Abstract
Human endogenous retroviruses (HERV) env proteins have been recently reported to be significantly up-regulated in certain cancers. Specifically, mRNA and protein levels of HERV-K (HML-2) are up-regulated in the blood plasma or serum of breast cancer patients. Here, we collected blood samples of 49 breast cancer patients and analyzed mRNA expressions of various HERVs env genes including HERV-R, HERV-H, HERV-K, and HERV-P by real-time PCR. The expression of env genes were significantly increased in the blood of primary breast cancer patients but were decreased in patients undergoing chemotherapy to a similar level with benign patients. When we compared the group currently undergoing chemotherapy and those patients undergoing chemotherapy simultaneously with radiotherapy, HERVs env genes were reduced more in the chemotherapy only group, suggesting that chemotherapy is more effective in reducing HERV env gene expression than is radiotherapy. Among chemotherapy groups, HERV env gene expression was the lowest in the taxotere- or taxol-treated group, suggesting that taxotere and taxol can reduce HERVs env expression. These data suggest the potential to use HERVs env genes as a diagnosis marker for primary breast cancer, and further studies are needed to identify the mechanism and physiological significance of the reduction of HERV env gene expression during chemotherapy.
Keywords: HERV, env, mRNA, realtime PCR, blood, gene expression
1. Introduction
Retroviruses, a form of mobile genetic elements, have important roles in both disease and primate evolution. Endogenous retroviruses (ERVs) was pathogenic, as well as being beneficial to a host in some cases. Furthermore, retroviruses also have played a key role in primate evolution that resulted in the incorporation of these elements into the human genome [1]. Human ERV (HERV) is now part of the human genome, occupying nearly 8% of the total genome, with approximately 98,000 ERV elements and fragments [2]. However, all HERVs appear to be defective due to major deletions or nonsense mutations, and no HERVs capable of replication had been identified, suggesting that most HERVs are merely traces of original viruses, having first integrated millions of years ago [3]. HERVs have been proposed to play a role in several forms of human cancer and autoimmune diseases, but conclusive evidence is still lacking [4,5,6]. Several reports have shown that HERVs are involved in several diseases including multiple sclerosis [7,8, ]schizophrenia [9], and HIV-infection [10].
Recent studies have suggested a strong relationship between HERVs and cancer based on the mRNA expression profile of HERVs in normal and cancer tissues [11,12,13,14,15,16]. One study compared the mRNA expression of HERVs in tumors to adjacent normal tissues and found high levels of HERV-K (HML-2) expression in testis tumor tissues, HERV-R (ERV3-1) in liver and lung tumor tissues, HERV-H in liver, lung, and testis tumor tissues, and HERV-P in colon and liver tumor tissues [16]. Another study analyzed multiple HERV-K (HML-2) surface envelope proteins in ovarian cancer using anti-HERV-K (HML-2)-specific antibody and found significantly increased expression in tumors with low malignant potential and low grade relative to expression in normal ovarian tissues. They also identified other classes of HERV env mRNAs, including HERV-R (ERV3-1) and HERV-E, which are expressed in these same ovarian cancer tissues. Anti-HERV antibodies including anti-ERV3 (30%), anti-HERV-E (40%) and anti-HERV-K (HML-2) (55%) were also detected in patients with ovarian cancer but not in normal female controls [17]. Furthermore, HERV-K (HML-2) mRNA and protein expressions in serum and blood were found to be significantly increased in cancer patients, and to be reduced by chemotherapy [18,19,20,21]. In addition, the HERV-K-MEL antigen, product of a pseudo-gene incorporated into the HERV-K env gene, was found to be significantly expressed in the majority of melanoma cells and other cancers [22,23].
We previously analyzed the HERV-R (ERV3-1) env protein in both adult human organs and in tumors using a tissue microarray and also compared the expression of HERV-R (ERV3-1) between normal and tumor tissues to study the relationship between HERV-R (ERV3-1) and tumor formation. HERV-R (ERV3-1) was highly expressed in normal organs and was significantly increased in various tumors, including lung adenocarcinoma, renal cell carcinoma, papillary carcinoma, hepatocellular carcinoma, and adenocarcinoma in the gastrointestinal tract [24]. Here, we analyzed and compared various HERV env gene mRNA expressions in the blood of 49 breast cancer patients to identify blood HERV env gene expression and to examine the potential use of HERVs as a breast cancer detection marker.
2. Results and Discussion
We collected blood samples of 10 healthy normal and 47 breast cancer patients containing benign tumors (Benign), primary breast cancers without chemotherapy (Primary no chemo), primary breast cancers undergoing chemotherapy (Primary chemo), and recurred and metastasized cancers. Recurred and metastasized cancers were divided into two groups: before chemotherapy (Meta prechemo) and undergoing chemotherapy (Meta chemo). However, Meta prechemo patients were treated with chemotherapy when they had primary tumors. The env genes expression of various HERVs including HERV-R (ERV3-1), HERV-H, HERV-K (HML-2), and HERV-P were analyzed by real-time PCR.
