Summary
PTEN dysfunction plays a crucial role in the pathogenesis of hereditary and sporadic cancers. Here we show that PTEN homo-dimerizes, and in this active conformation exerts lipid phosphatase activity on PtdIns(3,4,5)P3. We demonstrate that catalytically inactive cancer-associated PTEN mutants hetero-dimerize with wild-type PTEN and constrain its phosphatase activity in a dominant-negative manner. To study the consequences of homo- and hetero-dimerization of wild-type and mutant PTEN in vivo, we generated Pten knock-in mice harboring two cancer-associated PTEN mutations (PtenC124S and PtenG129E). Heterozygous PtenC124S/+ and PtenG129E/+ cells and tissues exhibit increased sensitivity to PI3-K/Akt activation compared to wild-type and Pten+/- counterparts, while this difference is no longer apparent between PtenC124S/- and Pten-/- cells. Notably, PtenKI mice are more tumor-prone and display features reminiscent of complete Pten loss. Our findings reveal that PTEN loss and PTEN mutations are not synonymous, and define a new working model for the function and regulation of PTEN.
Introduction
Phosphatase and Tensin Homolog Deleted on Chromosome Ten (PTEN) is a tumor suppressor frequently lost or mutated in human cancers and in a number of tumor syndromes, referred to as “PTEN hamartoma tumor syndromes” (PHTS), which include Cowden Disease (CD) and Bannayan-Zonana syndrome (Hollander et al., 2011).
To study the consequences of Pten loss in vivo, we and others generated animal models with partial and total loss of Pten (Di Cristofano et al., 1998; Podsypanina et al., 1999; Suzuki et al., 1998). Total Pten loss was found to lead to embryonic lethality, and additional investigations in a hypomorphic allelic series of mice with sequentially lower Pten expression revealed that even small reductions in Pten doses can elicit cancer phenotypes (Alimonti et al., 2010; Trotman et al., 2003). Conversely, systemic elevation of Pten through transgenic overexpression results in a constitutively augmented tumor-suppressive state (Garcia-Cao et al., 2012).
PTEN functions as a dual-specificity protein phosphatase (DSP) with predominant enzymatic activity on phosphoinositides (Maehama and Dixon, 1998). As a phospholipid phosphatase, PTEN catalyzes the hydrolysis of the second messenger PtdIns (3,4,5)P3 (PIP3) and counteracts the activation of the PI3K/AKT pathway, thus regulating cellular growth, proliferation and metabolism (Maehama and Dixon, 1998).
In line with its protein phosphatase function, PTEN has been shown to dephosphorylate phospho-peptides in vitro (Myers et al., 1998), and reported phospho-protein targets include the focal adhesion kinase, c-SRC, as well as PTEN itself (Tamura et al., 1999; Tibarewal et al., 2012; Zhang et al., 2011).
Heterozygous deletion of Pten in mice faithfully phenocopies biological features found in many human tumors with partial loss of PTEN (Di Cristofano et al., 1998). However, reports indicate that genetic loss of PTEN and mutations leading to PTEN loss-of-function may not be equivalent. For instance, Marsh et al. reported a genotype-phenotype correlation in patients diagnosed with CD who developed several tumors, including breast tumors. Importantly, they found that patients harboring missense PTEN mutations in the phosphatase core developed higher numbers of lesions than patients with truncating mutations (Marsh et al., 1998). This led us to hypothesize that expression of catalytically inactive mutant PTEN enzyme may be more unfavorable than PTEN protein loss.
Regulation of PTEN function occurs through various post-translational modifications implicated in PTEN membrane recruitment, sub-cellular localization, or protein-protein interactions (Wang and Jiang, 2008). Structurally, PTEN belongs to the Class I Cys-based protein tyrosine phosphatase (PTP), and more specifically to the VH1-like family (Alonso et al., 2004). PTEN contains an N-terminal phosphatase domain with a conserved active site; a C-terminal C2 domain followed by two PEST sequences, and a PDZ-binding domain (Lee et al., 1999). It has been reported that PTEN interacts with a number of PDZ-domain bearing proteins to achieve higher levels of complex formation (Sotelo et al., 2012; Vazquez et al., 2001). We therefore hypothesized, and have here demonstrated, that PTEN can interact with itself. We show that dimer PTEN is active toward its phosphoinositide substrate PIP3, and thereby inhibits the activation of the PI3K/AKT signaling pathway. Critically, we find that in a dimeric conformation, cancer-associated missense mutations have dominant negative consequences over wild-type protein function, with ensuing implications for tumorigenesis.
Results
PTEN exists in a dimeric complex
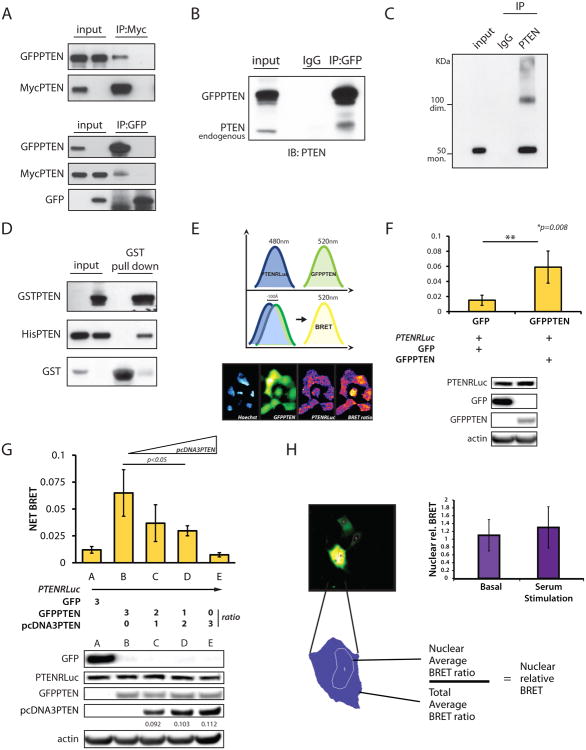
Given that VH1-like phosphatases are known to exist in higher order complexes/dimers (Koksal and Cingolani, 2011), we examined whether PTEN could form similar complexes. For this, we performed co-immunoprecipitation (co-IP) experiments using the PTEN-null cell line PC3. By co-transfecting differentially tagged PTEN variants we were able to reciprocally co-IP GFPPTEN and MycPTEN (Figure 1A). Moreover, in 293T cells overexpressing GFPPTEN, we were able to co-IP endogenous PTEN (Figure 1B).
Figure 1. PTEN exists in a dimeric complex.
(A) Co-IPs from total lysate of PC3 cells. IP of MycPTEN (top panel), and IP of GFPPTEN (bottom panel) with specific tag antibodies revealed reciprocal PTEN-PTEN interaction.
(B) Co-IPs from total lysate of 293T cells, transfected with GFPPTENWT and IP with anti-GFP antibody. Western blot was probed with a PTEN antibody.
(C) NIH3T3 cell lysates were IP with anti-rabbit PTEN antibody; native elution and Western blots show monomer and dimer of the protein using mouse anti-PTEN antibody.
(D) In bacteria, GST-PTEN specifically pulls down His-PTEN.
(E) Renilla and GFP emissions are detected at the indicated wavelengths. Administration of Coelenterazine induces Renilla excitation which generates BRET signal when in proximity to the GFP. Bottom panel: images of fluorescent signals generated by the different chimeras.
(F) Detection of PTEN-PTEN interaction by BRET in PC3 cells transfected with the indicated plasmids. Bottom panel shows expression levels of the different chimeras. Mean values with associated SD are shown.
(G) Competition assays performed by BRET. Bottom panel, expression levels of the indicated chimeras. Quantification of Un-tagged PTEN cloned in the pcDNA3.1 vector is normalized to β-actin. Mean values with associated SD are shown.
(H) In PC3, BRET signal is collected in a single cell fashion and nuclear emission normalized over average of total emission. Mean values with relative SD are shown.
Monomeric PTEN has a molecular weight of 50-55 KDa in denaturing conditions. If a dimeric PTEN existed, it would migrate at double that size in non-reducing and non-denaturing SDS-PAGE. Accordingly, IP and native elution of MycPTEN from transfected PC3 cells revealed the presence of two bands consistent with the monomeric and dimeric status of the protein (100KDa circa) (Figure S1A). To control for over-expression artifacts, we repeated this assay by pulling down endogenous Pten in NIH3T3 and confirmed the presence of two bands (Figure 1C).
Next, we investigated the contribution of covalent and non-covalent interactions to dimer formation, and found that disulfide bonds are not a major requirement for the stabilization of this interaction (Figure S1B). We also verified PTEN homo-dimerization in vitro by co-expressing GST-PTEN and His-PTEN in bacteria. In these experiments we successfully recovered His-PTEN in GST-PTEN pull-downs (Figure 1D).
