofectamine 2000 (Life Technologies, Carlsbad, CA). The TCRα/β-deficient T cell hybridoma 5KC-73.8.20 was transduced with retroviruses expressing the TCR constructs using spinfection (1000 × g, 2hrs) (37). T cell hybridoma transfectants were selected in 1mg/mL neomycin or 0.8μg/mL puromycin for 1 week and stained with anti-TCRβ antibody (H57-597) to ensure equivalent levels of TCR expression.
T cell hybridoma activation assay
105 T cell hybridomas were incubated with 3 × 104 fibroblasts that express MHC, B7.1 and ICAM-1 for 24 hrs, and their supernatants were screened for IL-2 content using an HT-2 cell based bioassay (19). In some experiments, T cell hybridomas were incubated with MHC-expressing fibroblasts presenting titrating amounts of soluble peptides, or with 106 spleen cells expressing various MHC alleles. The IL-2 assays were performed three times and had a maximum value of IL-2 release of 1,000Units/mL. EC50 values were calculated by fitting to a log(agonist) vs response variable slope (four parameter) curve (GraphPad, Prism).
Production of soluble and membrane bound MHC-peptide and TCR proteins
Soluble and biotinylated pMHC and TCRs that were used in the MHC display library experiments were produced using the baculovirus expression system and purified as previously described (21, 38). Multivalent fluorescent TCRs were prepared as previously described (20). Briefly, biotinylated anti-Cα mAb ADO304 was complexed with AlexaFluor647-streptavidin. The complex was purified by size exclusion chromatography using a superdex200 column. Fluorescently labeled ADO304 was mixed with a concentration of soluble TCRs that saturated the antibody binding sites. The membrane bound IAb-3K variant baculoviruses have been previously described (20, 21). Soluble TCRs used for SPR experiments and crystallography were expressed in E. coli and refolded as previously described (27, 39). The TCRs produced in E. coli were murine TCR Vα and TCR Vβ domains fused to the human Cα and Cβ domains carrying an engineered disulfide bond to improve folding yield and protein stability. TCRα and TCRβ chains were expressed as inclusion bodies in the Rosetta2 (DE3) strain of E. coli (EMD Millipore, Darmstadt, Germany). Soluble TCRs were produced by refolding mixtures of denatured TCRα and TCRβ chains isolated from inclusion bodies (21). The refolded TCRs were further purified by FPLC size exclusion and ion exchange chromatography.
SF9 cells expressing IAb-3K variants with TCR multimers: Staining, analysis and peptide identification
Fluorescently labeled soluble TCR multimers were used to stain SF9 cells expressing IAb-3K, IAb-3K variants or IAb-library peptides as previously described (20, 21). SF9 cells were infected with baculovirus expressing membrane bound IAb-peptide complexes for 3 days using an MOI of 1–3. Following three days, 105 virus infected cells were washed and then stained with 20μg/ml TCR staining reagent and anti-IAb (mAb 17–227) for 2 hours at 27°C. Stained cells were then washed and immediately analyzed by flow cytometry. Each staining experiment was performed three independent times. To maximize the reproducibility of TCR multimer staining fluorescence, the experiments were repeated on subsequent days using the same prep of TCR multimers and the same flow cytometer and settings. For each TCR:IAb-3K pair, a standard curve relating the MFI of TCR multimer staining to the SPR measured equilibrium affinity was created using the least squares method of curve fitting with a Boltzmann sigmoidal fit (GraphPad, Prism) (21). The standard curves were then used to generate ΔΔG data for the interaction of TCRs with residues of IAb-3K. Staining of less than 5% of WT staining was considered non-binding and assigned the maximum ΔΔG value, as determined by SPR.
To identify novel IAb-peptide complexes recognized by Vα2+ Y31A substituted TCRs, SF9 cells were infected with an IAb-random peptide library at a multiplicity of infection of <1. Fluorescently labeled soluble TCR multimers carrying the Vα2+ Y31A substitution were used to stain the infected SF9 cells. SF9 cells that were positively stained with the TCR multimers were FACS cell sorted, and then incubated with additional SF9 cells to expand the virus secreted from the sorted cells. A second round of library enrichment was performed, followed by cloning the baculovirus by limiting dilution. SF9 cells infected with expanded viral clones were tested for I-Ab expression and TCR recognition. The peptide sequence encoded in baculovirus clones that were recognized by the TCRs of interest were determined by PCR sequencing.
Surface Plasmon Resonance
Equilibrium affinity and binding kinetics for TCRs binding to IAb-3K and APLs were obtained by surface plasmon resonance on a BIAcore 3000 instruments (BIAcore AB, Uppsala, Sweden). For equilibrium affinity experiments, approximately 1,000–8,000 RU of soluble, biotinylated pMHC was captured on the surface of a biosensor flowcell by via streptavidin linker. Soluble TCR was injected for 120 sec. at various concentrations in the range between 0.39 - 100μM. All samples reached equilibrium binding within 60 sec. The complex was allowed to dissociate for 5 min between injections, followed by a 60sec pulse of 0.1% SDS to remove any TCR still bound to the flow cell. Data points were collected at 0.5 sec intervals. Raw data were corrected for the bulk signal from buffer and TCR by performing identical injections through a flow cell in which an irrelevant H2-Db + peptide complex was immobilized. The data were analyzed with BIAcore BIAeval software. Scatchard analyses of the equilibrium data were used to determine the dissociation constant (Kd) for 14.C6, J809.B5, and amino acid substituted TCR and pMHC. No detectable binding was observed for some TCRs binding various IAb-3K peptide variants. In these cases the minimum possible Kd was estimated as (>250μM) assuming a maximum possible equilibrium signal of 50RU with the highest concentration of TCR variants tested (100μM) and an Rmax of 5000 RU, i.e. the maximum predicted signal if complete binding of IAb-3K to TCR occurred.
The kinetics of soluble, monomeric TCRs binding to immobilized pMHC complexes were analyzed by surface plasmon resonance using a BIAcore 3000 instrument and analyzed with BIAcore BIAeval 4.1 software (BIAcore AB, Uppsala, Sweden). Approximately 500 RU of soluble pMHC was captured on the surface of a biosensor flowcell by via streptavidin linker. TCRs were injected at 20μL/min for 60sec through the biosensor flow cell at concentrations that ranged from 3–50μM. All samples reached equilibrium binding within 10 sec. The complex was allowed to dissociate for 60 sec between injections. As a control for bulk fluid phase refractive index the TCRs were also injected through a flow cell with an immobilized irrelevant H2-Db + peptide complex. Data points were collected at 0.1 sec intervals and fit to a 1:1 Langmuir binding model to determine the dissociation rate (koff) and t1/2 of the TCR-pMHC complex. The kinetic data were used to determine the dissociation rate and the association rate (ka) was calculated from the KD and kd (ka=kd/KD).
Crystallization and Data Collection
TCR:MHC complexes were preformed by mixing the proteins at equimolar concentrations at 12mg/mL. Crystallization trials for all complexes were carried out using the hanging-drop vapor-diffusion method with a 1:1 protein to buffer ratio at room temperature. Single crystals were obtained for the J809.B5-IAb3K complexes with 100mM sodium cacodylate, 100mM sodium citrate (pH 5.8) and 10% (w/v) PEG4000, conditions similar to our previously reported lower resolution J809.B5-IAb3K structure. Crystals of the J809.B5 Y31A-IAb3K complexes were formed in 100mM sodium cacodylate, 100mM sodium citrate (pH 5.4) and 8% (w/v) PEG4000, and crystals of the the 14.C6-IAb3K complex formed in 100mM sodium cacodylate, 200mM sodium citrate (pH 5.5) 10% (w/v) PEG 4000 and 0.5% (w/v) n-octyl-β-D-glucoside. J809.B5-IAb3K and 14.C6-IAb3K crystals generally formed in 2–3 days, while the J809.B5 Y31A-IAb3K crystals formed within a four weeks. For data collection, crystals were transferred to crystallization buffer containing 25% (w/v) glycerol and were flash-cooled by plunging into liquid nitrogen. X-ray diffraction data were collected from a single crystal at 100°K with 1.10 Å radiation at the National Synchrotron Source X25 undulator beamline at Brookhaven National Laboratory. Diffraction data were indexed, integrated, and scaled with HKL2000 (40). Unit cell parameters and data collection statistics are shown (Table II). A crystal structure for the 809.B5-IAb3K complex (PDB 3RDT) has been previously reported for lower resolution data (21).
Table II.
Data collection and refinement statistics (molecular replacement)
| 14.C6:IAb3K | J809.B5:IAb3K | J809.B5Y31A:IAb3K | |
|---|---|---|---|
| Data collection | |||
| Space group | P1 21 1 | C 1 2 1 | C 1 2 1 |
| Cell dimensions | |||
| a, b, c (Å) | 66.007, 74.142, 259.289 | 239.121, 73.514, 65.73 | 242.123, 73.429, 65.68 |
| a, b, g (°) | 90.00, 92.00, 90.00 | 90.00, 90.58, 90.00 | 90, 90.34, 90 |
| Resolution (Å) | 50–3.25(3.31–3.25) | 41–2.24(2.28–2.24) | 42–2.85(2.95–2.85) |
| Rmerge | 15.5 (53) | 8.2(46.2) | 13.7 (53) |
| I/sI | 12.9(3.8) | 18.14(3.43) | 11.6(3.63) |
| Completeness (%) | 99.1(97.9) | 97.48(92.10) | 100.0(99.7) |
| Redundancy | 6.6(3.8) | 5.8(5.3) | 6.7(6.6) |
| Refinement | |||
| Resolution (Å) | 47–3.26(3.38–3.26) | 39–2.24(2.32–2.24) | 42–2.85(2.95–2.85) |
| No. reflections | 39080 (3819) | 53984 (5046) | 25527 (2335) |
| Rwork/Rfree | 18.63/23.62 | 16.43/21.21 | 18.13/24.67 |
| No. atoms | 6736 | ||
| Protein | 12366 | 6377 | 6273 |
| Ligand/ion | |||
| Water | 359 | ||
| B-factors | 44.2 | ||
| Protein | 65.4 | 44.0 | 60.1 |
| Ligand/ion | |||
| Water | 47.4 | ||
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.003 | 0.007 | 0.008 |
| Bond angles (°) | 0.666 | 1.06 | 1.18 |
| Ramachandran favored (%) | 95.55 | 98 | 94 |
| Allowed region | 4.13 | 2.0 | 6.0 |
| Outlier region | 0.32 | 0.0 | 0.0 |
X-ray diffraction data were collected from single crystals
Structure determination
Initial phases were obtained by molecular replacement using Phaser (41) and the lower resolution J809.B5:IAb3K structure 3RDT as the search model. One MHC and one TCR per asymmetric unit were found with the J809.B5 and the J809.B5Y31A complexes, while 2 TCRs and MHCs per asymmetric unit were found for the 14.C6 complex, consistent with the expected unit cell composition using a Matthews analysis. Initial NCS averaged maps of the 14.C6 complex were generated using RESOLVE. Refinements for 14.C6 constrained the same NCS operators in RESOLVE: pMHC α1β1, MHC α2β2, TCR-Vαβ, TCR-Cα, and TCR-Cβ but was relaxed in later cycles of refinement (42).
