Abstract
A lipoproteoplex comprised of an engineered supercharged coiled-coil protein (CSP) bearing multiple arginines and the cationic lipid formulation FuGENE HD (FG) was developed for effective condensation and delivery of nucleic acids. The CSP was able to maintain helical structure and self-assembly properties while exhibiting binding to plasmid DNA. The ternary CSP•DNA(8:1)•FG lipoproteoplex complex demonstrated enhanced transfection of β-galactosidase DNA into MC3T3-E1 mouse preosteoblasts. The lipoproteoplexes showed significant increases in transfection efficiency when compared to conventional FG and an mTat•FG lipopolyplex with a 6- and 2.5-fold increase in transfection, respectively. The CSP•DNA(8:1)•FG lipoproteoplex assembled into spherical particles with a net positive surface charge, enabling efficient gene delivery. These results support the application of lipoproteoplexes with protein engineered CSP for non-viral gene delivery.
Keywords: coiled-coil protein, cationic lipid, lipoproteoplexes, gene delivery, supercharge
1. Introduction
A central challenge for gene therapy is the effective delivery of highly labile nucleic acids that are susceptible to nucleases [1]. While there are examples of successful nucleic acid delivery in vitro and in vivo by viral and non-viral vectors, achieving high transfection efficiency while maintaining low toxicity remains a significant challenge [2, 3]. Although virus-mediated vehicles are very efficient in gene transduction [4–6], they exhibit severe immunogenic properties and can cause detrimental mutagenic responses rendering them problematic [7, 8].
Considerable effort has been made to develop non-viral vectors such as cationic lipids [9, 10], cationic polymers [3, 11], and cell-penetrating peptides (CPPs) [12, 13]. Cationic lipids form non-covalent complexes with nucleic acids to generate lipoplexes. However, as the condensation ability of lipids alone is not effective, the resulting lipoplexes do not protect genes against nucleases in vivo [14, 15]. Cationic polymers such as polyethylenimine (PEI) [16–18], poly (L-lysine) (PLL) [19, 20], polyamidoamine (PAMAM) dendrimers [21] and polymethacrylates [22] form particulate complexes with DNA producing polyplexes that can deliver genes [16, 23]. Although such polyplexes demonstrate higher transfection ability, they exhibit high cytotoxicity. Moreover, further chemical modifications to the cationic polymers are required to reduce their cytotoxicity. The resultant chemically modified polymers demonstrate decreased transfection ability [24, 25]. While CPPs have been explored for their ability to deliver nucleic acids, delivery remains a major challenge [13] due to entrapment into endocytic vesicle and lysosomal degradation [26, 27]. Recently, lipopolyplexes composed of a cationic lipid and cationic peptide-based ternary complex have been introduced to enhance transfection of nucleic acids [28–34]. While lipopolyplexes have been successfully employed for gene delivery, it depends on the development of branched systems carrying a net positive charge [35]; in such cases, identifying optimal branching, charge and sequence will require various synthetic design strategies.
Protein engineered systems have emerged as an alternative to synthetic counterparts due to the unique advantages of programmed specificity in terms of structure and assembly, environmentally friendly production, non-toxic contaminants and biodegradability [36, 37]. In this study, we have engineered supercharged coiled-coil protein (CSP), derived from cartilage oligomeric matrix protein coiled-coil (COMPcc). The solvent exposed residues are mutated into arginine for effective binding to plasmid DNA and cationic lipids are introduced in conjunction with CSP to produce what we term “lipoproteoplexes” for enhanced gene delivery (Fig. 1). The CSP is expressed, purified and assessed for its secondary structure and binding ability to DNA. The optimal ratio of FG to CSP isdeveloped for in vitro delivery of β-galactosidase gene into MC3T3-E1 mouse preosteoblasts. The CSP and lipoproteoplex are evaluated for cytotoxicity against MC3T3-E1 cells. Also, CSP•DNA complex and lipoproteoplexes are further characterized for their size, surface charge and morphology.
Fig. 1.
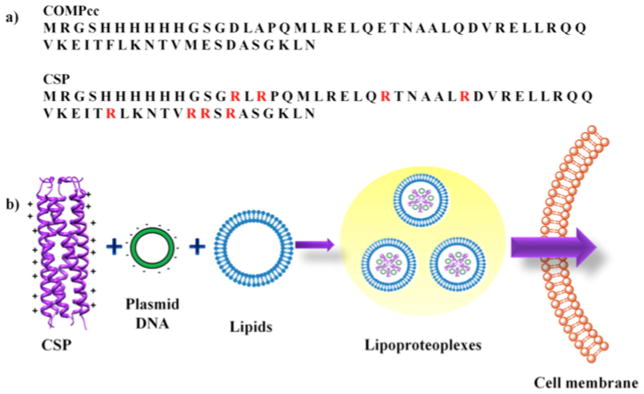
a) Aligned sequences of COMPcc and CSP with mutated arginine residue positions shown in red. b) Schematic of CSP complexation with plasmid DNA and a ternary complex with cationic lipids to form lipoproteoplexes for gene delivery.
