Abstract
Background
The Radiation Therapy Oncology Group 98-11 clinical trial demonstrated the superiority of standard 5FU/mitomycin-C over 5FU/cisplatin in combination with radiation in the treatment of anal squamous cell cancer. Tumor size (>5cm) and lymph node metastases are associated with disease progression. There may be key molecular differences (e.g. DNA methylation changes) in tumors at high-risk for progression.
Objectives
The objectives of this study were to determine if there are differences in DNA methylation at individual CpG sites and within genes among locally advanced anal cancers, with large tumor size and/or nodal involvement, compared to those that are less advanced.
Design
Case-case study among 121 patients defined as high-risk (tumor size>5cm and/or nodal involvement; n=59) or low-risk (≤5cm, node negative; n=62) within the mitomycin-C arm of RTOG98-11 trial. DNA methylation was measured using the Illumina HumanMethylation450 Array.
Settings
Tertiary care cancer center in collaboration with a national clinical trials cooperative group.
Patients
The patients consisted of 74 women and 47 men with a median age of 54 years (minmax 25-79).
Main Outcome Measures
DNA methylation differences at individual CpG sites and within genes between low and high-risk patients were compared using Mann-Whitney test (p-value<0.001).
Results
A total of 16 CpG loci were differentially methylated (14 increased and 2 decreased) in high vs. low-risk cases. Genes harboring differentially methylated CpG sites included known tumor suppressor genes and novel targets.
Limitations
This study only included patients in mitomycin-C arm with tumor tissue; however, this sample was representative of the trial.
Conclusions
This is the first study to apply genome-wide methylation analysis to anal cancer. Biologically relevant differences in methylated targets were found to discriminate locally advanced from early anal cancer. Epigenetic events likely play a significant role in the progression of anal cancer and may serve as potential biomarkers.
Keywords: anal cancer, methylation, genome-wide array, epigenetic, locally advanced anal cancer
INTRODUCTION
Anal cancer accounts for 4% of all lower gastrointestinal tract malignancies in the United States1 and the incidence of anal cancer continues to rise by 2.6% per year.2 In fact, the incidence rates of anal cancer have increased significantly in the past 30 years, jumping 160% in men and 78% in women. 3,4 The overall 5-year survival rate for anal cancer is 65.6%; however, this varies considerably by stage of diagnosis (80% for local disease; 60% for regional disease and 31% for distant disease at diagnosis).5
Radiation, 5-fluorouracil (5-FU), and mitomycin-C have remained the components of standard combined modality therapy over the last several decades despite the completion of several clinical trials.6-9 Of these, the Radiation Therapy Oncology Group (RTOG 98-11) evaluated two chemoradiation regimens for the treatment of anal canal carcinoma (standard mitomycin-based regimen versus an experimental cisplatin-based regimen). This trial included 644 patients and reported a significantly better 5-year disease-free survival in the mitomycin-C arm versus cisplatin arm (68% vs. 58%, p=0.006).6,7 While this treatment is effective, there are significantly associated morbidities and alternative dosing or novel targeted treatments are needed to reduce morbidity and improve outcomes for some patients.
Tumor size and nodal status are strong clinical prognostic factors for anal cancer.10 Within RTOG 98-11, patients with more advanced cancer at diagnosis, e.g. large tumors (>5cm diameter) and/or positive lymph nodes, had poorer disease-free survival outcomes than those with less advanced disease.7 However, both early and locoregionally advanced tumors have heterogeneous outcomes. We hypothesized that there are underlying biological differences between larger size and/or node-positive tumors that put tumors at high risk of progressing and not responding to treatment. These differences may explain some of the variability in patient outcomes given the same treatment.
One such biological alteration important to carcinogenesis is DNA methylation, an epigenetic modification.11,12 Epigenetic alterations encompass changes in chromatin structure, histone modification, and DNA methylation and play an important role in gene expression. Methylation of DNA occurs specifically at discrete sites termed CpG dinucleotides, where a cytosine (C) precedes a guanine (G). These CpG sites or loci generally reside in clusters known as CpG islands and are often associated with gene promoter regions.12 Dense DNA methylation in CpG islands can result in the epigenetic silencing of tumor suppressor genes and are an important part of the process of carcinogenesis. 13 Epigenetic gene silencing has been demonstrated in HPV-associated cervical cancer and commonly occurred in pathways that important in the carcinogenesis.13,14 For example, aberrant activity of the well described oncogenic Wnt/β-catenin pathway is prominent in numerous cancer types and genes encoding several key regulators of this pathway, such as CDH1, APC and WIF1, are frequently silenced via dense methylation of CpG islands in cervical cancer.15-17
Although likely important, the role of DNA methylation in the development of anal cancer remains very poorly studied. 18,19 Zhang et al. provided the first evidence of aberrant methylation in anal cancer among 11 candidate genes compared to normal tissue.18 Subsequently using an array-based assay analyzing >1500 CpG sites, we observed differences in DNA methylation patterns in 20 genes in the progression from normal anal mucosa, carcinoma in situ, to invasive anal carcinoma in a small set of cases.19 To date, a broad high-throughput characterization of methylation events in advanced anal cancer is clearly lacking. The objectives of this study were to determine if there are differences in DNA methylation at individual CpG sites and clusters within genes among locally advanced anal cancers, with large tumor size and/or nodal involvement, compared to those that are less advanced.
