Abstract
Exosomes and microvesicles are two classes of submicroscopic vesicle released by cells into the extracellular space. Collectively referred to as extracellular vesicles, these membrane containers facilitate important cell-cell communication by carrying a diverse array of signaling molecules, including nucleic acids, proteins, and lipids. Recently, the role of extracellular vesicle signaling in cancer progression has become a topic of significant interest. Methods to detect and target exosomes and microvesicles are needed to realize applications of extracellular vesicles as biomarkers and, perhaps, therapeutic targets. Detection of exosomes and microvesicles is a complex problem as they are both submicroscopic and of heterogeneous cellular origins. In this Minireview, we highlight the basic biology of extracellular vesicles, and address available biochemical and biophysical detection methods. Detectible characteristics described here include lipid and protein composition, and physical properties such as the vesicle membrane shape and diffusion coefficient. In particular, we propose that detection of exosome and microvesicle membrane curvature with lipid chemical probes that sense membrane shape is a distinctly promising method for identifying and targeting these vesicles.
Keywords: phospholipids, vesicles, membrane curvature, exosome, curvature sensing/binding peptide
1. Introduction
1.1 Ferrying cargo with extracellular vesicles
Microvesicles and exosomes, collectively referred to as extracellular vesicles in this Minireview, are submicron sized lipid containers released by cells. Both types of extracellular vesicles have an aqueous, cargo-containing core surrounded by a roughly spherical bilayer membrane. Arrival of the vesicles at a distant site and fusion with targeted cells allows transport of cargo as diverse as nucleic acids (DNA, mRNA, and microRNA), proteins, and lipids, facilitating important cell-cell communications.[1,2] Packaging the cargo in lipid vesicles protects the contents from extracellular degradation and allows for more precise targeting of the contents.[1,3] Packaging also provides temporal signaling control through the simultaneous delivery of an array of signaling molecules. Binding of vesicles to target cells is a specific interaction that is likely dependent on receptor mediated association, e.g., microvesicles originating from platelets interact with monocytes but not neutrophils.[4,5] A recent increase in understanding of the role extracellular vesicles play in normal, and pathologic, intercellular communication processes has made them a topic of intense interest.[6–11] The list of normal physiologic roles for microvesicles and exosomes continues to grow: microvesicles and exosomes have already been implicated in horizontal genetic transfer, cytokine release, angiogenesis, transfer of receptors between cells, and metalloproteinase release.[4]
1.2 Extracellular vesicle release, lipid composition, and shape
Although microvesicles and exosomes are structurally similar, they differ in cellular origin, lipid composition, and size. At present, precise definitions of the distinctions and the terminology used to describe these extracellular vesicles are still developing.[12] For example, the hematology research community often uses the term microparticles instead of microvesicles.[13,14] In general, microvesicles are described as being formed by outward budding and fission of the plasma membrane, and as having a particle diameter of 100 – 1000 nm.[15] The lipid composition of microvesicles is by and large similar to that of the cell membrane, but lacks the asymmetric distribution of lipids normally seen across the two leaflets of the plasma membrane. In particular, the aminophospholipids, phosphatidyl-serine and -ethanolamine, are no longer sequestered to the inner leaflet of the membrane, and are instead homogenously distributed across the microvesicle bilayer membrane.[16,17]