2. Materials and method
2.1. Materials
Primers were purchased from Eurofin MWG Operon (Huntsville, AL), pfu Ultra DNA polymerase from Stratagene (Santa Clara, CA) and DpnI restriction enzyme from New England Biolabs (Ipswich, MA). Tris base, isopropyl β-D-1-thiogalactopyranoside (IPTG), tryptone, ampicillin, sodium chloride, imidazole and urea were obtained from VWR. Ni-NTA beads were purchased from Sigma-Aldrich, β-galactosidase plasmid DNA from Genlantics (San Diego, CA) and Beta-Glo assay kit from Promega, (Madison, WI). Gibco alpha minimal essential medium (αMEM), Gibco fetal bovine serum (FBS), 5000 U/mL penicillin and 5000 ug/mL streptomycin were purchased from Invitrogen (Carlsbad, CA). FG was obtained from Roche (Branchburg, NJ) and the HIV-1-Tat (RKKRRQRRRR) modified (mTat) with ten histidine residues and two cysteine residues (C-5H-Tat-5H-C) was purchased from Biomatik Corporation (Cambridge, Canada) [38]. The MC3T3-E1, subclone 14 (CRL-2594) mouse preosteoblasts was gift from Dr. Mani Alikhani (New York University College of Dentistry).
2.2. Site-directed mutagenesis (SDM) and PCR assembly
The COMPcc gene in pQE9 vector was used as a template to perform multiple mutations. The residues at D28, A30, E39, Q45, F60, M66, E67 and D69 were mutated to arginine by using following primers and their complementary sequences. D28R and A30R/5′-CAT CAC GGA TCC GGT CGT CTG CGT CCG CAG ATG-3′; E39R/5′-GAA CTG CAG CGT ACC AAC-3′; Q45R/5′-GCG CTG CGT GAC GTT CG-3′; F60R/5′-GAA ATC ACC CGT CTG AAA-3′; M66R, E67R and D69R/5′-C ACC GTT CGT CGT TCT CGT GCG TCT GGT AAG CTT AAT TAG-3′
The DNA fragments with required mutations were synthesized by PCR by using forward primer of one mutant and reverse primer of the following mutant [39, 40]. The resulting gene bearing all 24 base pairs mutations was used as a megaprimer for mutagenesis of pQE9/COMPcc [41, 42] to produce pQE9/CSP. Site directed mutagenesis SDM was performed using a standard protocol and the resulting sample was digested with DpnI enzyme (New England Biolabs) for 3 hours at 37 °C. The DpnI digested sample was transformed into XL-1 blue cells.
2.3. Protein expression and purification
To express the CSP and COMPcc proteins, the E. coli strains AF-IQ [43, 44] and XL-1 blue were used for transformation of CSP and COMPcc, respectively. Starter culture of CSP and COMPcc were made in 5 mL of LB containing ampicillin (200 μg/mL) and chloramphenicol (35 μg/mL) and LB containing ampicillin (200 μg/mL), respectively and incubated overnight at 37 °C and 350 rpm. The starter cultures were used to innoculate 800 mL of LB media with the aforementioned respective antibiotics and incubated for 6 hours at 37 °C and 250 rpm for large scale expression. After 6 hours, OD600 was measured (≈0.8 to 1.0) and the protein expression was induced by the addition of 200 μg/mL IPTG and incubated under the same conditions for 3 hours. Cells were harvested after overexpression by centrifugation and stored at −80 °C until purification. Overexpression was confirmed by SDS-PAGE (Fig. S1a). The cells pellets were thawed and resuspended into 50 mM Tris-HCl buffer pH 8.0 with 0.5 M NaCl, 20 mM imidazole and 6 M urea and lysed via sonication. Whole cell lysates were clarified by centrifugation and purified under native condition using Ni-NTA beads. Purification was performed in a 10 mL gravity column (Thermo Scientific). The beads were washed with the buffer composed of 50 mM Tris-HCl, 0.5 M NaCl and 20 mM imidazole and the protein was eluted with increase concentration of imidazole from 200 mM, 500 mM and 1 M imidazole. The purity of the proteins was confirmed by SDS-PAGE (Fig. S1b and Fig. S1c). The proteins were dialyzed against 50 mM Tris-HCl buffer pH 8.0 with 0.5 M NaCl to remove the imidazole.
2.4. CD spectroscopy
The secondary structure of CSP/COMPcc was analyzed using a Jasco J-815 spectrometer at 10 μM protein concentration in 50 mM Tris-HCl buffer pH 8.0. The wavelength scans were performed at 4 °C over a range of 200–250 nm with a 1 nm step size. Temperature scans were performed at 222 nm from 20 °C to 85 °C with temperature ramp of 1 °C/min. The observed ellipticity value (Θ) was converted into mean residue ellipticity (MRE) using the standard equation ΘMRE = Θ/(10cpl) where c is the molar concentration of the protein, p is the path length in centimeters and l is the number of amino acids [17]. The fraction folded was derived using equation F = (ΘA − ΘU)/(ΘN − ΘU), where, ΘA is the MRE observed at given temperature, ΘU is MRE value for completely unfolded protein and ΘN is the MRE value of completely folded protein that is considered at 25 °C. The first derivative of fraction folded was used to calculate melting temperature (Tm) of protein [44]. All data were represented as an average of three trials.
2.5. Electrophoretic mobility shift assay and lipoproteoplex preparation
Plasmid DNA encoding β-galactosidase (gWiz β-galactosidase) under the control of the cytomegalovirus promoter/enhancer was used to investigate protein binding and also acted as a reporter for successful transfection. The β-galactosidase plasmid DNA (5.1 kb) at a concentration of 50 ng was mixed with different concentrations of CSP and COMPcc and incubated at room temperature for 30 minutes. The mixtures were run on 1% agarose gel (stained with ethidium bromide) and imaged under UV light using ImageQuant (GE healthcare). The lipoproteoplex was prepared by mixing the FG at 4:1 w/w ratio of lipid•DNA with an already formed condensed mixture of plasmid DNA and CSP/COMPcc and the ternary complex was incubated for 15 minutes at room temperature.