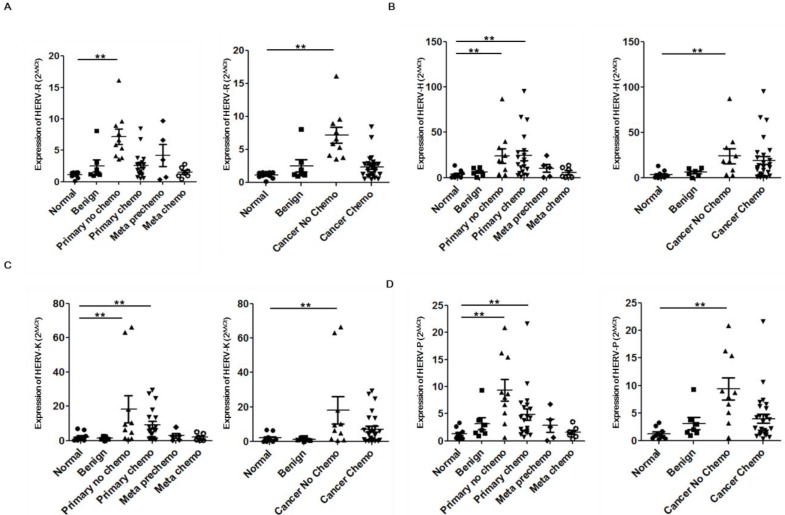
All patients’ information and expression score of each patient was described in Table 1. The expression of HERV-R (ERV3-1) env was significantly increased in the blood of Primary no chemo samples but reduced in patients undergoing or previously treated with chemotherapy to similar level with normal or benign patients (Figure 1A). HERV-H env expression was up-regulated in both the Primary no chemo and Primary chemo groups but was reduced in the Meta Prechemo and Meta chemo groups comparing with primary cancer patients. When we compared cancers having undergone chemotherapy and cancers not subjected to chemotherapy, we found that HERV-H env expression was also reduced in groups with chemotherapy (Figure 1B). The expression of HERV-K (HML-2) env in the Primary no chemo group was highly over-expressed (8.2-fold increase) compared to normal or benign group. HERV-K (HML-2) env expression was slightly reduced in the Primary chemo group but still showed a significant increase compared to normal or benign group. When we compared cancers with and without chemotherapy, we found that HERV-K (HML-2) expression, similar to HERV-R (ERV3-1), was reduced in groups with chemotherapy (Figure 1C). HERV-P env expression also showed a similar expression pattern to HERV-R (ERV3-1) env, including an increase in Primary no chemo group, and decrease in chemotherapy groups (Figure 1D). These data suggest that HERV env genes are significantly increased in the blood of primary breast tumor patients but this induction is reduced by chemotherapy.
Table 1.
Patients information and expressions of HERV env genes.
| Patient No. | Gender | Age | Status | Stage | CTx | Rx | Expression (2−ΔΔCt) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| HERV-R | HERV-H | HERV-K | HERV-P | |||||||
| 1 | Female | 25 | Normal | NA | − | − | 1.54806924 | 1.46936213 | 0.9275391 | 0.99967427 |
| 2 | Female | 31 | Normal | NA | − | − | 1.471890146 | 2.46791037 | 1.37724001 | 1.14023406 |
| 3 | Male | 25 | Normal | NA | − | − | 1.538228683 | 4.95879643 | 2.59526166 | 1.73282985 |
| 4 | Male | 38 | Normal | NA | − | − | 1.512746241 | 8.0583809 | 6.39294198 | 2.66203585 |
| 5 | Female | 25 | Normal | NA | − | − | 1.071157036 | 13.4086306 | 6.87891009 | 3.32217944 |
| 6 | Male | 31 | Normal | NA | − | − | 1.414870488 | 1.85316328 | 1.16342644 | 1.00558149 |
| 7 | Female | 25 | Normal | NA | − | − | 0.633655233 | 0.28319938 | 0.13652906 | 0.4591158 |
| 8 | Male | 26 | Normal | NA | − | − | 1.061327569 | 0.51662523 | 0.23777682 | 0.54624876 |
| 9 | Male | 25 | Normal | NA | − | − | 0.155951572 | 0.60970784 | 0.11957436 | 0.27466215 |
| 10 | Male | 26 | Normal | NA | − | − | 0.898300474 | 2.82527245 | 1.488831 | 0.80934684 |
| 11 | Female | 65 | Benign | NA | − | − | 0.969426438 | 6.60804418 | 1.16919319 | 2.01306906 |
| 12 | Female | 54 | Benign | NA | − | − | 1.316648275 | 10.5628214 | 2.48689685 | 3.75981932 |
| 13 | Female | 47 | Benign | NA | − | − | 1.131003453 | 7.46317751 | 2.59460214 | 1.52509241 |