Unlike microvesicles, exosomes are produced within the cell and are released through an exocytosis event. Exosome production begins when the bilayer membrane of late endosomes buds inward, filling the luminal space of the endosome with small luminal vesicles or exosomes.[18,19] The vesicle filled endosome, now called a multivesicular body (MVB), then fuses with the plasma membrane through a calcium dependent mechanism, releasing the exosomes into the extracellular space (Figure 1).[20] Similar to microvesicles, exosome membranes also contain an increased level of aminophospholipids compared to the outer leaflet of the plasma membrane.[21] In addition, exosome membranes also contain the lipid ceramide. Ceramide is produced by the hydrolysis of sphingomyelin by sphingomyelinases within the endosome, and its production is an essential step in the sorting and production of exosomes.[22] Exosomes are smaller and more homogenously distributed in size than microvesicles, commonly being described as having a particle diameter of 30 – 100 nm.[22,23] When prepared for and viewed with an electron microscope, exosomes typically display a cup-like shape.[24]
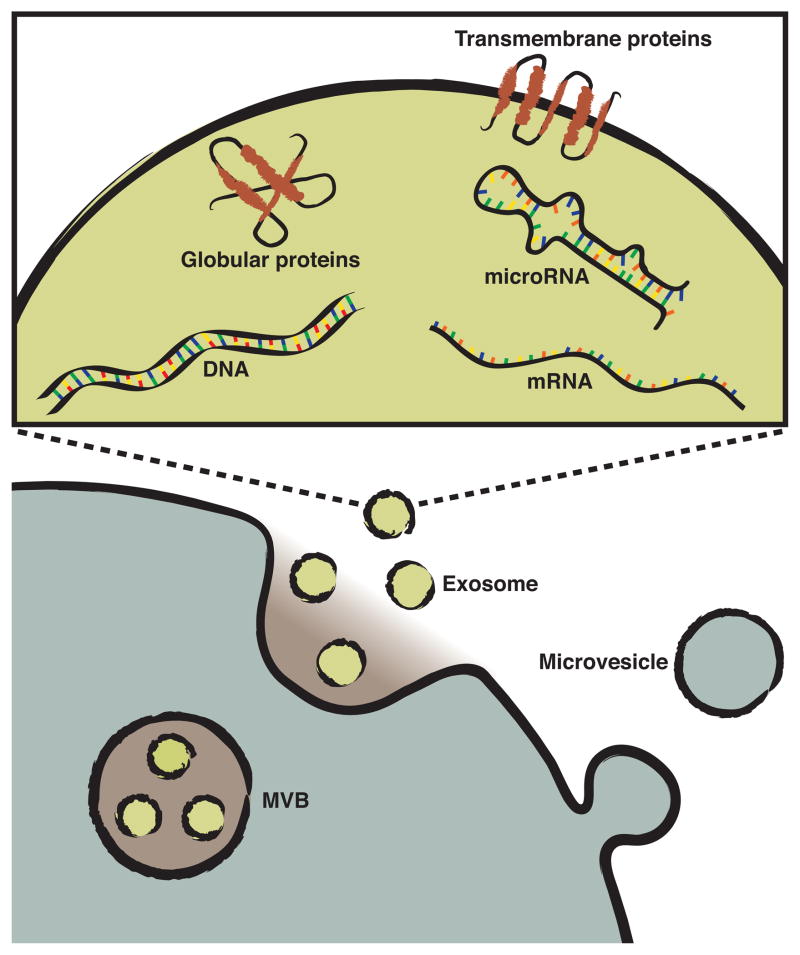
Figure 1.
Extracellular vesicle release and contents. Microvesicles bud directly from the plasma membrane. Exosomes, formed by inward budding of late endosomes, are held in multivesicular bodies (MVB) within the cell. Fusion of the MVBs with the cell membrane releases the exosomes into the extracellular space. Microvesicles and exosomes contain a diverse array of cargo, including lipids, transmembrane and globular proteins, DNA, mRNA, and microRNA. All represented membranes are phospholipid bilayers.
A significant feature of extracellular vesicles is their highly curved membrane surface. Highly curved membrane shapes, where curvature is defined as the reciprocal of the membrane radius, are commonly found within cells.[25] These membranes organize and compartmentalize organelles of complex shape such as the Golgi, and are the cargo of intracellular motor proteins like kinesin. In the extracellular space, highly curved membranes are less common. As most cells have diameters > 8 μm, submicron sized extracellular vesicles represent uniquely curved lipid surfaces.[10] Curvature affects lipid ordering within the membrane. Mismatch between the curvature of the membrane and the shape, or spontaneous curvature, of the lipids induces lipid-packing defects.[26–28] The holes created by the lipid-packing defects expose the hydrophobic core of the bilayer membrane to the aqueous phase (Figure 2). The defects are short-lived, but result in a net reduction of lipid density in the curved region. Along with absolute size, these membrane-packing defects are a characteristic feature of exosomes and microvesicles in the extracellular space, and one that will be further explored as a targetable or detectable feature in this Minireview.
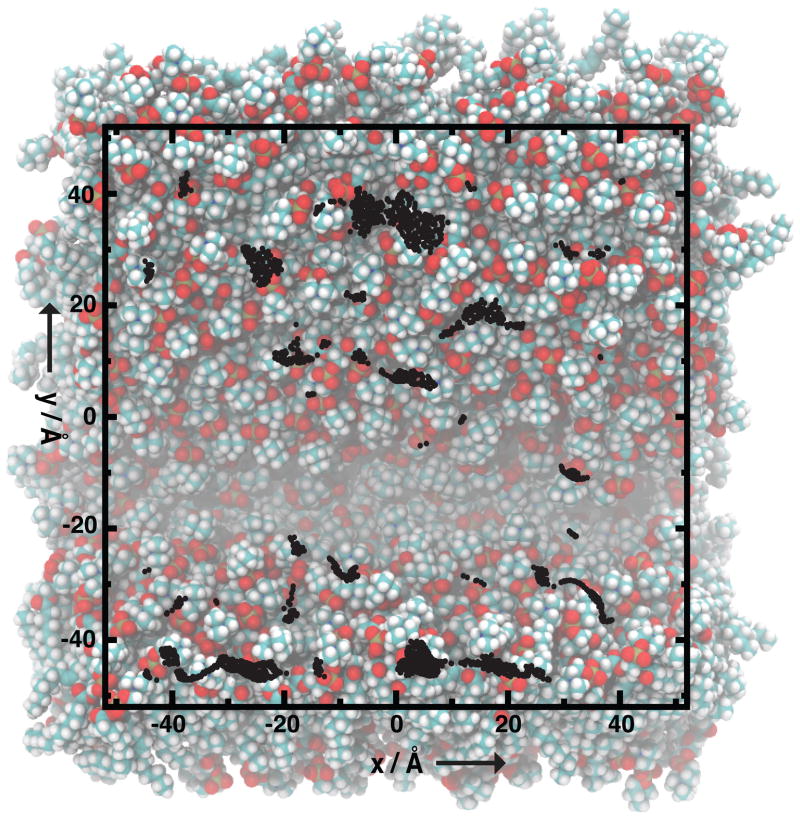
Figure 2.
