Successful drug treatment in human disease requires an adequate therapeutic index reflecting the treatment’s specific effects on target cells and its lack of clinically significant effects on the host. In cancer, the therapeutic goal is to trigger tumor-selective cell death. The mechanisms responsible for such death are of obvious importance in determining the efficacy of specific treatments. With the discovery that distinct death pathways exist in biology, and that certain of these are evolutionarily selected and highly efficient, came an explosion of interest in connecting such pathways to the pathophysiology of cancer. Thus, the biology of neoplasia has expanded to incorporate not only lesions that cause dysregulated growth, but also those that lead to inefficient cell death.
Apoptosis represents a universal and exquisitely efficient cellular suicide pathway. As our understanding of its vital role in normal development has deepened, we have identified numerous genes that encode apoptotic regulators, some of which represent familiar oncogenes or tumor-suppressor genes. Because of this convergence between cancer biology and cell death regulation in development, cancer biologists now frequently find themselves focusing on molecular pathways whose endpoint — death — coincides with the goal of successful treatment.
Apoptosis as a therapeutic goal offers advantages over nonapoptotic death mechanisms only if the therapeutic index or the availability of compounds that induce it is greater. In drug-curable malignancies, such as common pediatric leukemias and certain solid tumors, apoptosis is a prominent (if not the exclusive) mechanism associated with the induction of tumor remission. In addition, the expression of apoptotic modulators within a tumor appears to correlate with its sensitivity to traditional cancer therapies. No strict correlation between the induction of apoptosis and a patient’s long-term prognosis has emerged, perhaps in part because the ability to achieve initial remission alone does not adequately predict long-term outcome.
Routes to tumor-cell killing.
A drug that activates apoptosis might achieve a suitable therapeutic index in several ways. First, it might activate a death cascade via a drug target that is uniquely expressed in a cancer cell. Alternatively, it might be delivered to the target tissue in a manner that is selective for the cancer cell. These 2 strategies have proved frustratingly difficult to achieve in any broad clinical sense during the past decades. A third — and perhaps more promising — approach is to exploit a pathway that is activated by oncogenes, in order to provoke apoptosis selectively in cancer cells. It is now clear that oncoproteins can interact with apoptotic regulatory pathways. Thus, overexpression of Myc sensitizes cells to a wide assortment of apoptotic triggers (1), probably reflecting the role of apoptosis in the intracellular immunity that prevents normal cells from persisting in the body once they acquire cancer-causing genetic defects. However, many human tumors that overexpress Myc are highly resistant to apoptotic triggers, probably owing to a variety of downstream lesions that blunt the death pathway. Still, the recognition that oncogenes can sensitize cells to proapoptotic treatments suggests that if such lesions can be circumvented, drugs that induce cell death could prove highly selective for cancer cells.
In certain circumstances, normal cells may be impervious to the effects of therapy. For example, many normal cells are terminally differentiated and quiescent. Drugs that inhibit cell-cycle progression may be relatively nontoxic to such cells. Likewise, angiogenesis might occur preferentially in tumors rather than in most tissues in the adult organism. Angiogenesis inhibitors (see Keshet and Ben-Sasson in this Perspective series) might display relatively little toxicity toward normal cells, but all cells in the body appear to be susceptible to apoptosis, so drugs that target cancer by promoting this pathway must target molecules upon which the cancer cell has developed a particular reliance. The process of unraveling the mechanistic details of apoptosis has revealed evidence for a core machinery involved in execution of the later steps, as well as multiple regulatory networks. These regulatory paths frequently respond to environmental signals such as growth factor signaling or stress, and subsequently feed into the core apoptotic machinery at a variety of positions whose identities are emerging in an exciting wave of current investigations.
The core apoptotic machinery.
Apoptotic cell death is triggered by intracellular cues such as DNA damage and osmotic stress, and extracellular cues including growth factor withdrawal, matrix detachment, and direct cytokine-mediated killing. Two central pathways are involved in the process of apoptotic cell death, one involving the activation of the caspase proteases and a second, mitochondrial, pathway (Figure 1). A common feature of this machinery and of the signaling pathways that impinge on this machinery is that at nearly every level, the action of proapoptotic molecules is opposed by sets of inhibitors.
Figure 1.
