Abstract
Little is known about the catalytic mechanism of the recently discovered ThyX family of flavin-dependent thymidylate synthases that are required for thymidylate (deoxythymidine 5′-monophosphate) synthesis in a large number of microbial species. Using a combination of site-directed mutagenesis and biochemical measurements, we have identified several residues of the Helicobacter pylori ThyX protein with crucial roles in ThyX catalysis. By providing functional evidence that the active site(s) of homotetrameric ThyX proteins is formed by three different subunits, our findings suggest that ThyX proteins have evolved through multimerization of inactive monomers. Moreover, because the active-site configurations of ThyX proteins, present in many human pathogenic bacteria, and of human thymidylate synthase ThyA are different, our results will aid in the identification of compounds specifically inhibiting microbial growth.
Thymidylate (deoxythymidine 5′-monophosphate or dTMP) is a key metabolite required for the accurate replication of DNA genomes in all cellular organisms. Until recently, thymidylate synthase ThyA was thought to correspond to the sole enzyme catalyzing the formation of dTMP de novo. However, using in silico and experimental approaches, we recently identified a family of thymidylate synthases (ThyXs) (1). Both ThyA (EC 2.1.1.45) and ThyX (EC 2.1.1.148) proteins catalyze the methylation of deoxyuridine 5′-monophosphate (dUMP) to dTMP. However, the residues essential for catalysis in ThyA are absent in ThyX proteins (1), and no sequence nor structural similarity exists between the homodimeric ThyA and homotetrameric ThyX proteins (2-4). Whereas in the ThyA-catalyzed reaction, methylenetetrahydrofolate (CH2H4folate) provides a methylene group and functions as reductant (5), ThyX proteins contain a tightly bound FAD (1, 4). Our observation that ThyX activity is substantially increased when flavin nucleotides are added to the reaction mixtures indicated that FAD-independent (ThyA) and FAD-dependent (ThyX) molecular mechanisms for thymidylate synthesis have been established during the evolution of DNA as genetic material. The observation that two thymidylate synthases use different reductive mechanisms has, in addition to understanding the mechanisms of thymidylate synthesis, additional implications for other pathways of intermediate metabolism that depend on reduced folates (6, 7).
Although over the years many elegant studies addressed the role of conserved amino acid residues in ThyA catalysis (for reviews, see refs. 5 and 8), the amino acid residues that allow the transfer and reduction of the donating methylene moiety of CH2H4folate in ThyX proteins are unknown. In this work, using site-directed mutagenesis, biochemical analyses, and mass spectrometric measurements, we have performed a detailed study of the functional properties of a subset of conserved amino acid residues in ThyX proteins. This combined approach has allowed the attribution of specific roles in ThyX catalysis to several residues of previously unknown function. Our results have revealed that residues from three different monomers of the homotetrameric ThyX complex are required for catalysis, suggesting that the active homooligomeric ThyX complex evolved through multimerization of “inactive” ThyX monomers. Moreover, together with the presence of ThyX proteins in numerous pathogenic Bacteria (1, 9, 10), our findings will also aid in identifying specific ThyX inhibitors, which in consequence do not have to rely on subtle mechanistic or structural differences in the active sites of human ThyA and ThyX proteins.
Materials and Methods
Bacterial Strains and Molecular Genetic Techniques. Bacterial strains and plasmids used in this study are indicated in Table 3, which is published as supporting information on the PNAS web site. Escherichia coli strains were grown using LB medium or M9 minimal medium (Difco) supplemented with 2 mM MgSO4/0.1 mM CaCl2/40 μg/ml leucine and proline/0.4% glucose as described. When necessary, 100 μg/ml ampicillin or 0.2% arabinose where added for plasmid maintenance or protein induction, respectively (1). Complementation tests were performed as described (1).
The pGL2 plasmid containing Helicobacter pylori thyX gene (HP1533) in pBAD-Topo (Invitrogen) was used as template for site-directed mutagenesis reactions that were performed with QuickChange Site-directed and Multi site-directed mutagenesis kits (Stratagene). The sequences of the mutagenic primers [carrying 5′-phosphorylations when appropriate (Sigma Genosys)] are listed in Table 3. All plasmid constructs were confirmed by DNA sequencing before further analysis.