2.6. Transfection studies
The transfection studies were performed using β-galactosidase plasmid DNA. The MC3T3-E1 were seeded in 96-well plate at a density of 1 × 104 cells/well in αMEM with 10% FBS, 5000 U/mL penicillin and 5000 ug/mL streptomycin for 24 hours prior to experiment. The proteoplex and lipoproteoplexes were prepared as explained above at different CSP•DNA w/w ratios of 5:1 and 8:1. The mixtures were added to different wells and incubated for 24 hours at 37 °C at 5% (v/v) CO2. As a negative and positive control, plasmid DNA alone and FG•DNA(4:1) were also prepared, respectively. Based on previous studies using mTat•DNA(10:1)•FG lipopolyplexes, the optimized component mixture was also used as a positive control [38]. The expression of β-galactosidase was confirmed using a standard Beta-Glo assay kit provided by manufacturer. The luminescent signal obtained after reaction of β-galactosidase with Beta-Glo reagent was observed using the microplate reader Synergy™ HT (BioTek Instruments, Winooski, VT). Data were expressed as mean β-galactosidase activity (relative light units, RLU) per well ± standard deviation from quadruplicates. This detection system is designed to measure directly the expression level using 96-well plates [45].
2.7. Cell viability studies
The cells were seeded in 96-well plate at a density of 1 × 106 cells/well in αMEM with 10% FBS and incubated at 37°C with 5% CO2 overnight. The cytotoxicity of different complexes was evaluated by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrasodium bromide, MTT assay. Approximately 10 μL of MTT reagent from 5 mg/mL stock was added to each well and incubated for 4 hours at 37 °C with 5% CO2. After incubation, the entire medium was aspirated and 100 μL of dimethyl sulfoxide was added to each well. The change in absorbance of colored solution was observed using a microplate reader Synergy HT at 570 nm. The percent cell viability was calculated by normalizing the observed absorbance value to that of the control cells without any treatment. The data was represented as the average of quadruplicates ± standard deviation.
2.8. Zeta potential
Zeta potential of protein, plasmid DNA and complex was determined using Zetasizer Nano ZS90 (Malvern Instruments, UK) with a laser source of 630 nm. The instrument calculates the zeta potential based on the measured electrophoretic mobility and this is fit into a Smoluchowski equation [46]. The plasmid DNA alone was prepared at 90 ng/μL and for other mixtures the DNA was kept at a final concentration of 2.5 ng/μL with 8 times higher concentrations of CSP/COMPcc and 4 times the concentration of FG. For protein and FG alone, the concentrations were at 59.7 ng/μL and 18.7 ng/μL, respectively. All samples were prepared in 0.22 μm filtered dH2O and the ionic strength was kept constant for all solutions with approximate concentration of buffer as 2.7 mM with 27.6 mM of NaCl. Three trials were performed and within each each sample, three measurements were taken where each measurement consisted of 20 runs. The error bars represented standard deviation.
2.9. Transmission Electron Microscopy studies
The morphology of the protein•DNA complex and lipoproteoplex were studied using transmission electron microscopy (TEM). The complexes were formed as explained above and 3 μL of solution was applied on formvar carbon coated 400 mesh copper grids and incubated for 1 minute. The excess solution was blotted out and washed with 2 × 3 μL of dH2O and the sample was stained using 3 μL of 1% uranyl acetate solution for 1 minute and excess solution was blotted off. TEM was performed using Phillips CM-100 transmission electron microscope. The sizes of the complexes were measured using ImageJ [47].
2.10. Statistical Analysis
Statistics were computed for each experimental method to summarize the mean expression levels and associated standard deviations. A Student’s t-test was used to compare different experimental conditions. In each statistical analysis, a p value less than 0.05 was considered significant [48].
3. Results
3.1. Expression and purification of protein
The solvent exposed residues at b and c position of the heptad repeat of abcdefg of COMPcc were mutated into arginine by PCR assembly to generate the supercharged protein CSP (Fig 1). Notably, CSP showed higher expression in E. coli relative to the parent COMPcc, consistent with previous supercharged proteins [49] (Fig. S1a). While a specific purification protocol was developed to stabilize the highly charged (theoretical charge of +53) CSP, sufficient quantities of protein were produced (Fig. S1b, c).
3.2. Secondary structure studies
Circular dichroism (CD) spectroscopy was conducted on CSP and its parent COMPcc to assess the effects of arginine mutations on the secondary structure (Fig. 2a). Wavelength scans demonstrated that both CSP and COMPcc were indeed alpha-helical. Surprisingly, CSP exhibited enhanced helical structure when compared to the parent COMPcc (Fig. 2a). The supercharging did not negatively affect the structure; however, CSP exhibited a 10 °C decrease in the melting temperature (Tm) in comparison to the parent COMPcc (Fig. S2).
Fig. 2.