The core apoptotic machinery. The core apoptotic machinery comprises the caspases, members of the Bcl-2 family, and Apaf-1. Caspase 3 and other effector caspases carry out the process of cellular destruction that accompanies apoptotic signals. Effector caspases are cleaved and activated by initiator caspases, such as caspases 8 and 9. Caspase 8 is a target of activation by death signaling pathways. Bcl-2 family members regulate both mitochondrial damage and caspase activation. Certain antiapoptotic proteins can directly inhibit caspase 9 activation by Apaf-1, and in turn are antagonized by proapoptotic Bcl-2 family members such as tBid. The cleavage of Bid to tBid by caspase 8 and the inhibition of caspase 9 by Bcl-2 family members are examples of interconnections between the caspase pathway and the mitochondrial pathway of apoptosis.
Caspases are cysteine proteases that cleave proteins at aspartic acid residues contained within a tetrapeptide recognition motif. These enzymes are produced as inactive zymogens and are activated during apoptosis. They are divided into effector caspases and initiator caspases. When activated, effector caspases assist in carrying out the process of cellular death, either by directly cleaving proteins such as lamin (found in the nuclear membrane) or by activating other degradation enzymes such as CAD (a DNA degradation enzyme). Initiator caspases, on the other hand, transmit apoptotic signals by promoting the cleavage and activation of the effector caspases.
Caspase activity is regulated by adaptor molecules that promote or inhibit caspase activation. Activators promote caspase activation either by increasing the local concentration of caspases, allowing autoactivation to occur, or by inducing a conformational change that leads to caspase activation. Activators (such as the adaptor protein FADD) promote the formation of a death-inducing signaling complex. Their actions are opposed by multiple inhibitor proteins (IAPs) that block activation of both effector and initiator caspases.
A number of death pathways converge on the caspase cascade (Figures 1 and 2). These pathways begin when a death ligand such as TNF or FasL interacts with its cognate receptor, TNF-R or Fas (CD95), inducing the trimerization of the receptors. Receptors in this family (TNF-R1, Fas, DR3, DR4, DR5, and DR6) all contain an intracellular “death domain,” and receptor trimerization recruits adaptor proteins such as FADD to this domain. These adaptor molecules then recruit and activate caspase 8, although in certain instances caspase 8 may be recruited directly to the receptor. The death receptors are countered by a number of decoy receptors, which lack the death domain and may antagonize death receptor activation (2).
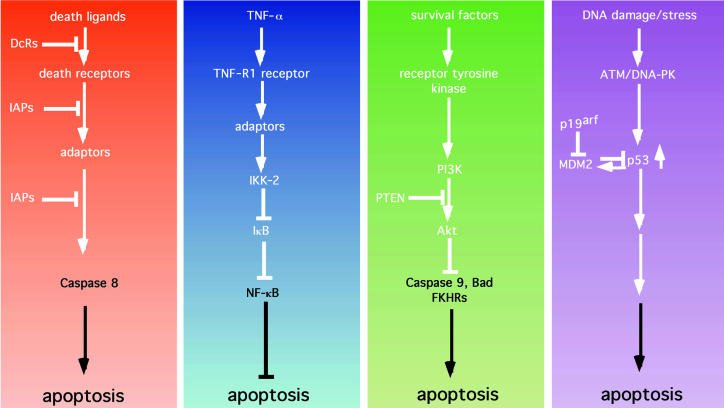
Figure 2.