| 14 | Female | 62 | Benign | NA | − | − | 1.235136082 | 11.1308356 | 2.20403429 | 1.42355345 |
| 15 | Female | 23 | Benign | NA | − | − | 3.067726784 | 5.04920645 | 0.92465046 | 2.9775588 |
| 16 | Female | 72 | Benign | NA | − | − | 1.958437833 | 3.11879775 | 0.72987096 | 0.95265781 |
| 17 | Female | 53 | Benign | NA | − | − | 8.088114008 | 0.20038179 | 0.03046887 | 9.30321907 |
| 18 | Female | 45 | Primary | 3c | − | − | 5.319792596 | 18.2529364 | 6.3774514 | 8.69446145 |
| 19 | Female | 64 | Primary | 2a | − | − | 16.14262545 | 39.6795383 | 10.1711671 | 15.4610172 |
| 20 | Female | 69 | Primary | 2a | − | − | 9.608447428 | 8.99396124 | 4.93020169 | 8.97988312 |
| 21 | Female | 48 | Primary | 1 | − | − | 8.913363856 | 87.2262221 | 66.7816577 | 20.8662098 |
| 22 | Female | 57 | Primary | 1 | − | − | 3.813819767 | 18.7491939 | 9.5227615 | 7.94857735 |
| 23 | Female | 58 | Primary | 4 | − | − | 4.12807734 | 3.28552366 | 1.48206974 | 3.14537058 |
| 24 | Female | 75 | Primary | 2a | − | − | 3.549551305 | 3.70504226 | 0.99168007 | 0.61249218 |
| 25 | Female | 49 | Primary | 0 | − | − | 6.627064679 | 33.2770737 | 63.1836773 | 16.2480724 |
| 26 | Female | 41 | Primary | 1 | − | − | 7.63980951 | 24.3444893 | 18.1808364 | 6.60914356 |
| 27 | Female | 64 | Primary | 2a | − | − | 5.867326733 | 2.70049398 | 0.86049974 | 5.10306258 |
| 28 | Female | 44 | Primary chemo | 2b | FEC | + | 0.583229932 | 3.42132858 | 1.35070567 | 0.70656621 |
| 29 | Female | 59 | Primary chemo | 1 | FEC | − | 1.922348628 | 19.9588497 | 13.4846546 | 2.12642504 |
| 30 | Female | 89 | Primary chemo | 2b | FEC | − | 2.655413743 | 1.21267252 | 0.32439272 | 1.91732734 |
| 31 | Female | 78 | Primary chemo | 2a | CMF | + | 6.824878361 | 9.60170093 | 4.13870667 | 4.4970014 |
| 32 | Female | 51 | Primary chemo | 1 | FEC | + | 1.27006588 | 6.65339183 | 1.08423689 | 7.41885563 |
| 33 | Female | 46 | Primary chemo | 1 | FAC | − | 1.703344207 | 28.0725848 | 5.5797997 | 7.00473271 |
| 34 | Female | 46 | Primary chemo | 3c | AC-taxol | + | 0.730760217 | 2.65103674 | 0.43893524 | 1.69693347 |
| 35 | Female | 52 | Primary chemo | 2a | AC | + | 3.813907886 | 22.6455656 | 5.9942531 | 6.20184155 |
| 36 | Female | 50 | Primary chemo | 1 | FEC | + | 2.83884353 | 14.931874 | 6.69126972 | 6.64835153 |
| 37 | Female | 61 | Primary chemo | 1 | FEC | + | 8.470829981 | 63.937481 | 24.7538325 | 21.6310231 |
| 38 | Female | 53 | Primary chemo | 1 | CMF | + | 2.113845596 | 12.3478675 | 3.84667871 | 3.80772688 |
| 39 | Female | 46 | Primary chemo | 2a | FEC | + | 2.873687797 | 11.3739122 | 8.0083961 | 5.84659504 |
| 40 | Female | 48 | Primary chemo | 2a | FEC | + | 2.776120133 | 25.2437908 | 17.7988649 | 10.6208697 |
| 41 | Female | 64 | Primary chemo | 2a | TA | + | 0.544990631 | 4.74241966 | 1.46403157 | 1.24584967 |
| 42 | Female | 56 | Primary chemo | 1 | FEC | + | 3.620787059 | 16.8844506 | 13.754324 | 4.36202085 |
| 43 | Female | 53 | Primary chemo | 2a | CMF | − | 1.035985157 | 2.41299234 | 1.01158204 | 1.64714017 |
| 44 | Female | 49 | Primary chemo | 2b | FEC | + | 1.265233213 | 45.0438009 | 6.23611126 | 1.15212453 |