Membrane curvature induces lipid-packing defects in regions of positive curvature. Top view of a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine (POPC) bilayer membrane induced to curve by gradual lateral compression during molecular dynamics simulation. Lipid packing defects, as measured by solvent accessible surface area, are mapped to the surface as black dots. Lipid head groups are shown in orange and red, and hydrocarbon tails in white and blue. The region of negative curvature, in the area of y ≈ −20, shows few packing defects. Figure adapted with permission.[27]
1.3 Extracellular vesicles promote cancer progression
Cancer cells have long been known to gain a competitive advantage by releasing factors that modulate the extracellular environment to promote cancer growth and metastasis. There is growing evidence that these factors are released via extracellular vesicles. Cultured glioblastoma multiforme, melanoma, and breast cancer cells have all shown significantly elevated release of microvesicles or exosomes.[29,30] Exosomes released from melanoma cells have been shown to promote angiogenesis, matrix remodeling, and anergy in lymph nodes, creating an optimal environment for metastatic cancer cells.[31] Colorectal cancer cells have been shown to shed Fas ligand laden vesicles that reduce the ability of T-cells to induce cancer cell apoptosis.[32]
Increased microvesicle and exosome levels have also been found in the blood of cancer patients. Elevated levels of microRNA-containing microvesicles have been found in the serum of patients with prostate cancer, and increased exosome levels were found in the plasma of patients with lung adenocarcinoma and melanoma.[33–35] Higher levels of circulating vesicles have also been correlated with a poorer prognosis in patients with gastric cancer.[36] Increasing evidence of the role of extracellular vesicles in the progression of cancer suggests that methods to detect, quantify, and modulate exosomes and microvesicles may potentially have prognostic, diagnostic, and therapeutic value. With this goal in mind, we discuss here current methods used to detect and target extracellular vesicles, and address the applicability of the emerging field of membrane curvature and lipid sensing peptides to this function.
2. Sensing the submicroscopic
2.1 Methods for detecting extracellular vesicles
Widely applied technologies used for detection of ultramicroscopic particles like exosomes and microvesicles include electron microscopy, flow cytometry, dynamic light scattering (DLS), and nanoparticle tracking analysis (NTA). Depending on the method used, the reported absolute number of extracellular vesicles in a liter of blood can vary by as much as five orders of magnitude.[37]
Each method mentioned above has advantages and drawbacks when used to detect extracellular vesicles. Electron microscopy can directly show that vesicles exist in a sample, but cannot provide quantitative data, and the fixation process can alter vesicle shape and size.[38] Flow cytometry is typically limited to the identification of particles greater than 300 nm, preventing the detection of smaller microvesicles and all exosomes.[39] Recently developed flow cytometry protocols have lowered this limit, but the detection of sub 100 nm particles, like exosomes, still remains an outstanding problem.[40] Challenges of detecting extracellular vesicles with DLS include: 1) the low refractive index of vesicles, and 2) a bias towards detection of larger particles when used with heterogeneous solutions.[38] This makes it problematic to distinguish between microvesicles (>100 nm) and exosomes (<100 nm) in mixed solution.[41] NTA is perhaps the most promising method because it can identify both microvesicles and exosomes and is not dependent on the refractive index of the vesicles. However, without a fluorescently labeled antibody directed towards a vesicle surface marker, or without use of a vesicle isolation method to reduce polydispersity of the sample, there can be considerable intra-assay count variability.[38,42]
