Abstract
Helix-hairpin-helix DNA glycosylases are typically small proteins that initiate repair of DNA by excising damaged or mispaired bases. An invariant aspartic acid in the active site is involved in catalyzing the excision reaction. Replacement of this critical residue with an asparagine severely reduces catalytic activity but preserves enzyme stability and structure. The Arabidopsis DEMETER (DME) gene encodes a large 1,729-aa polypeptide with a 200-aa DNA glycosylase domain. DME is expressed primarily in the central cell of the female gametophyte. DME activates maternal allele expression of the imprinted MEDEA (MEA) gene in the central cell and is required for seed viability. We mutated the invariant aspartic acid at position 1304 in DME to asparagine (D1304N) to determine whether the catalytic activity of the DNA glycosylase domain is required for DME function in vivo. Transgenes expressing wild-type DME in the central cell rescue seed abortion caused by a mutation in the endogenous DME gene and activate maternal MEA:GFP transcription. However, transgenes expressing the D1304N mutant DME do not rescue seed abortion or activate maternal MEA:GFP transcription. Whereas ectopic expression of the wild-type DME polypeptide in pollen is sufficient to activate ectopic paternal MEA and MEA:GUS expression, equivalent expression of the D1304N mutant DME in pollen failed to do so. These results show that the conserved aspartic acid residue is necessary for DME to function in vivo and suggest that an active DNA glycosylase domain, normally associated with DNA repair, promotes gene transcription that is essential for gene imprinting.
DNA glycosylases are typically low-molecular-weight (200-300 aa) enzymes responsible for recognizing base lesions in the genome and initiating base excision repair. These proteins excise mispaired or damaged (e.g., oxidized, alkylated, deaminated, or methylated) bases (1, 2). After base excision, the DNA is further processed by the concerted action of an endonuclease, a DNA polymerase, and a DNA ligase. DNA glycosylases have been conserved during evolution and four structural families have been identified based on similarity to a uracil DNA glycosylase (UDG) family, an alkyladenine DNA glycosylase (AAG) family, a bacterial 8-oxoguanine DNA glycosylase (MutM/Fpg) family, and a HhH family that share a HhH active-site motif (3). The mispaired or damaged bases that are repaired by DNA glycosylases arise from the inherent chemical instability of DNA, from errors arising from the activity of DNA polymerase during replication, and from exposure to DNA-damaging agents present in the environment (4). Such damage has the potential to cause mutations and cell death, and bacterial strains deficient in DNA glycosylases (Ung, MutY, and MutM) show a mutator phenotype (1, 5). However, whereas DNA glycosylase-deficient mice show marked sensitivity to the DNA-damaging agents at the cellular level, they do not display any developmental abnormalities (6, 7). Therefore, little is known about the function of DNA glycosylases during eukaryote development.
DEMETER (DME) encodes a large polypeptide (1,729 aa) with a nuclear localization signal and a 200-aa DNA glycosylase domain. The DME DNA glycosylase domain is most closely related to the HhH family of DNA glycosylases (8). Within the DME DNA glycosylase domain are conserved amino acid residues essential for DNA glycosylase activity. DME is required for maternal allele expression of the imprinted genes MEDEA (MEA), a Polycomb group gene, and FWA, a transcription factor gene, in the central cell of the female gametophyte and in the endosperm of Arabidopsis (8, 9). The central cell gives rise to the endosperm, an embryo-nourishing tissue, on fertilization. MEA function is required for seed viability; a seed inheriting a mutant maternal mea allele aborts regardless of genotype of the silent paternal allele (10-13). Seed viability also depends solely on the maternal DME allele, and seeds that inherit a mutant maternal dme allele abort regardless of the paternal DME allele genotype. DME is primarily expressed in the central cell of the female gametophyte, where it activates maternal MEA allele expression. DME and MEA are not expressed in the stamens, which produce the male gametophyte or pollen. After fertilization of the central cell, DME expression is greatly reduced. Therefore, only the maternal MEA allele, not the paternal MEA allele, is exposed to DME activity. Thus, DME establishes MEA imprinting (maternal allele expressed, paternal allele not expressed) in the endosperm.
The crystal structures of four proteins in the HhH family of DNA glycosylases (EndoIII, AlkA, MutY, and hOGG1) revealed a conserved HhH motif followed at a fixed distance by a glycine-proline-rich loop and an invariant aspartic acid residue in the active site (3, 14-21). Mutation of the invariant aspartic acid residue to asparagine reduces the catalytic activity of DNA glycosylases in vitro; depending on the enzyme, in vitro activity is either abolished or reduced by ≈65-fold. Biochemical and crystallographic analyses indicate that this mutation preserves the DNA glycosylase structure and stability but reduces enzymatic activity (19, 20), establishing an essential role for the invariant aspartic acid in the catalysis of base excision.