To further validate the direct binding between PTEN molecules in a eukaryotic system, we employed Bioluminescence Resonance Energy Transfer (BRET). To this end, we used Renilla luciferase-PTEN (PTENRluc) as energy donor, and GFPPTEN as energy acceptor (Figure 1E); coelenterazine was used as substrate for the luciferase. Co-expression of PTENRluc with GFPPTEN generated a significant increase in the total BRET signal compared to empty GFP, with GFP emission only occurring when in close proximity (less than 100 Å) to the luminescent PTENRluc (Figure 1F). We also performed competition assays. Co-expression of donor and acceptor proteins with increasing doses of untagged PTEN showed a reduction of net BRET, providing further evidence of direct PTEN-PTEN interaction (Figure 1G).
Finally, we sought to determine whether PTEN dimerization occurs in both the nucleus and cytoplasm. Utilizing BRET, we found no variations in nuclear emission over the total BRET signal, neither in basal conditions nor upon serum stimulation, suggesting that PTEN dimer likely exists in both compartments (Figure 1H). This finding was confirmed in co-IPs performed upon nuclear vs. cytoplasmic fractionation (Figures S1C and S1D). Thus, we have demonstrated by multiple approaches that PTEN can exist in a homo-dimeric complex.
PTEN dimer is catalytically active
To identify the protein domains responsible for PTEN dimer formation, we generated GST-fusion proteins with PTEN N-terminus (GST-PTENΔC-terminus) and PTEN C-terminus domains (GST-PTENΔN-terminus) (Figure 2A). Co-expression of GST-PTEN full length (FL) with His-PTENFL confirmed the interaction between the two FL proteins (Figure 2B). Notably, His-PTENFL was pulled down by both GST-PTEN-domains in bacteria (Figure 2B). We then tested the binding by co-IPs in PC3 cells and confirmed that in eukaryotic cells PTEN uses multiple interfaces along the entire protein to achieve a dimeric conformation (Figures S2A and S2B).
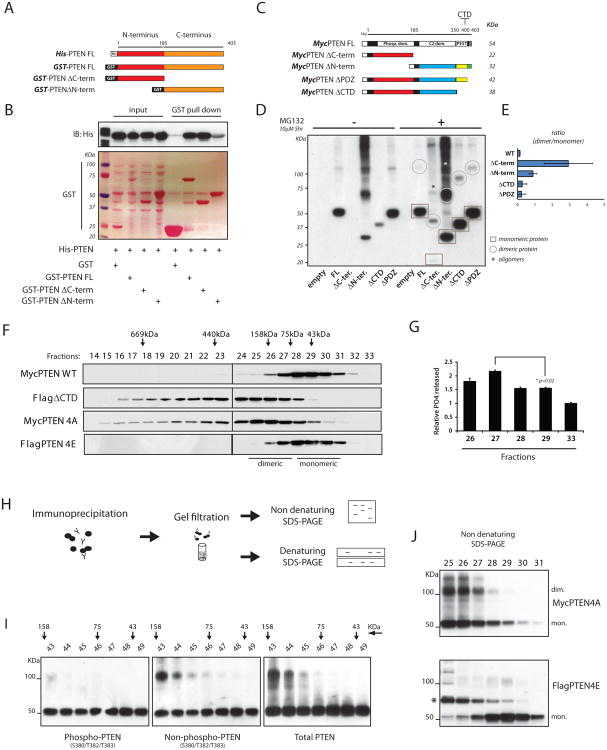
Figure 2. Dimerization defines a pool of catalytically active PTEN.
(A) Diagram of recombinant proteins showing PTEN full length (FL) and deletion mutants. In red, N-terminus domain, amino-acid (a.a.) 1-185; in orange, C-terminus domain, a.a. 186-403.
(B) GST-PTENFL and domains were purified (Ponceau-S staining); His-PTEN pulled down is detected by Western blot.
(C) Schematic representing series of Myc-tagged PTENFL and deletion mutant vectors. Predicted molecular weights are indicated.
(D) PC3 cells were transfected with the indicated expression vectors. Total lysates were resolved by non-reducing SDS-PAGE and probed with an anti-Myc antibody. Circles and squares indicate monomeric and dimeric PTEN conformations, respectively. Asterisks indicate oligomers of PTEN domains.
(E) Ratio between PTEN dimer/monomer in PTEN FL and deletion mutant series. Mean values with associated SD are shown.
(F) Lysates from HEK293 cells transfected with the indicated PTEN vectors were separated by gel filtration. Fractions were resolved by SDS-PAGE and probed with specific tag antibodies.
(G) Fractions containing different conformations of PTEN and collected as in (F) were tested for their activity toward PIP3, and normalized over protein levels. Mean values from triplicate wells with associated SD are shown.
(H) Experimental flow chart: cell lysates were subjected to IP with an anti-Myc antibody. Immuno-complexes were eluted in native conditions and separated by tandem-column gel filtration (See also Figure S2H). Collected fractions were resolved by reducing (Figure S2H) and non-reducing SDS-PAGE (Figure 2I).
(I) Fractions collected as in (H) were resolved by non-reducing SDS-PAGE and probed with the indicated PTEN antibodies.
(J) Non-reducing SDS-PAGE of eluted fractions generated as in (F) were blotted with anti-Myc (top panel) and anti-Flag (bottom panel) antibodies. Asterisk indicates a shift in FlagPTEN4E probably due to post-translational modifications.
To corroborate these findings, we tested the ability of different PTEN domains to homo-dimerize in vivo (Figure 2C). In non-reducing SDS-PAGE, total lysates from PC3 cells transfected with MycPTENFL, MycPTENΔN-terminus, or MycPTENΔPDZ revealed the appearance of upper bands at twice the size of the respective monomeric proteins. Strikingly, upon stabilization of MycPTENΔC-terminus and MycPTENΔCTD with the protease inhibitor MG132, we observed dimeric bands with all PTEN variants tested (Figure 2D), with the MycPTENΔC-terminus showing the highest propensity to self-associate (Figure 2E). Notably, the MycPTENΔC-terminus and MycPTENΔN-terminus exhibited a potential to oligomerize, as evidenced by additional higher molecular weight bands (Figure 2D, asterisks).
We also studied binding directionality, and found that while in a closed head-to-tail monomeric conformation PTEN is inactive toward its lipid substrate (Leslie and Foti, 2011), through head-to-head inter-molecular binding, it achieves a more active enzymatic conformation (Figure S2C-S2F).
We next tested the catalytic activity of antibody-purified PTEN dimers from PC3 cells. PTENFL displayed different affinities toward PTEN deletion mutant versions (Figure S2B), and higher levels of dimerization between PTENFL and PTEN domains correlated with higher levels of free phosphate release (Figure S2E).
We likewise performed gel filtration of various PTEN species followed by phosphatase assays using PIP3 as substrate. Total lysates of HEK293 cells with exogenous MycPTEN expression were fractioned and a low molecular weight range (30-to-160 KDa) identified by Western blot analysis (Figure 2F, top panel; and S2G). Importantly, eluted fractions corresponding to dimeric PTEN (fractions 26-27) generated higher phosphate release than fractions of a lower molecular weight which contained monomeric PTEN (fractions 28-29) (Figure 2G). Thus we conclude that PTEN dimerization defines a more active complex with respect to the less active PTEN monomer.
Phosphorylation of the PTEN tail regulates PTEN dimerization
Next we studied the conformational status of PTEN by gel filtration analyses. FLAG-tagged PTEN without the C-terminal tail (FlagPTENΔCTD) was expressed in HEK293 cells and total lysates were fractioned. Western blot analysis revealed a striking upward shift of tail-less PTEN compared to PTENFL, indicating an enriched dimeric status (Figure 2F).
Because the physiologic role of the PTEN tail is mediated by its phosphorylation status (Vazquez et al., 2000), we performed gel filtration analyses by using phospho-mimetic and phospho-dead mutant-tail versions of PTEN. We focused specifically on amino-acids S380, T382, T383, and S385. By expressing non-phosphorylatable PTEN (PTEN S380A, T382A, T383A, S385A: PTEN4A) we functionally mimicked deletion of the PTEN C-terminal tail. Notably, the PTEN4A presented the same upward shift we found with expression of PTENΔCTD (Figure 2F). In contrast, expression of a phospho-mimetic PTEN (PTEN4E) prevented this shift, indicating a possible role for PTEN tail-phosphorylation in governing its dimeric status (Figure 2F).
Next we assessed the phosphorylation level of PTEN dimer. To this end, we immuno-purified PTEN and eluted the complex in non-reducing conditions. After tandem-column gel filtrations we analyzed the immuno-complex by reducing and non-reducing SDS-PAGE (Figure 2H). In reducing SDS-PAGE, immuno-purified PTENFL appeared in two peaks corresponding to a “low” and a “high-molecular weight complex” (Figure S2H). We then tested the phospho/non-phospho status of the dimeric PTEN in non-reducing SDS-PAGE. First, with a total PTEN antibody we confirmed the presence of PTEN as a monomer and dimer (Figure 2I, right panel). Importantly, we found that while a specific “phospho-PTEN” antibody only recognizes the monomeric form, the relative “non-phospho” PTEN showed the appearance of both monomeric and dimeric protein (Figure 2I, left and middle panels).