| 45 | Female | 36 | Primary chemo | 3b | TA | + | 1.342234427 | 30.3275419 | 8.30499903 | 1.51382314 |
| 46 | Female | 50 | Primary chemo | 2b | CMF | + | 3.154118071 | 95.4643386 | 29.5362207 | 2.34984131 |
| 47 | Female | 64 | Primary chemo | 1 | FEC | + | 2.795558755 | 66.8485023 | 27.5099193 | 4.24480025 |
| 48 | Female | 33 | Meta no chemo | 4 | FEC | + | 0.746425408 | 0.45963376 | 0.07435361 | 0.68133206 |
| 49 | Female | 47 | Meta no chemo | 4 | CMF | + | 3.49050715 | 24.6256517 | 3.02817098 | 2.55660628 |
| 50 | Female | 43 | Meta no chemo | 4 | CMF | + | 0.397355807 | 1.72938211 | 0.38039759 | 0.10406547 |
| 51 | Female | 49 | Meta no chemo | 4 | CMF | + | 9.724135523 | 10.9923149 | 2.68548004 | 6.7503548 |
| 52 | Female | 56 | Meta no chemo | 4 | CMF | + | 6.626911563 | 13.408011 | 7.89577019 | 3.87500096 |
| 53 | Female | 49 | Meta chemo | 4 | CMF | + | 1.383737227 | 0.53792987 | 0.12805599 | 1.64584674 |
| 54 | Female | 53 | Meta chemo | 4 | FEC | + | 2.838056544 | 0.50303244 | 0.26561588 | 0.84934936 |
| 55 | Female | 47 | Meta chemo | 4 | AC | − | 0.973399092 | 1.9376706 | 0.39927279 | 3.47905785 |
| 56 | Female | 59 | Meta chemo | 4 | CMF | + | 1.757514591 | 8.90854611 | 5.13375104 | 2.23178286 |
| 57 | Female | 60 | Meta chemo | 4 | FEC | + | 0.976259554 | 11.4398006 | 4.19425139 | 1.56341054 |
CTx: Chemotherapy; Rx: Radiotherapy; CMF: six cycles of cyclophsphamide, methotrexate, 5-FU; FEC: six cycles of 5-FU, epirubicin, cyclophosphamide; TA: six cycles of taxotere and adriamycin.
Figure 1.
Expressions of HERVs env genes in the blood of breast cancer patients. Expression of the HERV-R (ERV3-1) (A); HERV-H (B); HERV-K (HML-2) (C); and HERV-P (D) env gene in the blood of breast cancer patients were analyzed by real-time PCR. Real-time PCR reactions were performed with blood total RNA of breast cancer patients including benign tumors (Benign), primary breast cancers without chemotherapy (Primary no chemo), primary breast cancers undergoing chemotherapy (Primary chemo), recurred and metastasized cancers before chemotherapy (Meta prechemo), and metastasized cancers undergoing chemotherapy (Meta chemo). Analyzed data were also compared to benign tumors (Benign), breast cancers without chemotherapy (Cancer No chemo), and breast cancers with chemotherapy (Cancer Chemo). Meta prechemo patients were ruled out in both Cancer No chemo and Cancer Chemo group because they are pre chemo stage but had experience of chemotherapy when they had primary cancer. Real-time PCR reactions were coupled to melting-curve analysis to confirm the amplification specificity. Non-template controls were included for each primer pair to check for any significant levels of contaminants. Real-time PCR was performed in three independent experiments with three different samples per group. The mRNA expression levels are presented relative to the control with calculated mean values and 95% confidence intervals. Statistical significance of differences between groups was determined using a two-tailed Student’s t-test. ** p < 0.01.