Apoptotic and antiapoptotic signaling pathways. (a) Death pathways. Death ligands typified by FasL, TNF-α, Apo2L, and Apo3L bind to and activate death receptors (e.g., Fas). This activation can be blocked by the presence of nonfunctioning decoy receptors (DcRs). Receptor trimerization and activation leads to the recruitment of adaptor molecules (such as FADD) that can recruit and activate caspase 8. Activation of caspase 8 can be blocked by inhibitors of apoptosis such as cIAP-1 and cIAP-2, as well as TRAF-1 and TRAF-2. (b) The NF-κB survival pathway. In addition to eliciting a death signal, TNF-α transmits a survival signal. Here, binding to the TNF-R1 receptor leads to the recruitment of adaptors that are capable of indirectly activating IκB kinase 2 (IKK-2), which phosphorylates IκB, leading to ubiquitin-mediated proteolysis of IκB. Loss of IκB allows NF-κB to localize to the nucleus and to induce the expression of antiapoptotic factors such as the IAPs, the TRAFs, and Bfl-1. (c) The PI3K/Akt pathway. Soluble growth and survival factors such as IL-3 and IGFs bind to and activate transmembrane receptor tyrosine kinases. Activation and phosphorylation of the receptors allows the recruitment and activation of PI3K. PI3K phosphorylates phosphatidylinositol 3,4-bisphosphate (PI3,4P2) to generate PIP3. This step is antagonized by the product of the tumor-suppressor gene PTEN, which acts as a lipid phosphatase. PIP3 activates downstream targets, such as the serine-threonine kinase Akt. Akt, in turn, phosphorylates and inactivates a number of substrates including the proapoptotic proteins Bad, caspase 9, and FKHR, and thereby inhibits apoptosis. (d) The p53 pathway. Cellular stresses — particularly DNA damage —are sensed by proteins such as ATM and DNA-PK, which phosphorylate and stabilize p53. p53 induces apoptosis by an as-yet undefined mechanism. The action of p53 is opposed by Mdm2, which binds to and inactivates p53. Mdm2, in turn, is inhibited by p19arf, the product of the INK4A tumor-suppressor gene.
A second, mitochondrial pathway to apoptosis is the likely province of the Bcl-2 family of proteins. The antiapoptotic protein Bcl-2, originally cloned as an oncogene from the t(14:18) breakpoint found in low-grade lymphomas, is the founding member of a large family that consists of proapoptotic factors such as Bax, Bak, Bcl-XS, and Bad, and antiapoptotic factors such as Bcl-2, Bcl-XL, and Bcl-W.
In response to apoptotic signals, proapoptotic Bcl-2 family members translocate to and alter the permeability of the mitochondrial membrane. These proteins are thought to either form channels in the outer mitochondrial membrane or to alter the activity of existing channels, leading to changes in the mitochondrial membrane potential, cytochrome c release, and the production of reactive oxygen species. Antiapoptotic members of the Bcl-2 family (such as Bcl-2 itself) reside in the outer mitochondrial membrane and counter these effects. Certain of the proapoptotic family members, including Bcl-XS, Bid, and Bad, may act by binding and antagonizing the antiapoptotic members of the family, rather than by any direct effects on mitochondrial permeability (see Figure 1).
The mitochondrial and caspase apoptotic pathways are intimately connected. For example, caspase 8 cleaves the Bcl-2 family member known as Bid to produce truncated Bid (tBid). In turn, tBid, in cooperation with Bad, can induce cytochrome c release, causing the caspase adaptor Apaf-1 to activate caspases 9 and 3. This interconnection leads to signal amplification involving a caspase-mitochondrial loop (see Figure 1). In addition, Bcl-XL binds and inactivates Apaf-1, whereas proapoptotic members can displace Bcl-XL from Apaf-1, allowing Apaf-1 to activate caspase 9. Thus, these Bcl-2 family members can directly influence the caspase pathway.
Drug targets in the core apoptotic machinery.
A number of genes that encode components of the apoptotic machinery are directly targeted by activating or inactivating genetic lesions in cancer. These genetic changes, which may inform future drug development programs, presumably highlight proteins or pathways upon which the tumor is critically dependent. Several of the genes have been identified only recently, so the frequency and extent of lesions at these loci are not well known. However, Bcl-2, which was originally cloned by its association with a chromosomal translocation common among follicular B-cell lymphomas, is overexpressed in a number of malignancies.
Targeting the core apoptotic machinery is bound to be a tricky business. Successful therapies will need to intervene very precisely in the pathway to avoid unrestricted cell killing. Direct activation of caspase 3, for example, would probably kill any cell. On the other hand, Bcl-2 is sometimes altered in tumors, and therefore must either be required at critical steps in tumorigenesis, or must significantly enhance cancer cell proliferation. Surprisingly, follicular lymphomas in which Bcl-2 is misexpressed represent some of the most highly apoptotic tumors, and they not infrequently undergo spontaneous remission, although eventual relapse is very common. Inhibition of Bcl-2 has been attempted through antisense oligonucleotides, antisense mRNA, and ribozyme constructs primarily in tumor cell lines.