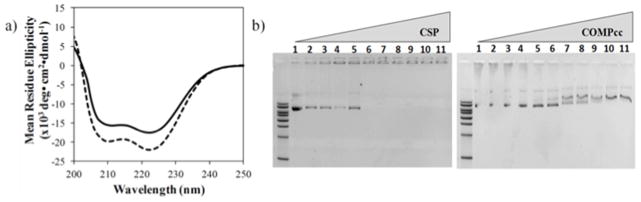
In vitro studies on CSP and COMPcc. a) CD wavelengths scan of CSP (dashed line) and COMPcc (solid line) at 4 °C at 10 μM concentration. Scans represent an average of three trials. b) Plasmid DNA binding to protein with increasing w/w ratio evaluated through mobility shift assay on 1% agarose gel of CSP binding to DNA (left) and COMPcc binding to DNA (right). In both gels L- 1kb DNA ladder, 1- plasmid DNA alone. Proteoplexes prepared at different protein to DNA w/w ratio: 2 – 0.5:1, 3 – 1:1, 4 – 2:1, 5 – 3:1, 6 – 5:1, 7 – 8:1, 8 – 10:1, 9 – 13:1, 10 – 16:1, 11 – 18:1.
3.3. Optimization of plasmid DNA and protein binding ratio
The ability of CSP to complex plasmid DNA was evaluated by electrophoretic mobility shift assay on agarose gel (Fig. 2b). A fixed concentration of β-galactosidase plasmid DNA was mixed with varying amount of CSP or COMPcc at certain w/w ratios. The CSP, bearing a theoretical charge of +53 as a pentamer, exhibited efficient binding to plasmid DNA through non-covalent interactions with a complete shift at a protein to DNA w/w ratio of 5:1 (Fig. 2b). By contrast, COMPcc (theoretical charge −6) demonstrated a negligible DNA shift even at higher ratios of 18:1 (Fig. 2b), suggesting that the engineered positive charge indeed enabled DNA complexation.
3.4. Lipoproteoplex leads to enhanced transfection
To determine whether the lipoproteoplexes could indeed deliver DNA, the transfection of the β-galactosidase gene was assessed in presence and absence of the proteins plus cationic lipid, FG. As a control, DNA alone was treated with cells, yielding little evidence of transfection (Fig. 3a). FG in presence of DNA revealed a modest 3-fold increase in transfection relative to naked DNA. COMPcc•DNA(8:1)•FG demonstrated nearly similar transfection to FG•DNA(4:1) (Fig. 3a). Compared to the FG positive control, the ternary lipoproteoplex complex CSP•DNA(8:1)•FG outperformed it, with a 6-fold increase in transfection (Fig. 3a). The efficacy of the CSP lipoproteoplex was compared to a previously studied mTat•DNA(10:1)•FG mixture, yielding a 2.5 fold improved transfection [38]. Notably, superior transfection efficiency was observed with CSP•DNA(8:1)•FG lipoproteoplex (Fig. 3b).
Fig. 3.
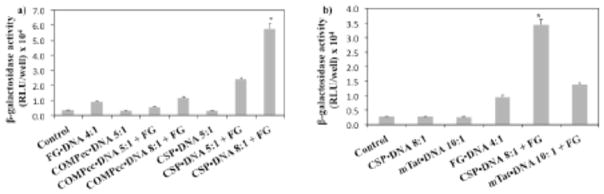
In vitro transfection efficiency for β-galactosidase DNA. FG•DNA(4:1), COMPcc•DNA (5:1), COMPcc•DNA(5 or 8:1)•FG, CSP•DNA(5:1), CSP•DNA(5 or 8:1)•FG, mTat•DNA(10:1) and mTat•DNA(10:1)•FG. a) * indicates p < 0.0001, comparison of control (DNA only), FG•DNA(4:1) etc, b)* indicates p < 0.0001 comparision of control (DNA only), CSP•DNA(8:1) etc. Data represents the mean β-galactosidase activity (relative light units, RLU)/well with a standard deviation obtained from quadruplicates.
3.5. Cytotoxicity of lipoproteoplexes
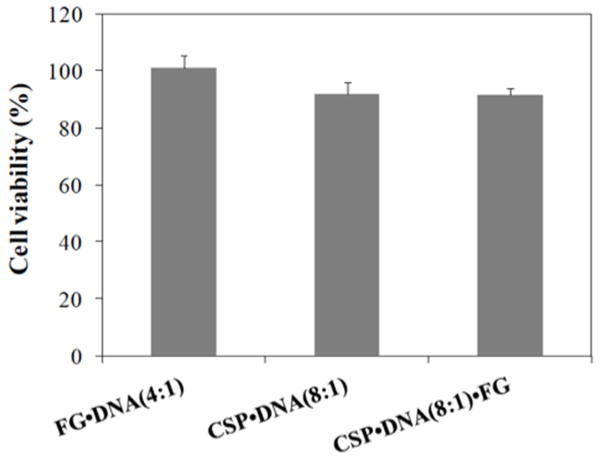
Cellular viability was measured by the MTT assay of cells treated with the plasmid DNA complexed with CSP and FG. The proteoplex of CSP•DNA(8:1)•FG illustrated 91.8 ± 4.3 % cell viability. The ternary lipoproteoplex also showed similar cell viability (91.6 ± 2.26%) to the CSP•DNA(8:1) treatment. All of the constructs were essentially non-toxic in which no significant difference was observed between CSP•DNA(8:1) and FG•DNA(4:1) or CSP•DNA(8:1)•FG and FG•DNA(4:1) (p < 0.05) (Fig. 4).
Fig. 4.
Cell viability evalauted by MTT assay after treatment of CSP and FG complexed with plasmid DNA and liproteoplex complex with MC3T3-E1 cell. Data is shown as the mean ± standard deviation obtained from quadruplicates and compared to the mean of control group. The control group consist of non-transfected cells (100% viability). No significant difference was observed between CSP•DNA(8:1) and FG•DNA(4:1) or CSP•DNA(8:1)•FG and FG•DNA(4:1) (p < 0.05).