The function of the predicted DME DNA glycosylase domain in the activation of gene transcription in the central cell is not known. One possibility is that the DNA glycosylase domain is required to activate transcription. Alternatively, the DME DNA glycosylase domain might not be needed and other DME domains are responsible for activation of transcription. To distinguish between these alternatives, we mutated the conserved aspartic acid at position 1304 in DME to asparagine (D1304N) and compared the in vivo function of wild-type DME and mutant DME(D1304N) polypeptides in Arabidopsis plants. We show that the invariant aspartic acid residue D1304 is necessary for DME function in the central cell, suggesting that an active DME DNA glycosylase is essential to regulate MEA gene imprinting and seed viability. Thus, the DNA glycosylase domain that is usually associated with DNA repair can also function during plant reproduction as a regulator of gene transcription, imprinting, and endosperm development.
Materials and Methods
Plant Materials and Microscopy. Wild-type and mutant dme alleles (Landsberg er ecotype) are as described (8). Methods for growing plants, fixing tissues, photography, β-glucuronidase (GUS) activity localization, and fluorescence microscopy were as described (22).
Generation of Transgenic Lines. To mutate the conserved aspartic acid at position 1304 to asparagine, JHDME1 primer (CCCTGTTaACACGAATGTTGGAAGGATAGC; codon for asparagine is underlined, mutagenized base is lowercase) and SKEN-5 (8) primer were used in a PCR to amplify a 1.3-kb DME (D1304N) cDNA. A 0.5-kb DNA fragment spanning the D1304 codon was excised from the wild-type DME cDNA clone by digestion with HincII and XmaI restriction endonucleases and was replaced with the 0.5-kb HincII and XmaI DNA fragment with the D1304N mutation, creating the full-length DME(D1304N) cDNA clone. The DME promoter ligated to DME cDNA [DME:DME (8)] and the DME promoter ligated to a mutagenized DME cDNA where aspartic acid at position 1304 is changed to asparagine [DME:DME(D1304N)] transgenes were generated by ligating 3.4 kb of DME 5′ flanking sequences to the wild-type full-length DME cDNA and DME(D1304N) cDNA, respectively. Transgenes were inserted into the Agrobacterium vector, pBI-GFP(S65T), and transgenic wild-type (Landsberg er ecotype) Arabidopsis lines were generated (8). Four independent DME:DME(D1304N) T1 transgenic lines were pollinated with DME/dme-2 pollen to generate F1 lines heterozygous DME/dme-2 and hemizygous DME:DME(D1304N). Likewise, four independent DME:DME T1 transgenic lines were pollinated with DME/dme-2 pollen to generate F1 plants, heterozygous DME/dme-2 and hemizygous DME:DME. Identification of these plants was facilitated by the fact that the dme-2 mutant allele is due to insertion of a pSKI015 T-DNA (23) with a BAR gene that confers resistance to glufosinate ammonium herbicide (Basta, Crescent Chemical, Islandia, NY). Plants bearing the DME:DME or DME:DME(D1304N) transgenes were identified by PCR amplification of a 460-bp DNA by using 3′ RACE1(5′-GCCTACA AGCCAGTGGGATAG-3′) and SKB4 (5′-GGATGGACTCGAGCACTGGG-3′) primers.
Plants homozygous for a MEA promoter ligated to GFP cDNA (MEA:GFP) transgene (8), heterozygous DME/dme-2, DME(D1304N) transgenes were generated by standard genetic crosses. Plants bearing the MEA:GFP transgene were identified by PCR amplification of an 860-bp DNA by using UCB3-F2 (5′-AGGAATTTAACCCGTATATATGTC-3′) and 5′ sGFPrev (5′-GAACTTGTGGCCGTTCACGTCGCC-3′) primers.
The cauliflower mosaic virus promoter (CaMV) was ligated to a full-length DME cDNA to create a CaMV:DME transgene as described (8). To generate a CaMV promoter ligated to mutant DME(D1304N) cDNA [CaMV:DME(D1304N)] transgene, a 2.3-kb BsrGI and SmaI DNA spanning the D1304N mutation was excised and used to replace the 2.3-kb BsrGI and SmaI DNA from a full-length DME cDNA, creating the CaMV:DME(D1304N) cDNA clone.
Pollen Collection and RNA Analysis. Pollen were isolated and processed by using procedures modified from Preuss et al. (24). About 30 open flowers were harvested in an Eppendorf tube on ice with 750 μl of tobacco pollen germination medium (20 mM Mes-KOH, pH 6.0/0.07% Ca(NO3)2/0.02% MgSO4·7H2O/0.01% KNO3/0.01% H3BO3/2% sucrose/15% PEG 4000). Tubes were vortexed, and then the pollen was sedimented at 9,000 rpm for 30 s. After supernatant and flower debris were removed, pollen was resuspended by vortex mixing in 50 μl of ice-cold pollen germination medium. Pollen from ≈600 flowers was pooled into a single tube, sedimented at 9,000 rpm for 30 s, resuspended by vortex mixing in 800 μl of ice-cold Arabidopsis pollen germination medium (17% sucrose/2 mM CaCl2/1.625 mM boric acid, pH 7.5, with 4 M KOH), and incubated for 1 h at room temperature to induce germination. Pollen was then sedimented at 3,000 rpm for 5 min. RNA was isolated with 1 ml of TRIzol (Invitrogen), and RT-PCRs were carried out as described (25). Primers for amplifying DME were cDNA-5 (CAGAAGTGTGGAGGGAAAGCGTCTGGC) and SKEN-5 (8). Primers for MEA were as described (13).