Protein Expression, Purification, and Spectroscopic Analyses. The wild-type enzyme and mutant ThyX proteins of H. pylori were expressed in E. coli strain BL21 [F-, ompT, hsdS (rB-, mB-) gal dcm] at 37°C in 800 ml of LB medium containing 100 mg/liter ampicillin. Protein expression was induced by adding 0.2% l-arabinose to early exponential-phase cultures (OD600 ≈ 0.5) for 2 h. 6xHis-tagged proteins were purified by gravity-flow chromatography on Ni-NTA agarose (Qiagen, Valencia, CA) and obtained protein samples were stored at -80°C in 50 mM Hepes (pH 7.0), supplemented with 10% glycerol.
Visible absorption spectroscopy was used to quantify the relative binding of FAD to mutant ThyX proteins. Spectra between 350 and 600 nm were recorded at room temperature with a CARY 50 spectrophotometer (Varian) with a total volume of 50 μl. Absorption changes were normalized to protein concentrations.
Optimization of the Assay Conditions for H. pylori ThyX Proteins. The tritium release assay for H. pylori ThyX activity measurements in vitro (1) was systematically optimized by using different concentrations of dUMP, CH2H4folate, MgCl2, β-NADH, β-NADPH, and FAD. [5-3H]dUMP [Moravek Biochemicals, Brea, CA (specific activity, 16.6 Ci/mmol)] was used in all experiments. CH2H4folate was either obtained directly from Merck Eprova (Schaffhausen, Switzerland), or it was formed chemically by incubating 2 mM H4folate (Sigma) in 50 mM Hepes/10% glycerol/96 mM 2-mercaptoethanol/42 mM formaldehyde at room temperature. The highest deprotonation activity was observed under single-turnover conditions. The reactions were started by adding enzyme (120 μg/ml), and incubations were stopped after 15 min of incubation time at 37°C. (Under these conditions, the observed activity was linearly proportional to incubation time and protein quantity.) One milliliter of activated charcoal [10% (wt/vol), Norit A] in 2% trichloroacetic acid was used to remove radioactive nucleotides from the reaction mixtures. Kinetic parameters were determined by using nonlinear regression as described in the graphpad software package. Radioactivity remaining in the supernatant was measured by using Ecolune (ICN) scintillation liquid as described (1).
Matrix-Assisted Laser Desorption-Ionization-Time-of-Flight (MALDI-TOF) MS. All spectra were acquired in positive-ion mode on a Voyager DE-STR MALDI-TOF mass spectrometer (Applied Biosystems) equipped with a 337-nm nitrogen laser. Determination of the molecular masses of the wild-type and mutant proteins were performed in linear mode (accelerating voltage, 25 kV; grid voltage, 93%; guide wire, 0.3%; delay, 600 ns) with close external calibration. Protein solutions were diluted to 30 μM with 30% acetonitrile/0.3% trifluoroacetic acid. Of these solutions 1.5 μl was mixed with 1.5 μl of saturated solution of sinapinic acid in 30% acetonitrile/0.3% trifluoroacetic acid, and 1.5 μl of this premix were then deposited on the sample plate and allowed to dry at room temperature.
Before digestion, 2.3 nmol of wild-type and S84C proteins were reduced with 5 mM DTT and carbamidomethylated with 15 mM iodoacetamide in 50 mM NH4HCO3/6.2 M urea buffer. They were subsequently purified by RP-HPLC on a C4 column (300 Å, 2.1 mm i.d. × 150 mm; Vydac, Hesperia, CA) with a linear gradient of 0-70% acetonitrile in 0.1% trifluoroacetic acid for 30 min at 0.3 ml/min. Collected fractions were pooled and dried under vacuum in a SpeedVac concentrator. Purified samples were reconstituted in 100 mM NH4HCO3 (estimated concentration, 0.8 mg/ml) and digested overnight at 37°C with 0.027 mg/ml sequencing grade endoproteinase Glu-C (Roche Diagnostics). After acidification with 10% trifluoroacetic acid, 1.5 μl were mixed with 1.5 μl of a saturated solution of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile/0.3% trifluoroacetic acid. Of this premix 1.5 μl were deposited on the sample plate. Peptide mass fingerprints were recorded in reflector mode (accelerating voltage, 20 kV; grid voltage, 73%; guide wire, 0.002%; delay, 200 ns) with close external calibration covering the range 750-4,000 Da.