3.6. Surface properties and morphology of complexes
To assess the surface characteristics of the proteins, proteoplexes and lipoproteoplexes, the surface charge and size of the individual components and complexes were determined via zeta potential measurements. As expected, plasmid DNA revealed a negative zeta potential of −49.70 ± 1.28 mV, while CSP alone was positive with a value of +25.51 ± 1.28 mV (Table 1, Fig. S3). The CSP•DNA(8:1) complex exhibited a slightly less positive zeta potential relative to CSP alone with a value of +24.00 ± 0.27 mV; with the addition of FG, the zeta potential did not change significantly, demonstrated by the value of +26.58 ± 0.10 mV (Table 1, Fig. S3). FG alone possessed a positive zeta potential of +24.83 ± 0.27 mV; after mixing it with negatively charge DNA, the zeta potential significantly decreased to +17.17 ± 0.68 mV (Table 1, Fig. S3).
Table 1.
Zeta potentials measurements
| Zeta Potential (mV)* | |
|---|---|
| DNA | − 49.70 ± 1.28 |
| CSP | + 25.51 ± 1.28 |
| FG | + 24.83 ± 0.27 |
| CSP•DNA | + 24.00 ± 0.36 |
| CSP•DNA•FG | + 26.58 ± 0.10 |
| DNA•FG | + 17.17 ± 0.68 |
The data represents the average of three individual trials with standard deviation.
TEM analysis was performed for the CSP•DNA(8:1), FG•DNA(4:1) and the ternary lipoproteoplex complex. At 8:1 w/w ratio, the CSP•DNA(8:1) showed aggregate structures with an average feret diameter of 270.4 ± 84.7 nm (Fig. 5a, Table S1). A drastic change in shape, size and overall morphology of the complex was observed in presence of FG; the FG•DNA(4:1) and CSP•DNA (8:1)•FG showed small spherical particles with a feret diameter of 354.8 ± 58.4 nm and 317 ± 47 nm, respectively (Fig. 5b, c, Table S1).
Fig. 5.
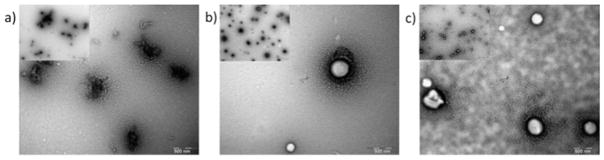
TEM images of different complexes a) CSP•DNA(8:1), b) FG•DNA(4:1), c) CSP•DNA(8:1)•FG. The scale bars in the image is 500 nm and the insets has 2 μm.
4. Discussion
The primary aim of this study was to develop lipoproteoplexes comprised of an engineered CSP for effective condensation of negatively charged DNA in conjunction with cationic lipid, FG, for enhanced gene delivery. While supercharging the protein did not negatively affect its structure, CSP exhibited a 10 °C decrease in the melting temperature (Tm) in comparison to the parent COMPcc (Fig. 2a, S2). This may be due to changes in the ionic environment of the CSP as the arginine residues presumably impact the solvation of the protein and electrostatic interactions [50]. The COMPcc, which possessed a more negatively charged surface, demonstrated poor binding ability to plasmid DNA, while CSP exhibited a pronounced positively charged surface that bound plasmid DNA at lower w/w ratio as confirmed by electrophoretic mobility shift assay (Fig. 2b). The in vitro transfection studies showed that the CSP with cationic lipid, FG, significantly enhanced the transfection relative to FG and CSP alone. More importantly, it outperformed another cell penetrating peptide mTat [38] as shown via a significant increase in β-galactosidase activity (Fig. 3).
Although we do not know the exact reason for the enhanced transfection of the CSP•DNA(8:1)•FG complex, it is believed that cell entry is governed by the lipid character of the lipoproteoplex since studies using poly-L-lysine [51] and protamine sulfate [52] and PEI [53–55] as a DNA condensing agent have shown increased transfection efficiency by 2 to 28 fold. The CSP acts as a DNA condensing agent and much like CPPs, it may be poorly interacting with cell membrane; however, in presence of FG, the ternary CSP•DNA(8:1)•FG lipoproteoplex is likely interacting more effectively with the cell membrane for greater transfection efficiency.
Although many cationic polymer and cationic lipid-based systems have been developed for enhanced transfection, cytotoxicity is always a major concern for in vitro and in vivo studies [56]. Our cell viability assay on MC3T3-E1 cells show that the CSP•DNA(8:1) with or without FG did not contribute any significant cytotoxic effect when compared to FG•DNA(4:1) (Fig. 4). Although cell viability is compromised to a minimal extent with the lipoproteoplex as compared to the FG•DNA(4:1), it demonstrates a significant increase in transfection.
The zeta potential studies demonstrate that CSP has better DNA condensation ability relative to FG alone as CSP•DNA(8:1) reveals a more positive zeta potential than the FG•DNA(4:1) complex (Table 1). Although the condensation of ability of CSP is higher than FG, the particles are polydisperse with irregular size as supported by literature [57]. By contrast, both FG•DNA(4:1) and CSP•DNA(8:1)•FG complex exhibit spherical particles (Fig. 5). For effective gene delivery, the particle size is critical [35] and the ternary lipoproteoplex reveals a more uniform particle size leading to enhanced transfection.