2.2 Brownian motion of extracellular vesicles
Both DLS and NTA rely on the relationship between particle size and diffusion coefficient to determine the size of the extracellular vesicles in solution. This is described quantitatively for a spherical particle in a low Reynolds number fluid by the Stokes-Einstein equation
where D is the diffusion coefficient, kB is Boltzmann’s constant, T is the temperature, η is viscosity of the fluid, and r is the radius of the particle. The estimated diffusion coefficients for microvesicles and exosomes in the extracellular environment are shown in Figure 3A. It is important to note that the relative differences in the diffusion coefficient between different vesicle sizes becomes more difficult to resolve as the vesicles become larger. For example, going from a vesicle 30 nm in diameter to 130 nm in diameter changes the diffusion coefficient by ~12 μm2/s, but from 900 nm to 1000 nm the diffusion coefficient changes by only ~0.05 μm2/s. For experiments using NTA, it is therefore important to consider if the minimum track length (how long a particle must be followed before the diffusion coefficient is calculated) is long enough to properly resolve such differences in vesicle diffusion coefficients. In general, and depending on the settings of the NTA instrument, particles with diameters in the range of 1 μm or greater move too slowly to be accurately sized using NTA.[38]
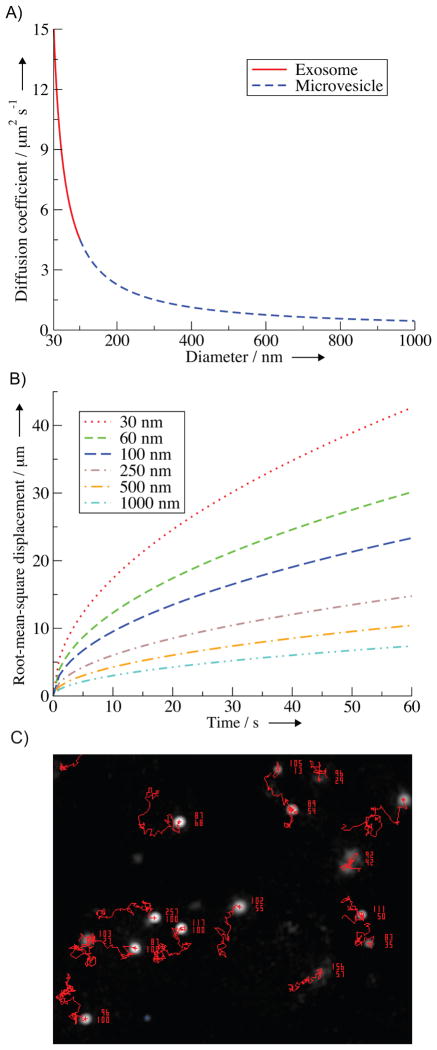
Figure 3.
Extracellular vesicle size dependent diffusion and displacement. A) Diffusion coefficients of exosomes (solid red) and microvesicles (dashed blue) as given by the Stokes-Einstein equation. B) Root-mean-square displacement of exosomes (dashed only lines) and microvesicles (dashed with dotted lines) through an aqueous extracellular environment. C) Representative image of ~100 nm particles visualized using a NTA instrument (NanoSight LM14). Red lines are the diffusional displacement of the particles tracked by the NTA software. Each particle is associated with two numbers, the upper number represents the particle size as calculated using the Stokes-Einstein equation and the lower number represents the particle’s track length.
The calculated diffusion coefficients can also be used to estimate the average distance a vesicle diffuses from its origin in a given time. The root-mean-square displacement (xrms) for diffusion in the absence of an external force is given by
where t is time.[43] The behavior and presence of exosomes and microvesicles is arguably best studied in blood.[44] Vesicles in the blood are subjected to large external forces from the pulsatile flow of the circulation system that trump in scale any diffusive processes. Cases of smaller external forces, where we can make the approximation of using the root-mean-square displacement equation, do exist, such as paracrine signaling between nearby cells or in the sample chamber of an NTA instrument (Figure 3C). For these cases, Figure 3B illustrates that smaller exosomes are able to diffuse greater distances in a given time, making them the faster traveling and equilibrating vesicle.