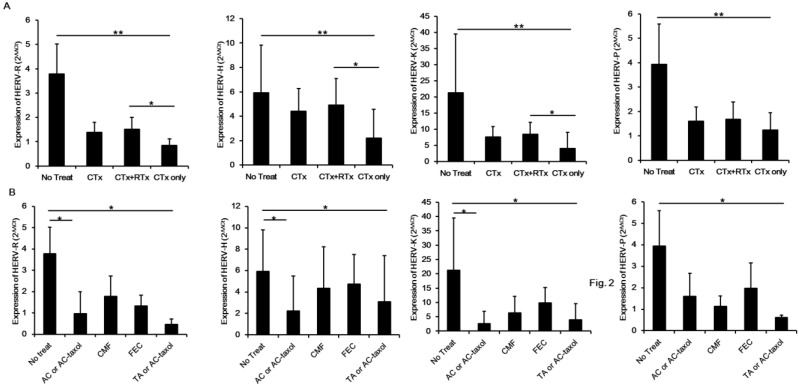
In order to identify which treatment has a critical effect on reducing the expression of HERVs env genes, we analyzed data by classifying the patients into treatment groups. As shown in Figure 2A, the expression of HERV env genes was the lowest in the chemotherapy only group. Even the chemotherapy and radiotherapy group showed a higher level of HERV env gene expression. These data suggest that chemotherapy has a significant effect on reducing HERV env gene expression, and radiotherapy has no effect or may up-regulate the reduced expression of HERV env genes in chemotherapy-treated patients. We also analyzed the expression pattern of HERV env genes by classifying the chemotherapy methods. As shown in Figure 2B, taxotere- or taxol-containing therapy has the maximum effect on reducing HERV env gene expression in blood of breast cancer patients. This finding suggests that taxotere or taxol may reduce the expression of HERV env genes, although further studies are needed to identify the action mechanism and physiological roles of taxol on the reduction of HERV env genes.
Figure 2.
Analysis of HERV env genes expression according to chemotherapy protocol. Effect of chemotherapy and co-treated with radiotherapy on the reduction of HERV env genes in the blood of breast cancer patients (A); Expression of HERV env genes was compared by chemotherapy method including no chemotherapy (No Treat), chemotherapy included (CTx), chemotherapy accompanying radiotherapy (CTx+RTx), and chemotherapy only (CTx only) groups. Comparing the HERV env genes reduction according to chemotherapy protocol (B). The effect of each chemotherapy protocol on the reduction of HERVs env gene in the blood of breast cancer patients was analyzed. CMF: six cycles of cyclophsphamide, methotrexate, 5-FU; FEC: six cycles of 5-FU, epirubicin, cyclophosphamide; TA: six cycles of taxotere and adriamycin. * p < 0.05, ** p < 0.01.
The relationship between HERV-derived protein and breast cancer was first identified through the finding of the expression of HERV-K-MEL env gene in various tumors including breast cancer [23]. In addition, very high titers of HERV-K (HML-2) RNA in the plasma of patients with lymphomas and breast cancer measured by either reverse transcriptase PCR or nucleic acid sequence-based amplification [18]. That study also reported that the mRNA expression of HERV-K (HML-2) decreased dramatically with cancer treatment, and the presence of HERV-K (HML-2) virus-like particles in the plasma of lymphoma patients suggests that elements of the endogenous retrovirus HERV-K (HML-2) can be found in the blood of modern-day humans with certain cancers [18]. Other studies have since been conducted that suggest that HERV-K (HML-2) env could be a novel candidate prognostic marker and immunotherapeutic target for breast cancer [19,20]. HERV-K (HML-2) reverse transcriptase also has been reported to be a breast cancer prognostic marker [25]. An RT-PCR approach demonstrated that the HERV-R (ERV3-1) env gene was expressed in several human tissues (brain, prostrate, testis, kidney, placenta, thymus, and uterus) and cancer cells (RT4, BT-474, MCF7, OVCAR-3, LOX-IMVI, and AZ521). In addition, our previous reports showed HERV-R (ERV3-1) was highly expressed in certain tumors [24]. In case of the HERV-H env gene in human tissues, the env fragments were detected in the mRNA of several tissues (placenta, skeletal muscle, spleen, and thymus) and various cancer cells (RT4, BT-474, HCT-116, TE-1, UO-31, Jurkat, HepG2, A549, MCF7, OVCAR-3, MIA-PaCa-2, PC3, LOX-IMVI, AZ521, 2F7, U-937, and C-33A) by RT-PCR analysis [26]. Transcripts of HERV-P structural genes have also be detected in various human tissues and cancer cells, suggesting a potential role in carcinogenesis [27]. Quantitative real-time RT-PCR analysis between the cancer tissues and adjacent normal tissues in env genes of HERV-R, HERV-H, HERV-K, and HERV-P indicated that high levels of expression were detected in various cancer tissues compared to the adjacent normal tissues [16]. A recent study also confirmed the potential of HERV-K (HML-2) as a diagnostic marker for detecting early-stage breast cancer by detecting HERV-K (HML-2) protein and mRNA in the serum of breast cancer patient through ELISA and real-time PCR analysis, respectively [21].
Here, we first analyzed various HERV env mRNAs including HERV-R (ERV3-1), HERV-H, HERV-K (HML-2), and HERV-P in whole blood samples from breast cancer patients and compared the mRNA expression patterns of each HERV env gene. The expressions of HERV-K (HML-2) and other HERV env genes were significantly increased in the blood of breast cancer patients and decreased in the patient groups undergoing chemotherapy. These data agree with previous studies and also suggest other HERV candidates as diagnostic markers or therapeutic targets for breast cancer.