One strategy for promoting tumor cell apoptosis would be to mimic the action of proapoptotic molecules. Thus, overexpression of antiapoptotic proteins such as Bcl-2 in tumor cells might serve to sequester and inactivate Apaf-1, elevating the cell’s resistance to proapoptotic molecules such as Bax. Peptides that mimic the domain of Bax, which mediates binding to Bcl-2 (the so-called BH3 domain), would be predicted to disrupt the interaction of Bcl-2 with Apaf-1, which suppresses caspase 9 activity. For this reason, BH3 domains and peptides have been used to induce apoptosis in cells, but whether they preferentially kill tumor cells or cells that contain amplified Bcl-2, rather than normal cells, is not known (3).
Alternatively, one could activate the death pathways directly, using soluble death ligands. Thus far, this approach has been problematic, because administered TNF-α and FasL are each broadly cytotoxic. Whether differential expression of other death receptors or decoy receptors in tumor and normal cells will allow for a therapeutic window for the more recently described death ligands such as Apo2L remains to be seen.
NF-κB and the regulation of apoptosis.
NF-κB is a transcription factor composed of dimers of the NF-κB/Rel family of proteins (4). Family members are defined by the presence of a so-called Rel homology domain, which mediates DNA binding and homo- or heterodimerization, as well as nuclear localization and binding to inhibitory proteins of the IκB family. NF-κB activity is induced in response to a large number of stimuli, including cytokines such as TNF-α and IL-1, and viral infection. In most cell types, NF-κB is sequestered by its interaction with IκB proteins in the cytoplasm and is consequently inactive (see Figure 2). IκB binding to the Rel homology domain of NF-κB blocks the nuclear localization signal of NF-κB. Upon cytokine stimulation, IκB is phosphorylated and subsequently ubiquitinated and degraded, releasing NF-κB. The ubiquitination process itself is constitutive — degradation is regulated primarily at the level of phosphorylation. Phosphorylation of IκB is mediated by a protein complex containing 2 kinases, IκB kinase 1 and IκB kinase 2 (IKK-1 and IKK-2; see Figure 2, first panel).
NF-κB has been implicated in a number of cellular functions, including cell-cycle regulation and immunoglobulin gene expression. Emerging data from the study of knockout animals suggest that, at least in some cells, NF-κB mediates a critical antiapoptotic signal. Specifically, RelA–/– mice, which lack the 65-kDa subunit of NF-κB, die during embryogenesis of hepatocellular apoptosis, and their fibroblasts and macrophages are particularly sensitive to TNF-α–mediated cell killing (5). The liver phenotype is the same in mice that lack IKK-2, and this phenotype is suppressed in animals with reduced expression of TNF-R1 (6).
NF-κB is activated by TNF-α, a cytokine that can also induce apoptosis. Thus, 1 model that has emerged is that TNF-α elicits 2 signals, 1 apoptotic and 1 antiapoptotic. Inhibition of NF-κB appears to eliminate the latter signal, leading to deregulated cell death. The mechanism by which NF-κB inhibits apoptosis is unknown. One possibility is that certain critical transcriptional targets act to block apoptotic signals. Indeed, targets of NF-κB regulation include the IAPs (inhibitors of apoptosis) as well as TRAF1 and TRAF2, which are thought to suppress caspase 8 activation (7). Recently, Bfl-1/A1, an antiapoptotic member of the Bcl-2 family, was cloned in a search for NF-κB–responsive genes (8).
Multiple human tumors and an avian retrovirus have evolved mechanisms for dysregulating the NF-κB pathway, suggesting that NF-κB participates in cellular transformation (9). The cellular REL gene is found in birds as a retroviral oncogene (v-rel), and T cell–specific expression of this oncogene gives rise to T-cell leukemias in transgenic mice. In chronic lymphocytic leukemia, a rare translocation activates Bcl-3, a member of the IκB family that, unlike other family members, localizes to the nucleus, where it stimulates the transactivation function of NF-κB. Overexpression of this protein in tumors might therefore enhance NF-κB function. Similarly, mutations of IκB-α that are predicted to dysregulate NF-κB activity have been reported in Hodgkin’s disease. Finally, direct alterations of the NF-κB2 locus have also been noted in T-cell lymphomas.
Drug targets in the NF-κB pathway.