Density for all MHC and TCR domains and peptide was observed clearly in the composite omit maps using CNS (43), and used as a guide for model building. Cycles of automated rigid body, coordinate position, atomic displacement factor, and TLS refinement using Phenix (44) were alternated with manual model building using Coot (45). Analysis of the J809.B5Y31-IAb3K dataset using Xtriage (Phenix) revealed an estimated 10% twin fraction (Britton and H Test) with twin law -h, -k, l. The dataset used for refinement was altered taking the twin operator into account using Detwin (CCP4i). Molecular replacement and model refinement followed similar steps as outlined for the J809.B5 and 14.C6 IAb3K complexes. The final models include coordinates for TCR extracellular domains (alpha3-201, beta2-239), MHC IAb extracellular domains (alpha 1–179, beta 6–191) and all 13 residues of the 3K peptide. No density was observed in any of the crystals for the linker (GGGGSLVPRGSGGGGS) tethering the peptide to the N-terminus of the beta subunit. Final refinement statistics are shown in (Table II).
Structure analysis
The two molecules within the 14.C6 asymmetric unit are highly similar (RMSD of 0.853 or 0.492 if the TCR C-doamins are removed), especially within the binding interface including the CDR3 loops. The only observable difference between the two structures within the binding interface occurs at residue R50. In molecule one and molecule two, two different rotomers are found. In molecule one, the R50 side chain interacts with the MHCβ residue E69 (<4.0Å between the two side chains). In molecule two, the R50 side chain does not appear to interact with this side chain, the R50 side chain is >4.0 Å distance away from any E69 side chain atoms. However, alanine scanning experiments show that energetically, the MHCβ E69A substitution coordinates approximately1.0 kcal/mol free energy (see Fig. S1). This indicates that the TCR R50-MHC E69A interactions are important for binding. Therefore, we used molecule one for generating the figures in the manuscript. Structural alignments were done using LSQMAN (46). The structure of the 14.C6 complex was aligned with the J809.B5 complex using coordinates of the MHC αβ helical domains and the 3K peptide. The angle of engagement was calculated from the p5 Cα atom to the center of mass (Pymol, Delano Scientific) for each TCR. Surface and interaction areas were calculated using the PISA server (47). Coordinate statistics, and atom-atom distances were evaluated using Phenix and NCONT from the CCP4i Suite (48).
Accession Numbers
Coordinates and structure factors for the J809.B5 TCR-IAb-3K, J809.B5 Y31A TCR-IAb-3K, and the 14.C6 TCR-IAb-3K complexes will be available from the Protein DataBank under accession numbers 4P23, 4P46, and 4P5T (www.rcsb.org/).
Results
CDR3α residues can affect the geometry in which TCRs bind pMHC
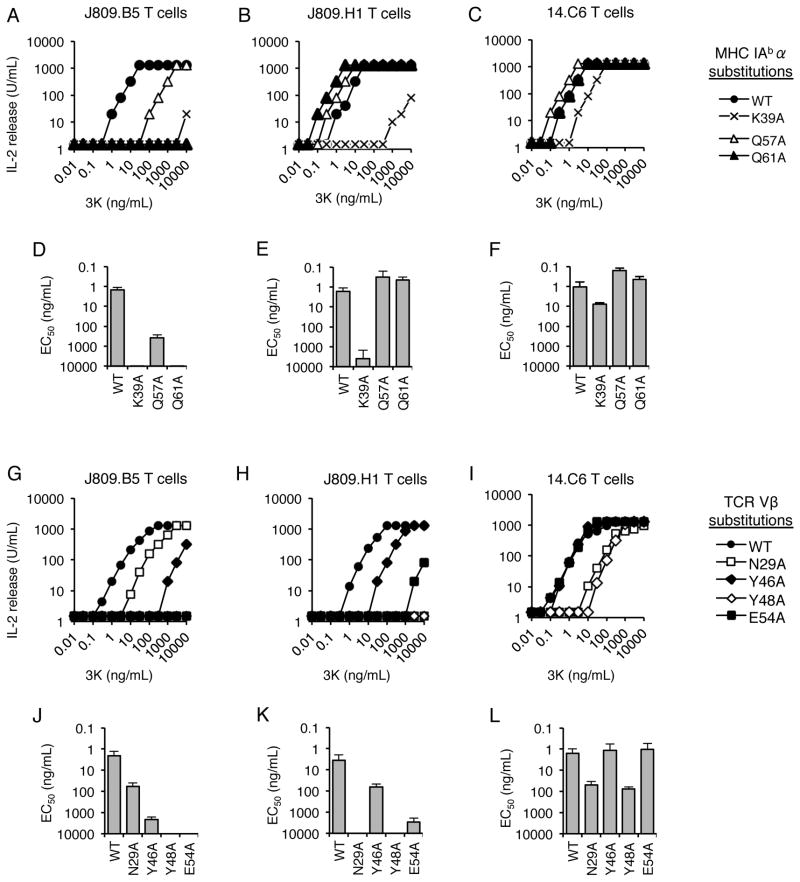
To examine the influence of CDR3α sequences on TCR-MHC interactions, the IAb-3K-reactive TCRs J809.B5, J809.H1 and 14.C6 were studied. These three TCRs carry the identical TCRβ and TCR Vα2.8 sequences, differing only in the CDR3α sequence (Table I). Alanine scanning mutagenesis experiments indicate that these TCRs differentially require several MHC residues for binding and for T cell expressing these receptors to undergo activation (Fig 1, Fig. S1 and Table S1, S2). These MHC residues include potential sites of TCRβ-MHC contact (21, 26, 27, 49). For example, the activation threshold (EC50 value) of J809.B5 T cells responding to the 3K peptide is reduced 100-fold and greater than 1,000-fold when APCs express IAb carrying alanine substitutions at the IAbαQ57 and αQ61 residues, respectively. In contrast, the response of J809.H1 and 14.C6 T cells was minimally changed when the IAb MHC molecules express these alanine substitutions (Fig. 1A–F). Similarly, J809.B5, J809.H1 and 14.C6 T cells differentially require TCR CDR1β and CDR2β residues to be activated by APC presenting IAb-3K (Fig. 1G–L). Importantly, differences in requirements of these TCR Vβ and MHC residues were not predicated on the TCRs having different equilibrium affinities for IAb-3K, as the J809.B5 and 14.C6 TCRs bind IAb-3K with similar affinity (KD = 6μM and 5μM, respectively) (Fig. S1).
Table I.
Sequence of TCRs
| TCR | Vα | CDR1α | CDR2α | CDR3α | Vβ | CDR1β | CDR2β | CDR3β |
|---|---|---|---|---|---|---|---|---|
| J809.B5 | 2.8 | ENSAFDY | AIRSVSDK | CAASKGADRLTFG | 8.2 | TNNHNN | YSYGAGSTEKGD | CASGDFWGDTLYFG |
| J809.H1 | 2.8 | ENSAFDY | AIRSVSDK | CAASKSSGQKLVFG | 8.2 | TNNHNN | YSYGAGSTEKGD | CASGDFWGDTLYFG |
| 14.C6 | 2.8 | ENSAFDY | AIRSVSDK | CAASRDSGQKLVFG | 8.2 | TNNHNN | YSYGAGSTEKGD | CASGDFWGDTLYFG |
| YAe62 | 4.12 | STTGYPT | QVTTANNK | CAANSGTYQRFG | 8.2 | TNNHNN | YSYGAGSTEKGD | CASGDFWGDTLYFG |
| 2W1S20.4 | 2.9 | EDSTFDY | AIRPVSNK | CASSDNRIFFG | 8.2 | TNNHNN | YSYGAGSTEKGD | CASGDAWGYEQYFG |
| 75–55 | 4.11 | STTWYPT | KVTTANNK | CALAGGSNAKLTFG | 8.2 | TNNHNN | YSYGAGSTEKGD | CASGEAWGGRYFG |
| 13.B1 | 2.3 | ENSAFDY | SILSVSDK | CAASDDNNRIFFG | 8.2 | TNNHNN | YSYGAGSTEKGD | CASGDFWGDTLYFG |
| 13.D5 | 2.3 | ENSAFDY | SILSVSDK | CAASDGNNRIFFG | 8.2 | TNNHNN | YSYGAGSTEKGD | CASGDFWGDTLYFG |
| J809.G3 | 2.9 | EDSTFDY | AIRPVSNK | CAASKDGGADRLTF | 8.2 | TNNHNN | YSYGAGSTEKGD | CASGDFWGDTLYFG |
| 14.A6 | 2.9 | EDSTFDY | AIRPVSNK | CAASKGGQKLVFG | 8.2 | TNNHNN | YSYGAGSTEKGD | CASGDFWGDTLYFG |
FIGURE 1.