Results
The Invariant Aspartic Acid Is Essential for Seed Viability. We mutated the conserved aspartic acid at position 1304 to asparagine (D1304N) to determine whether DNA glycosylase activity is critical for DME function. The full-length cDNA clone with the mutation, DME(D1304N), as well as the control wild-type full-length DME cDNA clone, was ligated to 3.4-kb of DME 5′ flanking sequences that activate transcription in the central cell of the female gametophyte (8). We transformed Arabidopsis plants with DME:DME(D1304N) and obtained four independently isolated transgenic lines designated DME:DME(D1304N)-1, -2, -3, and -4. We also transformed Arabidopsis plants with the control DME:DME transgene and obtained four independently isolated transgenic lines designated DME:DME-1, -2, -3, and -4. Transgenes were crossed into a dme-2 heterozygous background so that we could compare the function of the DME(D1304N) and DME proteins during seed development.
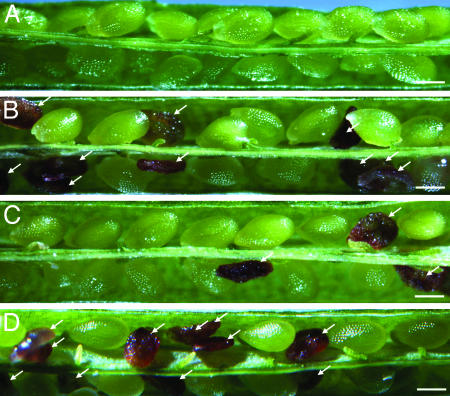
Wild-type Arabidopsis seeds rarely abort (Fig. 1A). Self-pollinated heterozygous DME/dme-2 siliques display 50% seed abortion (Fig. 1B and Table 1) because seed viability depends on the presence of a wild-type maternal DME allele (8). Seeds that inherit a wild-type maternal DME allele are viable, and seeds that inherit the mutant maternal dme-2 allele abort. Self-pollinated heterozygous DME/dme-2 siliques that are also hemizygous for the DME:DME transgene show a reduced level of seed abortion. Two transgenic lines (DME:DME-3 and DME:DME-4) displayed 25% seed abortion (Table 1 and Fig. 1C). In these two lines inheritance of a mutant dme-2 allele and a transgene produced a viable seed, indicating that the DME:DME transgene had suppressed the dme-2 allele and rescued seed viability. Two other transgenic lines (DME:DME-1 and DME:DME-2) displayed ≈30% seed abortion and therefore partially suppressed the dme-2 mutation (Table 1). By contrast, four independent lines heterozygous for DME/dme-2 and hemizygous for a mutant DME:DME(D1304N) transgene had siliques with 50% seed abortion (Table 1 and Fig. 1D), indicating that seeds with a dme-2 allele and the mutant transgene were not viable, indicating that the DME:DME(D1304N) transgene had not suppressed the dme-2 mutant allele. These results show that the conserved aspartic acid is essential for DME function in developing seeds.
Fig. 1.
Effect of the D1304N mutation on seed viability. Siliques in A-D were dissected and photographed 14 days after self-pollination. (Scale bars = 0.5 mm.) Arrows indicate aborted seeds. (A) Wild-type silique. (B) Heterozygous DME/dme-2 silique. (C) Silique is heterozygous DME/dme-2 and hemizygous for a DME:DME-4 transgene. (D) Silique is heterozygous DME/dme-2 and hemizygous for a DME:DME(D1304N)-3 transgene.
Table 1. Effect of the D1304N mutation on ratios of viable and aborted seeds.
| Genotype | n* | %† | P for 1:1‡ | P for 3:1§ |
|---|---|---|---|---|
| DME/dme-2 | 762 | 51 | 0.5 | — |
| DME/dme-2, DME:DME(D1304N)-1 | 181 | 45 | 0.2 | — |
| DME/dme-2, DME:DME(D1304N)-2 | 986 | 49 | 0.6 | — |
| DME/dme-2, DME:DME(D1304N)-3 | 1,588 | 48 | 0.1 | — |
| DME/dme-2, DME:DME(D1304N)-4 | 764 | 50 | 0.8 | — |
| DME/dme-2, DME:DME-1 | 699 | 30 | — | <0.005 |
| DME/dme-2, DME:DME-2 | 525 | 33 | — | <0.005 |
| DME/dme-2, DME:DME-3 | 322 | 23 | — | 0.5 |
| DME/dme-2, DME:DME-4 | 1,041 | 25 | — | 0.6 |
Number of seeds checked.
Percentage of aborted seed.
Probability that deviation from a 1:1 segregation of viable aborted seeds is due to chance.
Probability that deviation from a 3:1 segregation of viable aborted seeds is due to chance.