Finally, we tested total lysates from HEK293 cells expressing PTEN4A and PTEN4E. Here we found that while PTEN4E only runs as a single monomeric band (Figure 2J, bottom panel), PTEN4A showed monomeric and dimeric PTEN conformations (Figure 2J, top panel), in agreement with the shift found by gel filtration. Thus we demonstrate that phosphorylation of the C-terminal tail maintains PTEN as a monomer, whereas the absence of phosphorylation is associated with a dimeric conformation.
PTEN cancer-associated mutations exert dominant negative effects over wild-type protein
We then asked if mutant PTEN protein could form dimers, and whether mutations altered dimer formation or activity. The most studied cancer-associated PTEN mutations are the Cys-124 to Ser (C124S) and the Gly-129 to Glu (G129E). C124S mutation generates a catalytically dead PTEN variant that is associated with endometrial cancer and is reported to completely ablate PTEN phosphatase activity (Bonneau and Longy, 2000; Myers et al., 1997), whereas the G129E mutation is associated with CS and abrogates the phosphoinositide phosphatase function but retains activity towards phospho-peptides (Liaw et al., 1997; Myers et al., 1998). Both mutations lie in the PTEN catalytic core.
To test our hypothesis, we first analyzed the ability of MycPTENC124S and MycPTENG129E to homo-dimerize. In non-reducing conditions we found that both mutants appear as monomers and dimers, like the wild-type protein (Figure 3A). PTEN mutants were also able to form hetero-dimers with wild-type PTEN as found in our co-IP experiments (Figure 3B). BRET experiments further demonstrated that PTENWT-Rluc interacted with either GFPPTEN-C124S or G129E (Figure 3C).
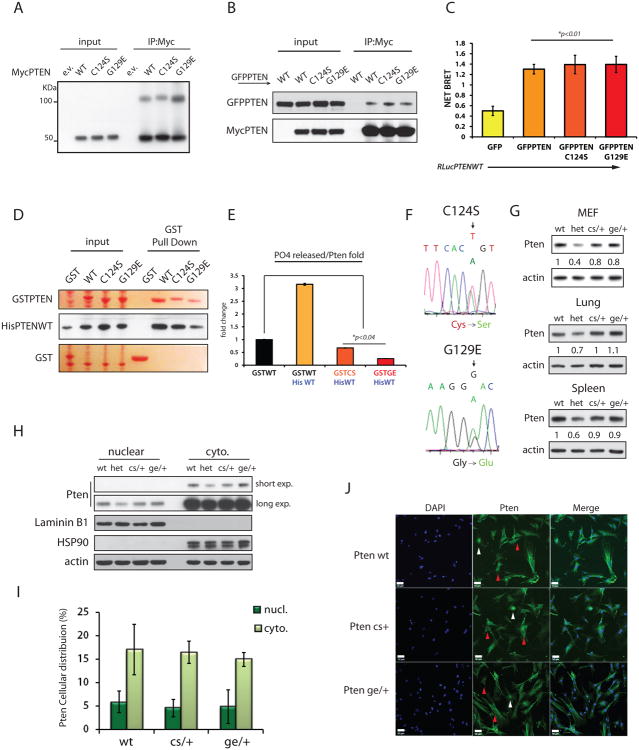
Figure 3. PTENC124S and PTENG129E mutations hetero-dimerize with PTEN wild-type and inhibit its phosphatase function.
(A) Non-reducing SDS-PAGE of lysates from PC3 cells transfected with MycPTENWT or mutant vectors, and IP with an anti-Myc antibody.
(B) IPs from PC3 cell lysates transfected with the indicated vectors, were performed and Western blots probed with an anti-PTEN antibody.
(C) BRET analysis on PC3 cells complemented with the indicated vectors. Mean values with associated SD are shown.
(D) GST-pull down (in Ponceau-S staining) reveals binding between prokaryotic PTEN WT and mutant versions. Western blots are probed with anti-His antibody.
(E) Fractions of PTEN dimers purified as in (D), plus GST-PTENWT alone were purified. Catalytic activity of PTEN dimers was tested in phosphatase assays using PIP3 as substrate. PO4 released is normalized over levels of His-PTEN pulled down, and is shown relative to the PO4 released by GST-PTENWT alone. Mean values of triplicate wells with associated SD are shown.
(F) Sequencing of Pten-exon5 amplified from DNA of targeted ES cells. Electropherograms show expression of both Ptenwt and Pten mutated alleles (Top panel C124S, T-to-A; bottom panel G129E, G-to-A).
(G) Analysis of Pten protein levels. Total lysates of MEFs and adult tissues (lung and spleen) derived from Pten+/+, Pten+/-, PtenC124S/+, and PtenG129E/+ mice were resolved by SDS-PAGE. Pten quantification is normalized to β-actin and relative to wt band.
(H) Nuclear vs. Cytoplasmic fractionation of MEFs. Comparable distribution of Pten was observed across the different genotypes.
(I) Pten IF in MEFs: 120 cells per field (200X magnification) were scored on average. Percentage of Pten accumulation in the nucleus vs. cytoplasm (at the membrane and in the cytosol) is shown with associated SD.
(J) Representative IF showing endogenous nuclear (white arrow-heads) and cytoplasmic (red arrow-heads) distribution of Pten in MEFs. Scale bar is 50μm.
In bacteria, GST pull-down experiments confirmed that His-PTENWT binds both GST-tagged PTEN mutants (Figure 3D). Importantly, purified hetero-dimers were tested for phosphatase activity on PIP3. In control experiments GST-PTENWT alone or a His-PTENWT:GST-PTENWT mix produced effective free phosphate release. However, hetero-dimers comprised of His-PTENWT:GSTPTENC124S or His-PTENWT:GSTPTENG129E had reduced capacity to hydrolyze PIP3 compared to GST-PTENWT (Figure 3E), leading us to consider the possibility that in a heterodimeric state, the catalytically inactive mutations inhibited activity of the wild-type PTEN protein towards PIP3, as we next demonstrated in vivo.
Generation and characterization of Pten knock-in mutant mice
To investigate the physiological consequences of PTEN-PTEN-mutant heterodimers we generated mouse models expressing PTENC124S and PTENG129E (Figures S3A and S3B). For the PtenC124S/+ and PtenG129E/+ mouse models, we substituted T to A at position 370 or G to A in position 386 of Pten exon5, respectively (Figure 3F).
In performing the in vivo characterization of these new PtenKI mice, we also aimed to investigate two critical aspects of PTEN regulation: stability and localization. First, as many PTEN missense point mutations render PTEN unstable (Georgescu et al., 1999), we measured the mutant Pten proteins levels in our PtenKI mice. We observed that PtenC124S/+ and PtenG129E/+ express a total level of Pten that is comparable to levels in Pten wild-type mice (Figure 3G). Second, to exclude the contribution of defective cellular distribution to our analysis, we performed cell-fractionation and immunofluorescence (IF) analyses. Here we found that PtenC124S/+ and PtenG129E/+ MEFs (Figures 3H-3J) displayed comparable Pten localization patterns to wild type cells.
In vivo confirmation of the dominant negative functions of mutant Pten
Since homozygous Pten loss is lethal in embryogenesis, we investigated the possibility of generating homozygous PtenKI mice. 3 independent crosses per mutation failed to yield live pups with a homozygous mutant genotype, indicating that the lipid phosphatase activity of Pten is an essential function during embryogenesis (Figures S4A and S4B).
To evaluate the consequences of harboring loss-of-function Pten mutants in heterozygosity, we established cohorts of PtenC124S/+, PtenG129E/+, and Pten+/- mice as controls. Since Pten heterozygosity initiates neoplastic transformation of epithelial tissues, and leads to severe lympho-proliferation (Di Cristofano et al., 1999), we collected a number of organ samples for histological analysis. We found that young mice between 8 and 16 weeks of age showed hyperplastic changes in the lymph nodes, with expansion of T-cells (data not shown). Surprisingly, unlike Pten+/- mice, a significant number of PtenC124S/+, PtenG129E/+ mice developed lympho-proliferation features by 16 weeks (Figure S3E). Accordingly, older PtenKI mice had increased spleen weights, compared to Pten+/- mice, owing to severe extra-medullary hematopoiesis and lymphoid hyperplasia; we also found extended expansion of white blood cells in the peripheral blood (Figure S3F). In solid tissues, the tumor spectrum of PtenKI mice was indistinguishable from Pten+/- mice, with lesions developing in several glands and organs (Figures S3C and S3D). However, histological assessments at different time points revealed that a greater proportion of PtenKI mice developed adenomas of the thyroid, adrenal, and gall bladder, with 2 out of 29 PtenC124S/+, and 3 out of 37 PtenG129E/+ male mice also developing invasive adenocarcinoma of the thyroid. In addition, 3 PtenC124S/+ mice developed lung adenomas, and 4 out of 30 PtenG129E/+ female mice developed liver adenomas, a lesion never observed in our Pten+/- mice (Figure S3C and S3D). In mammary tissues, 68.4% of our Pten+/-mice between 9 and 12 months developed hyperplastic lesions (13 out of 29, 44.8%) or small adenocarcinomas (7 out of 29, 24.1%). Lesions developed in 66% and 67% of PtenC124S/+and PtenG129E/+ mice, respectively; however, along with a number of hyperplasia cases and small adenocarcinomas (9 out of 27, 33.3%, for PtenC124S/+, and 7 out of 24, 29.1%, for PtenG129E/+) we also found that 33.3% of PtenC124S/+ mice (9 out of 27) and 37.5% of PtenG129E/+ mice (9 out of 24) developed large invasive adenocarcinomas (Figures 4A and 4B). Thoracic and inguinal fat pads presented massive expansion of epithelial cells and connective tissue, with central necrotic areas likely due to tumor size, which reached 0.6 cm2 on average (Figures 4C and S4C).