It is likely that a therapeutic inhibitor of NF-κB activation would prove useful in controlling lymphoid malignancies, but the cellular target of such an agent would have to be chosen with care. In most normal cells, except mature B cells, NF-κB is sequestered in the cytoplasm and is inactive. Therefore, it seems that additional inhibition of this pathway would be of little consequence to normal cells. On the other hand, RelA–/– mice and IKK-2–/– mice die during embryogenesis from massive hepatocellular apoptosis, and if adult hepatocytes are equally sensitive to the loss of NF-κB, the inhibition of this pathway could produce intolerable liver toxicity.
A number of the genetic alterations in this pathway bypass upstream regulatory mechanisms and lead directly to the production of activated NF-κB, so one may be left with trying to target the transcription factor itself. Still, the kinase IKK-2 has been identified as a necessary regulator of IκB whose inhibition should block the disruption of the IκB/NF-κB complex, keeping NF-κB sequestered in the cytoplasm. The growing number of successful kinase inhibitors might make IKK-2 a reasonable target. Downstream deregulation of this pathway could render such a maneuver futile, but if cytokine or autocrine loops become a recognized indispensable component of Rel-dependent transformation, IKK-2 inhibition might still be effective.
A number of other strategies can be envisioned for this pathway. In particular, adenoviral mediated delivery of IκB-α, a potent inhibitor of NF-κB, could be advantageous, because many normal cells already carry a stable IκB/NF-κB complex in their cytoplasm. Another strategy would be to target NF-κB at the level of transcriptional activation. NF-κB is thought to activate transcription through the p300/CBP family of coactivators, so molecules that disrupt this complex could prove therapeutic.
Survival signaling through phosphoinositide-3 kinase.
The phosphoinositide-3 kinase (PI3K)/Akt pathway shown in Figure 2 (second panel) is a potent mediator of cell survival signals (10). Extracellular survival signals delivered as soluble factors or through cell attachment can inhibit apoptosis by activating this pathway. Upon growth factor binding, transmembrane receptor tyrosine kinases undergo auto- or transphosphorylation, creating binding sites for PI3K in their cytoplasmic domains, thus recruiting active PI3K to the inner surface of the plasma membrane. Here, PI3K acts on membrane phosphoinositides to generate phosphatidylinositol 3,4-bisphosphate and phosphatidylinositol 3,4,5-trisphosphate (PIP3). These phospholipids serve as foci for recruiting and activating a number of signaling proteins to the membrane. Among these proteins are the serine-threonine kinases of the Akt family (Akt-1, -2, and -3, collectively referred to as Akt), which have emerged as critical regulators of apoptosis. Overexpression of Akt can confer cell survival, and Akt phosphorylates and inactivates a number of substrates that are intimately connected to apoptosis, including caspase 9 and BAD. In addition, genetic studies of the Caenorhabditis elegans aging pathway have linked PI3K and Akt to the regulation of a member of the forkhead transcription factor family. Based upon this work, 3 related mammalian forkhead genes have been identified as proapoptotic Akt targets: FKHR, FKHRL1, and AFX. Finally, Akt can also inhibit cytochrome c release from mitochondria. Thus, Akt can potentially affect the apoptotic machinery at multiple points (see Figure 2).
PI3K activity is also regulated by cellular interactions with other cells or with the extracellular matrix. In certain epithelial cells, expression of oncogenic Ras, working through Akt, can block the process of detachment-induced apoptosis (anoikis), permitting these cells to survive in the absence of the normal adhesive interactions (11).
A role for the PI3K/Akt pathway in transformation was first suspected with the identification of PI3K as a target of the polyoma viral oncoprotein middle T antigen (12). Subsequently, both Akt-1 and the gene encoding the catalytic subunit of PI3K were identified as transforming retroviral oncogenes. Furthermore, the protein product of the recently cloned PTEN tumor-suppressor gene functions as a lipid phosphatase and specifically antagonizes the action of PI3K. Loss of PTEN in cell lines and in tumors leads to deregulation of the PI3K pathway and unregulated activation of Akt. As a result, PTEN-deficient tumor cells may be protected from apoptosis. In keeping with this notion, reconstitution of PTEN to certain PTEN null cells induces apoptosis in an Akt-dependent manner. Somatic mutation of PTEN is found in a variety of malignancies, and amplification of Akt-2 has been reported in certain breast and pancreatic cancers. Therefore it is likely that deregulated PI3K/Akt contributes to tumorigenesis in a significant number of human tumors.
Drug targets in the PI3K/Akt pathway.