2.3 Protein markers of extracellular vesicles
Up to this point, we have focused on identifiable biophysical characteristics of extracellular vesicles. In addition to fundamental properties such as diffusion coefficient, exosomes and microvesicles also display surface markers that can be used for quantification and detection. The distinction between markers specific to exosomes or microvesicles is muddled by variations in literature terminology previously mentioned, and challenges isolating and identifying the separate populations of vesicles. Universal markers commonly used to identify exosomes are better characterized, and include transmembrane proteins like tetraspanins (CD9, CD63, CD81, and CD82) and MHC class I and II, and cytosolic proteins like heat shock proteins (HSP-70 and HSP-90).[18,45,46] Source specific markers that represent the proteome of the cell of origin can also be used for exosome identification. For example, urinary exosomes of patient’s with non-small cell lung cancer were found to carry proteins representative of their primary tumor.[47]
Detection of extracellular vesicle proteins is relatively straight forward using analytical techniques like western blot or ELISA. However, soluble antigens may also be detected, and it is not possible to distinguish vesicle sizes or concentrations with these techniques.[38] Additionally, interindividual differences in exosome tetraspanin expression levels, in particular CD63, may reduce the value of comparative studies.[48]
2.4 Targeting extracellular vesicle lipids
The final component of the extracellular vesicle surface is the bilayer lipid membrane. As discussed earlier, the membranes of exosomes and microvesicles both contain rare lipids (e.g. ceramide and aminophospholipids) and have highly curved surfaces that contain lipid-packing defects. In the extracellular environment, these are unique traits that can be targeted by membrane binding proteins. Here we will briefly discuss these proteins and their applications to extracellular vesicle detection. For a broader and more comprehensive summary of membrane binding proteins, see these other excellent reviews.[49–51]
Membrane binding proteins encompass a variety of proteins, from random coils to those with complex secondary structures. Compared to a protein-protein binding interface that can have complex three-dimensional tertiary and quaternary structure, the membrane-protein interface can be thought of as planar regions of discrete interaction regimes: the hydrophobic core, the electrostatically charged phosphate and lipid head groups, and the hydrophilic solvent. Three interaction regime regions, and two interfaces between them, leave open the possibility for many membrane recognition mechanisms.
The most conventional protein-membrane interaction mechanism is the recognition of rare lipid head groups by a folded protein domain in a lock and key like mechanism.[50,52] A well known example is annexin V, which recognizes the lipid phosphatidylserine using folded domain repeats that bind the lipid head group through coordinated Ca2+ ions.[52] Membrane binding through coordinated divalent cations, Ca2+ or Zn2+, is a common mechanism for folded protein domains.[52] Among the lipids found in the extracellular environment, phosphatidylserine carries an uncommon net negative charge. Although most other lipids have ionic character, they are zwitterionic and do not have a net charge.[21] Fluorescently labeled annexin V is commonly used to detect the exposure of phosphatidylserine on the outer membrane leaflet of apoptotic cells.[53] As discussed earlier, extracellular vesicles also contain phosphatidylserine in their outer membrane leaflet. Annexin V has been used in methods to quantify exosome and microvesicle phosphatidylserine levels, for a filtration and flow cytometry based microvesicle counting assay, and to inhibit exosome signaling by blocking exosome-cell membrane fusion.[18,54,55]
2.5 Probes for extracellular vesicle membrane curvature
The next examples include proteins that can sense membrane curvature. Membrane curvature sensors are proteins that bind the membrane with varying affinity based on the membrane’s shape. Curvature sensing occurs through many motifs, including, but not limited too: electrostatic interaction with clustered anionic lipids, well structured protein-membrane shape matching, insertion of hydrophobic residues or loops into membrane defects, and curved membrane induced protein folding.[49,51] Proteins/peptides can also actively induce membrane curvature, altering the shape of the membrane. The distinction between a protein that induces membrane curvature and one that senses it is likely as dependent on the concentration of the protein as it is on its physical properties.[50] The outer membrane of extracellular vesicles is positively curved, or convex, so we focus here on examples of sensors for these surfaces.
Three recent cases show how truncation or structural modification of a known membrane binding protein can lead to a curvature sensing peptide. Myristoylated alanine-rich C kinase substrate (MARCKS) is an intracellular protein that normally functions to sequester free PIP2 lipids on the interior leaflet of the plasma membrane.[56] In its active form, the lysine rich effector domain of MARCKS binds PIP2 through a non-specific electrostatic interaction.[56] Truncation of MARCKS to just its effector domain results in a 25 amino acid unstructured peptide.[57] This peptide, MARCKS-ED, preferentially binds phosphatidylserine enriched and exosome sized (< 100 nm diameter) synthetic liposomes and isolated extracellular vesicles.[57] Electrostatic attraction between positively charged peptide lysine residues and negatively charged phosphatidylserine lipid head groups, as well insertion of five bulky hydrophobic phenylalanine residues into lipid-packing defects, likely explains this behavior.[57] Phenylalanine insertion would be entropically favorable; it would exclude the solvent from the membrane defect and remove the need for a solvation shell to form around the hydrophobic peptide residues.