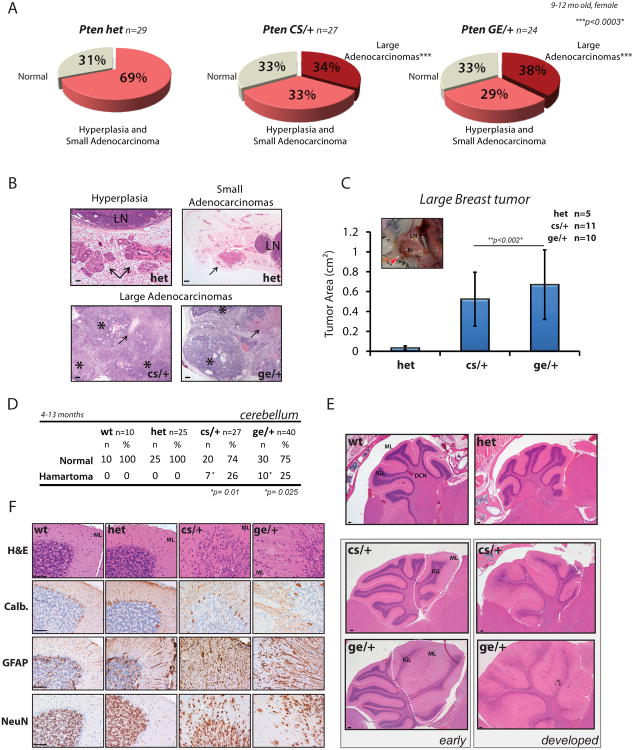
Figure 4. Dominant negative effect of PTEN cancer-associated mutations leads to enhanced tumorigenesis and development of hamartoma features.
(A) Pie-charts showing breast tumor distribution in Pten+/-, PtenC124S/+ and PtenG129E/+ female mice between 9 and 12 months of age.
(B) H&E staining of tumoral lesions found in breast tissues of Pten mouse lines. Top panels, arrows point to hyperplasia (left) and small adenocarcinomas (right) found in the 68.5% of Pten+/- mice (n=20 out of 29); LN, lymph-nodes (See also Figures S3E and S3F). Bottom panels, sections of large adenocarcinomas developing in 34% of PtenC124S/+ mice (n=9 out of 27), left, and 37.5% of PtenG129E/+ mice (n=9 out of 24), right. Asterisks indicate areas of epithelial expansion surrounded by connective tissue. Arrows point to areas of necrotic tissue found in the middle core of the tumors. Scale bar is 1mm in hyperplasia sample, top left; and 500μm in the remaining panels.
(C) Tumor area of large breast adenocarcinomas found in PtenKI mice. Inset, picture of large breast tumor (red arrow-head) developing in the inguinal fat-pad of a 9-month old PtenC124S/+ female mouse. Histogram shows average tumor area measured on paraffin blocks samples (side x side). Br, breast: LN, lymph-node. Mean values with associated SD are shown.
(D) Lhermitte-Duclos disease develops in the cerebellum of PtenKI mice. Table shows penetrance of disease in PtenC124S/+ (6 females and 1 male) and PtenG129E/+ mice (3 females and 7 males) (See also Figures S4D and S4G).
(E) H&E staining of cerebellar sections from Pten mouse lines. Top panel, histologically normal cerebellum from Pten+/+ and Pten+/- mice displaying: molecular layer (ML), internal granule cell layer (IGL), and deep cerebellar nuclei (DCN). Bottom panel, Lhermitte-Duclos disease in PtenKI mice present in small areas of early lesions, left, or developing through the entire cerebellum, right (See also Figure S4D). Scale bar is 1mm.
(F) Immunohistochemistry (IHC) of cerebellar sections from 10-month old Pten mouse lines. From the top: H&E staining shows disorganized granule cells spreading into the ML. Calbindin immunostaining shows dispersion of Purkinje cells due to granule cells overgrowth (GFAP immunostaining). NeuN immunostaining identifies presence of scattered neurons. Scale bar is 25 μm.
As PTEN loss also leads to defects in neuronal development, we assessed the status of different neuronal populations (Fraser et al., 2004). Histo-pathological analysis revealed that only the pituitary gland presented signs of tumorigenesis, with adenomas developing primarily in the anterior lobe (Figure S4E) (Bai et al., 2006). However, in the cerebellum, we found that while Pten+/- mice had histologically normal brains, both PtenKI cohorts developed features of Lhermitte-Duclos disease, or “dysplastic gangliocytoma” (Figures 4D and 4E, and Figures S4D and S4G) (Backman et al., 2001; Kwon et al., 2001). An average of 25% of PtenC124S/+ and PtenG129E/+ mice had enlarged areas in the cerebellum, with disorganized spreading of granule cells into the molecular layer, increased thickness of the molecular layer itself, and the presence of neurons expressing the neuronal marker, NeuN (Figure 4F). Immuno-staining confirmed the expansion of glial cells (mostly astrocytes, GFAP positive, Figure 4F), together with an increased number and size of glial fibers, as shown by positivity to the proliferation marker PCNA (Figure S4F). Affected areas reshaped the structure of the cerebellar lobes and led to dispersion of the Purkinje cells in the molecular layer (Calbindin staining in Figure 4F). These “PHTS” features were found as early as 4 months of age, and presented in small localized areas, as well as in more developed lesions that pervaded the whole cerebellum (Figure 4E). Notably, conditional loss of Pten in the brain, under Gfap-driven Cre expression, leads to similar findings (Backman et al., 2001; Kwon et al., 2001). Taken together, our in vivo data reveal that PtenKI mice have an exacerbated tumor spectrum and PHTS features compared to Pten+/- mice.
PtenKI mice show higher sensitivity to growth factor stimulation and increased Akt activation
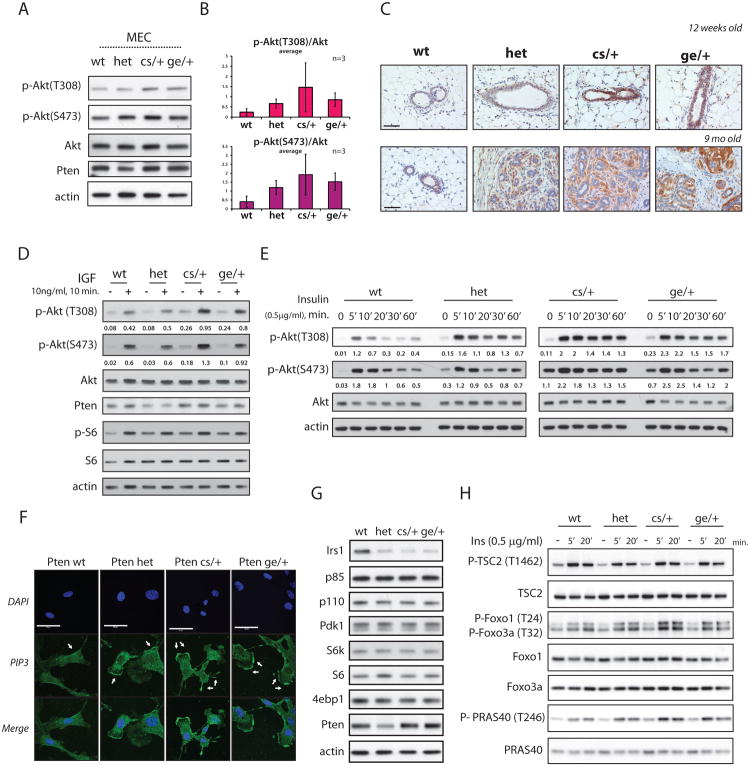
To investigate the molecular basis of the augmented tumorigenesis observed in PtenKIs vs. Pten+/- mice, we monitored downstream effectors of Pten. For this, we isolated primary mammary epithelial cells (MECs) from young Pten+/+, Pten+/-, PtenC124S/+ and PtenG129E/+ female mice to examine levels of Akt phosphorylation. Importantly, while Pten+/- derived MECs had increased phospho-Akt levels compared to wild-type cells, lysates from PtenC124S/+ and PtenG129E/+ derived MECs displayed levels of Akt activation consistently higher than those found in Pten+/- (Figures 5A and 5B). We next examined Akt phosphorylation in pre-onset mammary glands of 10-to-12-week old mice by immunohistochemical staining (IHC) and found that PtenKI mice presented the strongest signal (Figure 5C), which persisted in tumor lesions of older mice (Figure S3G and S4C).