CDR3α Sequences Can Alter CDR1β and CDR2β Interactions with MHC. (A–F) The dose response and EC50 values of the (A, D) J809.B5, (B, E) J809.H1 and (C, F) 14.C6 T cell hybridomas responding to titrating concentrations of 3K peptide presented by fibroblasts expressing IAb, or IAb in which the IAbα side chains αK39, αQ57, or αQ61 have been substituted with alanine (labeled WT, K39A, Q57A or Q61A). EC50 values are log10-based half-maximal responses in ng/mL of 3K peptide. The dose response and EC50 value of T cell hybridomas expressing either the (G, J) J809.B5, (H, K) J809.H1 and (I, L) 14.C6 TCR, or each TCR carrying alanine substitutions at the TCR residues CDR1β N29, CDR2β Y46, CDR2β Y48 or CDR2β E54, responding to titrating concentrations of 3K peptide presented by IAb expressing fibroblasts. EC50 values are log10-based half-maximal responses in ng/mL of 3K peptide. Error bars represent the SEM of three independent experiments.
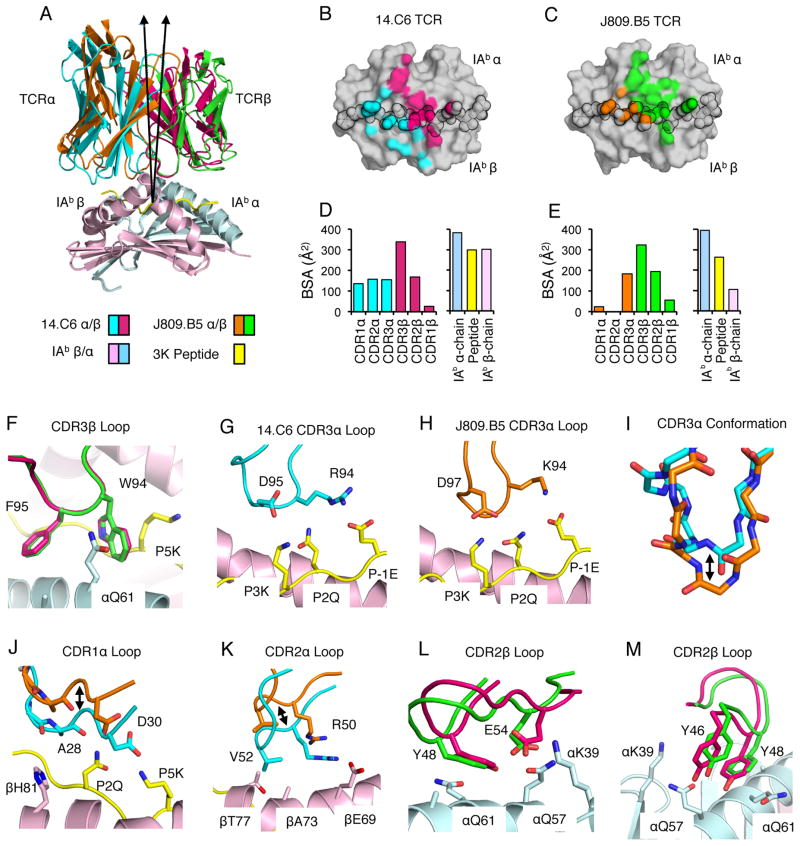
To gain insight into how CDR3α sequences can influence CDR1 and CDR2 interactions with MHC, we determined the 3.2Å co-crystal structure of the 14.C6 TCR bound to IAb-3K, and compared it to a new 2.3Å structure of the J809.B5 TCR bound to the same ligand. A lower-resolution J809.B5 - IAb-3K structure has been reported previously (21). The J809.B5 TCR and the 14.C6 TCR engage IAb-3K similarly but with a marked change in orientation, such that there is a 7.5° difference in the tilt of the TCR relative to pMHC (Fig. 2A, Table II and Fig. S2). This change in TCR-pMHC binding geometry is confirmed using an R-free analysis of the TCR V domains. The 14.C6:IAb-3K complex has an R-free of 23% whereas if the 14.C6 V domain if forced into the J809.B5 docking geometry, the resulting complex would have an R-free of 42%. This change in tilt allows the 14.C6 TCR to form an increased number of contacts with the IAbβ-chain, as compared to the J809.B5 TCR (Fig. 2B, C). In addition, the total buried surface area (BSA) for the 14.C6 complex (approximately 1970 Å2) was greater than that for the J809.B5 complex (approximately 1540 Å2), primarily stemming from changes to the BSA of the CDR1α and CDR2α loops and the IAb β-chain (Fig. 2D, E). In contrast to these changes, CDR3β loop conformations and the placement of CDR3 βW94 and βF95 residues within these structures are similar (Fig. 2F).
FIGURE 2.
CDR3α Sequence Can Alter the Positioning of CDR1 and CDR2 Loop Atop MHC. (A) Overlay of the 14.C6 and J809.B5 TCRs binding IAb-3K. The 14.C6 TCR is colored red (TCRβ) and blue (TCRα); the J809.B5 TCR is colored green (TCRβ) and orange (TCRα). IAb-3K is colored cyan (IAbα chain), yellow (peptide), and magenta (IAbβ chain). A 7.5% difference in the tilt of the TCR docking geometry atop IAb-3K is observed based on a vector from the center of mass of each TCR to the α-carbon of the p5 peptide residue. (B) Projection of the 14.C6 TCR or (C) J809.B5 TCR binding onto IAb-3K. 14.C6 TCRα contacts are colored blue, 14.C6 TCRβ contacts are colored red. The J809.B5 TCRα contacts are colored orange and the TCRβ contacts are colored green. The peptide residues are outlined in black. (D) The amount of buried surface area (BSA) contributed by the 14.C6 or (E) J809.B5 TCRα, TCRβ, peptide or MHC chains for the binding reaction with IAb-3K. (F) The 14.C6 CDR3β loop (magenta) and the J809.B5 CDR3β loop (green) are in a similar conformation and make similar contacts with IAb-3K. (G) The 14.C6 CDR3α loop residues R94 and D95 interact with the peptide residues, P-1E, P2Q and P3K. (H) The J809.B5 CDR3α loop residues K94 and D97 interact with the peptide residues, P-1E, P2Q and P3K. (I) The 14.C6 CDR3α loop (blue) and the J809.B5 CDR3α loop (orange) are in a different conformation when bound to IAb-3K. (J) 14.C6 CDR1α residues A28 and D30 (blue) make extensive contacts with the P2Q and P5K residues of the peptide, and the IAbβ chain residue H81. The J809.B5 CDR1α loops residues A28 and D30 (orange) are shifted approximately 4 Å, allowing only the A28 residue to make minimal contacts with peptide, and no contact with MHC. (K) The 14.C6 TCR CDR2α residues R50 and V52 (blue) interact with IAb β-chain residues βE69, βA73 and βT77, contacts that are not present in the J809.B5:IAb-3K structure (orange) due to a 4 Å rigid body shift. (L, M) The main-chain of CDR2β loop at the βY48 position in the 14.C6 structure (green) is shifted approximately 1Å as compared to the J809.B5 structure (magenta), and some rotamer differences are observed for the CDR2β residues E54 and Y46. Figures were made with PyMol (76).
CDR3α sequences can modulate CDR1 and CDR2 interactions with pMHC
To evaluate how CDR3α sequences can affect TCR interactions with pMHC, we first compared the 14.C6 and J809.B5 CDR3α loop conformations when bound to IAb-3K. In both structures the 14.C6 and J809.B5 CDR3α loops make similar types of contacts with the peptide (Fig. 2G, H). Despite the 14.C6 CDR3α loop being one amino acid longer, the J809.B5 CDR3α loop extends 2.5Å further towards the peptide, relative to the 14.C6 CDR3α loop (Fig. 2I). Because of the different CDR3α loop conformations, a steric clash would occur between the J809.B5 CDR3α loop and the peptide if this TCR bound IAb-3K with the same geometry as that of the 14.C6 TCR. Thus, the differences in CDR3α loop conformation likely underlie why the J809.B5 and 14.C6 TCRs bind IAb-3K with different tilts. The most prominent differences in how the J809.B5 and 14.C6 TCRα chains engage IAb-3K involve CDR1α and CDR2α residues. The 14.C6 TCR docking geometry allows its CDR1α loop to make extensive contacts with the P2 and P5 positions of the 3K peptide, as well as the IAbβ chain residue βH81 (Fig. 2J). In contrast, the J809.B5 CDR1α loop is positioned such that it makes only minimal contacts with the 3K peptide and no contacts with MHC (Fig. 2J). Likewise, the 14.C6 TCR CDR2α residues interact with IAbβ chain, whereas these contacts are absent in the J809.B5:IAb-3K structure due to the 4 Å rigid body shift (Fig. 2K).
Difference in the requirements of TCR Vβ and IAbα residues for J809.B5 and 14.C6 T cells to undergoactivation, shown in Figure 1 and Supplementary Figure S1, arise from less dramatic structural changes. The main-chain of the J809.B5 and 14.C6 CDR2β loops are shifted approximately 1Å at the CDR2 βY48 position. In addition, some rotamer differences are observed for the CDR2β residues, βY46 and βE54; while the IAb αQ57 and αQ61 side chain rotamers are similarly orientated (Fig. 2L, M). Thus, the requirements of TCR and MHC side chains for T cell activation can be strongly affected by modest shifts in CDR loop placement and side chain rotamer differences.
CDR3α sequences can regulate the specificity of a TCR Vβ-MHC interaction
Across different TCR Vβ gene families, the amino acid expressed at the CDR2 β48 position is variable (50). Nevertheless, analyses of different TCR-pMHC co-crystal structures have shown that the CDR2 β48 residue is often located in close proximity to MHC residues (28). Thus, different amino acid residues can be used at the TCR Vβ CDR2 β48 position to bind MHC. Because of the variance in how the J809.B5 and 14.C6 TCR CDR2β loops engage MHC residues, we hypothesized that CDR3α sequences may influence which amino acids can be expressed at a TCR Vβ position to facilitate MHC binding.