Results
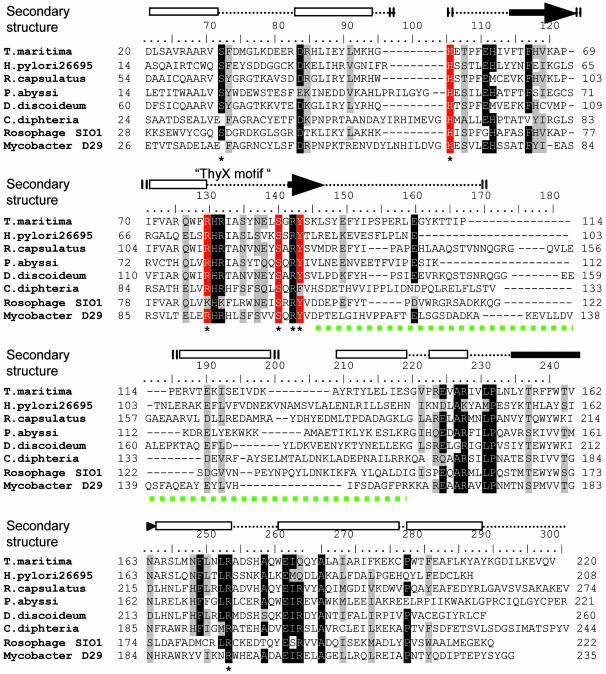
In Silico Identification of Putative Catalytic Residues of H. pylori ThyX. We identified a sequence motif RHRX7S (ThyX motif) common to the ThyX family in the conserved amino-terminal domain of these proteins (1). Moreover, ThyX orthologs contain highly conserved residues in their carboxyl-terminal domains, whereas their central regions show little sequence similarity (Fig. 1). Because our earlier studies indicated an essential role of FAD in mediating hydride transfer during ThyX catalysis (1), based on our sequence alignments (Fig. 1) and the 3D structure of the Thermotoga maritima ThyX/Thy1 protein (4), we consequently postulated that conserved amino acid residues located within 7 Å of the redox-active N5 atom of the isoalloxazine ring system of the FAD molecule (these residues are marked with an asterisk in Fig. 1; see also Fig. 5C) should have a crucial role in ThyX catalysis.
Fig. 1.
Sequence alignment of a diverse set of ThyX homologs is shown. The alignment was obtained with the use of clustalx program using default settings (19) followed by manual modifications. Conserved residues within 7 Å of the redox-active N5 atom of an FAD molecule are indicated by the asterisk. The secondary structure elements (cylinders, α-helices; arrows, β-strands) of T. maritima ThyX proteins are indicated above the alignment (4). The residues investigated in this study are indicated by the red background. The green dotted line indicates a central variable region of ThyX proteins that participates in the stabilization of ThyX tertiary structure (see also Fig. 5B).
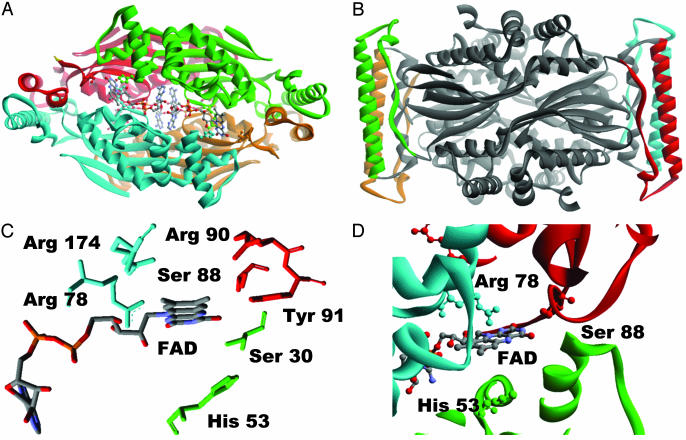
Fig. 5.
(A) ThyX tetramer (4) where the different subunits are shown with the different colors. FAD, S88, H53, and R78 (corresponding to active-site residues of H. pylori ThyX identified in this work) are represented with balls and sticks. (B) The central variable part of the ThyX protein indicated with the green dotted line in Fig. 1 is indicated by the different colors by monomer. (C) Only the conserved residues located within 7 Å of the redox-active N5 atom of the FAD molecule (only one of four FAD molecules of homotetramer is shown) are represented (see also Fig. 1). (D) The active-site configuration of T. maritima ThyX protein. A FAD molecule, Arg-78, Ser-88, and His-53 (T. maritima ThyX numbering) corresponding, respectively, to Ser-84, His-48, and Arg-74 in H. pylori protein are represented by balls and sticks. The different subunits of ThyX homotetramer are shown with the different colors. Note that the residues from the three different subunits participate in the formation of the single active site.