Another example, C2BL3C, is a 12 amino acid long peptide that was cyclized using “Click” chemistry.[58] The peptide is based on a Ca2+ dependent membrane inserting loop of the membrane fusion protein synaptotagmin-I. Truncation and cyclization of the membrane inserting loop created a more rigid peptide that selectively binds highly curved synthetic liposomes and blood derived exosomes in a calcium independent manner.[58] Although the mechanism of its curvature sensing remains unknown, the low net charge of C2BL3C suggests a lipid-packing defect stabilization mechanism.[58] The final modification example is a derivative of the cationic peptide bradykinin. In this case, a truncated bradykinin monomer showed modest curvature sensing; however, trimerization (conjugation of three monomers to a flexible linker backbone) significantly increased the peptides’ affinity for highly curved and charged surfaces.[59] A synergetic combination of electrostatic attraction and membrane structure scaffolding from the peptides’ claw-like shape may explain this curvature sensing behavior.[59]
These protein truncation, cyclization, and multimerization models illustrate how a simplified scaffold can maintain or gain new membrane curvature sensing properties relevant to extracellular vesicle detection. Compared to annexin V, short, easily synthesized curvature sensing peptides such as these are advantageous because they allow for easier chemical modification, conjugation, and preparation. Future in vivo or in vitro applications will require consideration of the peptides’ biological activity. For example, are the peptides present at concentrations sufficient to damage cells through induction of membrane curvature or other biological activity?
3. Outlook
In this Minireview, we have touched on the basic biology of extracellular vesicles, their importance as a signaling entity in normal and pathologic processes, and addressed bio-physical and -chemical detection methods. Major barriers to our growing understanding of exosome and microvesicle biology include finding reliable methods for quantification, isolation, and modulation. Identifying universal characteristics of submicroscopic particles of heterogeneous origin and content is a challenging task. Outlined characteristics include lipid and protein composition and physical properties such as diffusion coefficient and membrane curvature. As physical properties represent a sum of the vesicle parts, they are arguably more universal. Membrane curvature sensing peptides are a unique class of molecules that can sense the physical state of the membrane. Along with the peptides mentioned, many potential future scaffolds exist for extracellular vesicle curvature sensing probes. These include naturally occurring proteins that sense, and in some cases induce, membrane curvature, such as the BAR domains, the ALPS motif of ArfGAP1, or α-synuclein.[60–63] The development of new tools for extracellular vesicle detection will help address basic questions, such as how exosomes and microvesicles signal their target cells, and will also provide scaffolds for new biotechnological tools. Potential biomedical applications include quantifying circulating vesicles as a novel cancer biomarker and inhibiting the extracellular vesicle dependent signaling pathways that promote cancer growth and metastasis.
Acknowledgments
This work was supported by the National Institutes of Health (R01GM103843 to H.Y. and F30CA180249 to N.K.).
References
- 1.Rak J. Semin Thromb Hemost. 2010;36:888–906. doi: 10.1055/s-0030-1267043. [DOI] [PubMed] [Google Scholar]
- 2.Kahlert C, Kalluri R. J Mol Med. 2013;91:431–437. doi: 10.1007/s00109-013-1020-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El-Hefnawy T. Clin Chem. 2004;50:564–573. doi: 10.1373/clinchem.2003.028506. [DOI] [PubMed] [Google Scholar]
- 4.Camussi G, Deregibus MC, Bruno S, Cantaluppi V, Biancone L. Kidney Int. 2010;78:838–848. doi: 10.1038/ki.2010.278. [DOI] [PubMed] [Google Scholar]
- 5.Lösche W, Scholz T, Temmler U, Oberle V, Claus RA. Platelets. 2004;15:109–115. doi: 10.1080/09537100310001649885. [DOI] [PubMed] [Google Scholar]