Figure 5. PtenC124S/+ and PtenG129E/+ associated tumorigenesis is driven by Akt hyper-activation.
(A) Total lysates of mammary epithelial cells (MECs) derived from 10-to-12 weeks old female mouse lines. MECs were starved overnight and stimulated for 10min. with full serum (See Experimental Procedures).
(B) Phospho-Akt levels are normalized over total protein levels. Average of 3 independent experiments is shown.
(C) IHC of Phospho-Akt (S473) in breast tissue of 12-weeks-old female mice, top panels; and 9 months old female mice, bottom panels. Scale bar is 50μm.
(D) Activation of PI3K pathway in MEFs. Total lysates were resolved by SDS-PAGE and probed with the indicated antibodies. Phospho-AKT levels are normalized over total Akt
(E) MEFs were starved for 3hrs and stimulated with 0.5μg/ml of insulin for the indicated time-points. Phospho-AKT levels are normalized over total Akt.
(F) PIP3 immuno-staining in MEFs. MEFs were starved for 3hrs, stimulated with 0.5μg/ml of insulin for 1min., and fixed with 4% PFA (See Experimental Procedure). Arrows point to accumulation of PIP3 at the leading edges of membrane projections. Scale bar is 50μM.
(G) Total lysates of MEFs were resolved by SDS-PAGE and probed to detect protein levels of members of the PI3K/Akt pathway.
(H) Activation of Akt targets in Pten derived MEFs. MEFs were starved for 3hrs and stimulated for the indicated time-points. Total lysates were resolved by SDS-PAGE and probed with the indicated antibodies.
To further test the propensity of Pten mutations to induce Akt hyper-activation, we generated mouse embryonic fibroblasts (MEFs). Insulin-like growth factor (IGF) stimulation led to acute Akt phosphorylation in PtenKI-derived MEFs, again much higher than in Pten+/- cells (Figure 5D). Similar results were observed upon insulin stimulation (Figure S5A). In addition, by monitoring the activation profile over time, we found that after 1 hour of insulin stimulation, PtenKI MEFs sustained higher levels of phospho-Akt (Figure 5E).
We next examined cellular levels of PIP3, which is mainly found on the leading edges of filipodia and lamellipodia, to stimulate cell migration and invasion (Kolsch et al., 2008). In MEFs, IF experiments showed that one minute of insulin stimulation led to PIP3 accumulation at the membrane of all samples, with PtenKI MEFs showing the strongest signal (Figure 5F).
Next, we monitored activation levels of Akt isoforms. Out of the three, Akt1 and Akt2 are more widely expressed, and their contributions to tumorigenesis have been better characterized (Gonzalez and McGraw, 2009). Thus, we performed IP-Western blot analysis on Akt1 and Akt2, and found that both proteins were hyper-phosphorylated in PtenKI MEFs (Figures S5C and S5D). We also examined expression levels of key components of the PI3K pathway, and found no changes in total levels of its upstream or downstream components (Figure 5G); nor did we find alteration in the formation of the PI3K complex (p85:p110 binding) across the Pten genotypes (Figure S5B).
Then we monitored the activation of several Akt targets, including the mTORC1 complex. We found that upon insulin stimulation PtenKI MEFs displayed increased levels of PRAS40 phosphorylation compared to Pten+/- cells, resulting in mTORC1 activation and faster accumulation of phospho-S6, its downstream substrate (Figure 5H and Figures S5F and S5G). PtenKI MEFs also displayed reduced levels of IRS1, probably due to negative feedback loops driven by mTORC1activation (Figure 5G) (Shah et al., 2004). We also monitored activation of Akt targets including TSC2, Foxo1 and Foxo3a, but while both Foxo proteins were more phosphorylated in PtenKI MEFs compared to Pten+/-, the status of phospho TSC2 remained unchanged (Figure 5H).
Finally, we tested the status of proposed Pten protein substrates and established their potential contribution to Pten loss driven tumorigenesis. (Tamura et al., 1999; Zhang et al., 2011). In our experimental conditions, however, while IGF stimulation led to Akt hyper-activation, the phosphorylation status of Fak and Src were not significantly affected across the Pten genotypes (Figure S5E).
Dimerization and membrane recruitment identify an active pool of PTEN in vivo which is out-competed by mutant PTEN
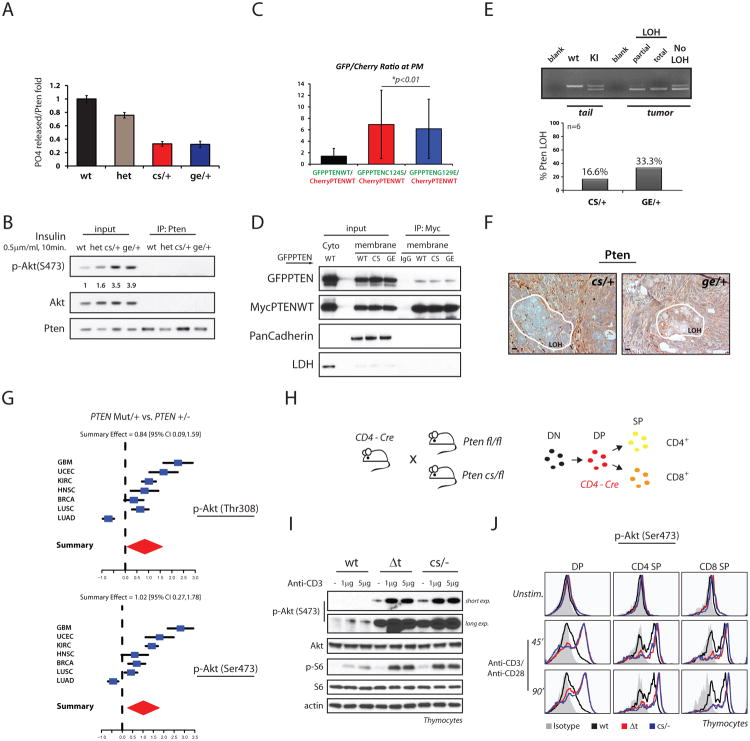
We then immuno-purified Pten from Pten+/+, Pten+/-, PtenC124S/+ and PtenG129E/+ MEFs and tested the respective phosphatase activity toward PIP3. We found that free phosphate production by the hetero-dimeric complexes was lower than that found under Pten heterozygous conditions. Accordingly, reduced PIP3 hydrolysis led to increased Akt phosphorylation (Figures 6A and 6B).
Figure 6. Akt hyper-activation leads to faster tumor formation.
(A) Analysis of Pten catalytic activity. Total lysates of MEFs were IP with an anti-Pten antibody, immuno-complexes eluted in native conditions and relative phosphatase activity normalized over levels of Pten IP. Mean values from triplicate wells with associated SD are shown.
(B) Levels of Akt phosphorylation found in MEFs treated as in (A).
(C) PC3 cells were transfected with mCherry-PTENWT alongside GFP-PTENWT, or mutants. Translocation to PM was assayed through generation of ratiometric images. Average ratios were calculated based on fluorescent intensities of mCherry and GFP species at the PM vs. intensities found in the cytosol during serum reactivation. Quantification of PTEN distribution is shown.
(D) Co-IPs from membrane or cytosolic fractions of PC3 cells transfected with the indicated PTEN vectors. PanCadherin and LDH are membrane and cytosolic protein markers, respectively
(E) Laser Capture Micro-dissection (LCM) analysis performed on large breast adenocarcinoma samples of PtenKI mice. Top panel: representative PCRs of Pten exon5 locus showing levels of wild-type and targeted Pten alleles found in micro-dissected breast samples. Bottom panel, percentage of LOH in breast tumor samples, n=6 per genotype.
(F) IHC of large breast adenocarcinomas from 10-month old PtenKI female mice. Pten expression is maintained in the majority of the breast tumor, with exception of localized areas with reduced intensity (surrounded in white). LOH, area with loss of heterozygosity.
(G) Association study between PTEN status and AKT phosphorylation in human cancer, (See Experimental Procedures). Samples harboring PTEN mutation are associated with higher levels of phospho-AKT(T308), top panel, and phospho-AKT(S473) bottom panel, compared to samples with PTEN heterozygosity.
(H) Ptenfl/fl and PtenC124S/fl mice were crossed with CD4- driven Cre transgenic mice (left panel). Pten deletion specifically occurs at the Double Positive (DP) stage of thymocyte development: DN, Double Negative thymocytes; SP, single Positive (right panel).