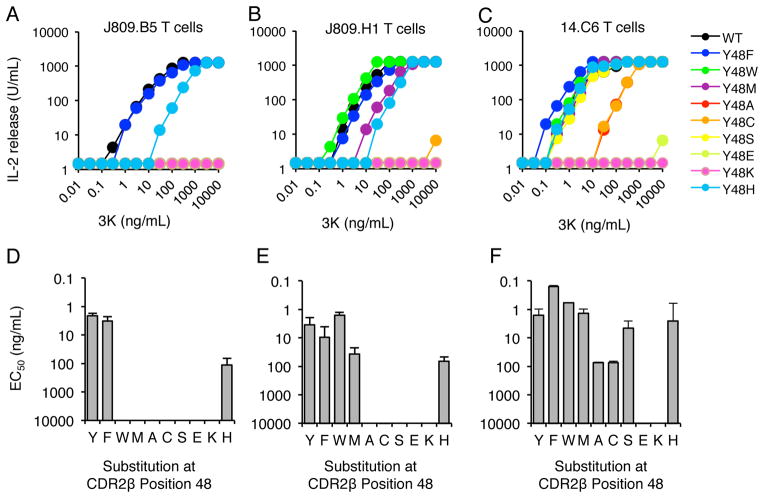
To test the hypothesis that CDR3α sequences can modulate the specificity of a TCR Vβ-MHC interaction, J809.B5, J809.H1 and 14.C6 T cell hybridomas were created that carried different amino acid substitutions at the CDR2 β48 position. These T cell hybridomas were then tested for the ability to respond to APC presenting the 3K peptide. Consistent with studies of other Vβ8.2+ T cells (22, 23, 27, 51), the J809.B5, J809.H1 and 14.C6 T cells lost or had a reduced response to 3K peptide when the CDR2 β48 position carried an alanine (A). In addition, T cells carrying the conservative tyrosine to phenylalanine (F) substitution maintained complete 3K peptide reactivity (Figure 3). When non-conserved side chains were expressed at the CDR2 β48 position, differing 3K peptide reactivity patterns were observed. J809.B5 T cells retained some response to 3K peptide when a histidine (H) was expressed, but not a tryptophan (W), methionine (M), cysteine (C), serine (S), glutamic acid (E), or lysine (K) (Fig. 3A). In contrast, J809.H1 T cells responded to 3K peptide when the CDR2 β48 position carried a W, M and H, but not an C, S, E or K, while 14.C6 T cells maintained strong 3K responses when the CDR2 β48 position carried a W, M, S and H (Figure 3B, C).
FIGURE 3.
CDR3α Sequences Can Regulate the Specificity of the Vβ8.2 CDR2 β48 Interaction with pMHC. IL-2 production of T cell hybridomas expressing the (A, D) J809.B5, (B, E) J809.H1 or (C, F) 14.C6 TCRs, or TCRs carrying amino acid substitutions at the TCR CDR2β residue Y48, in response to titrating concentrations of 3K peptide presented by IAb expressing fibroblasts. IL-2 secretion is reported as Units/mL. (D–F) The EC50 values for 3K dose responses shown in panels A–C and error bars represent the SEM of three independent experiments.
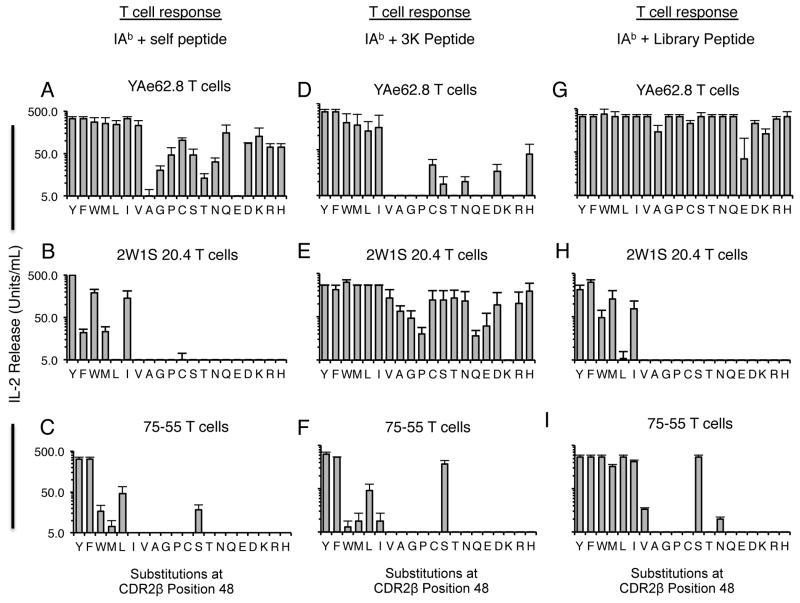
Similar studies of the self-reactive and IAb-3K-reactive T cells, YAe62, 2W1S 20.4 and 75–55 were consistent with the hypothesis that the TCR CDR3 sequences and TCRαβ chain pairing can modulate the specificity of a TCR V gene-MHC interaction. YAe62 T cells were activated by self-peptides when the CDR2 β48 position carried most amino acid sequences, although not an alanine (Fig. 4A), while the 2W1S 20.4 and 75–55 T cells required a more limited set of amino acids to be expressed at the CDR2 β48 position (Fig. 4B, C). In contrast to the anti-self responses, YAe62 T cells required a more restricted set of amino acids at the CDR2 β48 position to respond to APCs presenting 3K peptide (Fig. 4D). The peptide sequence was directly observed to influence the specificity of this CDR2 β48-MHC interaction, as YAe62 T cells maintained reactivity to a different peptide ligand, FMRKA, with most amino acids expressed at the CDR2 β48 position (Fig. 4G). Peptide-dependent effects on the requirements of the Vβ8.2 CDR2 β48 position were also observed for the 2W1S 20.4 T cells, though less so for 75–55 T cells (Fig. 4E, F, H and I).
FIGURE 4.
Vβ8.2+ T cells can have distinct requirements at CDR2 β48 Position when Responding to Self-peptides, and Different Foreign-peptides. IL-2 production of T cell hybridomas expressing the (A, D, G) YAe62.8, (B, E, H) 2W1S 20.4 or the (C, F, I) 75–55 TCRs substituted with all 20 amino acid at the CDR2β 48 position following challenge with (A–C) H2b expressing splenocytes, (D–F) IAb-3K expressing fibroblasts, or (G–I) an alternate IAb-foreign peptide ligand expressed on fibroblasts. For the YAe62 T cells, the alternate peptide is FMRKA. For the 2W1S 20.4 and 75–55 T cells, the alternate foreign peptide is RCKST. IL-2 secretion is reported as Units/mL. Error bars represent the SEM of three independent experiments.
Collectively, these studies demonstrate that the side chain requirements of a TCR Vβ-MHC interaction can be distinct for different T cell clonotypes. Furthermore, the resulting change in TCR V gene-MHC specificity can be regulated by the CDR3α sequence.
A CDR1 residue distal to the TCR-pMHC binding site can contribute to antigen recognition
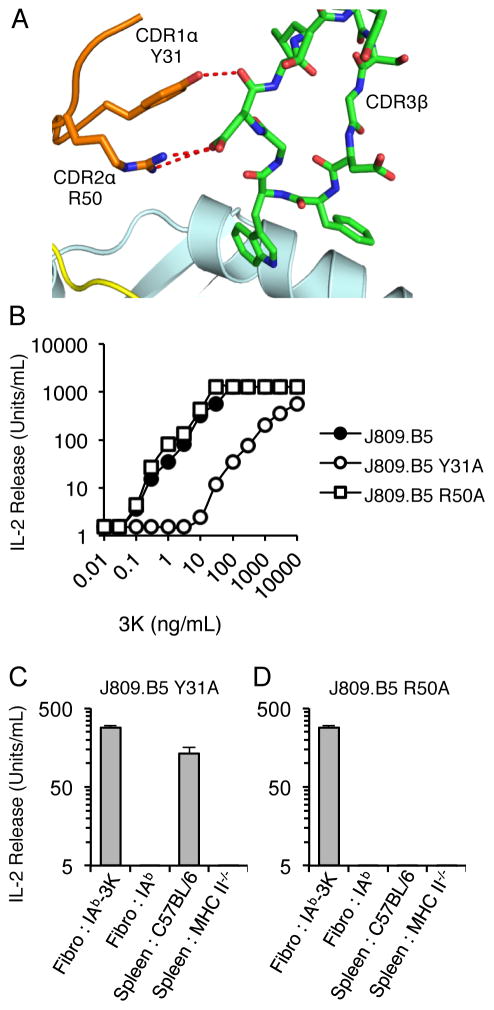
With the assembly of complete TCRs, interactions occur between the TCR Vα and Vβ domains and between different CDR loops (52, 53). The residues involved in these inter- and intra-chain interactions can be derived from conserved sequences of the TCR framework, or from variable sequences in the CDR loops. The inter- and intra-chain interactions that involve CDR loops can occur distal or proximal to the pMHC binding site. For example, when the J809.B5 TCR binds IAb-3K, the CDR1 αY31 forms contacts with the CDR3β loop at a site that is distal to where the CDR3 loop contacts the peptide, while the CDR2 αR50 residues forms contacts with the CDR3β loop at a site nearer to the pMHC interface (Fig. 5A, S2) (21). Having observed that CDR3α residues can create indirect effects that influence the specificity of TCR V gene-MHC interactions, we wondered if TCR Vα-Vβ interactions outside of the binding interface might also create indirect effects that influence ligand specificity.
FIGURE 5.