- 6.Andaloussi SEL, Mäger I, Breakefield XO, Wood MJA. Nat Rev Drug Discov. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 7.Raposo G, Stoorvogel W. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harding CV, Heuser JE, Stahl PD. J Cell Biol. 2013;200:367–371. doi: 10.1083/jcb.201212113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Biochim Biophys Acta. 2012;1820:940–948. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 10.György B, Szabó TG, Pásztói M, Pál Z, Misják P, Aradi B, László V, Pállinger É, Pap E, Kittel Á, et al. Cell Mol Life Sci. 2011;68:2667–2688. doi: 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tetta C, Ghigo E, Silengo L, Deregibus MC, Camussi G. Endocrine. 2013;44:11–19. doi: 10.1007/s12020-012-9839-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Couzin J. Science. 2005;308:1862–1863. doi: 10.1126/science.308.5730.1862. [DOI] [PubMed] [Google Scholar]
- 13.Lacroix R, Dubois C, Leroyer AS, Sabatier F, Dignat-George F. J Thromb Haemost. 2013;11:24–35. doi: 10.1111/jth.12268. [DOI] [PubMed] [Google Scholar]
- 14.Geddings JE, Mackman N. Blood. 2013;122:1873–1880. doi: 10.1182/blood-2013-04-460139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Nedawi K, Meehan B, Rak J. Cell Cycle. 2009;8:2014–2018. doi: 10.4161/cc.8.13.8988. [DOI] [PubMed] [Google Scholar]
- 16.Scott S, Pendlebury SA, Green C. Biochem J. 1984;224:285–290. doi: 10.1042/bj2240285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hugel B. Physiology. 2005;20:22–27. doi: 10.1152/physiol.00029.2004. [DOI] [PubMed] [Google Scholar]
- 18.Heijnen HF, Schiel AE, Fijnheer R, Geuze HJ, Sixma JJ. Blood. 1999;94:3791–3799. [PubMed] [Google Scholar]
- 19.Harding C, Heuser J, Stahl P. J Cell Biol. 1983;97:329–339. doi: 10.1083/jcb.97.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Savina A, Furlán M, Vidal M, Colombo MI. J Biol Chem. 2003;278:20083–20090. doi: 10.1074/jbc.M301642200. [DOI] [PubMed] [Google Scholar]
- 21.Laulagnier K, Motta C, Hamdi S, Roy S, Fauvelle F, Pageaux JF, Kobayashi T, Salles JP, Perret B, Bonnerot C. Biochem J. 2004;380:1–11. doi: 10.1042/BJ20031594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brügger B, Simons M. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 23.Denzer K, Kleijmeer MJ, Heijnen HF, Stoorvogel W, Geuze HJ. J Cell Sci. 2000;113(Pt 19):3365–3374. doi: 10.1242/jcs.113.19.3365. [DOI] [PubMed] [Google Scholar]
- 24.Théry C, Amigorena S, Raposo G, Clayton A. Curr Protoc Cell Biol. 2006;Chapter 3(Unit 3.22) doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 25.Zimmerberg J, Kozlov MM. Nat Rev Mol Cell Bio. 2005;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 26.Lyman E, Cui H, Voth GA. Biophys J. 2010;99:1783–1790. doi: 10.1016/j.bpj.2010.06.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Jesus AJ, Kastelowitz N, Yin H. RSC Adv. 2013;3:13622. doi: 10.1039/C3RA42332H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hatzakis NS, Bhatia VK, Larsen J, Madsen KL, Bolinger PY, Kunding AH, Castillo J, Gether U, Hedegård P, Stamou D. Nat Chem Biol. 2009;5:835–841. doi: 10.1038/nchembio.213. [DOI] [PubMed] [Google Scholar]
- 29.Suetsugu A, Honma K, Saji S, Moriwaki H, Ochiya T, Hoffman RM. Adv Drug Deliver Rev. 2012:1–8. doi: 10.1016/j.addr.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 30.Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO, Skog J. Nat Commun. 2011;2:180–9. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hood JL, San RS, Wickline SA. Cancer Res. 2011;71:3792–3801. doi: 10.1158/0008-5472.CAN-10-4455. [DOI] [PubMed] [Google Scholar]
- 32.Huber V, Fais S, Iero M, Lugini L, Canese P, Squarcina P, Zaccheddu A, Colone M, Arancia G, Gentile M, et al. Gastroenterology. 2005;128:1796–1804. doi: 10.1053/j.gastro.2005.03.045. [DOI] [PubMed] [Google Scholar]
- 33.Bryant RJ, Pawlowski T, Catto JWF, Marsden G, Vessella RL, Rhees B, Kuslich C, Visakorpi T, Hamdy FC. Brit J Cancer. 2012;106:768–774. doi: 10.1038/bjc.2011.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rabinowits G, Gerçel-Taylor C, Day JM, Taylor DD, Kloecker GH. Clin Lung Cancer. 2011;10:42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 35.Logozzi M, De Milito A, Lugini L, Borghi M, Calabrò L, Spada M, Perdicchio M, Marino ML, Federici C, Iessi E, et al. PLoS ONE. 2009;4:e5219. doi: 10.1371/journal.pone.0005219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim HK, Song KS, Park YS, Kang YH, Lee YJ, Lee KR, Kim HK, Ryu KW, Bae JM, Kim S. Eur J Cancer. 2003;39:184–191. doi: 10.1016/s0959-8049(02)00596-8. [DOI] [PubMed] [Google Scholar]