Transmission of a maternal mutant dme-2 allele is a more sensitive assay to compare DME and DME(D1304N) function during seed development. None of the viable F1 progeny inherit the maternal mutant dme-2 allele when a heterozygous DME/dme-2 plant is pollinated with wild-type pollen (Fig. 2), confirming the importance of the wild-type maternal DME allele during seed development. When a plant that is heterozygous DME/dme-2 and hemizygous for a DME:DME-4 transgene is pollinated with wild-type pollen, viable F1 progeny are detected that inherit both the maternal mutant dme-2 allele and the transgene (Fig. 2), indicating that the DME:DME-4 transgene suppressed the dme-2 mutant allele. However, when a heterozygous DME/dme-2, hemizygous DME:DME(D1304N)-3 plant is pollinated with wild-type pollen, the frequency of F1 progeny that inherit the maternal mutant dme-2 allele and the transgene is reduced ≈50-fold (Fig. 2). These results suggest that the D1304N mutation severely reduces but does not completely abolish DME activity during seed development. This is consistent with the finding that some glycosylases retain slight activity after mutation of the aspartic acid to asparagine (19, 21).
Fig. 2.
Effect of the D1304N mutation on transmission of the maternal mutant dme-2 allele. No transgene, heterozygous DME/dme-2 plant was pollinated with wild-type pollen and no F1 progeny with the dme-2 allele were detected (790 checked); DME:DME(D1304N), plant heterozygous DME/dme-2 and hemizygous for a DME:DME(D1304N)-3 transgene was pollinated with wild-type pollen, and three F1 progeny with the dme-2 allele and DME:DME(D1304N)-3 transgene were detected (379 checked); DME:DME, plant heterozygous DME/dme-2 and hemizygous for a DME:DME-4 transgene was pollinated with wild-type pollen and 36 F1 progeny with the dme-2 allele and the DME:DME-4 transgene were detected (97 checked).
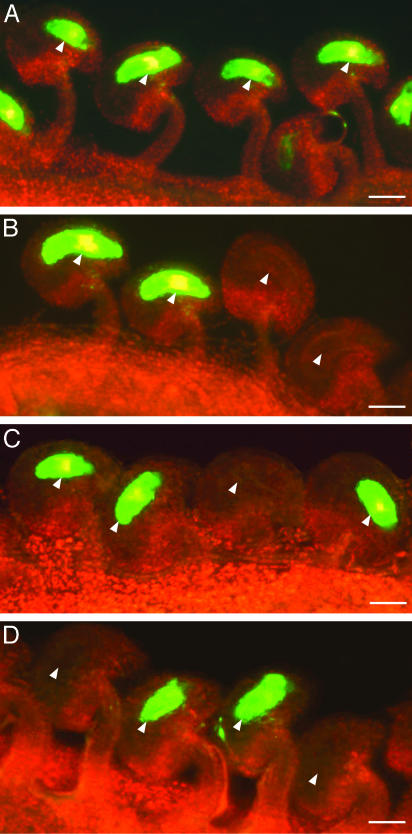
The Invariant Aspartic Acid Is Required for Activation of Maternal MEA:GFP Transcription in the Central Cell. Essentially all prefertilization ovules (153 of 155 checked) from MEA:GFP homozygous plants showed strong GFP fluorescence in the central cell nucleus and cytoplasm (Fig. 3A). DME is necessary for the transcription of a MEA:GFP transgene in the central cell of the female gametophyte (8, 26), and plants homozygous for MEA:GFP and heterozygous for DME/dme-2 display a 1:1 segregation ratio of fluorescent to nonfluorescent ovules (113:108, χ2 = 0.1, P > 0.8) (Fig. 3B). To determine whether DNA glycosylase activity is critical for MEA gene activation, we analyzed the effect of the D1304N mutation on transcription of the MEA:GFP transgene. In plants homozygous for a MEA:GFP transgene, heterozygous for DME/dme-2, and hemizygous for a DME:DME-4 transgene, we detected a 3:1 segregation ratio of fluorescent to nonfluorescent ovules (557:169, χ2 = 1.1, P > 0.4) (Fig. 3C). These results suggest that female gametophytes inheriting dme-2 and the DME:DME transgene transcribed MEA:GFP and that the DME:DME transgene suppressed the dme-2 mutant allele. By contrast, in plants homozygous for a MEA:GFP transgene, heterozygous DME/dme-2, and hemizygous for a DME:DME(D1304N)-3 transgene, we detected a 1:1 segregation ratio of fluorescent to nonfluorescent ovules (378:369, χ2 = 0.1, P > 0.8) (Fig. 3D). This result suggests that the female gametophytes inheriting dme-2 and the DME:DME(D1304N) transgene did not transcribe the MEA:GFP transgene, and DME:DME(D1304N) did not suppress the dme-2 mutant allele. Thus, the conserved aspartic acid D1304 is essential for activation of MEA:GFP transcription by DME.
Fig. 3.