To investigate the functional role of these residues in ThyX catalysis, we have created several site-specific single and/or double mutations of H. pylori ThyX residues H48, R84, S87, and Y91 (these residues are highlighted in red in Fig. 1; see also Table 3). Here, we have limited our experimental efforts to this subset of conserved residues, in particular, because they approach an FAD molecule from three different subunits of the ThyX homotetramer, suggesting unexpected complexity in the active-site configuration of ThyX proteins. Although earlier structural data have not been able to propose a catalytic role for the conserved residues H48, S87, and Y91, the structure for the cocrystal of dUMP (or its analogs) and the T. maritima ThyX homotetramer have indicated a direct interaction of R84 with a hydroxyl group of dUMP ribose, suggesting a functional role of the positively charged NH2 group of this arginine residue in dUMP binding.
Identification of Essential Amino Acid Residues for ThyX Function in Vivo. By using an arabinose-inducible promoter system (11), wild-type and mutant H. pylori ThyX proteins were expressed in E. coli strain χ2913(ΔthyA; see ref. 1), and the ability of H. pylori thyX mutant alleles to functionally replace the E. coli thyA gene was scored on defined growth medium in the presence and absence of arabinose after 3-4 days. Our results indicate that the substitutions of the residues H48, R74, and S84, with the exception S84Y, abolished the arabinose-dependent complementation activity observed for the wild-type allele, thus confirming their indispensable roles for ThyX function. The substitutions S84A [eliminating a potential nucleophilic hydroxyl group (ref. 1 and Table 1)] and S84C [changing a hydroxyl group to a thiol group, used by ThyA proteins as essential nucleophile (Fig. 2)] both abolished thymidylate synthase activity in vivo, whereas the mutant S84Y carrying a hydroxyl group still was able to confer thymidine-independent growth to a ΔthyA E. coli strain (Table 1). These results clearly indicate that a hydroxyl group at position 84 is required for complementing activity of H. pylori ThyX protein. Despite the high level of conservation of Y87, eliminating its hydroxyl group by changing this tyrosine residue to phenylalanine had no effect on in vivo complementation. Consequently, the hydroxyl group of residue Y87 does not have a physiologically relevant catalytic role in the ThyX reaction.
Table 1. Genetic and biochemical characterization of H. pylori ThyX mutant proteins.
| Kinetic parameters* deprotonation activity
|
||||||
|---|---|---|---|---|---|---|
| Complementation activity
|
FAD binding, %
|
kcat/Km
|
||||
| H. pylori thyX allele |
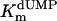 , μM , μM |
kcat, min−1 | min−1·μM−1 | %† | ||
| Wild-type | +++ | 100 | 7.2 ± 2.52 | 0.46 ± 0.02 | 0.064 | 100 |
| H48Q | − | 77.9 | ND | ND | ND | ND |
| H48Q/S84A | − | 10.3 | ND | ND | ND | ND |
| H48Q/S84C | − | 5.9 | ND | ND | ND | ND |
| R74A | − | 12.2 | 37.0 | 0.10 | 0.003 | 4.7 |
| R74K | − | 7.7 | 7.8 | 0.07 | 0.009 | 14.1 |
| S84A | − | 56.9 | 38.4 | 0.22 | 0.006 | 9.4 |
| S85A | +++ | 107.8 | NM | NM | NM | NM |
| S84A/S85A | − | 22.8 | ND | ND | ND | ND |
| S84C | − | 53.7 | 14.0 (n = 1) | 0.05 | 0.004 | 6.3 |
| S84Y | +++ | 113.1 | 9.7 (n = 1) | 0.72 | 0.074 | 115.6 |
| Y87F | +++ | 158.3 | 3.8 (n =1) | 0.48 | 0.126 | 196.9 |
Standard deviations shown for Km and kcat values obtained for wild-type protein were calculated by using five different data sets. Unless otherwise indicated, all other values are the averages of two independent measures. ND, nondetectable activity; NM, not measured.
100% refers to kcat/Km value of 0.064 min−1·μM−1 measured for wild-type protein.
Fig. 2.
The ability of wild-type or mutated H. pylori thyX genes to permit thymidine-independent growth of E. coli thymidine auxotroph χ2913 (ΔthyA) was scored after 3 days on M9 minimal agar lacking thymidine in the presence and absence of 0.2% l-arabinose.