The Vα2 CDR1 αY31 side chain contributes to the self-tolerance and IAb-3K reactivity of J809.B5 T cells. (A) The Vα2.8 CDR1α and CDR2α loops interact with the CDR3β loops of the J809.B5 TCR when bound to IAb-3K. Note the CDR1 αY31 is located 7 Å from the nearest peptide or MHC residue. (B) IL-2 production of T cell hybridomas expressing the J809.B5 TCR or J809.B5 TCRs carrying alanine substitutions at the CDR1 residue αY31, or the CDR2 residue αR50, in response to titrating concentrations of 3K peptide (ng/mL) presented by IAb expressing fibroblasts; or (C, D) in response to fibroblasts expressing IAb-3K, fibroblasts expressing IAb, C57BL/6 splenocytes expressing H2b MHC molecules or splenocytes from C57BL/6 MHC II−/− mice. IL-2 secretion is reported as Units/mL. Error bars represent the SEM of three independent experiments.
To determine whether CDR residues that are located outside of the pMHC interface contribute to peptide specificity, J809.B5 T cell hybridomas were created that express alanine substitution at the CDR1 αY31 residue (αY31A), or the CDR2 αR50 residues (αR50A). 3K peptide titration experiments show that T cell hybridomas expressing the J809.B5 αY31A TCR are 260-fold less reactive to 3K peptide presented by IAb-transfected fibroblasts. In contrast, no change in 3K-reactivity was observed for J809.B5 T cells carrying the αR50A substitution (Fig. 5B). T cell hybridomas expressing the J809.B5 αY31A TCR also become reactive to C57BL/6 splenocytes presenting IAb + self-peptides, inferring that the CDR1 αY31 side chain contributes to the peptide specificity of the J809.B5 TCR (Fig. 5C). Thus, the CDR1 αY31 side chain of the J809.B5 TCR influences 3K reactivity, as noted by a change in sensitivity to titrating concentrations of 3K peptide, and peptide specificity, as evidenced by the acquisition of self-reactivity even though it is positioned outside of the TCR-pMHC binding site.
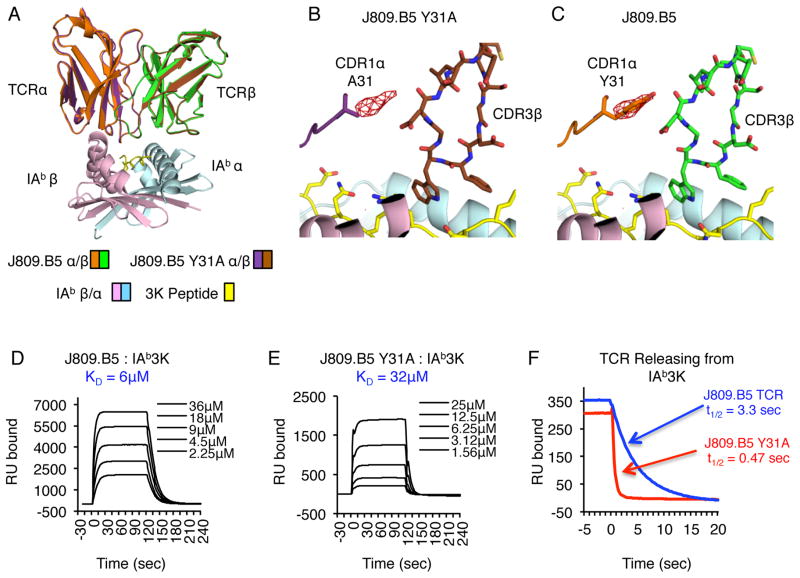
The J809.B5 CDR1 αY31 side chain stabilizes the IAb-3K binding conformation
To identify how the J809.B5 CDR1 αY31 side chain contributes to 3K peptide reactivity, we first determined the 2.9Å crystal structure of the J809.B5 αY31A TCR bound to IAb-3K and compared it to the 2.3Å structure of the J809.B5 TCR bound to the same ligand. The overall structures of the J809.B5 and J809.B5 αY31A TCRs bound to IAb-3K are extremely similar, including the conformation of the CDR loops and the total BSA (approximately 1540 Å2) (Fig. 6, Table II and Fig. S2). To confirm the high degree of similarity between the two models, we calculated an isomorphous difference map from the two x-ray datasets to identify the relative differences in electron density. The largest difference in density was localized to the CDR1 α31 position and can accommodate a phenol group that is lost with the tyrosine to alanine substitution (Figure 6B, C). Thus, the CDR1 αY31 side chain is not required for the J809.B5 TCR to adopt its parental IAb-3K binding conformation.
FIGURE 6.
The Vα2 CDR1 αY31 side chain stabilizes the J809.B5 TCR binding reaction with IAb-3K through indirect effects. (A) Overlay of J809.B5 and J809.B5 αY31A TCRs binding IAb-3K. The J809.B5 TCRβ is colored green, TCRα is orange; the J809.B5 αY31A TCRβ is colored purple, TCRα is brown. IAb-3K is colored cyan (IAbα chain), yellow (peptide) and magenta (IAbβ chain). (B, C) Isomorphous difference map indicates that the only major structural changes (5.5 sigma peak) are located in immediate proximity of the Vα2 αY31 residue, centered on the phenyl ring. Note the next highest peak occurs at a sigma of 3.8. (D) Soluble J809.B5 TCR or (E) J809.B5 αY31A TCRs were analyzed for equilibrium affinity binding to immobilized IAb-3K via SPR. Listed KD are the average of three independent experiments, sensograms are representative analyses. (F) Dissociation of J809.B5 TCR (blue) and J809.B5 αY31A TCR (red) from immobilized IAb-3K at a flow rate of 20μL/min at 25 °C. Data for multiple TCR concentrations were collected at 0.1-s intervals and fit to a 1:1 Langmuir binding model to determine the dissociation rate (koff) and t1/2 of the TCR-pMHC complex. Dissociation plots are examples of three independent experiments.
Despite the similar TCR loop conformations, the reduced 3K reactivity of T cell hybridomas expressing the J809.B5 αY31A TCR indicated that the TCR-pMHC binding reaction had changed. Therefore, surface plasmon resonance experiments were performed to identify the contribution of the J809.B5 CDR1 αY31 side chain to the binding of IAb-3K. Whereas the J809.B5 TCR binds IAb-3K with a KD = 6μM, the J809.B5 αY31A TCR binds IAb-3K with a KD = 32μM (Fig. 6D, E). The reduced binding affinity for the J809.B5 αY31A TCR is primarily the result of the interaction having a 7.5-fold faster off-rate (koff) and thus a shorter half-life (t1/2) (Fig. 6F). These data indicate that the CDR1 αY31 side chain contributes to setting the T cell activation threshold by stabilizing the J809.B5 TCR - IAb-3K binding reaction.
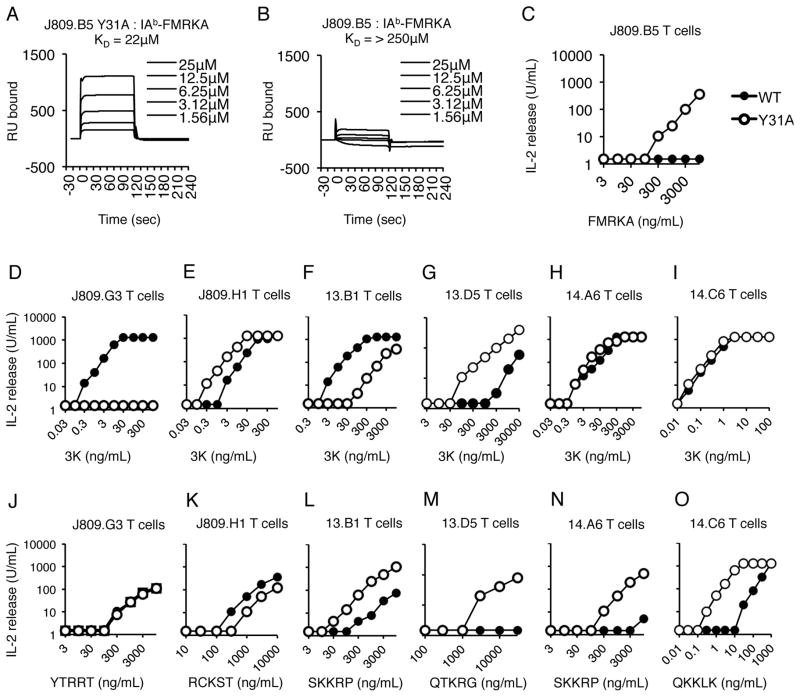
The Vα2 CDR1 αY31 side chain regulates the peptide specificity of several TCRs
The self-reactivity of T cells carrying the J809.B5 CDR1 αY31A substitution implies that the CDR1 αY31 side chain contributes to peptide specificity. To formally test this, an IAb-random peptide baculovirus library was probed to identify peptides that are recognized by J809.B5 αY31A T cells (54). Using this method, an IAb-peptide ligand, IAb-FMRKA, was identified which binds the J809.B5 αY31A TCR with a KD = 22μM. In addition, J809.B5 αY31A T cell hybridomas are activated by APC presenting the FMRKA peptide. The parent J809.B5 TCR, however, does not bind IAb-FMRK (KD > 250μM) and J809.B5 T cell hybridoma is not activated by the FMRKA peptide (Fig. 7A–C). The data formally demonstrate that the CDR1 αY31 side chain contributes to the peptide specificity of the J809.B5 TCR.
FIGURE 7.