5. Conclusion
We demonstrate that the CSP lipoproteoplex increases the transfection ability relative to CSP or FG alone and it outperforms a well-studied complex of mTat•DNA(10:1)•FG [38]. Our findings suggest that a self-assembling coiled-coil protein can be engineered with positively charged residues to effectively complex with plasmid DNA. Surprisingly, the engineered coiled-coil protein also maintains its self-assembling properties into a stable homopentamer. Since, the efficient gene delivery through non-viral vectors is a major challenge, the highly positively charged CSP based lipoproteoplexes represents a new avenue for gene delivery using protein engineered system. Further studies to explore gene delivery in vivo as well as with simultaneous gene/drug delivery are underway.
Supplementary Material
Acknowledgments
This work was supported by ARO W911NF-10-1-0228 and W911NF-11-1-0449 (J.K.M.), AFOSR FA-9550-08-1-0266 (J.K.M.) and in part by the NSF MRSEC Program under Award Number DMR-0820341, NSF GK-12 fellow grant DGE-0741714 (J.A.F.) and NYU CTSA grant UL1TR000038 from the National Center for Advancing Translational Sciences (NCATS), NIH (J.K.M.).
Footnotes
Supplementary data can be found in the online version at doi: 10.1016/j.biomaterials.2014.02.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Elsabahy M, Nazarali A, Foldvari M. Non-viral nucleic acid delivery: key challenges and future directions. Curr Drug Deliv. 2011;8:235–44. doi: 10.2174/156720111795256174. [DOI] [PubMed] [Google Scholar]
- 2.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4:581–93. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 3.Tros de Ilarduya C, Sun Y, Duzgunes N. Gene delivery by lipoplexes and polyplexes. Eur J Pharm Sci. 2010;40:159–70. doi: 10.1016/j.ejps.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 4.Schaffer DV, Koerber JT, Lim KI. Molecular engineering of viral gene delivery vehicles. Annu Rev Biomed Eng. 2008;10:169–94. doi: 10.1146/annurev.bioeng.10.061807.160514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mah C, Byrne BJ, Flotte TR. Virus-based gene delivery systems. Clin Pharmacokinet. 2002;41:901–11. doi: 10.2165/00003088-200241120-00001. [DOI] [PubMed] [Google Scholar]
- 6.Kay MA, Glorioso JC, Naldini L. Viral vectors for gene therapy: the art of turning infectious agents into vehicles of therapeutics. Nat Med. 2001;7:33–40. doi: 10.1038/83324. [DOI] [PubMed] [Google Scholar]
- 7.Lowenstein PR, Mandel RJ, Xiong WD, Kroeger K, Castro MG. Immune responses to adenovirus and adeno-associated vectors used for gene therapy of brain diseases: the role of immunological synapses in understanding the cell biology of neuroimmune interactions. Curr Gene Ther. 2007;7:347–60. doi: 10.2174/156652307782151498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniel R, Smith JA. Integration site selection by retroviral vectors: molecular mechanism and clinical consequences. Hum Gene Ther. 2008;19:557–68. doi: 10.1089/hum.2007.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin B, Sainlos M, Aissaoui A, Oudrhiri N, Hauchecorne M, Vigneron JP. The design of cationic lipids for gene delivery. Curr Pharm Des. 2005;11:375–94. doi: 10.2174/1381612053382133. [DOI] [PubMed] [Google Scholar]
- 10.Bhattacharya S, Bajaj A. Advances in gene delivery through molecular design of cationic lipids. Chem Commun (Camb) 2009:4632–56. doi: 10.1039/b900666b. [DOI] [PubMed] [Google Scholar]
- 11.De Smedt SC, Demeester J, Hennink WE. Cationic polymer based gene delivery systems. Pharm Res. 2000;17:113–26. doi: 10.1023/a:1007548826495. [DOI] [PubMed] [Google Scholar]
- 12.Veldhoen S, Laufer SD, Restle T. Recent developments in peptide-based nucleic acid delivery. Int J Mol Sci. 2008;9:1276–320. doi: 10.3390/ijms9071276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fonseca SB, Pereira MP, Kelley SO. Recent advances in the use of cell-penetrating peptides for medical and biological applications. Adv Drug Deliv Rev. 2009;61:953–64. doi: 10.1016/j.addr.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 14.Xu Y, Szoka FC., Jr Mechanism of DNA release from cationic liposome/DNA complexes used in cell transfection. Biochemistry. 1996;35:5616–23. doi: 10.1021/bi9602019. [DOI] [PubMed] [Google Scholar]
- 15.Bally MB, Harvie P, Wong FM, Kong S, Wasan EK, Reimer DL. Biological barriers to cellular delivery of lipid-based DNA carriers. Adv Drug Deliv Rev. 1999;38:291–315. doi: 10.1016/s0169-409x(99)00034-4. [DOI] [PubMed] [Google Scholar]
- 16.Moghimi SM, Symonds P, Murray JC, Hunter AC, Debska G, Szewczyk A. A two-stage poly(ethylenimine)-mediated cytotoxicity: implications for gene transfer/therapy. Mol Ther. 2005;11:990–95. doi: 10.1016/j.ymthe.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 17.Godbey WT, Wu KK, Mikos AG. Poly(ethylenimine) and its role in gene delivery. J Control Release. 1999;60:149–60. doi: 10.1016/s0168-3659(99)00090-5. [DOI] [PubMed] [Google Scholar]