We also analyzed the patterns of HERV env gene reduction by chemotherapy. Among chemotherapy groups including chemotherapy and radiotherapy group and a chemotherapy only group, the expressions of all HERV env genes were the lowest in the chemotherapy only group, indicating that radiotherapy has no effect or increases the expression of HERV env genes. This data agrees with our previous report showing that radiation induces HERV-R (ERV3-1) env expression by epigenetic modulation [28]. Among the chemotherapy methods, we found that taxotere- or taxol-containing therapy has the maximum effect on reducing expression of all HERV env genes. Contreras-Galindo et al. found virus-like particles in the plasma of lymphoma patients [18] and suggested HERV-K env expression comes from these viral particles. However, it is also possible that this expression comes from the contamination of debris nucleic acids from lymphocytes, circulating tumor cells, or exosomes. In this study, we collected whole blood to include whole possibilities including virus, lymphocytes, circulating tumor cells, or exosomes but excluded the possibility of genomic DNA contamination by treating DNase before constructing cDNA from purified total RNA. It is also an important issue whether the decreased expression of HERV env genes correlates with remission or simply just the chemotherapy treatment. The Meta Pre Chemo and Meta Chemo group consisted mainly of patients having recurred breast cancers, which is much more malignant than primary tumor. These groups had treated or have been treating with chemotherapy and showed the significant low level of all types of HERV env genes comparing with No Chemo group, suggesting the possibility that chemotherapy reduces the expression of HER env genes (Figure 1).
It is possible that these viruses up-regulate the expression of HERV env genes in the blood of breast cancer patients but real viral particles were not yet defined. Circulating tumor cells also can be candidates up-regulating HERVs env levels in blood but still needs direct evidences to probe it. The mechanism of reduction of HERV env genes by chemotherapy is also unclear and requires further study.
3. Experimental Section
3.1. Blood Samples of Patients
Blood samples were collected from 10 normal and 47 breast cancer patients including 7 benign tumors (Benign), 10 primary breast cancers without chemotherapy (Primary no chemo), 20 primary breast cancers undergoing chemotherapy (Primary chemo), 5 recurred and metastasized cancers before chemotherapy (Meta prechemo), and 5 metastasized cancers undergoing chemotherapy (Meta chemo). All primary and metastasized tumors were invasive ductal carcinoma. Both the collection and analysis of all samples were approved by the Institutional Review Board of Kosin University Gospel Hospital. Informed consent was obtained from each patient enrolled in the study (IRB approval number: KUCM IRB 12-003).
3.2. Real-Time PCR Analysis
We purified total RNA from whole blood which contains all possible HERV env gene RNA from particles in plasma or serum, lymphocytes, and circulating tumors. We treated blood samples with RBC lysis buffer (BioLegend, San Diego, CA, USA) to remove red blood cells and added Trizol reagent (Invitrogen, Carlsbad, CA, USA) to purify total RNA according to the manufacturer’s guidelines. All purified RNA from all samples was treated with DNase before constructing cDNA to remove genomic DNA contamination. One microgram of total RNA was subjected to reverse transcription using the AffinityScrip™ Multiple Temperature cDNA synthesis kit (Stratagene, La Jolla, CA, USA). Relative quantitative real-time PCR was performed in a Applied Biosystems 7500 Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA) using the SYBR® Green PCR Master Mix (Applied Biosystems, Carlsbad, CA, USA) according to the manufacturer’s instructions with the primer sets we used in previous study [16] and described in Table 2. All samples were performed in duplicate and related to the expression of an appropriate housekeeping gene (GAPDH), as confirmed in previous work [16].
Table 2.
Primers of HERVs env genes for realtime PCR analysis.
| Target Gene | Primers | GenBank Accession No. | |
|---|---|---|---|
| Forward | Reverse | ||
| HERV-R env | 5'-CATGGGAAGCAAGGGAACT-3' | 5'-CTTTCCCCAGCGAGCAATAC-3' | AC073210 from Chr.7q11.21 |
| HERV-H env | 5'-TTCACTCCATCCTTGGCTAT-3' | 5'-CGTCGAGTATCTACGAGCAAT-3' | AJ289711 from Chr.2q24.3 |
| HERV-K env | 5'-CACAACTAAAGAAGCTGACG-3' | 5'-CATAGGCCCAGTTGGTATAG-3' | AC074261 from Chr12q14.1 |
| HERV-P env | 5'-CAAGATTGGGTCCCCTCAC-3' | 5'-CCTATGGGGTCTTTCCCTC-3' | DQ247958 from Chr.14q32.12 |
All PCR reactions were coupled to melting-curve analysis to confirm the amplification specificity (Figure S1). Non-template controls were included for each primer pair to check for any significant levels of contaminants. Real-time PCR was performed in three independent. The mRNA expression levels of HERV env genes in breast cancer patients are presented relative to the benign patients group as control with the calculated mean values and 95% confidence intervals. Statistical significance of the differences between groups was determined using a two-tailed Student’s t-test. p values less than 0.05 were considered statistically significant.