The Vα2 CDR1 αY31 side chain contributes to the antigen specificity of multiple Vα2+ T cells. (A) Soluble J809.B5 αY31A or (B) J809.B5 TCR TCRs were analyzed for equilibrium affinity binding to immobilized IAb-FMRKA via SPR. Listed KD are the average of three independent experiments, sensograms are representative analyses. (C) IL-2 production of T cell hybridomas expressing the J809.B5 (filled squares) or J809.B5 αY31A (open circles) TCR in response to titrating concentrations of FMRKA peptide (ng/mL) presented by IAb expressing fibroblasts. IL-2 production of T cell hybridomas expressing the (D, J) J809.G3, (E, K) J809.H1, (F, L) 13.B1, (G, M) 13.D5, (H, N), 14.A6 or (I, O) 14.C6 TCRs (filled squares) or TCRs carrying alanine substitutions at the CDR1 residue αY31 (open circles), in response to (D–I) titrating concentrations of 3K peptide presented by IAb expressing fibroblasts, or (J–O) titrating concentrations of alternative peptide identified using baculovirus display library presented by IAb expressing fibroblasts. IL-2 secretion is reported as Units/mL. Data are the average of three independent experiments. See (Table S2) for EC50 and SEM values.
Six additional Vα2+ IAb-3K-reactive TCRs were studied to assess whether the CDR1 αY31 residue contributes to cognate peptide recognition and peptide specificity of other receptors. We observed that T cell hybridomas expressing CDR1 αY31A substituted TCRs could have an increase, decrease or unchanged reactivity to 3K peptide (Fig. 7D–I). In response to additional peptide ligands identified using an IAb-random peptide baculovirus library, a variety of peptide reactivity patterns were observed (Fig. 7J–O). T cell hybridomas expressing the 14.A6 αY31A TCR and 14.C6 αY31A TCR have an increased response to the SKKRP and QKKLK peptide, respectively, even though the response to 3K peptide is unchanged. In contrast, T cell hybridomas expressing the J809.G3 αY31A TCR have an equivalent response to the YTRRT peptide as compared to the parent J809.G3 TCR, yet lose all reactivity to APC presenting the 3K peptide. In addition, T cell hybridomas expressing the 13.B1 αY31A TCR have an approximate 80-fold loss in 3K reactivity, even though they show an approximate 140-fold increase in reactivity to APC presenting SKKRP peptide as compared to T cell hybridomas expressing the parent TCR (see Table S2 for EC50 values). Furthermore, T cell hybridomas expressing the 13.D5 αY31A TCR acquire reactivity to splenocytes presenting self-peptides (not shown). Collectively, these data indicate that the Vα2 CDR1 αY31 residue contributes to the peptide specificity of multiple MHC class II-restricted T cells.
Discussion
To provide protective immunity, T cell development creates a mature T cell repertoire that has the ability to respond to a broad range of foreign-antigens bound to host-MHC molecules. The T cell repertoire accomplishes this by being comprised of T cell clonotypes that express TCRs that have distinct binding requirements at potential sites of TCR-peptide and TCR-MHC interaction (1, 11). Much of the specificity of the TCR is derived from unique amino acid sequences present within the pMHC binding site. Data presented here demonstrate that the process of TCR combinatorial diversity and V(D)J rearrangements also create a collection of indirect effects that regulate the ligand specificity of TCRs. These effects allow CDR3α residues to modulate the geometry in which TCRs engage pMHC ligands, governing whether and how individual TCR residues interact with MHC. In addition, a TCR CDR1α residue that is located distal to the TCR-pMHC binding interface is shown to contribute to the peptide specificity of T cells.
Unlike immunoglobulin genes, TCRs do not undergo somatic hypermutation. Therefore, the only way TCRs can be created with reactivity for antigens presented by host-MHC alleles is through the combinatorial pairing of different TCR V gene segments and the creation of random CDR3 sequences through V(D)J recombination. The major force in creating TCRs with diverse antigen reactivity patterns occurs from V(D)J recombination, as this process determines both the length of the CDR3 loops and the identity of the amino acids present within the central portion of the pMHC binding site. Sequence differences amongst the Vα and Vβ gene segments also contribute to creating TCRs with a diversity of ligand specificities. Differential TCR V gene pairing creates TCRs that express different amino acid sequences at potential sites of pMHC contact. In addition, CDR1 and CDR2 loops can adopt one of a set of unique canonical structures, which allow different TCR Vα and Vβ pairings to create TCRs with a modest range of tertiary structures (55–57).
Several groups have analyzed structures of TCRs carrying the same TCR Vα or TCR Vβ gene sequences or TCRα or TCRβ subunit bound to the same MHC ligand. These studies have shown a spectrum of changes in how the identical TCR sequences can engage MHC. Studies of the 2C TCR and a high affinity variant M6 bound to H2-Ld, as well three Vβ8.2+ TCRs recognizing IAu, all show minimal differences in TCR V gene-MHC contacts (26, 58). Comparisons of the 2B4 and 226 TCRs bound to I-Ek show similar TCR V gene-MHC interactions, but with some modest shifts and side chain rotamer differences in the TCR Vβ3 CDR loop interactions with the IEkα-chain (59). Modest shifts in TCRβ CDR loop placements and differences side chain rotamer usage are also observed for the binding of the YAe62, 2W1S20.4, J809.B5 and 14.C6 Vβ8.2 chains with IAb(21, 27). Although variations in the placements of these TCRβ CDR loops atop MHC and side chain rotamer usage can appear subtle, these structural differences were associated with T cells having distinct requirements of TCR V gene residues and MHC residues for activation (see Figures 1, 3 and 4). Relatively subtle changes in MHC structure have also been shown to determine whether a T cell can recognize an allo-MHC ligand (60).
In contrast to the subtle variations outlined above, the comparisons of some TCRs carrying similar TCR V gene segments bound to the identical MHC can show more dramatic difference in CDR loop placements. Studies of the TCR Vβ CDR1 and CDR2 loops of the A6 versus B7 TCRs bound HLA-A2 demonstrate that different TCR V gene residues can be involved in the binding reactions (61). Differences in CDR loop placements are also observed when human Vα24+ and Vα24− TCRs bind CD1d MHC-like molecules presenting aGalCer (62). A comparison of the 2C TCR and the YAe62 TCR bound to H2-Kb ligands further show that the identical Vβ8.2 CDR2 loops can engage different positions atop the MHC (15). For the J809.B5 and 14.C6 TCRs structures bound to IAb-3K studied here, CDR3α sequences are shown to regulate the extent in which the CDR1α and CDR2α loops contribute to the binding reaction. The range of TCR sequence-dependent changes in TCR V gene-MHC interactions are mirrored by observations that the MHC allele and the bound peptide or lipid sequence can influence how TCR CDR1 and CDR2 loops engage MHC residues (21, 62–69). These findings may explain how very modest changes in TCR sequence, including TCRs with only a single amino acid change in the CDR3α loop, can distinguish TCRs expressed on CD4+ T cells recognizing MHC-II ligands from TCRs on CD8+ T cells recognizing MHC-I ligands (70). Collectively, the ability of CDR3 sequences and peptide sequences to modulate how TCR V gene residues engage MHC residues provides a mechanism whereby the TCR-MHC binding properties of the mature T cell repertoire can be tailored during T cell selection to recognize antigens displayed by host-MHC molecules (3, 71–73).
Germline encoded CDR sequence variations extend beyond the positions that most often contact the pMHC and into the core of the TCR V domain (2, 50). Due to the organization of CDR loops within TCRs, some of these variable CDR1 and CDR2 residues, as well as CDR3 residues, often abut other CDR loops (52, 53). By constructing TCRs through combinatorial diversity, TCRs express amino acid residues at potential sites of contact between the CDR loops based on the TCR V gene and CDR3 sequences used. Borg et. al. demonstrated that contacts between a CDR1α and CDR3α loop are required for the LC13 TCR to bind its cognate pMHC ligand (33), indicating that TCR intra-chain interactions can contribute to cognate ligand recognition. Through studying a set of MHC class II-restricted Vα2.3+, Vα2.8+ and Vα2.9+ TCRs, as well as a re-analyzing MHC class II-restricted TCRs carrying these Vα2 chains bound to their ligand (21, 26, 27) (Figure S2), we observed that the CDR1 αY31 residue often forms contacts with the CDR3 loops at a site outside of the TCR-pMHC interface. The CDR-CDR contacts made by the CDR1 αY31 residue in each Vα2+ TCR structures are unique. Nevertheless, the alanine substitution of the CDR1 αY31 residue modulated the ability of a set of Vα2.3+, Vα2.8+ and Vα2.9+ T cells to respond to their cognate ligand. Because in vivo CD4 T cell activation to viral epitopes are highly sensitive to the dwell time of TCR-pMHC interactions, the 100-fold changes in in vitro dose response can determine whether a CD4 T cell productively enters into an immune response (74). A result of the amino acid expressed at the CDR1α 31 position being variable (50), is that this CDR position may induce a variety of indirect effects based on the particular V gene segment.
To identify how the Vα2 CDR1 αY31 side chain contributes to the ability of TCRs to bind their cognate ligand, we compared the co-crystal structures of the J809.B5 TCR and J809.B5 αY31A TCRs bound to IAb-3K. The CDR1 αY31A substitution does not change the conformation of the J809.B5 TCR in complex with IAb-3K. Rather, by slowing the off-rate, the CDR1 αY31 side chain stabilizes the TCR conformation that binds IAb-3K. Through analyzing multiple Vα2+ T cells, we observed that the CDR1 αY31 side chain does not contribute uniform effects on either cognate peptide recognition or the ability of T cells to recognize alternate peptides. The CDR1 αY31A substitution also caused two of the Vα2+ T cells to acquire self-reactivity, indicating that this CDR1 residue can contribute to the ability of T cells to distinguish foreign-antigens from self-antigens. These findings argue that combinatorial diversity allows germline encoded TCR V gene residues to contribute to the specificity of TCRs in multiple ways. CDR1 and CDR2 residues can make direct contacts with the pMHC ligand. In addition, CDR residues that do not contact the pMHC can affect the ability of TCRs to adopt or stabilize different pMHC binding conformations. The indirect effects created by TCR inter- and intra-chain interactions that regulate antigen specificity from positions outside of the binding interface can be seen as analogous to the allosteric regulation of protein function, that is controlled by V(D)J recombination and TCR V gene pairing (75). The common occurrence of interactions between CDR loops suggests that this is a general feature of TCRs, and that one of the functions of combinatorial diversity and T cell development is to equip mature T cells with TCRs that stabilize CDR loop conformations that specifically recognize antigens presented by host MHC.