- 18.Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci USA. 1995;92:7297–301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishikawa M, Takemura S, Takakura Y, Hashida M. Targeted delivery of plasmid DNA to hepatocytes in vivo: optimization of the pharmacokinetics of plasmid DNA/galactosylated poly(L-lysine) complexes by controlling their physicochemical properties. J Pharmacol Exp Ther. 1998;287:408–15. [PubMed] [Google Scholar]
- 20.Wagner E, Ogris M, Zauner W. Polylysine-based transfection systems utilizing receptor-mediated delivery. Adv Drug Deliv Rev. 1998;30:97–113. doi: 10.1016/s0169-409x(97)00110-5. [DOI] [PubMed] [Google Scholar]
- 21.Leng Q, Mixson AJ. Modified branched peptides with a histidine-rich tail enhance in vitro gene transfection. Nucleic Acids Res. 2005;33:e40. doi: 10.1093/nar/gni040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dubruel P, Christiaens B, Rosseneu M, Vandekerckhove J, Grooten J, Goossens V. Buffering properties of cationic polymethacrylates are not the only key to successful gene delivery. Biomacromolecules. 2004;5:379–88. doi: 10.1021/bm034438d. [DOI] [PubMed] [Google Scholar]
- 23.Benns JM, Choi JS, Mahato RI, Park JS, Kim SW. pH-sensitive cationic polymer gene delivery vehicle: N-Ac-poly(L-histidine)-graft-poly(L-lysine) comb shaped polymer. Bioconjug Chem. 2000;11:637–45. doi: 10.1021/bc0000177. [DOI] [PubMed] [Google Scholar]
- 24.Maheshwari A, Han S, Mahato RI, Kim SW. Biodegradable polymer-based interleukin-12 gene delivery: role of induced cytokines, tumor infiltrating cells and nitric oxide in anti-tumor activity. Gene Ther. 2002;9:1075–84. doi: 10.1038/sj.gt.3301766. [DOI] [PubMed] [Google Scholar]
- 25.Maheshwari A, Mahato RI, McGregor J, Han S, Samlowski WE, Park JS. Soluble biodegradable polymer-based cytokine gene delivery for cancer treatment. Mol Ther. 2000;2:121–30. doi: 10.1006/mthe.2000.0105. [DOI] [PubMed] [Google Scholar]
- 26.Richard JP, Melikov K, Vives E, Ramos C, Verbeure B, Gait MJ. Cell-penetrating peptides. A reevaluation of the mechanism of cellular uptake. J Biol Chem. 2003;278:585–90. doi: 10.1074/jbc.M209548200. [DOI] [PubMed] [Google Scholar]
- 27.Al-Taei S, Penning NA, Simpson JC, Futaki S, Takeuchi T, Nakase I. Intracellular traffic and fate of protein transduction domains HIV-1 TAT peptide and octaarginine. Implications for their utilization as drug delivery vectors. Bioconjug Chem. 2006;17:90–100. doi: 10.1021/bc050274h. [DOI] [PubMed] [Google Scholar]
- 28.Zuhorn IS, Engberts JB, Hoekstra D. Gene delivery by cationic lipid vectors: overcoming cellular barriers. Eur Biophys J. 2007;36:349–62. doi: 10.1007/s00249-006-0092-4. [DOI] [PubMed] [Google Scholar]
- 29.Leng Q, Scaria P, Ioffe OB, Woodle M, Mixson AJ. A branched histidine/lysine peptide, H2K4b, in complex with plasmids encoding antitumor proteins inhibits tumor xenografts. J Gene Med. 2006;8:1407–15. doi: 10.1002/jgm.982. [DOI] [PubMed] [Google Scholar]
- 30.Chen QR, Zhang L, Stass SA, Mixson AJ. Branched co-polymers of histidine and lysine are efficient carriers of plasmids. Nucleic Acids Res. 2001;29:1334–40. doi: 10.1093/nar/29.6.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y, Sen J, Bathula SR, Yang Q, Fittipaldi R, Huang L. Novel cationic lipid that delivers siRNA and enhances therapeutic effect in lung cancer cells. Mol Pharm. 2009;6:696–705. doi: 10.1021/mp800136v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan Y, Whitmore M, Li S, Frederik P, Huang L. LPD nanoparticles--novel nonviral vector for efficient gene delivery. Methods Mol Med. 2002;69:73–81. doi: 10.1385/1-59259-141-8:073. [DOI] [PubMed] [Google Scholar]
- 33.Jenkins RG, Herrick SE, Meng QH, Kinnon C, Laurent GJ, McAnulty RJ. An integrin-targeted non-viral vector for pulmonary gene therapy. Gene Ther. 2000;7:393–400. doi: 10.1038/sj.gt.3301095. [DOI] [PubMed] [Google Scholar]
- 34.Hart SL, Arancibia-Carcamo CV, Wolfert MA, Mailhos C, O’Reilly NJ, Ali RR. Lipid-mediated enhancement of transfection by a nonviral integrin-targeting vector. Hum Gene Ther. 1998;9:575–85. doi: 10.1089/hum.1998.9.4-575. [DOI] [PubMed] [Google Scholar]
- 35.Welser K, Campbell F, Kudsiova L, Mohammadi A, Dawson N, Hart SL. Gene delivery using ternary lipopolyplexes incorporating branched cationic peptides: the role of Peptide sequence and branching. Mol Pharm. 2013;10:127–41. doi: 10.1021/mp300187t. [DOI] [PubMed] [Google Scholar]
- 36.More HT, Yang CY, Montclare JK. Posttranslational modification of proteins Incorporating nonnatural amino acids. In: Theato P, Klok H-A, editors. Functional polymers by post-polymerizarion modificiation: Concepts, guidelines, and applications. Germany: Wiley-VCH Verlag GmbH & Co. KGaA; 2013. pp. 291–331. [Google Scholar]