Effect of the D1304N mutation on maternal MEA:GFP transcription in the central cell. Fluorescence micrographs of ovules harvested from stage 12 flowers (46) are shown. GFP and chlorophyll fluorescence were converted to green and red, respectively. Arrows point to central cells. (Scale bars = 0.04 mm.) (A) Ovules from a wild-type flower that is homozygous for a MEA:GFP transgene. (B) Ovules from a flower that is heterozygous DME/dme-2 and homozygous for a MEA:GFP transgene. (C) Ovules from a flower that is heterozygous DME/dme-2, hemizygous for a DME:DME-4 transgene, and homozygous for a MEA:GFP transgene. (D) Ovules from a flower that is heterozygous DME/dme-2, hemizygous for a DME:DME(D1304N)-3 transgene, and homozygous for a MEA:GFP transgene.
The Invariant Aspartic Acid Is Required for Ectopic Paternal MEA Allele Expression. Ectopic DME expression in the leaf and endosperm is sufficient to induce MEA and paternal MEA allele expression, respectively (8). These results suggest that restriction of DME expression to the central cell of the female gametophyte is responsible, at least in part, for MEA imprinting in the endosperm. Consistent with this model, DME and MEA paternal allele RNA is not detected in the male gametophyte, pollen (Fig. 4), and GUS staining is not detected in DME:GUS (8) or MEA promoter ligated to GUS cDNA (MEA:GUS) (Fig. 5A) transgenic pollen. Thus, the pollen provides an opportunity to compare the ectopic activation of MEA and MEA:GUS expression by DME and DME(D1304N) in a cellular environment that is free from endogenous DME and MEA expression.
Fig. 4.
Effect of the D1304N mutation on ectopic paternal MEA allele expression. CaMV:DME-4 and CaMV:DME-5 represent two independently isolated transgenic lines that ectopically express the wild-type DME cDNA (8). CaMV:DME(D1304N)-1 and CaMV:DME(D1304N)-3 represent two independently isolated transgenic lines that ectopically express the mutant D1304N form of DME. Total RNA was isolated from pollen harvested from open flowers, and the approximate level of MEA and DME RNA was determined by semiquantitative RT-PCR. (A) CaMV:DME transgenes activate paternal MEA allele gene expression in pollen. (B) CaMV:DME(D1304N) transgenes do not activate paternal MEA allele gene expression in pollen.
Fig. 5.
Effect of the D1304N mutation on ectopic paternal MEA:GUS gene transcription. Light micrographs were taken 12 h after staining for GUS activity. (A) Stamen is hemizygous for a MEA:GUS transgene. (B) Stamen is hemizygous for a CaMV:DME-4 transgene and hemizygous for a MEA:GUS transgene. (C) Stamen is hemizygous for a CaMV:DME(D1304N)-1 transgene and hemizygous for a MEA:GUS transgene. (Scale bars = 0.005 mm.)
We generated CaMV:DME transgenic lines (8) where transcription of the wild-type DME cDNA is under the control of the CaMV promoter (27). Here, we show that both DME and MEA RNAs were present in pollen from independently isolated CaMV:DME transgenic lines (Fig. 4A). This result indicates that expression of wild-type DME in pollen is sufficient to activate paternal MEA expression and supports the model that preventing DME expression in the male gametophyte is necessar y for MEA gene imprinting. By contrast, DME(D1304N) RNA, but not MEA RNA, was detected in pollen harvested from multiple independently isolated CaMV:DME(D1304N) lines that express the mutant DME(D1304N) protein (Fig. 4B). The level of mutant DME(D1304N) and wild-type DME RNAs were similar in pollen harvested from the DME:DME(D1304N) and DME:DME lines, respectively, suggesting that the failure to induce MEA gene expression in the DME:DME(D1304N) lines is due to the replacement of aspartic acid by asparagine in the DME(D1304N) DNA glycosylase active site. We also crossed the CaMV:DME-4 and CaMV:DME(D1304N)-1 transgenes into a MEA:GUS genetic background. We detected GUS staining in pollen grains from plants hemizygous for a CaMV:DME-4 transgene and hemizygous for a MEA:GUS transgene (Fig. 5B). By contrast, no GUS staining was detected in plants hemizygous for CaMV:DME(D1304N)-1 and MEA:GUS transgenes (Fig. 5C). These results suggest that active DME DNA glycosylase is essential for activation of MEA gene transcription in pollen.
Discussion
Most DNA glycosylases are low-molecular-weight enzymes that catalyze the first step in the base-excision DNA repair pathway by excising damaged or mispaired bases. These lesions are mutagenic and believed to play a role in cancer and aging, so the mechanism of base-excision DNA repair has been studied in great detail, and the mechanism of base excision is well understood at the atomic level (1, 3). Elucidation of DNA glycosylase 3D structures has led to a detailed understanding of their lesion recognition and catalysis mechanisms. However, the role of these DNA glycosylases in controlling development is largely unknown.