Supplementary Material
Acknowledgments
This work was supported by a Searle Scholars Award to E.S.H. and NIH grants to E.S.H. (RO1-DK095077), NIH grants to L.J.S. (R01-AI-38996), and a NIH training grant to B.D.S. (T32 AI 007349). E.S.H. is a member of the UMass DERC (grant DK32520). Data for this study were measured at beamline X25 of the National Synchrotron Light Source, for which financial support comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and from the National Center for Research Resources (P41RR012408) and the National Institute of General Medical Sciences (P41GM103473) of the National Institutes of Health
The authors would like to thank Frances Crawford and John Kappler for the IAb-random peptide baculovirus display library, and Rebecca Smith for assistance with some experiments.
Footnotes
There are two Supplementary Figures and two Tables associated with this manuscript.
References
- 1.Jenkins MK, Chu HH, McLachlan JB, Moon JJ. On the composition of the preimmune repertoire of T cells specific for Peptide-major histocompatibility complex ligands. Annu Rev Immunol. 2010;28:275–294. doi: 10.1146/annurev-immunol-030409-101253. [DOI] [PubMed] [Google Scholar]
- 2.Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 3.Bevan MJ. In a radiation chimaera, host H-2 antigens determine immune responsiveness of donor cytotoxic cells. Nature. 1977;269:417–418. doi: 10.1038/269417a0. [DOI] [PubMed] [Google Scholar]
- 4.Hogquist KA, Tomlinson AJ, Kieper WC, McGargill MA, Hart MC, Naylor S, Jameson SC. Identification of a naturally occurring ligand for thymic positive selection. Immunity. 1997;6:389–399. doi: 10.1016/s1074-7613(00)80282-4. [DOI] [PubMed] [Google Scholar]
- 5.Hu Q, Bazemore Walker CR, Girao C, Opferman JT, Sun J, Shabanowitz J, Hunt DF, Ashton-Rickardt PG. Specific recognition of thymic self-peptides induces the positive selection of cytotoxic T lymphocytes. Immunity. 1997;7:221–231. doi: 10.1016/s1074-7613(00)80525-7. [DOI] [PubMed] [Google Scholar]
- 6.Lo WL, Felix NJ, Walters JJ, Rohrs H, Gross ML, Allen PM. An endogenous peptide positively selects and augments the activation and survival of peripheral CD4+ T cells. Nat Immunol. 2009;10:1155–1161. doi: 10.1038/ni.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebert PJ, Jiang S, Xie J, Li QJ, Davis MM. An endogenous positively selecting peptide enhances mature T cell responses and becomes an autoantigen in the absence of microRNA miR-181a. Nat Immunol. 2009;10:1162–1169. doi: 10.1038/ni.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kappler JW, Roehm N, Marrack P. T cell tolerance by clonal elimination in the thymus. Cell. 1987;49:273–280. doi: 10.1016/0092-8674(87)90568-x. [DOI] [PubMed] [Google Scholar]
- 9.Palmer E, Naeher D. Affinity threshold for thymic selection through a T-cell receptor-co-receptor zipper. Nat Rev Immunol. 2009;9:207–213. doi: 10.1038/nri2469. [DOI] [PubMed] [Google Scholar]
- 10.Venturi V, Rudd BD, Davenport MP. Specificity, promiscuity, and precursor frequency in immunoreceptors. Curr Opin Immunol. 2013;25:639–645. doi: 10.1016/j.coi.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Morris GP, Allen PM. How the TCR balances sensitivity and specificity for the recognition of self and pathogens. Nat Immunol. 2012;13:121–128. doi: 10.1038/ni.2190. [DOI] [PubMed] [Google Scholar]
- 12.Zerrahn J, Held W, Raulet DH. The MHC reactivity of the T cell repertoire prior to positive and negative selection. Cell. 1997;88:627–636. doi: 10.1016/s0092-8674(00)81905-4. [DOI] [PubMed] [Google Scholar]
- 13.Merkenschlager M, Graf D, Lovatt M, Bommhardt U, Zamoyska R, Fisher AG. How many thymocytes audition for selection? J Exp Med. 1997;186:1149–1158. doi: 10.1084/jem.186.7.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stritesky GL, Xing Y, Erickson JR, Kalekar LA, Wang X, Mueller DL, Jameson SC, Hogquist KA. Murine thymic selection quantified using a unique method to capture deleted T cells. Proc Natl Acad Sci U S A. 2013;110:4679–4684. doi: 10.1073/pnas.1217532110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin L, Scott-Browne J, Kappler JW, Gapin L, Marrack P. T cells and their eons-old obsession with MHC. Immunol Rev. 2012;250:49–60. doi: 10.1111/imr.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia KC. Reconciling views on T cell receptor germline bias for MHC. Trends Immunol. 2012;33:429–436. doi: 10.1016/j.it.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Laethem F, Tikhonova AN, Singer A. MHC restriction is imposed on a diverse T cell receptor repertoire by CD4 and CD8 co-receptors during thymic selection. Trends Immunol. 2012;33:437–441. doi: 10.1016/j.it.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gras S, Burrows SR, Turner SJ, Sewell AK, McCluskey J, Rossjohn J. A structural voyage toward an understanding of the MHC-I-restricted immune response: lessons learned and much to be learned. Immunol Rev. 2012;250:61–81. doi: 10.1111/j.1600-065X.2012.01159.x. [DOI] [PubMed] [Google Scholar]
- 19.Huseby ES, White J, Crawford F, Vass T, Becker D, Pinilla C, Marrack P, Kappler JW. How the T cell repertoire becomes peptide and MHC specific. Cell. 2005;122:247–260. doi: 10.1016/j.cell.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 20.Huseby ES, Crawford F, White J, Marrack P, Kappler JW. Interface-disrupting amino acids establish specificity between T cell receptors and complexes of major histocompatibility complex and peptide. Nat Immunol. 2006;7:1191–1199. doi: 10.1038/ni1401. [DOI] [PubMed] [Google Scholar]
- 21.Stadinski BD, Trenh P, Smith RL, Bautista B, Huseby PG, Li G, Stern LJ, Huseby ES. A role for differential variable gene pairing in creating T cell receptors specific for unique major histocompatibility ligands. Immunity. 2011;35:694–704. doi: 10.1016/j.immuni.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scott-Browne JP, White J, Kappler JW, Gapin L, Marrack P. Germline-encoded amino acids in the alphabeta T-cell receptor control thymic selection. Nature. 2009;458:1043–1046. doi: 10.1038/nature07812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rubtsova K, Scott-Browne JP, Crawford F, Dai S, Marrack P, Kappler JW. Many different Vbeta CDR3s can reveal the inherent MHC reactivity of germline-encoded TCR V regions. Proc Natl Acad Sci U S A. 2009;106:7951–7956. doi: 10.1073/pnas.0902728106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blackman M, Yague J, Kubo R, Gay D, Coleclough C, Palmer E, Kappler J, Marrack P. The T cell repertoire may be biased in favor of MHC recognition. Cell. 1986;47:349–357. doi: 10.1016/0092-8674(86)90591-x. [DOI] [PubMed] [Google Scholar]
- 25.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 26.Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon’. Nat Immunol. 2007;8:975–983. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- 27.Dai S, Huseby ES, Rubtsova K, Scott-Browne J, Crawford F, Macdonald WA, Marrack P, Kappler JW. Crossreactive T Cells spotlight the germline rules for alphabeta T cell-receptor interactions with MHC molecules. Immunity. 2008;28:324–334. doi: 10.1016/j.immuni.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marrack P, Scott-Browne JP, Dai S, Gapin L, Kappler JW. Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu Rev Immunol. 2008;26:171–203. doi: 10.1146/annurev.immunol.26.021607.090421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia KC, Adams JJ, Feng D, Ely LK. The molecular basis of TCR germline bias for MHC is surprisingly simple. Nat Immunol. 2009;10:143–147. doi: 10.1038/ni.f.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Laethem F, Tikhonova AN, Pobezinsky LA, Tai X, Kimura MY, Le Saout C, Guinter TI, Adams A, Sharrow SO, Bernhardt G, Feigenbaum L, Singer A. Lck availability during thymic selection determines the recognition specificity of the T cell repertoire. Cell. 2013;154:1326–1341. doi: 10.1016/j.cell.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collins EJ, Riddle DS. TCR-MHC docking orientation: natural selection, or thymic selection? Immunol Res. 2008;41:267–294. doi: 10.1007/s12026-008-8040-2. [DOI] [PubMed] [Google Scholar]
- 32.Holland SJ, Bartok I, Attaf M, Genolet R, Luescher IF, Kotsiou E, Richard A, Wang E, White M, Coe DJ, Chai JG, Ferreira C, Dyson J. The T-cell receptor is not hardwired to engage MHC ligands. Proc Natl Acad Sci U S A. 2012;109:E3111–3118. doi: 10.1073/pnas.1210882109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borg NA, Ely LK, Beddoe T, Macdonald WA, Reid HH, Clements CS, Purcell AW, Kjer-Nielsen L, Miles JJ, Burrows SR, McCluskey J, Rossjohn J. The CDR3 regions of an immunodominant T cell receptor dictate the ‘energetic landscape’ of peptide-MHC recognition. Nat Immunol. 2005;6:171–180. doi: 10.1038/ni1155. [DOI] [PubMed] [Google Scholar]