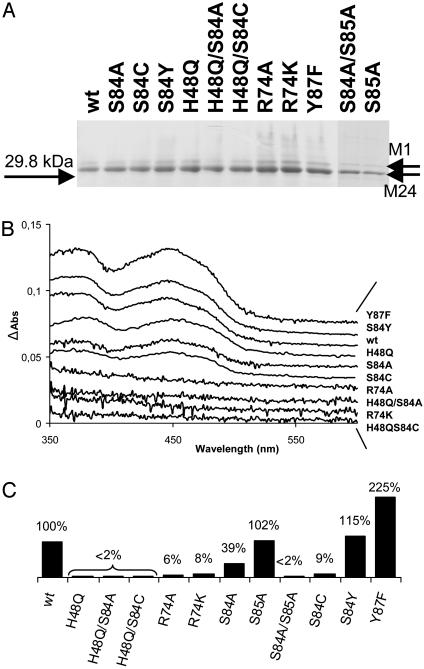
Purification and Spectroscopic Characterization of H. pylori ThyX Mutant Proteins. To address their molecular defect in detail, we purified the H. pylori mutant proteins by their carboxyl-terminal oligohistidine epitope (see Materials and Methods). All isolated H. pylori ThyX proteins were found to be stable and were expressed in amounts similar to wild-type protein (Fig. 3A). Amino-terminal protein sequencing and analyses by MS (MALDI-TOF) of pure protein samples indicated that H. pylori ThyX protein is preferentially expressed from the methionine residue at position 24 of the originally annotated ORF HP1533 (12), explaining why pure protein samples run as a doublet on SDS/polyacrylamide gels (Fig. 3A). Consequently, the residue numbering for H. pylori ThyX protein used throughout this work is based on the revised protein sequence of this protein (accession no. GI:14916856).
Fig. 3.
(A) SDS/10% PAGE of H. pylori wild-type (wt) and mutated ThyX proteins by using ≈1 μg of purified protein stained with Coomassie Brilliant Blue. M1 and M24 refer to the alternative start codons of H. pylori HP1533. (B) Spectroscopic analysis of H. pylori ThyX wild-type and mutant proteins (1). (C) “Deprotonation” activity of H. pylori ThyX proteins with 6.25 μM dUMP at 37°C as described in Materials and Methods. Complete reaction mixtures contained 50 mM Hepes (pH 7.0), 10% glycerol, 0.9 mM CH2H4folate, 10 mM MgCl2, 2 mM NADPH, 1 mM NADH, 0.5 mM FAD, and 6.25 μM [5-3H]dUMP (specific activity, 2.55 Ci/mmol). The specific activity for the wild-type protein corresponding to 0.95 nmol of 3H2O formed per min per mg of protein was arbitrarily chosen to correspond to 100%.
To investigate whether the loss of functional complementation resulted from a defect in FAD binding, the relative amounts of tightly bound FAD in pure protein samples were quantified by using visible absorption spectroscopy (Fig. 3B and Table 1). The spectra obtained revealed two distinct groups of mutant proteins that (i) either easily lose considerable amounts of bound FAD (R74A, R74K, H48Q/S84A, and H48Q/S84C) or (ii) contain quantities of tightly bound FAD similar to wild-type protein (Y87F, S84Y, H48Q, S84A, and S84C). Note that the effects on FAD binding for the double mutants H48Q/S84A and H48Q/S84C are additive (see Table 1).
H48 and S84 of H. pylori ThyX Are Essential for Catalysis. To address the effect of the described amino acid substitutions on ThyX activity, we measured ThyX activity in vitro through the loss of proton from [5-3H]dUMP for wild-type and mutant ThyX proteins in the presence of an excess of FAD. This activity is directly linked to the formation of dTMP (1). Deprotonation activity of H. pylori ThyX protein was undetectable for mutant proteins carrying the H48Q substitution in 15 min of incubation time under limiting dUMP concentrations, whereas the R74A and R74K mutations showed 5-10% of the wild-type activity (Fig. 3C; note that values for specific activities are shown). These results indicate that the imidazole side chain of H48 is essential for ThyX catalysis, whereas R74 is not absolutely required for ThyX activity in vitro. Unexpectedly, despite the S84A mutation being clearly nonfunctional in our complementation tests, the corresponding purified protein still has 40% of the wild-type activity. However, the primary sequence of H. pylori ThyX contains an additional nonconserved serine residue at position 85 (Fig. 1), raising the possibility that the hydroxyl group of S85 could function as nonphysiologically relevant catalytic residue in the absence of S84. To test this possibility, two additional mutant alleles (carrying substitutions S85A and S84A/S85A) were constructed. Although the S85A replacement had no effect on H. pylori ThyX activity either in vivo or in vitro, the double mutant S84A/S85A was completely inactive in both assays, indicating that S85 can partially replace S84 as active-site residue in vitro. Finally, the complementing mutant protein carrying the S84Y substitution shows an in vitro activity similar to wild type, whereas, at least under limiting dUMP concentrations, the mutant Y87F demonstrates increased specific activity.