4. Conclusions
In conclusion, various HERV genes including HERV-R (ERV3-1), HERV-H, and HERV-P env show the same pattern of regulation as HERV-K (HML-2) and may be potent diagnostic markers and therapeutic targets for breast cancer patients.
Acknowledgments
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (No. 2008-0062611).
Supplementary Files
Supplementary Information (PDF, 489 KB)
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Hayes M., Whitesell M., Brown M.A. Pathological and evolutionary implications of retroviruses as mobile genetic elements. Genes. 2013;4:573–582. doi: 10.3390/genes4040573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belshaw R., Pereira V., Katzourakis A., Talbot G., Paces J., Burt A., Tristem M. Long-term reinfection of the human genome by endogenous retroviruses. Proc. Natl. Acad. Sci. USA. 2004;101:4894–4899. doi: 10.1073/pnas.0307800101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belshaw R., Dawson A.L., Woolven-Allen J., Redding J., Burt A., Tristem M. Genomewide screening reveals high levels of insertional polymorphism in the human endogenous retrovirus family HERV-K(HML2): Implications for present-day activity. J. Virol. 2005;79:12507–12514. doi: 10.1128/JVI.79.19.12507-12514.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannert N., Kurth R. Retroelements and the human genome: New perspectives on an old relation. Proc. Natl. Acad. Sci. USA. 2004;101:14572–14579. doi: 10.1073/pnas.0404838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nelson P.N., Carnegie P.R., Martin J., Davari Ejtehadi H., Hooley P., Roden D., Rowland-Jones S., Warren P., Astley J., Murray P.G. Demystified. Human endogenous retroviruses. Mol. Pathol. 2003;56:11–18. doi: 10.1136/mp.56.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh S.K. Endogenous retroviruses: Suspects in the disease world. Future Microbiol. 2007;2:269–275. doi: 10.2217/17460913.2.3.269. [DOI] [PubMed] [Google Scholar]
- 7.Mameli G., Astone V., Arru G., Marconi S., Lovato L., Serra C., Sotgiu S., Bonetti B., Dolei A. Brains and peripheral blood mononuclear cells of multiple sclerosis (MS) patients hyperexpress MS-associated retrovirus/HERV-W endogenous retrovirus, but not Human herpesvirus 6. J. Gen. Virol. 2007;88:264–274. doi: 10.1099/vir.0.81890-0. [DOI] [PubMed] [Google Scholar]
- 8.Serra C., Mameli G., Arru G., Sotgiu S., Rosati G., Dolei A. In vitro modulation of the multiple sclerosis (MS)-associated retrovirus by cytokines: Implications for MS pathogenesis. J. Neurovirol. 2003;9:637–643. doi: 10.1080/13550280390246462. [DOI] [PubMed] [Google Scholar]
- 9.Yolken R. Viruses and schizophrenia: A focus on herpes simplex virus. Herpes. 2004;11:83A–88A. [PubMed] [Google Scholar]
- 10.Garrison K.E., Jones R.B., Meiklejohn D.A., Anwar N., Ndhlovu L.C., Chapman J.M., Erickson A.L., Agrawal A., Spotts G., Hecht F.M., et al. T cell responses to human endogenous retroviruses in HIV-1 infection. PLoS Pathog. 2007;3:e165. doi: 10.1371/journal.ppat.0030165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyd M.T., Bax C.M., Bax B.E., Bloxam D.L., Weiss R.A. The human endogenous retrovirus ERV-3 is upregulated in differentiating placental trophoblast cells. Virology. 1993;196:905–909. doi: 10.1006/viro.1993.1556. [DOI] [PubMed] [Google Scholar]
- 12.Takeuchi K., Katsumata K., Ikeda H., Minami M., Wakisaka A., Yoshiki T. Expression of endogenous retroviruses, ERV3 and lambda 4–1, in synovial tissues from patients with rheumatoid arthritis. Clin. Exp. Immunol. 1995;99:338–344. doi: 10.1111/j.1365-2249.1995.tb05555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsson E., Venables P.J., Andersson A.C., Fan W., Rigby S., Botling J., Oberg F., Cohen M., Nilsson K. Expression of the endogenous retrovirus ERV3 (HERV-R) during induced monocytic differentiation in the U-937 cell line. Int. J. Cancer. 1996;67:451–456. doi: 10.1002/(SICI)1097-0215(19960729)67:3<451::AID-IJC23>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 14.Andersson A.C., Venables P.J., Tonjes R.R., Scherer J., Eriksson L., Larsson E. Developmental expression of HERV-R (ERV3) and HERV-K in human tissue. Virology. 2002;297:220–225. doi: 10.1006/viro.2002.1428. [DOI] [PubMed] [Google Scholar]