- 34.Bulek AM, Cole DK, Skowera A, Dolton G, Gras S, Madura F, Fuller A, Miles JJ, Gostick E, Price DA, Drijfhout JW, Knight RR, Huang GC, Lissin N, Molloy PE, Wooldridge L, Jakobsen BK, Rossjohn J, Peakman M, Rizkallah PJ, Sewell AK. Structural basis for the killing of human beta cells by CD8(+) T cells in type 1 diabetes. Nat Immunol. 2012;13:283–289. doi: 10.1038/ni.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson J, Waller MJ, Fail SC, McWilliam H, Lopez R, Parham P, Marsh SG. The IMGT/HLA database. Nucleic Acids Res. 2009;37:D1013–1017. doi: 10.1093/nar/gkn662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huseby ES, Crawford F, White J, Kappler J, Marrack P. Negative selection imparts peptide specificity to the mature T cell repertoire. Proc Natl Acad Sci U S A. 2003;100:11565–11570. doi: 10.1073/pnas.1934636100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White J, Pullen A, Choi K, Marrack P, Kappler JW. Antigen recognition properties of mutant V beta 3+ T cell receptors are consistent with an immunoglobulin-like structure for the receptor. J Exp Med. 1993;177:119–125. doi: 10.1084/jem.177.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Govern CC, Paczosa MK, Chakraborty AK, Huseby ES. Fast on-rates allow short dwell time ligands to activate T cells. Proc Natl Acad Sci U S A. 2010;107:8724–8729. doi: 10.1073/pnas.1000966107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tynan FE, Reid HH, Kjer-Nielsen L, Miles JJ, Wilce MC, Kostenko L, Borg NA, Williamson NA, Beddoe T, Purcell AW, Burrows SR, McCluskey J, Rossjohn J. A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule. Nat Immunol. 2007;8:268–276. doi: 10.1038/ni1432. [DOI] [PubMed] [Google Scholar]
- 40.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 41.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 44.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Kleywegt GJ. Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr D Biol Crystallogr. 1996;52:842–857. doi: 10.1107/S0907444995016477. [DOI] [PubMed] [Google Scholar]
- 47.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 48.Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 49.Reinherz EL, Tan K, Tang L, Kern P, Liu J, Xiong Y, Hussey RE, Smolyar A, Hare B, Zhang R, Joachimiak A, Chang HC, Wagner G, Wang J. The crystal structure of a T cell receptor in complex with peptide and MHC class II. Science. 1999;286:1913–1921. doi: 10.1126/science.286.5446.1913. [DOI] [PubMed] [Google Scholar]
- 50.Arden B, Clark SP, Kabelitz D, Mak TW. Mouse T-cell receptor variable gene segment families. Immunogenetics. 1995;42:501–530. doi: 10.1007/BF00172177. [DOI] [PubMed] [Google Scholar]
- 51.Holler PD, Kranz DM. Quantitative analysis of the contribution of TCR/pepMHC affinity and CD8 to T cell activation. Immunity. 2003;18:255–264. doi: 10.1016/s1074-7613(03)00019-0. [DOI] [PubMed] [Google Scholar]
- 52.Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley DC. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 53.Garcia KC, Degano M, Stanfield RL, Brunmark A, Jackson MR, Peterson PA, Teyton L, Wilson IA. An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex. Science. 1996;274:209–219. doi: 10.1126/science.274.5285.209. [DOI] [PubMed] [Google Scholar]
- 54.Crawford F, Huseby E, White J, Marrack P, Kappler JW. Mimotopes for alloreactive and conventional T cells in a peptide-MHC display library. PLoS Biol. 2004;2:E90. doi: 10.1371/journal.pbio.0020090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arden B. Conserved motifs in T-cell receptor CDR1 and CDR2: implications for ligand and CD8 co-receptor binding. Curr Opin Immunol. 1998;10:74–81. doi: 10.1016/s0952-7915(98)80035-6. [DOI] [PubMed] [Google Scholar]
- 56.Garcia KC, Teyton L, Wilson IA. Structural basis of T cell recognition. Annu Rev Immunol. 1999;17:369–397. doi: 10.1146/annurev.immunol.17.1.369. [DOI] [PubMed] [Google Scholar]
- 57.Al-Lazikani B, Lesk AM, Chothia C. Canonical structures for the hypervariable regions of T cell alphabeta receptors. J Mol Biol. 2000;295:979–995. doi: 10.1006/jmbi.1999.3358. [DOI] [PubMed] [Google Scholar]
- 58.Colf LA, Bankovich AJ, Hanick NA, Bowerman NA, Jones LL, Kranz DM, Garcia KC. How a single T cell receptor recognizes both self and foreign MHC. Cell. 2007;129:135–146. doi: 10.1016/j.cell.2007.01.048. [DOI] [PubMed] [Google Scholar]
- 59.Newell EW, Ely LK, Kruse AC, Reay PA, Rodriguez SN, Lin AE, Kuhns MS, Garcia KC, Davis MM. Structural basis of specificity and cross-reactivity in T cell receptors specific for cytochrome c-I-E(k) J Immunol. 2011;186:5823–5832. doi: 10.4049/jimmunol.1100197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tynan FE, Burrows SR, Buckle AM, Clements CS, Borg NA, Miles JJ, Beddoe T, Whisstock JC, Wilce MC, Silins SL, Burrows JM, Kjer-Nielsen L, Kostenko L, Purcell AW, McCluskey J, Rossjohn J. T cell receptor recognition of a ‘super-bulged’ major histocompatibility complex class I-bound peptide. Nat Immunol. 2005;6:1114–1122. doi: 10.1038/ni1257. [DOI] [PubMed] [Google Scholar]
- 61.Ding YH, Smith KJ, Garboczi DN, Utz U, Biddison WE, Wiley DC. Two human T cell receptors bind in a similar diagonal mode to the HLA-A2/Tax peptide complex using different TCR amino acids. Immunity. 1998;8:403–411. doi: 10.1016/s1074-7613(00)80546-4. [DOI] [PubMed] [Google Scholar]
- 62.Lopez-Sagaseta J, Sibener LV, Kung JE, Gumperz J, Adams EJ. Lysophospholipid presentation by CD1d and recognition by a human Natural Killer T-cell receptor. EMBO J. 2012;31:2047–2059. doi: 10.1038/emboj.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adams JJ, Narayanan S, Liu B, Birnbaum ME, Kruse AC, Bowerman NA, Chen W, Levin AM, Connolly JM, Zhu C, Kranz DM, Garcia KC. T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex. Immunity. 2011;35:681–693. doi: 10.1016/j.immuni.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reiser JB, Darnault C, Gregoire C, Mosser T, Mazza G, Kearney A, van der Merwe PA, Fontecilla-Camps JC, Housset D, Malissen B. CDR3 loop flexibility contributes to the degeneracy of TCR recognition. Nat Immunol. 2003;4:241–247. doi: 10.1038/ni891. [DOI] [PubMed] [Google Scholar]
- 65.Borbulevych OY, Santhanagopolan SM, Hossain M, Baker BM. TCRs used in cancer gene therapy cross-react with MART-1/Melan-A tumor antigens via distinct mechanisms. J Immunol. 2011;187:2453–2463. doi: 10.4049/jimmunol.1101268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Felix NJ, Donermeyer DL, Horvath S, Walters JJ, Gross ML, Suri A, Allen PM. Alloreactive T cells respond specifically to multiple distinct peptide-MHC complexes. Nat Immunol. 2007;8:388–397. doi: 10.1038/ni1446. [DOI] [PubMed] [Google Scholar]
- 67.Deng L, Langley RJ, Wang Q, Topalian SL, Mariuzza RA. Structural insights into the editing of germ-line-encoded interactions between T-cell receptor and MHC class II by Valpha CDR3. Proc Natl Acad Sci U S A. 2012;109:14960–14965. doi: 10.1073/pnas.1207186109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mazza C, Auphan-Anezin N, Gregoire C, Guimezanes A, Kellenberger C, Roussel A, Kearney A, van der Merwe PA, Schmitt-Verhulst AM, Malissen B. How much can a T-cell antigen receptor adapt to structurally distinct antigenic peptides? EMBO J. 2007;26:1972–1983. doi: 10.1038/sj.emboj.7601605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu YC, Miles JJ, Neller MA, Gostick E, Price DA, Purcell AW, McCluskey J, Burrows SR, Rossjohn J, Gras S. Highly divergent T-cell receptor binding modes underlie specific recognition of a bulged viral peptide bound to a human leukocyte antigen class I molecule. J Biol Chem. 2013;288:15442–15454. doi: 10.1074/jbc.M112.447185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Correia-Neves M, Waltzinger C, Mathis D, Benoist C. The shaping of the T cell repertoire. Immunity. 2001;14:21–32. doi: 10.1016/s1074-7613(01)00086-3. [DOI] [PubMed] [Google Scholar]
- 71.Berg LJ, Pullen AM, Fazekas de St Groth B, Mathis D, Benoist C, Davis MM. Antigen/MHC-specific T cells are preferentially exported from the thymus in the presence of their MHC ligand. Cell. 1989;58:1035–1046. doi: 10.1016/0092-8674(89)90502-3. [DOI] [PubMed] [Google Scholar]
- 72.Scott B, Bluthmann H, Teh HS, von Boehmer H. The generation of mature T cells requires interaction of the alpha beta T-cell receptor with major histocompatibility antigens. Nature. 1989;338:591–593. doi: 10.1038/338591a0. [DOI] [PubMed] [Google Scholar]
- 73.Morris GP, Ni PP, Allen PM. Alloreactivity is limited by the endogenous peptide repertoire. Proc Natl Acad Sci U S A. 2011;108:3695–3700. doi: 10.1073/pnas.1017015108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vanguri V, Govern CC, Smith R, Huseby ES. Viral antigen density and confinement time regulate the reactivity pattern of CD4 T-cell responses to vaccinia virus infection. Proc Natl Acad Sci U S A. 2013;110:288–293. doi: 10.1073/pnas.1208328110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nussinov R, Tsai CJ. Allostery in disease and in drug discovery. Cell. 2013;153:293–305. doi: 10.1016/j.cell.2013.03.034. [DOI] [PubMed] [Google Scholar]
- 76.DeLano WL. Unraveling hot spots in binding interfaces: progress and challenges. Curr Opin Struct Biol. 2002;12:14–20. doi: 10.1016/s0959-440x(02)00283-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.