A number of considerations make the PI3K/Akt pathway an attractive target for therapeutic intervention. First, PTEN loss deregulates the pathway in a number of common malignancies. Second, the pathway involves 3 distinct classes of kinases, any 1 of which could be targeted. Third, the pathway is relatively inactive in resting cells, although it does play an important role in regulating glucose metabolism and appears to be critical for survival of lymphocytes and possibly endothelial cells or neurons. Thus, toxicity in these tissues could be problematic.
Targets in the pathway include the upstream tyrosine kinase receptors themselves, activated Ras, PI3K, and the downstream targets of PI3K, including Akt, PDK1, and SGK. In the absence of PTEN, downstream targets of PI3K appear to be constitutively active, even in the absence of serum, suggesting that the inhibition of growth factor receptors may be an ineffective strategy for blocking the proliferation of cells that have lost this tumor-suppressor activity. On the other hand, inhibitors to PI3K can induce apoptosis in certain tumor cell lines including cells that lack PTEN. In addition, because the PI3K multigene family governs a diverse array of cellular functions, it is possible that cell survival signals in tumors rely upon specific family members, which might provide a more restricted target for kinase inhibition.
Kinases activated by PI3K include Akt-1, Akt-2, Akt-3, PDK1, SGK, and BTK. Whether 1 or more of these kinases is necessary for transformation is not known. However, the fact that Akt is found as a retroviral oncogene, and that amplification of cellular AKT-1 and AKT-2 genes occurs in some tumors suggests that this may be a critical kinase for tumor cells. Thus, targeted inhibition of Akt may be effective. In support of this notion, dominant-negative Akt can reverse Ras-dependent survival signals (11), and antisense Akt can reduce growth of cell lines containing BCR-ABL (13; see also Druker and Lydon in this Perspective series). An alternative strategy for pathway inhibition is to restore PTEN to PTEN–/– tumor cells by adenoviral mediated gene delivery, a treatment that has little effect on PTEN-expressing cells. Finally, therapeutic inactivation of Ras is being attempted, using compounds that prevent membrane localization by blocking Ras farnesylation. Evidence linking Ras effector pathways to PI3K/Akt suggests that farnesyl transferase inhibitors could work, at least in part, through inhibition of this pathway. Again, because this is an upstream activating event, such a manipulation might fail in cells lacking PTEN.
Apoptosis and p53.
The p53 tumor-suppressor gene is the most commonly mutated gene in human cancer. It is a striking example of a factor whose tumor-selective effects could provide the basis of treatments with high therapeutic indices. Two biological functions are attributed to p53: regulation of cell cycle progression after stress, and regulation of apoptosis. After stresses such as DNA damage, a signaling cascade involving the DNA-dependent protein kinase and the ATM protein results in p53 phosphorylation and stabilization (see Figure 2, third panel). This p53 upregulation produces cell-cycle arrest in untransformed primary fibroblasts but leads to apoptosis in oncogene-transformed, but otherwise genetically matched fibroblasts (14). Conversely, p53-deficient cells display sluggish cell-cycle arrest, particularly involving G1/S and G2/M checkpoints (see Shapiro and Harper in this Perspective series). Also, oncogene-transformed, p53-deficient fibroblasts display dramatic resistance to apoptosis triggers to which p53-expressing transformed cells are exquisitely sensitive. Moreover, in normal cells, cell-cycle arrest creates a barrier to replication of a damaged genome. This behavior of p53 — provoking apoptosis in transformed cells and arrest in untransformed cells — therefore constitutes tumor-selective death regulation.
In addition to the mutation or deletion of the p53 gene in approximately 50% of human cancers, a number of additional lesions are found that appear to act downstream of p53. For example, 1 of p53’s transcriptional targets, Mdm2, feeds back to inhibit p53 function, at least in part by enhancing p53 degradation. MDM2 is amplified in a number of human cancers that lack p53 mutations. In addition, the alternative reading-frame product of the p16/INK4A locus (ARF) negatively regulates Mdm2 activity. Loss of ARF occurs in many cancers, including some, such as melanomas, in which p53 mutations are rare. Although the mechanistic details linking p53 to caspase activity remain to be fully elucidated, it is likely that mutations within the p53 pathway will be found in the vast majority of human cancers.