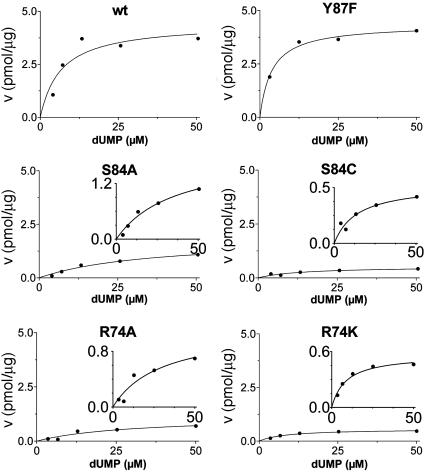
Additional activity measurements were performed under single-turnover conditions with a range of dUMP concentrations (for maximum specific activities, see Fig. 4; kcat and kcat/Km values for mutants are indicated in Table 1). dUMP concentrations only up to 100 μM were used in these experiments, because we have observed that higher, nonphysiological concentrations of dUMP resulted in substrate inhibition of the activity (data not shown). For the wild-type protein and the complementing mutants (S84Y and Y87F) similar kcat values (ranging from 0.46 to 0.72 min-1) were measured, whereas kcat and kcat/Km values, commonly used as measure for catalytic effectiveness of mutant proteins, of non-complementing mutants were at least one order of magnitude lower. 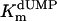 values for wild-type ThyX and the S84Y protein were 7.2 and 9.7 μM, respectively, whereas the Y87F mutant showed a slightly lower
values for wild-type ThyX and the S84Y protein were 7.2 and 9.7 μM, respectively, whereas the Y87F mutant showed a slightly lower 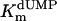 value of 3.8 μM (albeit, more experiments are needed to confirm the latter observation). Our measurements also indicate that the
value of 3.8 μM (albeit, more experiments are needed to confirm the latter observation). Our measurements also indicate that the 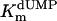 value for the R74A protein is significantly higher than observed for wild-type protein, indicating that the strongly basic guanino side group of R74 has a dual role in dUMP (Fig. 4) and FAD binding (Fig. 3B). Our measurements of an (unexpected) deprotonation activity of the S84A mutant protein demonstrate that the hydroxyl group of S84 participates, in addition to its catalytic role, in dUMP binding (Fig. 4 and Table 1).
value for the R74A protein is significantly higher than observed for wild-type protein, indicating that the strongly basic guanino side group of R74 has a dual role in dUMP (Fig. 4) and FAD binding (Fig. 3B). Our measurements of an (unexpected) deprotonation activity of the S84A mutant protein demonstrate that the hydroxyl group of S84 participates, in addition to its catalytic role, in dUMP binding (Fig. 4 and Table 1).
Fig. 4.
Maximum specific activities for ThyX (dUMP) wild-type (wt) and mutants were measured under single-turnover conditions. The reaction conditions are as described in Fig. 3C, except various dUMP concentrations were used.
In conclusion, our measurements indicate that H48, S84, and R74 play a crucial role in ThyX catalysis and suggest that the hydroxyl group of Y87 modulates ThyX activity, perhaps by a steric effect, which could regulate the access of various substrates to the active site.
Detection of a Covalent Reaction Intermediate Identifies S84 as Nucleophile. The canonical enzyme thymidylate synthase ThyA forms a stable covalent complex in the presence of fluorode-oxyuridine 5′-monophosphate (FdUMP) and CH2H4folate (13). Its formation is brought about by a nucleophilic attack of the catalytic cysteine on position C6 of dUMP (for a recent review on the mechanism of ThyA, see ref. 8). ThyX proteins do not contain conserved cysteine residues and we have been unable to detect a covalent complex of FdUMP with H. pylori ThyX. However, although the molecular masses of the mutant proteins H48Q and S84A are in agreement with their theoretical values (Table 2), MALDI-TOF analyses of the S84C mutant protein purified from E. coli cell-free extracts by using Ni-NTA agarose and RP-HPLC, under denaturing conditions, have revealed that this mutant protein carries a modification of ≈322 Da. Separation of the purified peptide fragments of the S84C proteins, followed by their MALDI-TOF analysis, demonstrated that the molecular mass of the peptide N-LSRHRIASLSVKCSRYTLRE-C [the highlighted cysteine corresponds to the S84C mutation (Table 2)] was 2,695.320 ± 0.216 Da. Strikingly, this value is, within the experimental error, identical with the predicted molecular mass of 2,695.3 Da for the covalent adduct of dTMP and the peptide carrying the S84C substitution. Together with our observations indicating that S84 makes functionally essential interactions with dUMP, the simplest explanation for these results is that the universally conserved hydroxyl group of S84 functions acts as nucleophile during ThyX catalysis.