(I) Thymocytes from 3-to-4 week old mice with the indicated genotypes were extracted and stimulated as indicated. Total lysates were resolved by SDS-PAGE and probed with the indicated antibodies.
(J) Representative flow-histogram of thymocyte populations stimulated with Anti-CD3/Anti-CD28 for the indicated time. Pten-/- and PtenC124S/- thymocytes show comparable levels of phospho-Akt.
Because PIP3 is produced in the inner leaflet of the plasma membrane, we next analyzed Pten membrane recruitment. To this end, we co-expressed GFPPTEN WT or mutants alongside untagged PTENWT in PC3 cells, and monitored the ability of the different GFPPTEN species to localize to the plasma-membrane (PM) (Figure S6A). We found that upon starvation and serum stimulation, GFPPTEN mutant proteins showed a 12% greater concentration (on average) at the membrane than GFPPTEN WT (Figure S6B).
To verify that PTEN is present at the membrane as homo- or hetero-dimers, we repeated this assay by co-expressing GFPPTEN variants with mCherry-tagged PTENWT. Co-expression of mCherryPTENWT with GFPPTENWT displayed a linear accumulation of GFP and mCherry signals at the PM, with less than 2% intrinsic variation, likely due to different backbone plasmids. However, in co-expressing C124S or G129E-GFP variants with mCherry-PTENWT, we observed that the GFP detection at the PM was only 4% more intense than the mCherry signal (Figures 6C and S6C), suggesting that although both mutants are recruited to the membrane more rapidly than PTEN WT, they are also able to promote enhanced mCherry-PTENWT membrane recruitment through dimerization.
In addition, co-IPs from PC3 cell-membrane fractions confirmed that MycPTENWT can bind GFPPTENWT and GFPPTEN mutants at the membrane (Figure 6D). Thus we propose that “inactive” hetero and homo-dimers may displace and out-compete the function of “active” PTEN WT homo-dimers.
Consequence of PTEN mutations on loss of heterozygosity and Akt activation in mouse models and humans
In the mouse, mono-allelic loss of Pten leads to tumor initiation while cancer progression often selects for loss of the functional Pten allele (Hollander et al., 2011). Thus we asked whether in PtenKI mice, the dominant negative action of mutant over wild-type Pten protein would trigger advanced tumorigenesis also in the absence of loss of heterozygosity (LOH). We therefore performed southern blot (SB) analysis and laser capture micro-dissection (LCM) on large breast adenocarcinoma samples from PtenKI mice. By SB we found that on average, 50% of the PtenKI breast samples fully retained the Pten wt allele (Figure S6D). To test a larger number of samples we then performed LCM and PCR amplification on DNA extracts and found mixed genomic profiles with only 1 out of 6 PtenC124S/+ samples (16%) displaying total LOH, and 2 out of 6 PtenG129E/+ samples (33%) presenting either partial or total LOH (Figure 6E).
Next, we examined the status of the Pten protein by IHC, and found predominantly positive staining, with areas of reduced intensity in agreement with the percentage of LOH found by SB and LCM (Figure 6F). This finding argues that the dominant negative effect of Pten mutations exerts their negative pro-tumorigenic effects even in the absence of LOH; however, when cancer arises, intra-tumoral variability and accumulation of additional mutations will eventually favor focal loss of Pten. Thus the consistent Akt hyper-activation, as found in large adenocarcinomas (Figure S6E), leads to accelerated cancer formation, while PTEN LOH, when observed, may further favor tumor progression.
To assess whether our observations were consistent with the molecular changes occurring in human tumorigenesis, we examined the association between PTEN mutational status and AKT activation in human cancers. For this, we analyzed datasets from The Cancer Genome Atlas available at the cBio portal (http://cbioportal.org), and matched the genetic status of PTEN with the reverse-phase protein arrays (RPPA) data for AKT. We found that samples with heterozygous mutations in PTEN (and no copy number alteration, including two PTENG129E mutant cases) were associated with higher levels of AKT phosphorylation (on both Thr308 and Ser473) compared to samples with heterozygous PTEN loss (and no additional mutations) (Figure 6G), independent of total Akt levels (Figure S6F). These data support the notion that PTEN mutations have greater consequence than heterozygous PTEN loss in promoting PI-3K/AKT hyper-activation, and may predict for higher sensitivity to AKT inhibition in human cancers.
Genetic assessment of the dominant negative role of mutant Pten
Finally, we reasoned that if our model was correct, then the observed differential signaling output between PtenKI cells vs. Pten+/- should be absent in PtenKI/- vs. Pten-/- cells. Unfortunately this could not be addressed in MEFs, since complete Pten loss is known to trigger cellular senescence in these cells (Chen et al., 2005); indeed, we found that PtenC124S/+ MEFs presented increased senescence over Pten+/- MEFs upon passaging (Figures S6G-S6I). To overcome this problem, we made use of a conditional knock-out strategy induced by the CD4-Cre recombinase, which is active in the double positive lymphocyte stage of thymus development (Figure 6H). Extracted thymocytes were cultured and their PI3K-signaling output analyzed by Western blot and flow cytometry analysis, respectively. Critically, in this setting we found no consistent differences in signaling output across the genotypes (Figures 6I and 6J), further supporting a model in which Pten mutations enhance PI3K/Akt oncogenic signaling by inhibiting its wt counterpart.
Functional evaluation of PTENR130G mutation
Since PTENC124S and PTENG129E mutations are only found in a subset of cancer and genetic syndromes, we next tested the functional role of three additional PTEN mutations more commonly found in human disease: PTENR130G, PTENR130X, and PTENR233X (Bonneau and Longy, 2000). PTENR130X and R233X nonsense mutations introduce stop codons that generate very unstable PTEN proteins that are almost undetectable (Figure S7A), and thus are functionally comparable to the PTEN heterozygous condition. The PTENR130G missense mutation however generates a stable protein which suffers loss of its lipid phosphatase function (Kato et al., 2000). In this respect PTENR130G phenocopies PTENC124S and PTENG129E, so we further investigated its potential dominant negative effect.
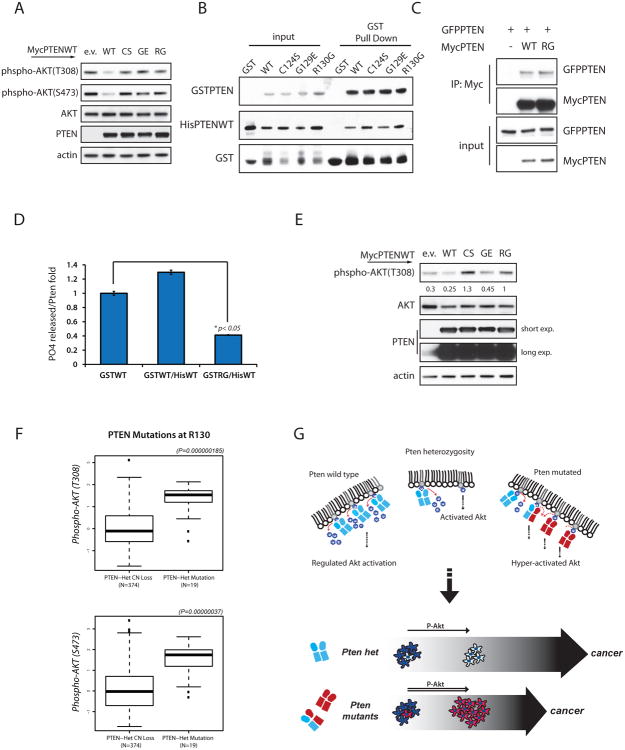
First we confirmed that this mutation has no suppressive effect on phospho-AKT levels in PTEN null cells (Figure 7A). We next determined that PTENR130G can interact with WT PTEN in bacteria as well as eukaryotic cells (Figures 7B and 7C). Then by testing the phosphatase function of HisPTENWT-GSTPTENR130G hetero-dimer purified from bacteria, we found that when bound together, PTENR130G limits the function of the WT protein to inhibit PIP3 dephosphorylation (Figure 7D). In addition, over-expression of PTEN mutations in PTEN-competent 293T cells displayed increased phospho-AKT on T308 than cells expressing empty vector, overcoming the function of endogenous PTEN (Figure 7E).
Figure 7. PTENR130G parallels PTENC124S and PTENG129E mutations.
(A) Total lysates of PC3 cells transfected with PTEN WT and mutant expression vectors. Western-blots were probed with the indicated antibodies.
(B) Bacteria were co-transformed with His-PTENWT vector alongside empty GST, or GST-PTENWT or GST-PTEN mutant vectors as indicated. Protein lysates were subjected to GST-pull down and SDS-PAGE blots probed with His-tag antibody.
(C) Co-IPs from total lysates of PC3 cells transfected with the indicated vectors, and IP with a Myc-tag antibody. Western-blot revealed interaction between PTENWT and PTENR130G mutant protein.