Compared with the well studied low-molecular-weight DNA glycosylases, the Arabidopsis DME protein has a distinct structure and function (8, 9). Embedded within the 1,729-aa DME polypeptide is a 200-aa domain related to the HhH family of DNA glycosylase. DME was discovered by a mutation that results in seed abortion. DME activates transcription of maternal alleles in the central cell of the female gametophyte, resulting in endosperm gene imprinting. We compared the in vivo function of wild-type DME and mutant DME(D1304N) polypeptides in Arabidopsis plants to understand the role of the DME DNA glycosylase domain in the activation of gene transcription in the central cell. We replaced the conserved aspartic acid residue with an asparagine residue because this mutation decreases DNA glycosylase activity without altering enzyme stability or structure in vitro (19, 20). We found that only the wild-type DME protein, not the mutant DME(D1304N) protein, rescued dme-mediated seed abortion (Figs. 1 and 2 and Table 1) and activated MEA transcription in the central cell (Fig. 3) and pollen (Figs. 4 and 5). These results are consistent with our finding that in vivo DME expression results in nicks in the MEA promoter that may be due to DME-mediated base excision (8). These experiments show that DME DNA glycosylase activity is essential for activation of imprinted gene transcription, a process that plays a critical role in reproductive development in plants.
Multiple Mechanisms for Regulation of Gene Transcription by DNA Glycosylases. Physical and functional linkages between DNA glycosylases and proteins that regulate gene transcription (e.g., transcription factors, receptors, and chromatin-remodeling proteins) have recently been discovered (28). Thymine DNA glycosylase has been reported to both activate and repress gene transcription by a variety of mechanisms, including binding to hormone receptors (29, 30), interacting with transcription factors (31), and associating with CBP/p300 acetylase, which remodels chromatin and activates transcription through histone acetyltransferase activity (32). Methylpurine DNA glycosylase has a synergistic effect on gene silencing by interacting with the methyl CpG-binding domain protein 1 (MBD1) transcriptional repressor (33) and 3-methyladenine DNA glycosylase interacts with estrogen receptor α to inhibit gene transcription (34). Thus, DNA glycosylases are linked to the process of gene regulation by physical interactions that modulate the activities of transcription factors, receptors, and chromatin-remodeling proteins. However, it is not known whether base-excision activity is a requirement for regulation of gene transcription by DNA glycosylases in addition to direct physical association. Indeed, in one case it was shown that inactive and wild-type thymine DNA glycosylases bound estrogen receptors and activated them with equal efficiency (30), suggesting that receptor binding, rather than DNA glycosylase activity, was responsible for modulating the rate of gene transcription.
We have shown that an active DNA glycosylase domain is necessary to induce MEA gene transcription in the central cell. We recently found that DNA methylation plays an important role in MEA imprinting and seed viability. These processes are controlled by an antagonism between the MET1 methyltransferase and the DME DNA glycosylase in the central cell of the female gametophyte (26). MET1 is the Arabidopsis ortholog of mammalian Dnmt1 methyltransferase, which maintains DNA methylation at CpG sites (35). One possibility is that DME initiates the replacement of 5-methylcytosine with cytosine, resulting in hypomethylation and activation of maternal MEA allele expression. In support of this model, ROS1 DNA glycosylase, an Arabidopsis protein related to DME, can excise 5-methylcytosine in vitro and represses DNA methylation-mediated transgene silencing (36). Also, an animal thymine DNA glycosylase can excise 5-methylcytosine in vitro, and its inhibition suppresses genome-wide hypomethylation, whereas overexpression causes promoter hypomethylation and activates transcription (37, 38). Alternatively, the antagonistic relationship between DME and MET1 may be indirect, whereas MEA gene transcription is promoted by DNA nicking associated with DME DNA glycosylase activity (8). This process may facilitate nucleosome sliding and alteration of chromatin structure (39).
DNA glycosylases may modulate gene transcription by two distinct mechanisms. One mechanism involves modulation by the association of DNA glycosylases with transcription factors and/or chromatin-remodeling proteins. A second mechanism invokes modification of the DNA by base-excision DNA glycosylase activity.
Possible Function for Other DME Protein Domains. Arabidopsis has numerous low-molecular-weight DNA glycosylases that are responsible for repairing DNA damage throughout the genome (40-45). By contrast, DME appears to acts at very distinct sites in the Arabidopsis genome to activate transcription of specific genes such as MEA and FWA in the central cell (8, 9, 26). Thus, DME DNA glycosylase activity plays a precise and critical role in plant development. What accounts for the highly restricted genome target specificity of DME? One possibility is that the DME DNA glycosylase domain binds to a factor that directs it to its target in the genome. Alternatively, other domains within the high-molecular-weight DME polypeptide may play a role. Comparing the Arabidopsis DME to a related rice protein has revealed multiple regions of significant amino acid sequence homologies that are outside the shared DNA glycosylase domains (8). Although the function of these domains is unknown, they might direct DME, or interact with other molecules that direct DME, to specific sites within the genome where the DNA glycosylase activity of DME is needed to promote maternal allele transcription in the central cell, a process required for endosperm imprinting and seed viability.
Acknowledgments
We thank M. Gehring, R. Pennell, T. Hazra, and A. Britt for critically reading this manuscript and S. Rashid for technical assistance with these experiments. This research was supported by U.S. Department of Agriculture Grant 2002-01400 and National Institutes of Health Grant GM069415 (to R.L.F.).