Table 2. Analyses by MALDI-TOF MS of H. pylori ThyX mutant proteins.
| Protein substitution | Predicted mass, Da | Observed mass, Da* |
|---|---|---|
| None (wild-type) | 27,169.9 | 27,168.2 |
| H48Q | 27,159.9 | 27,155.7 |
| S84A | 27,152.9 | 27,146.9 |
| S84C | 27,185.0 | 27,493.1 |
| Peptide (S84C) | 2,375.308† | 2,695.320 |
| LSRHRIASLSVKCSRYTLRE |
The precision of MALDI-TOF measurements was 0.05% for proteins and 0.008% for peptides.
The value shown is for a nonderivatized cysteine residue. Expected value for carbamidomethylated C84 would be 2,432.330.
Discussion
ThyX proteins represent a recently discovered family of flavoproteins, essential for DNA replication in at least 30% of the microbial genomes sequenced to date [the considerable biases in available genome sequences underestimate this number (14)]. During this work we have undertaken structure-function studies on the functional roles of conserved ThyX amino acid residues. Our results indicate that the strongly basic side chain of R74 participates in FAD and dUMP binding, in agreement with earlier structural data (15). We have also shown that the residues H48 and S84 of H. pylori ThyX, previously of unknown function, are essential residues for ThyX catalysis. Although the exact role of the imidazole group of H48, likely in folate activation and/or acid-base catalysis, remains to be determined in further experiments, our results have already indicated that S84 functions as a nucleophile. This latter conclusion is unexpected, because the very recent structure of T. maritima ThyX/Thy1 in complex with dUMP (15) has revealed that the Oγ of the equivalent serine residue (S88 in T. maritima) is located too far away to act in a nucleophilic attack at the C6 of the pyrimidine ring. The position of dUMP in the cocrystal is thus not in agreement with our biochemical measurements, possibly reflecting that conformational changes take place during ThyX catalysis or, alternatively, that the exact orientation of the dUMP molecule in the cocrystal results from a crystal-packing artifact. To definitely resolve this discrepancy, the structure of reduced and/or CH2H4folate-bound enzyme will be required, allowing direct attention to whether reorientation of dUMP in the active site of ThyX proteins is brought about by folate binding and/or reduction of FAD in the ThyX complex.
Strikingly, the functionally crucial residues H48, R74, and S84 (H. pylori numbering) identified in this work are contributed by three different subunits of the ThyX tetramer to form a single active site (Fig. 5D). This situation is analogous to what has been shown for tetrameric adenylsuccinate lyase (16) and raises the question of how these rare active sites at the interphase of three monomers have evolved. ThyX proteins could have evolved through the multimerization of “inactive” ThyX monomers or, alternatively, the Chlamydia and Thermoplasma ThyX proteins that consist of two fused “ThyX domains” on the same polypeptide could correspond to the ancestral enzyme. Because these duplicated ThyX genes likely represent independent fusion events (14), we favor the former possibility. Our functional studies have also clearly demonstrated that two independent mechanisms for thymidylate synthesis of modern-day DNA have been established during the evolution of DNA as genetic material. Strikingly, as obvious structural homologs for ThyA or ThyX monomers have not (yet) been detected, the evolutionary origins of thymidylate are still unknown. In addition, the three classes of ribonucleotide reductases, essential enzymes for converting RNA precursors to DNA precursors, are thought to have originated from a common ancestor (17, 18), whereas our studies on ThyX enzymes have indicated that a specific step of DNA precursor synthesis can be achieved by two distinct enzymatic mechanisms.
Our findings also have practical implications for inhibiting microbial growth in a large number of pathogenic Bacteria carrying the thyX gene. Our data suggest that the rational conception of ThyX inhibitors should focus on the identification of molecules that prevent the catalytic function of H48 and S84, possibly by directly binding to oxidized and/or reduced ThyX protein in the vicinity of these amino acid residues. Consequently, our future work will be orientated toward the identification of specific ThyX inhibitors, a task that will be aided by the drastically different active-site geometries and distinct enzymatic mechanisms of ThyX and the human ThyA protein.