(D) Phosphatase assay of PTEN dimers purified from bacteria. PO4 released is normalized over levels of His-PTEN pulled down, and shown relative to the PO4 released by GST-PTENWT alone. Mean value of triplicate wells with associated SD are shown.
(E) HEK293T transfected with the indicated MycPTEN vectors were starved overnight and stimulated with 10% serum for 10min. Total lysates were resolved by Western-blot and probed with the indicated antibodies. Phospho-AKT levels are normalized over total AKT.
(F) Association study between AKT activation and PTEN mutations at a.a. R130 in human cancers.
(G) Top panel: Pten heterozygosity leads to Akt activation and tumorigenesis. Heterozygous expression of cancer-associated PTEN mutations leads to formation of catalytically inactive hetero-dimers. Consequentially, increased PIP3 levels induce Akt hyper-activation and augmented tumorigenesis, bottom panel.
Finally, we found that samples harboring mutations on PTENR130 either mutated to Gly (R130G) or to Gln (R130Q) in a total of 19 samples from glioblastoma and endometrial cancers, which exhibited higher levels of AKT phosphorylation than samples with mono-allelic loss of PTEN (Figure 7F).
Discussion
In this study we have demonstrated that PTEN dimerization is critical for its lipid phosphatase function. We have also proposed that dimeric PTEN complexes are more active than PTEN monomer in dephosphorylating PIP3 and regulating PI3K signaling. By studying two PTEN cancer-associated mutations, we revealed that while disrupting PTEN activity in-cis, PTENC124S and PTENG129E inhibit the wt protein function in-trans in a dominant negative manner as a result of hetero-dimerization. In turn, this reduced Pten lipid-phosphatase activity leads to Akt hyper-activation and increased tumorigenesis in the mouse.
These findings allow us to reach several conclusions. Mechanistically, we have defined that PTEN exists as a dimer. Given that PTEN is part of the VH1-like family of DSPs, it is relevant that the prototypical VH1 phosphatase is known to exist in a dimeric-quaternary complex whose assembly is essential for the recognition of its substrate, STAT1 (Koksal and Cingolani, 2011). Moreover, phosphorylation of the PTEN tail is known to produce an inactive form of the enzyme in a closed conformation (Leslie and Foti, 2011). In agreement, we provide evidence that dephosphorylation of the PTEN-tail, while favoring a more open conformation, also allows subsequent dimerization and, possibly, oligomerisation, in view of the multiple interfaces found to mediate the PTEN-PTEN interaction.
The role of PTEN dimerization in cancer is particularly important when we consider the high frequency of PTEN mutations in sporadic tumors and inherited syndromes. In this study, we have analyzed in vivo and in vitro two cancer- and CD-associated missense mutations, and extended our analysis to a third mutation, PTENR130G. In the future, it will be important to study various PTEN mutations in a systematic manner. It is tempting to speculate that other PTEN mutants may affect the function of the PTEN wt protein through physical interactions, and perhaps at multiple levels, e.g. by affecting PTEN extracellular export (Hopkins et al., 2013; Putz et al., 2012).
Clinically, our study implies that patients harboring these, or similar loss-of-function missense mutations, may be more susceptible to malignant cancer, and may develop it more rapidly, than patients expressing reduced levels of PTEN, or expressing PTEN destabilizing mutations. Thus we propose that PTEN mutational status may be utilized to stratify patients who may benefit from earlier and more radical therapeutic intervention modalities, potentially leading to improved prognoses.
Although we cannot exclude the possibility that certain PTEN mutations may become competent toward targets not recognized by the wild-type protein in a “gain-of-function” scenario (Wang et al., 2010), our genetic analyses support a model in which the ability of mutant PTEN to interfere with wt protein function contributes to exacerbation of tumor spectrum compared to Pten heterozygosity (Fig. 7G). While these mutations induce acceleration of tumorigenesis, they specifically act by further lowering Pten activity rather than engaging alternative pathways. Accordingly, we found that PtenKI mice developed Lhermitte-Duclos disease, as previously reported in mice with total conditional Pten loss, while lesions of different histological origins not associated with Pten loss (such as sarcomas) were not observed.
Additionally, in monitoring the activation status of proposed PTEN phospho-protein targets we found no obvious changes upon IGF stimulation, or variation between tumor spectra of the two PtenKI models. This implies that even if cancer-relevant PTEN phospho-protein targets are present and deregulated in PtenC124S/+ mice, concomitant Akt hyper-activation may overcome their effects, perhaps due to increased cellular senescence (Figure S6G). While several reports have highlighted the possibility that PTEN protein phosphatase activity may be related to regulation of migration and invasion (Tibarewal et al., 2012), in our in vivo analysis of the PtenKI mice we did not observe metastasis. However, the general exacerbation of solid tumorigenesis in multiple organs and pronounced lympho-proliferation may have masked a possible metastatic phenotype in elder mutants.
In summary, we have identified new features of PTEN biology through the characterization of cancer associated PTEN missense mutations. Given the ongoing development of agents targeting the PI3K pathway including AKT-inhibitors, our findings may help to identify patients that may be sensitive to these agents due to high levels of AKT activation associated with a PTEN mutant state.
Experimental Procedures
Western blotting, Immunoprecipitation and In vitro binding
For details on Western blotting, Immunoprecipitation, and in vitro binding please refer to Extended Experimental Procedures.
Gel Filtration Chromatography
For Gel filtration experiments please refer to Extended Experimental Procedures.
Mice and Immunohistochemistry
Autopsy and histological analysis was performed on cohorts of female and male mice from 2-to-13 month of age. Mouse tissues were fixed in 4% PFA. Normal and tumor tissues were embedded in paraffin, sectioned, and H&E stained for pathological evaluation. Brain tissues were fixed in Bouin's solution. Please refer to Extended Experimental procedure for details on the generation of mouse lines. All mice were cared for according to NIH-approved institutional animal care guidelines and studies approved by the Institutional Committee at the Beth-Israel Deaconess Medical Center.
Studies with primary cells
Mouse Embryonic Fibroblasts (MEFs) were isolated at day E13.5 and maintained in culture as described (Todaro and Green, 1963). MEFs between passages 1 and 3 were used for all experiments. Senescence assays were performed as described (Chen et al., 2005).
Isolation of primary mammary epithelial cells (MECs) was performed as previously described (Song SJ, Cell 2013). Please refer to Extended Experimental Procedures for details.
PtdIns(3,4,5)P3 Phosphatase assay
PC3 cells and primary MEFs lysates from Pten lines were IP and subjected to native elution. For phosphatase assays, a solution with 25mM Tris-HCl (pH7.5), 140mM NaCl, 1mM DTT, 100μM diC8-PtdIns(3,4,5)P3 (Echelon) was prepared and assay ran at 37°C for 45min. Free phosphate release was measured with Green Reagent (Biomol) and according to the manufacturer's instructions.
Population BRET imaging
PC3 cells seeded on 35mm wells were transfected with indicated expression vectors. After 48hrs, cells were scraped and transferred into a white OptiPlate-96 (Perkin Elmer, Milltown). Luminescent emission was stimulated by adding 5μM Coelenterazine, and signal collected with the Victor3 plate reader (Perkin Elmer, Milltown). Please refer to Extended Experimental Procedures for details.
Bioinformatic analysis
We used tissue samples from The Cancer Genome Atlas (TCGA) project to assess relationships between PTEN mutation status, PTEN copy number status, and expression levels of AKT and pAKT in human cancer samples. Please refer to Extended Experimental Procedures for details.
Supplementary Material
Highlights.