Abbreviations: DME, DEMETER; MEA, MEDEA; GUS, β-glucuronidase; DME:DME, DME promoter ligated to DME cDNA; DME:DME(D1304N), DME promoter ligated to a mutagenized DME cDNA where aspartic acid at position 1304 is changed to asparagine; CaMV:DME, cauliflower mosaic virus promoter ligated to DME cDNA; CaMV:DME(D1304N), cauliflower mosaic virus promoter ligated to mutant DME(D1304N) cDNA; MEA:GFP, MEA promoter ligated to GFP cDNA; MEA:GUS, MEA promoter ligated to GUS cDNA; HhH, helix-hairpin-helix.
References
- 1.Scharer, O. D. & Jiricny, J. (2001) BioEssays 23 270-281. [DOI] [PubMed] [Google Scholar]
- 2.Krokan, H. E., Standal, R. & Slupphaug, G. (1997) Biochem. J. 325 1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fromme, J. C., Banerjee, A. & Verdine, G. L. (2004) Curr. Opin. Struct. Biol. 14 1-7. [DOI] [PubMed] [Google Scholar]
- 4.Lindahl, T. (1993) Nature 362 709-715. [DOI] [PubMed] [Google Scholar]
- 5.Minowa, O., Arai, T., Hirano, M., Monden, Y., Nakai, S., Fukuda, M., Itoh, M., Takano, H., Hippou, Y., Arburatani, H., et al. (2000) Proc. Natl. Acad. Sci. USA 97 4156-4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engelward, B. P., Weeda, G., Wyatt, M. D., Broekhof, J. L., Donker, I., Allan, M. M., Gold, B., Hoeijmakers, J. H. & Samson, L. D. (1997) Proc. Natl. Acad. Sci. USA 94 13087-13092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hang, B., Singer, B., Margison, G. P. & Elder, R. H. (1997) Proc. Natl. Acad. Sci. USA 94 12869-12874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi, Y., Gehring, M., Johnson, L., Hannon, M., Harada, J. J., Goldberg, R. B., Jacobsen, S. E. & Fischer, R. L. (2002) Cell 110 33-42. [DOI] [PubMed] [Google Scholar]
- 9.Kinoshita, T., Miura, A., Choi, Y., Kinoshita, Y., Cao, X., Jacobsen, S. E., Fischer, R. L. & Kakutani, T. (2004) Science 303 521-523. [DOI] [PubMed] [Google Scholar]
- 10.Grossniklaus, U., Vielle-Calzada, J.-P., Hoeppner, M. A. & Gagliano, W. B. (1998) Science 280 446-450. [DOI] [PubMed] [Google Scholar]
- 11.Kohler, C., Hennig, L., Spillane, C., Pien, S., Gruissem, W. & Grossniklaus, U. (2003) Genes Dev. 17 1540-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaudhury, A. M., Luo, M., Miller, C., Craig, S., Dennis, E. S. & Peacock, W. J. (1997) Proc. Natl. Acad. Sci. USA 94 4223-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiyosue, T., Ohad, N., Yadegari, R., Hannon, M., Dinneny, J., Wells, D., Katz, A., Margossian, L., Harada, J., Goldberg, R. B. & Fischer, R. L. (1999) Proc. Natl. Acad. Sci. USA 96 4186-4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bruner, S. D., Norman, D. P. & Verdine, G. L. (2000) Nature 403 859-866. [DOI] [PubMed] [Google Scholar]
- 15.Thayer, M. M., Ahern, H., Xing, D., Cunningham, R. P. & Tainer, J. A. (1995) EMBO J. 14 4108-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan, Y., Manuel, R. C., Arvai, A. S., Parikh, S. S., Mol, C. D., Miller, J. H., Lloyd, R. S. & Tainer, J. A. (1998) Nat. Struct. Biol. 5 1058-1064. [DOI] [PubMed] [Google Scholar]
- 17.Labahn, J., Scharer, O. D., Long, A., Ezaz-Nikpay, K., Verdine, G. L. & Ellenberger, T. E. (1996) Cell 86 321-329. [DOI] [PubMed] [Google Scholar]
- 18.Yamagata, Y., Kato, M., Odawara, K., Tokuno, Y., Nakashima, Y., Matsushima, N., Yasumura, K., Tomita, K., Ihara, K., Fujii, Y., et al. (1996) Cell 86 311-319. [DOI] [PubMed] [Google Scholar]
- 19.Norman, D. P., Chung, S. J. & Verdine, G. L. (2003) Biochemistry 42 1564-1572. [DOI] [PubMed] [Google Scholar]
- 20.Fromme, J. C., Banerjee, A., Huang, S. J. & Verdine, G. L. (2004) Nature 427 652-656. [DOI] [PubMed] [Google Scholar]