Supplementary Material
Acknowledgments
We thank A. Sodolescu, W. Abou-Jaoude, and J. Bullet for preliminary characterization of some mutant proteins; S. Skouloubris for critical reading of the manuscript; and Dr. R. Moser (Merck Eprova AG) for folate samples. Our work on ThyX proteins is supported by research funds from the French Ministry of Research (to U.L. and H.M.) and the BIOAVENIR program of the Institut National de la Santé et de la Recherche Medical (to H.M.). H.M. also received financial support from Fondation Bettencourt Schueller.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ThyX, thymidylate synthase ThyX; dUMP, deoxyuridine 5′-monophosphate; dTMP, deoxythymidine 5′-monophosphate; CH2H4folate, methylenetetrahydrofolate; MALDI-TOF, matrix-assisted laser desorption-ionization-time-of-flight.
References
- 1.Myllykallio, H., Lipowski, G., Leduc, D., Filee, J., Forterre, P. & Liebl, U. (2002) Science 297, 105-107. [DOI] [PubMed] [Google Scholar]
- 2.Hardy, L. W, Finer-Moore, J. S., Montfort, W. R., Jones, M. O., Santi, D. V. & Stroud, R. M. (1987) Science 235, 448-455. [DOI] [PubMed] [Google Scholar]
- 3.Murzin, A. G. (2002) Science 297, 61-62. [DOI] [PubMed] [Google Scholar]
- 4.Kuhn, P., Lesley, S. A., Mathews, I. I., Canaves, J. M., Brinen, L. S., Dai, X., Deacon, A. M., Elsliger, M. A., Eshaghi, S., Floyd, R., et al. (2002) Proteins 49, 142-145. [DOI] [PubMed] [Google Scholar]
- 5.Carreras, C. W. & Santi, D. V. (1995) Annu. Rev. Biochem. 64, 721-762. [DOI] [PubMed] [Google Scholar]
- 6.Matthews, R. G. (1996) in Escherichia coli and Salmonella, ed. Neidhardt, F. C. (Am. Soc. Microbiol. Press, Washington, DC), pp. 600-611.
- 7.Myllykallio, H., Leduc, D., Filee, J. & Liebl, U. (2003) Trends Microbiol. 11, 220-223. [DOI] [PubMed] [Google Scholar]
- 8.Finer-Moore, J. S., Santi, D. V. & Stroud, R. M. (2003) Biochemistry 42, 248-256. [DOI] [PubMed] [Google Scholar]
- 9.Galperin, M. Y. & Koonin, E. V. (2000) Nat. Biotechnol. 18, 609-613. [DOI] [PubMed] [Google Scholar]
- 10.Giladi, M., Bitan-Banin, G., Mevarech, M. & Ortenberg, R. (2002) FEMS Microbiol. Lett. 216, 105-109. [DOI] [PubMed] [Google Scholar]
- 11.Guzman, L. M., Belin, D., Carson, M. J. & Beckwith, J. (1995) J. Bacteriol. 177, 4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomb, J.-F., White, O., Kerlavage, A. R., Clayton, R. A., Sutton, G. G., Fleischmann, R. D., Ketchum, K. A., Klenk, H. P., Gill, S., Dougherty, B. A., et al. (1997) Nature 388, 539-547. [DOI] [PubMed] [Google Scholar]
- 13.Santi, D. V., McHenry, C. S. & Perriard, E. R. (1974) Biochemistry 13, 467-470. [DOI] [PubMed] [Google Scholar]
- 14.Leduc, D., Graziani, S., Meslet-Cladiere, L., Sodolescu, A., Liebl, U. & Myllykallio, H. (2004) Biochem. Soc. Trans. 32, 231-235. [DOI] [PubMed] [Google Scholar]
- 15.Mathews, I. I., Deacon, A. M., Canaves, J. M., McMullan, D., Lesley, S. A., Agarwalla, S. & Kuhn, P. (2003) Structure (London) 11, 677-690. [DOI] [PubMed] [Google Scholar]
- 16.Brosius, J. L. & Colman, R. F. (2002) Biochemistry 41, 2217-2226. [DOI] [PubMed] [Google Scholar]
- 17.Stubbe, J., Ge, J. & Yee, C. S. (2001) Trends Biochem. 26, 93-99. [DOI] [PubMed] [Google Scholar]
- 18.Torrents, E., Aloy, P., Gibert, I. & Rodriquez-Trelles, F. (2002) J. Mol. Evol. 55, 138-152. [DOI] [PubMed] [Google Scholar]
- 19.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 25, 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.