- 37.van der Pol E, Hoekstra AG, Sturk A, Otto C, van Leeuwen TG, Nieuwland R. J Thromb Haemost. 2010;8:2596–2607. doi: 10.1111/j.1538-7836.2010.04074.x. [DOI] [PubMed] [Google Scholar]
- 38.Dragovic RA, Gardiner C, Brooks AS, Tannetta DS, Ferguson DJP, Hole P, Carr B, Redman CWG, Harris AL, Dobson PJ, et al. Nanomedicine. 2011;7:780–788. doi: 10.1016/j.nano.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perez-Pujol S, Marker PH, Key NS. Cytometry. 2007;71A:38–45. doi: 10.1002/cyto.a.20354. [DOI] [PubMed] [Google Scholar]
- 40.van der Vlist EJ, Nolte-’t Hoen ENM, Stoorvogel W, Arkesteijn GJA, Wauben MHM. Nat Protoc. 2012;7:1311–1326. doi: 10.1038/nprot.2012.065. [DOI] [PubMed] [Google Scholar]
- 41.Lawrie AS, Albanyan A, Cardigan RA, Mackie IJ, Harrison P. Vox Sang. 2009;96:206–212. doi: 10.1111/j.1423-0410.2008.01151.x. [DOI] [PubMed] [Google Scholar]
- 42.Oosthuyzen W, Sime NEL, Ivy JR, Turtle EJ, Street JM, Pound J, Bath LE, Webb DJ, Gregory CD, Bailey MA, et al. J Physiol. 2013;591:5833–5842. doi: 10.1113/jphysiol.2013.264069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Howard J. Mechanics of Motor Proteins & the Cytoskeleton. Sinauer Associates Incorporated; 2001. [Google Scholar]
- 44.Cocucci E, Racchetti G, Meldolesi J. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 45.Caby MP. Int Immunol. 2005;17:879–887. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- 46.Lee TH, D’Asti E, Magnus N, Al-Nedawi K, Meehan B, Rak J. Semin Immunopathol. 2011;33:455–467. doi: 10.1007/s00281-011-0250-3. [DOI] [PubMed] [Google Scholar]
- 47.Li Y, Zhang Y, Qiu F, Qiu Z. Electrophoresis. 2011;32:1976–1983. doi: 10.1002/elps.201000598. [DOI] [PubMed] [Google Scholar]
- 48.Jørgensen M, Bæk R, Pedersen S, Søndergaard EKL, Kristensen SR, Varming K. J Extracell Vesicles. 2013;2:20920. doi: 10.3402/jev.v2i0.20920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bigay J, Antonny B. Dev Cell. 2012;23:886–895. doi: 10.1016/j.devcel.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 50.Antonny B. Annu Rev Biochem. 2011;80:101–123. doi: 10.1146/annurev-biochem-052809-155121. [DOI] [PubMed] [Google Scholar]
- 51.Cui Q, Zhang L, Wu Z, Yethiraj A. Curr Opin Solid State Mater Sci. 2013;17:164–174. [Google Scholar]
- 52.Lemmon MA. Nat Rev Mol Cell Bio. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 53.van Engeland M, Nieland LJ, Ramaekers FC, Schutte B, Reutelingsperger CP. Cytometry. 1998;31:1–9. doi: 10.1002/(sici)1097-0320(19980101)31:1<1::aid-cyto1>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 54.Grant R, Ansa-Addo E, Stratton D, Antwi-Baffour S, Jorfi S, Kholia S, Krige L, Lange S, Inal J. J Immunol Methods. 2011;371:143–151. doi: 10.1016/j.jim.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 55.Keller S, König AK, Marmé F, Runz S, Wolterink S, Koensgen D, Mustea A, Sehouli J, Altevogt P. Cancer Lett. 2009;278:73–81. doi: 10.1016/j.canlet.2008.12.028. [DOI] [PubMed] [Google Scholar]
- 56.Arbuzova A, Schmitz AAP, Vergères G. Biochem J. 2002;362:1–12. doi: 10.1042/0264-6021:3620001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morton LA, Yang H, Saludes JP, Fiorini Z, Beninson L, Chapman ER, Fleshner M, Xue D, Yin H. ACS Chem Biol. 2013;8:218–225. doi: 10.1021/cb300429e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saludes JP, Morton LA, Ghosh N, Beninson LA, Chapman ER, Fleshner M, Yin H. ACS Chem Biol. 2012;7:1629–1635. doi: 10.1021/cb3002705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saludes JP, Morton LA, Coulup SK, Fiorini Z, Cook BM, Beninson L, Chapman ER, Fleshner M, Yin H. Mol BioSyst. 2013;9:2005. doi: 10.1039/c3mb70109c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Madsen KL, Bhatia VK, Gether U, Stamou D. FEBS Lett. 2010;584:1848–1855. doi: 10.1016/j.febslet.2010.01.053. [DOI] [PubMed] [Google Scholar]
- 61.Beck R, Sun Z, Adolf F, Rutz C, Bassler J, Wild K, Sinning I, Hurt E, Brügger B, Béthune J, et al. Proc Natl Acad Sci USA. 2008;105:11731–11736. doi: 10.1073/pnas.0805182105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Drin G, Casella JF, Gautier R, Boehmer T, Schwartz TU, Antonny B. Nat Struct Mol Biol. 2007;14:138–146. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- 63.Pranke IM, Morello V, Bigay J, Gibson K, Verbavatz JM, Antonny B, Jackson CL. J Cell Biol. 2011;194:89–103. doi: 10.1083/jcb.201011118. [DOI] [PMC free article] [PubMed] [Google Scholar]