In addition, p53 status in human cancer may predict clinical outcome. Despite a large and not entirely consistent body of literature on this topic, several arguments suggest that the status of this pathway is of clinical importance. First, in some instances where p53 mutation fails to correlate with prognosis, genetic events acting elsewhere in the pathway could be identified that were consistent with the prognosis. Examples include the amplification of MDM2 in soft tissue sarcoma, and the presence of human papilloma virus E6 protein, which promotes ubiquitin-dependent proteolysis of p53 in cervical carcinomas. Other, still unrecognized factors that regulate caspases in response to p53 signaling may be aberrant in aggressive tumors that retain wild-type p53. Second, in genetically defined mouse models, p53 status correlates not only with apoptosis in vitro, but also with chemosensitivity and radiation sensitivity in solid tumors in animals (15). Third, cancers that are known to respond well to chemotherapy are far more likely to retain wild-type p53 expression than are other tumor types. In fact, currently the most curable cancers are among the minority of tumors in which p53 is not mutated.
Drug targets and p53 in cancer.
If p53 loss disables a major therapeutic death pathway, it is easy to understand how loss of this protein or other effector molecules would be selected during treatment, or even during tumorigenesis, when cells are under stress from growth factor deprivation, hypoxia, or acidosis (16). The restoration of this pathway to p53-deficient cells is therefore a promising approach for controlling tumors, and has been pursued by several strategies. First, gene-transfer vectors might be used to restore wild-type p53. This strategy, which is currently under study, must overcome numerous obstacles inherent to gene therapy technology, including the requirement to efficiently infect all (or nearly all) cancer cells. In addition, animal studies show that certain untransformed cell types, such as irradiated thymocytes, undergo apoptosis in response to p53 induction (17), so targeted delivery of transgenes is being used as well.
An interesting potential complication of restoring p53 to such mutant tumors involves the paradoxical potential for blocking apoptosis in treated cells. In several experimental systems, tumor lines containing wild-type p53 became more sensitive to DNA-damaging treatments after loss of endogenous p53 (18), perhaps because of the presence of additional lesions that block p53’s function in the apoptotic pathway, but that leave it active in controlling cell-cycle arrest. If the primary function of p53 is arrest (death having been inhibited downstream of p53), then reintroduction of wild-type p53 would enhance genomic fidelity after genotoxic stress, and thus enhance tumor cell survival. Thus, restoration of the wild-type p53 gene would be predicted to enhance apoptosis only in tumors in which the p53-apoptosis pathway is otherwise intact.
Another therapeutic strategy that targets p53 deficiency uses an engineered adenovirus, exploiting a requirement for p53 protein to prevent lytic viral infections (19). A deficiency of p53 results in a cell that is susceptible to lytic infection, although normal host cells are resistant to such infection. Still another strategy that targets p53 and apoptosis for therapeutic benefit aims to inhibit p53-dependent apoptosis: because drug toxicity in normal cells is mediated in many cases by p53-dependent apoptosis, an inhibitor of p53 could theoretically enhance the therapeutic index, not by sensitizing tumor cells, but by increasing resistance in normal cells. Both of these strategies await clinical analysis to determine in vivo efficacy. Finally, one might target p53-dependent apoptosis using the recently discovered homologues of p53. Proteins in this family appear to trigger apoptosis upon overexpression and might therefore serve as surrogates of p53. It remains to be fully determined whether these factors are also deficient in human cancers, but if not, strategies to upregulate their activity may act selectively to promote apoptosis in cancer cells.
The mechanisms by which p53 regulates apoptosis are poorly understood. In some experimental contexts, p53 appears to act through a transcription-dependent pathway (20), whereas in other systems, p53-dependent apoptosis can occur in the complete absence of new gene expression (21). Numerous potential targets of transcriptional regulation by p53 have been identified whose overexpression can produce apoptosis, but it is still uncertain if these molecules are primarily responsible for tumor cell death after induction of p53. Transcription-independent induction of apoptosis is also incompletely understood, and further work on both of these pathways is needed to identify intermediates that connect p53 to the caspase machinery in vivo. Once the biochemical events are elucidated, it may be possible to develop compounds that mimic p53 in sensitizing tumor cells selectively to apoptosis.