PTEN exists in an active, unphosphorylated dimeric conformation
PTEN missense mutations exert dominant negative effects over the wild-type protein
Pten mutations elicit Akt hyper-activation and augmented tumorigenesis in the mouse
Mutant PTEN associates with increased AKT phosphorylation in human cancer
Acknowledgments
Authors would like to thank Pandolfi lab members for critical comments, and Thomas Garvey for editing the manuscript. Authors are indebted to SuJung Song and Dimitrios Anastosiou for insightful discussion. A.P. was supported in part by the American-Italian Cancer Foundation Post-Doctoral Fellowship. M.B. and P.P. are supported by the Italian Association for Cancer Research (AIRC, grant number 14442). This work was supported by the NIH grant, U01-CA 141496 to P.P.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alimonti A, Carracedo A, Clohessy JG, Trotman LC, Nardella C, Egia A, Salmena L, Sampieri K, Haveman WJ, Brogi E, et al. Subtle variations in Pten dose determine cancer susceptibility. Nature genetics. 2010;42:454–458. doi: 10.1038/ng.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A, Sasin J, Bottini N, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Backman SA, Stambolic V, Suzuki A, Haight J, Elia A, Pretorius J, Tsao MS, Shannon P, Bolon B, Ivy GO, et al. Deletion of Pten in mouse brain causes seizures, ataxia and defects in soma size resembling Lhermitte-Duclos disease. Nature genetics. 2001;29:396–403. doi: 10.1038/ng782. [DOI] [PubMed] [Google Scholar]
- Bai F, Pei XH, Pandolfi PP, Xiong Y. p18 Ink4c and Pten constrain a positive regulatory loop between cell growth and cell cycle control. Molecular and cellular biology. 2006;26:4564–4576. doi: 10.1128/MCB.00266-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau D, Longy M. Mutations of the human PTEN gene. Human mutation. 2000;16:109–122. doi: 10.1002/1098-1004(200008)16:2<109::AID-HUMU3>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cristofano A, Kotsi P, Peng YF, Cordon-Cardo C, Elkon KB, Pandolfi PP. Impaired Fas response and autoimmunity in Pten+/- mice. Science. 1999;285:2122–2125. doi: 10.1126/science.285.5436.2122. [DOI] [PubMed] [Google Scholar]
- Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nature genetics. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- Fraser MM, Zhu X, Kwon CH, Uhlmann EJ, Gutmann DH, Baker SJ. Pten loss causes hypertrophy and increased proliferation of astrocytes in vivo. Cancer research. 2004;64:7773–7779. doi: 10.1158/0008-5472.CAN-04-2487. [DOI] [PubMed] [Google Scholar]
- Garcia-Cao I, Song MS, Hobbs RM, Laurent G, Giorgi C, de Boer VC, Anastasiou D, Ito K, Sasaki AT, Rameh L, et al. Systemic elevation of PTEN induces a tumor-suppressive metabolic state. Cell. 2012;149:49–62. doi: 10.1016/j.cell.2012.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgescu MM, Kirsch KH, Akagi T, Shishido T, Hanafusa H. The tumor-suppressor activity of PTEN is regulated by its carboxyl-terminal region. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:10182–10187. doi: 10.1073/pnas.96.18.10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez E, McGraw TE. The Akt kinases: isoform specificity in metabolism and cancer. Cell Cycle. 2009;8:2502–2508. doi: 10.4161/cc.8.16.9335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander MC, Blumenthal GM, Dennis PA. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nature reviews Cancer. 2011;11:289–301. doi: 10.1038/nrc3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins BD, Fine B, Steinbach N, Dendy M, Rapp Z, Shaw J, Pappas K, Yu JS, Hodakoski C, Mense S, et al. A Secreted PTEN Phosphatase that Enters Cells to Alter Signaling and Survival. Science. 2013 doi: 10.1126/science.1234907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Kato S, Kumabe T, Sonoda Y, Yoshimoto T, Han SY, Suzuki T, Shibata H, Kanamaru R, Ishioka C. Functional evaluation of p53 and PTEN gene mutations in gliomas. Clinical cancer research : an official journal of the American Association for Cancer Research. 2000;6:3937–3943. [PubMed] [Google Scholar]
- Koksal AC, Cingolani G. Dimerization of Vaccinia virus VH1 is essential for dephosphorylation of STAT1 at tyrosine 701. The Journal of biological chemistry. 2011;286:14373–14382. doi: 10.1074/jbc.M111.226357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolsch V, Charest PG, Firtel RA. The regulation of cell motility and chemotaxis by phospholipid signaling. Journal of cell science. 2008;121:551–559. doi: 10.1242/jcs.023333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon CH, Zhu X, Zhang J, Knoop LL, Tharp R, Smeyne RJ, Eberhart CG, Burger PC, Baker SJ. Pten regulates neuronal soma size: a mouse model of Lhermitte-Duclos disease. Nature genetics. 2001;29:404–411. doi: 10.1038/ng781. [DOI] [PubMed] [Google Scholar]
- Lee JO, Yang H, Georgescu MM, Di Cristofano A, Maehama T, Shi Y, Dixon JE, Pandolfi P, Pavletich NP. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99:323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- Leslie NR, Foti M. Non-genomic loss of PTEN function in cancer: not in my genes. Trends in pharmacological sciences. 2011;32:131–140. doi: 10.1016/j.tips.2010.12.005. [DOI] [PubMed] [Google Scholar]
- Liaw D, Marsh DJ, Li J, Dahia PL, Wang SI, Zheng Z, Bose S, Call KM, Tsou HC, Peacocke M, et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nature genetics. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. The Journal of biological chemistry. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- Marsh DJ, Coulon V, Lunetta KL, Rocca-Serra P, Dahia PL, Zheng Z, Liaw D, Caron S, Duboue B, Lin AY, et al. Mutation spectrum and genotype-phenotype analyses in Cowden disease and Bannayan-Zonana syndrome, two hamartoma syndromes with germline PTEN mutation. Human molecular genetics. 1998;7:507–515. doi: 10.1093/hmg/7.3.507. [DOI] [PubMed] [Google Scholar]
- Myers MP, Pass I, Batty IH, Van der Kaay J, Stolarov JP, Hemmings BA, Wigler MH, Downes CP, Tonks NK. The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:13513–13518. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers MP, Stolarov JP, Eng C, Li J, Wang SI, Wigler MH, Parsons R, Tonks NK. P-TEN, the tumor suppressor from human chromosome 10q23, is a dual-specificity phosphatase. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:9052–9057. doi: 10.1073/pnas.94.17.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podsypanina K, Ellenson LH, Nemes A, Gu J, Tamura M, Yamada KM, Cordon-Cardo C, Catoretti G, Fisher PE, Parsons R. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:1563–1568. doi: 10.1073/pnas.96.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putz U, Howitt J, Doan A, Goh CP, Low LH, Silke J, Tan SS. The tumor suppressor PTEN is exported in exosomes and has phosphatase activity in recipient cells. Science signaling. 2012;5:ra70. doi: 10.1126/scisignal.2003084. [DOI] [PubMed] [Google Scholar]
- Shah OJ, Wang Z, Hunter T. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Current biology : CB. 2004;14:1650–1656. doi: 10.1016/j.cub.2004.08.026. [DOI] [PubMed] [Google Scholar]
- Sotelo NS, Valiente M, Gil A, Pulido R. A functional network of the tumor suppressors APC, hDlg, and PTEN, that relies on recognition of specific PDZ-domains. Journal of cellular biochemistry. 2012;113:2661–2670. doi: 10.1002/jcb.24141. [DOI] [PubMed] [Google Scholar]
- Suzuki A, de la Pompa JL, Stambolic V, Elia AJ, Sasaki T, del Barco Barrantes I, Ho A, Wakeham A, Itie A, Khoo W, et al. High cancer susceptibility and embryonic lethality associated with mutation of the PTEN tumor suppressor gene in mice. Current biology : CB. 1998;8:1169–1178. doi: 10.1016/s0960-9822(07)00488-5. [DOI] [PubMed] [Google Scholar]
- Tamura M, Gu J, Takino T, Yamada KM. Tumor suppressor PTEN inhibition of cell invasion, migration, and growth: differential involvement of focal adhesion kinase and p130Cas. Cancer research. 1999;59:442–449. [PubMed] [Google Scholar]
- Tibarewal P, Zilidis G, Spinelli L, Schurch N, Maccario H, Gray A, Perera NM, Davidson L, Barton GJ, Leslie NR. PTEN protein phosphatase activity correlates with control of gene expression and invasion, a tumor-suppressing phenotype, but not with AKT activity. Science signaling. 2012;5:ra18. doi: 10.1126/scisignal.2002138. [DOI] [PubMed] [Google Scholar]
- Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. The Journal of cell biology. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotman LC, Niki M, Dotan ZA, Koutcher JA, Di Cristofano A, Xiao A, Khoo AS, Roy-Burman P, Greenberg NM, Van Dyke T, et al. Pten dose dictates cancer progression in the prostate. PLoS biology. 2003;1:E59. doi: 10.1371/journal.pbio.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Grossman SR, Takahashi Y, Rokas MV, Nakamura N, Sellers WR. Phosphorylation of the PTEN tail acts as an inhibitory switch by preventing its recruitment into a protein complex. The Journal of biological chemistry. 2001;276:48627–48630. doi: 10.1074/jbc.C100556200. [DOI] [PubMed] [Google Scholar]
- Vazquez F, Ramaswamy S, Nakamura N, Sellers WR. Phosphorylation of the PTEN tail regulates protein stability and function. Molecular and cellular biology. 2000;20:5010–5018. doi: 10.1128/mcb.20.14.5010-5018.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Karikomi M, Naidu S, Rajmohan R, Caserta E, Chen HZ, Rawahneh M, Moffitt J, Stephens JA, Fernandez SA, et al. Allele-specific tumor spectrum in pten knockin mice. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:5142–5147. doi: 10.1073/pnas.0912524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Jiang X. Post-translational regulation of PTEN. Oncogene. 2008;27:5454–5463. doi: 10.1038/onc.2008.242. [DOI] [PubMed] [Google Scholar]
- Zhang S, Huang WC, Li P, Guo H, Poh SB, Brady SW, Xiong Y, Tseng LM, Li SH, Ding Z, et al. Combating trastuzumab resistance by targeting SRC, a common node downstream of multiple resistance pathways. Nature medicine. 2011;17:461–469. doi: 10.1038/nm.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.