- 15.Andersson A.C., Yun Z., Sperber G.O., Larsson E., Blomberg J. ERV3 and related sequences in humans: Structure and RNA expression. J. Virol. 2005;79:9270–9284. doi: 10.1128/JVI.79.14.9270-9284.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahn K., Kim H.S. Structural and quantitative expression analyses of HERV gene family in human tissues. Mol. Cells. 2009;28:99–103. doi: 10.1007/s10059-009-0107-y. [DOI] [PubMed] [Google Scholar]
- 17.Wang-Johanning F., Liu J., Rycaj K., Huang M., Tsai K., Rosen D.G., Chen D.T., Lu D.W., Barnhart K.F., Johanning G.L. Expression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer. Int. J. Cancer. 2007;120:81–90. doi: 10.1002/ijc.22256. [DOI] [PubMed] [Google Scholar]
- 18.Contreras-Galindo R., Kaplan M.H., Leissner P., Verjat T., Ferlenghi I., Bagnoli F., Giusti F., Dosik M.H., Hayes D.F., Gitlin S.D., et al. Human endogenous retrovirus K (HML-2) elements in the plasma of people with lymphoma and breast cancer. J. Virol. 2008;82:9329–9336. doi: 10.1128/JVI.00646-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao J., Rycaj K., Geng S., Li M., Plummer J.B., Yin B., Liu H., Xu X., Zhang Y., Yan Y., et al. Expression of human endogenous retrovirus type K envelope protein is a novel candidate prognostic marker for human breast cancer. Genes Cancer. 2011;2:914–922. doi: 10.1177/1947601911431841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang-Johanning F., Rycaj K., Plummer J.B., Li M., Yin B., Frerich K., Garza J.G., Shen J., Lin K., Yan P., et al. Immunotherapeutic potential of anti-human endogenous retrovirus-K envelope protein antibodies in targeting breast tumors. J. Natl. Cancer Inst. 2012;104:189–210. doi: 10.1093/jnci/djr540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang-Johanning F., Li M., Esteva F.J., Hess K.R., Yin B., Rycaj K., Plummer J.B., Garza J.G., Ambs S., Johanning G.L. Human endogenous retrovirus type K antibodies and mRNA as serum biomarkers of early-stage breast cancer. Int. J. Cancer. 2014;134:587–595. doi: 10.1002/ijc.28389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cegolon L., Salata C., Weiderpass E., Vineis P., Palu G., Mastrangelo G. Human endogenous retroviruses and cancer prevention: Evidence and prospects. BMC Cancer. 2013;13:4. doi: 10.1186/1471-2407-13-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schiavetti F., Thonnard J., Colau D., Boon T., Coulie P.G. A human endogenous retroviral sequence encoding an antigen recognized on melanoma by cytolytic T lymphocytes. Cancer Res. 2002;62:5510–5516. [PubMed] [Google Scholar]
- 24.Kang Y.J., Jo J.O., Ock M.S., Chang H.K., Baek K.W., Lee J.R., Choi Y.H., Kim W.J., Leem S.H., Kim H.S., et al. Human ERV3–1 env protein expression in various human tissues and tumours. J. Clin. Pathol. 2014;67:86–90. doi: 10.1136/jclinpath-2013-201841. [DOI] [PubMed] [Google Scholar]
- 25.Golan M., Hizi A., Resau J.H., Yaal-Hahoshen N., Reichman H., Keydar I., Tsarfaty I. Human endogenous retrovirus (HERV-K) reverse transcriptase as a breast cancer prognostic marker. Neoplasia. 2008;10:521–533. doi: 10.1593/neo.07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yi J.M., Kim H.M., Kim H.S. Human endogenous retrovirus HERV-H family in human tissues and cancer cells: Expression, identification, and phylogeny. Cancer Lett. 2006;231:228–239. doi: 10.1016/j.canlet.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 27.Yi J.M., Schuebel K., Kim H.S. Molecular genetic analyses of human endogenous retroviral elements belonging to the HERV-P family in primates, human tissues, and cancer cells. Genomics. 2007;89:1–9. doi: 10.1016/j.ygeno.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 28.Lee J.R., Ahn K., Kim Y.J., Jung Y.D., Kim H.S. Radiation-induced human endogenous retrovirus (HERV)-R env gene expression by epigenetic control. Radiat. Res. 2012;178:379–384. doi: 10.1667/RR2888.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information (PDF, 489 KB)