- 37.Voloschuk N, Montclare JK. Incoproation of unnatural amino acids for synthetic biology. Mol BioSys. 2010;6:65–80. doi: 10.1039/b909200p. [DOI] [PubMed] [Google Scholar]
- 38.Yamano S, Dai J, Yuvienco C, Khapli S, Moursi AM, Montclare JK. Modified Tat peptide with cationic lipids enhances gene transfection efficiency via temperature-dependent and caveolae-mediated endocytosis. J Control Release. 2011;152:278–85. doi: 10.1016/j.jconrel.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 39.Xiong AS, Yao QH, Peng RH, Duan H, Li X, Fan HQ. PCR-based accurate synthesis of long DNA sequences. Nat Protoc. 2006;1:791–7. doi: 10.1038/nprot.2006.103. [DOI] [PubMed] [Google Scholar]
- 40.Shevchuk NA, Bryksin AV, Nusinovich YA, Cabello FC, Sutherland M, Ladisch S. Construction of long DNA molecules using long PCR-based fusion of several fragments simultaneously. Nucleic Acids Res. 2004;32:e19. doi: 10.1093/nar/gnh014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Angelaccio S, Bonaccorsi di Patti MC. Site-directed mutagenesis by the megaprimer PCR method: variations on a theme for simultaneous introduction of multiple mutations. Anal Biochem. 2002;306:346–49. doi: 10.1006/abio.2002.5689. [DOI] [PubMed] [Google Scholar]
- 42.Ke SH, Madison EL. Rapid and efficient site-directed mutagenesis by single-tube ‘megaprimer’ PCR method. Nucleic Acids Res. 1997;25:3371–72. doi: 10.1093/nar/25.16.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuvienco C, More HT, Haghpanah JS, Tu RS, Montclare JK. Modulating supramolecular assemblies and mechanical properties of engineered protein materials by fluorinated amino acids. Biomacromolecules. 2012;13:2273–78. doi: 10.1021/bm3005116. [DOI] [PubMed] [Google Scholar]
- 44.Gunasekar SK, Asnani M, Limbad C, Haghpanah JS, Hom W, Barra H. N-terminal aliphatic residues dictate the structure, stability, assembly, and small molecule binding of the coiled-coil region of cartilage oligomeric matrix protein. Biochemistry. 2009;48:8559–67. doi: 10.1021/bi900534r. [DOI] [PubMed] [Google Scholar]
- 45.Eguchi A, Akuta T, Okuyama H, Senda T, Yokoi H, Inokuchi H. Protein transduction domain of HIV-1 Tat protein promotes efficient delivery of DNA into mammalian cells. J Biol Chem. 2001;276:26204–10. doi: 10.1074/jbc.M010625200. [DOI] [PubMed] [Google Scholar]
- 46.Hunter RJ. Zeta potential in colloid science: principles and applications. London; New York: Academic Press; 1981. [Google Scholar]
- 47.Abramoff MDMP, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 48.du Prel JB, Hommel G, Rohrig B, Blettner M. Confidence interval or p-value?: part 4 of a series on evaluation of scientific publications. Dtsch Arztebl Int. 2009;106:335–9. doi: 10.3238/arztebl.2009.0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lawrence MS, Phillips KJ, Liu DR. Supercharging proteins can impart unusual resilience. J Am Chem Soc. 2007;129:10110–12. doi: 10.1021/ja071641y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sokalingam S, Raghunathan G, Soundrarajan N, Lee SG. A study on the effect of surface lysine to arginine mutagenesis on protein stability and structure using green fluorescent protein. PLoS One. 2012;7:e40410. doi: 10.1371/journal.pone.0040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao X, Huang L. Potentiation of cationic liposome-mediated gene delivery by polycations. Biochemistry. 1996;35:1027–36. doi: 10.1021/bi952436a. [DOI] [PubMed] [Google Scholar]
- 52.Li S, Huang L. In vivo gene transfer via intravenous administration of cationic lipid-protamine-DNA (LPD) complexes. Gene Ther. 1997;4:891–900. doi: 10.1038/sj.gt.3300482. [DOI] [PubMed] [Google Scholar]
- 53.Schafer J, Hobel S, Bakowsky U, Aigner A. Liposome-polyethylenimine complexes for enhanced DNA and siRNA delivery. Biomaterials. 2010;31:6892–900. doi: 10.1016/j.biomaterials.2010.05.043. [DOI] [PubMed] [Google Scholar]
- 54.Yamano S, Dai J, Hanatani S, Haku K, Yamanaka T, Ishioka M. Long-term efficient gene delivery using polyethylenimine with modified Tat peptide. Biomaterials. 2014;35:1705–15. doi: 10.1016/j.biomaterials.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 55.Chen JL, Wang H, Gao JQ, Chen HL, Liang WQ. Liposomes modified with polycation used for gene delivery: preparation, characterization and transfection in vitro. Int J Pharm. 2007;343:255–61. doi: 10.1016/j.ijpharm.2007.05.045. [DOI] [PubMed] [Google Scholar]
- 56.Lv H, Zhang S, Wang B, Cui S, Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J Control Release. 2006;114:100–9. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 57.Werth S, Urban-Klein B, Dai L, Hobel S, Grzelinski M, Bakowsky U. A low molecular weight fraction of polyethylenimine (PEI) displays increased transfection efficiency of DNA and siRNA in fresh or lyophilized complexes. J Control Release. 2006;112:257–70. doi: 10.1016/j.jconrel.2006.02.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.