References
- 1.Evan GI, et al. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 2.Pan G, et al. An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science. 1997;277:815–818. doi: 10.1126/science.277.5327.815. [DOI] [PubMed] [Google Scholar]
- 3.Holinger EP, Chittenden T, Lutz RJ. Bak BH3 peptides antagonize Bcl-xL function and induce apoptosis through cytochrome c-independent activation of caspases. J Biol Chem. 1999;274:13298–13304. doi: 10.1074/jbc.274.19.13298. [DOI] [PubMed] [Google Scholar]
- 4.Baeuerle PA, Baltimore D. NF-kappa B: ten years after. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 5.Beg AA, Baltimore D. An essential role for NF-kappaB in preventing TNF-alpha-induced cell death. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 6.Li Q, Van Antwerp D, Mercurio F, Lee K-F, Verma IM. Severe liver degeneration in mice lacking the IKappaB kinase 2. Science. 1999;284:321–325. doi: 10.1126/science.284.5412.321. [DOI] [PubMed] [Google Scholar]
- 7.Wang CY, Mayo MW, Korneluk RG, Goeddel DV, Baldwin AS., Jr NF-kappaB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science. 1998;281:1680–1683. doi: 10.1126/science.281.5383.1680. [DOI] [PubMed] [Google Scholar]
- 8.Zong WX, Edelstein LC, Chen C, Bash J, Gelinas C. The prosurvival Bcl-2 homolog Bfl-1/A1 is a direct transcriptional target of NF-kappaB that blocks TNFalpha-induced apoptosis. Genes Dev. 1999;13:382–387. doi: 10.1101/gad.13.4.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilmore TD, Koedood M, Piffat KA, White DW. Rel/NF-kappaB/IkappaB proteins and cancer. Oncogene. 1996;13:1367–1378. [PubMed] [Google Scholar]
- 10.Cantley LC, Neel BG. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the PI3K/AKT pathway. Proc Natl Acad Sci USA. 1999;96:4240–4245. doi: 10.1073/pnas.96.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khwaja A, Rodriguez-Viciana P, Wennstrom S, Warne PH, Downward J. Matrix adhesion and Ras transformation both activate a phosphoinositide 3-OH kinase and protein kinase B/Akt cellular survival pathway. EMBO J. 1997;16:2783–2793. doi: 10.1093/emboj/16.10.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitman M, Kaplan DR, Schaffhausen B, Cantley L, Roberts TM. Association of phosphatidylinositol kinase activity with polyoma middle-T competent for transformation. Nature. 1985;315:239–242. doi: 10.1038/315239a0. [DOI] [PubMed] [Google Scholar]
- 13.Skorski T, et al. Transformation of hematopoietic cells by BCR/ABL requires activation of a PI3K/Akt-dependent pathway. EMBO J. 1997;16:6151–6161. doi: 10.1093/emboj/16.20.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowe SW, Ruley HE, Jacks T, Housman DE. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 15.Lowe SW, et al. p53 status and the efficacy of cancer therapy in vivo. Science. 1994;266:807–810. doi: 10.1126/science.7973635. [DOI] [PubMed] [Google Scholar]
- 16.Schmaltz C, Hardenbergh PH, Wells A, Fisher DE. Regulation of proliferation-survival decisions during tumor cell hypoxia. Mol Cell Biol. 1998;18:2845–2854. doi: 10.1128/mcb.18.5.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowe SW, Schmitt EM, Smith SW, Osborne BA, Jacks T. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature. 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 18.Xu C, Meikrantz W, Schlegel R, Sager R. The human papilloma virus 16E6 gene sensitizes human mammary epithelial cells to apoptosis induced by DNA damage. Proc Natl Acad Sci USA. 1995;92:7829–7833. doi: 10.1073/pnas.92.17.7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bischoff JR, et al. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science. 1996;274:373–376. doi: 10.1126/science.274.5286.373. [DOI] [PubMed] [Google Scholar]
- 20.Attardi LD, Lowe SW, Brugarolas J, Jacks T. Transcriptional activation by p53, but not induction of the p21 gene, is essential for oncogene-mediated apoptosis. EMBO J. 1996;15:3693–3701. [PMC free article] [PubMed] [Google Scholar]
- 21.Ding HF, Fisher DE. Mechanisms of p53-mediated apoptosis. Crit Rev Oncog. 1998;9:83–98. doi: 10.1615/critrevoncog.v9.i1.60. [DOI] [PubMed] [Google Scholar]