- 21.Eichman, B. F., O'Rourke, E. J., Radicella, J. P. & Ellenberger, T. (2003) EMBO J. 22 4898-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yadegari, R., Kinoshita, T., Lotan, O., Cohen, G., Katz, A., Choi, Y., Katz, A., Nakashima, K., Harada, J. J., Goldberg, R. B., et al. (2000) Plant Cell 12 2367-2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weigel, D., Ahn, J. H., Blazquez, M. A., Borevitz, J. O., Christensen, S. K., Frankhauser, C., Ferrandiz, C., Kardailsky, I., Malancharuvil, E. J., Neff, M. M., et al. (2000) Plant Physiol. 122 1003-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Preuss, D., Lemieux, B., Yen, G. & Davis, R. W. (1993) Genes Dev. 7 974-985. [DOI] [PubMed] [Google Scholar]
- 25.Kinoshita, T., Yadegari, R., Harada, J. J., Goldberg, R. B. & Fischer, R. L. (1999) Plant Cell 11 1945-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao, W., Gehring, M., Choi, Y., Margossian, L., Pu, H., Harada, J. J., Goldberg, R. B., Pennell, R. I. & Fischer, R. L. (2003) Dev. Cell 5 891-901. [DOI] [PubMed] [Google Scholar]
- 27.Rogers, S. G., Klee, H. J., Horsch, R. B. & Fraley, R. T. (1987) Methods Enzymol. 153 253-277. [Google Scholar]
- 28.Mitra, S., Izumi, T., Boldogh, I., Bhakat, K. K., Hill, J. W. & Hazra, T. K. (2002) Free Radical Biol. Med. 33 15-28. [DOI] [PubMed] [Google Scholar]
- 29.Um, S., Harbers, M., Benecke, A., Pierrat, B., Losson, R. & Chambon, P. (1998) J. Biol. Chem. 273 20728-20736. [DOI] [PubMed] [Google Scholar]
- 30.Chen, D., Lucey, M. J., Phoenix, F., Lopez-Garcia, J., Hart, S. M., Losson, R., Lakjaya, B., Coombes, R. C., Chambon, P., Schar, P. & Ali, S. (2003) J. Biol. Chem. 278 38586-38592. [DOI] [PubMed] [Google Scholar]
- 31.Missero, C., Pirro, M. T., Simeone, S., Pischetola, M. & Di Lauro, R. (2001) J. Biol. Chem. 276 33569-33575. [DOI] [PubMed] [Google Scholar]
- 32.Tini, M., Benecke, A., Um, S.-J., Torchia, J., Evans, R. M. & Chambon, P. (2002) Mol. Cell 9 265-277. [DOI] [PubMed] [Google Scholar]
- 33.Watanabe, S., Ichimura, T., Fujita, N., Tsuruzoe, S., Ohki, I., Shirakawa, M., Kawasuji, M. & Nakao, M. (2003) Proc. Natl. Acad. Sci. USA 100 12859-12864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Likhite, V. A., Cass, E. I., Anderson, S. D., Yates, J. R. & Nardulli, A. M. (2004) J. Biol. Chem., 10.1074/jbc.M313155200. [DOI] [PubMed]
- 35.Finnegan, E. J. & Kovac, K. A. (2000) Plant Mol. Biol. 43 189-201. [DOI] [PubMed] [Google Scholar]
- 36.Gong, Z., Morales-Ruiz, T., Ariza, R. R., Roldan-Arjona, T., David, L. & Zhu, J.-J. (2002) Cell 111 803-814. [DOI] [PubMed] [Google Scholar]
- 37.Jost, J.-P., Oakeley, E. J., Zhu, B., Benjamin, D., Thiry, S., Siegmann, M. & Jost, Y.-C. (2001) Nucleic Acids Res. 29 4452-4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu, B., Benjamin, D., Zheng, Y., Angliker, H., Thiry, S., Siegmann, M. & Jost, J.-P. (2001) Proc. Natl. Acad. Sci. USA 98 5031-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langst, G. & Becker, B. (2001) Mol. Cell 8 1085-1092. [DOI] [PubMed] [Google Scholar]
- 40.The Arabidopsis Genome Initiative (2000) Nature 408 796-815. [DOI] [PubMed] [Google Scholar]
- 41.Ohtsubo, T., Matsuda, O., Iba, K., Terashima, I., Sekiguchi, M. & Nakabeppu, Y. (1998) Mol. Gen. Genet 259 577-590. [DOI] [PubMed] [Google Scholar]
- 42.Roldan-Arjona, T., Garcia-Ortiz, M.-V., Ruiz-Rubio, M. & Ariza, R. R. (2000) Plant Mol. Biol. 44 43-52. [DOI] [PubMed] [Google Scholar]
- 43.Garcia-Ortiz, M.-V., Ariza, R. R. & Roldan-Arjona, T. (2001) Plant Mol. Biol. 47 795-804. [DOI] [PubMed] [Google Scholar]
- 44.Gao, M.-J. & Murphy, T. M. (2001) Photochem. Photobiol. 73 128-134. [DOI] [PubMed] [Google Scholar]
- 45.Hayes, J. G. (2002) DNA Repair 1 579-600. [DOI] [PubMed] [Google Scholar]
- 46.Bowman, J. L. (1994) in Arabidopsis: An Atlas of Morphology and Development, ed. Bowman, J. L. (Springer, New York), pp. 135-145.