MATERIAL AND METHODS
RTOG 98-11 Trial
As reported,6,7 this US Gastrointestinal Intergroup trial RTOG 98-11 evaluated combinations of external beam irradiation (XRT) plus chemotherapy in a 2-arm phase III randomized trial comparing XRT plus concurrent 5-FU and mitomycin-C with induction 5-FU+cisplatin followed by concurrent XRT plus 5-FU+cisplatin.6 Patients were excluded if their primary diagnosis was T1 or M1, severe comorbid conditions (including AIDS), or prior malignancy within the last 5 years.6 Randomization was stratified by gender, clinical nodal status (positive or negative), and size of the primary tumor (2-5cm or >5cm). For anal cancer, clinical lymph node staging was conducted by physical examination and radiological imaging. All patients enrolled in RTOG 98-11 signed an IRB approved informed consent form. The use of de-identified tissues was IRB-approved by the University of South Florida IRB. This study includes patients from the Mitomycin-C arm of RTOG 98-11 that had archived tumor tissue available (N=186).
RTOG Tissues and DNA extraction
Archived formalin fixed paraffin embedded (FFPE) tumors were macrodissected and genomic DNA was isolated using the QIAamp DNA FFPE Tissue Kit (QIAGEN, Valencia, CA). DNA quality was evaluated using the qPCR Illumina FFPE QC kit (Illumina, San Diego, CA) in triplicate. Samples with sufficient DNA (≥250ng) that met criteria for inclusion by the QC assay (Illumina, San Diego, CA) were included.
Bisulfite Modification and Infinium Methylation Analysis
DNA methylation cannot be measured directly and requires treatment with sodium bisulfite which converts all unmethylated cytosines to uracil, while methylated cytosines are not altered. Thus, genomic DNA is first sodium bisulfite-modified using the EZ DNA Methylation kit (Zymo Research, Orange, CA). Bisulfite-modified DNA was prepared using the Infinium HD FFPE DNA Restore kit (Illumina, San Diego, CA). Methylation was interrogated using the Infinium HumanMethylation450K BeadChip following manufactures specifications throughout the whole-genome amplification, fragmentation, hybridization, base extension, counterstaining and scanning). A Tecan Liquid Handling robot with the Te-Flow apparatus was used for the single base extension and staining, and the chips were scanned on a single HiScanSQ System (Illumina Inc.). The Infinium HumanMethylation450K BeadChip incorporates both Infinium I (methylated and unmethylated beads per CpG locus) and Infinium II assays (one bead type with the methylated state determined at the single base extension step after hybridization) to evaluate DNA methylation status at 485,512 CpG loci, which covers 99% of annotated genes and 96% of defined CpG islands.20,21
Statistical Analysis
Patients were classified as low-risk with tumor diameter of less than 5 cm and node negative and advanced high-risk with tumor diameter of more than 5 cm and/or node positive). Patients were also grouped individually by tumor size (≤5cm vs. >5cm) or nodal status (negative vs. positive). Differences in patient characteristics were determined by chi-square, Fisher's exact, or t-test. Overall survival was defined as the time from randomization to death due to any cause. Disease-free survival was defined as the time from randomization to local, regional, or distant failure, second primary tumor, or death due to any cause.6 Differences in survival by clinical risk group were estimated univariately with the Kaplan-Meier method and the clinical risk groups were compared using the log-rank test.
Methylated CpG loci were defined on the Infinium array as a β-value, which was calculated for the 485,512 CpG-loci from unmethylated (U) and methylated (M) signal [M / (U+M+100)] and assigned a range between 0 and 1 (unmethylated to 100% methylated). β-values with a corresponding detection p-value>0.05 were set as missing. Methylation data were pre-processed using the R statistical software package and Bioconductor packages methylumi and wateRmelon.22 β-values were normalized using the normalizeViaControls function in the methylumi package. Chip-wide controls and Multi-Dimensional Scaling plots were used to visualize data quality. β-values were analyzed as continuous variables. To quantitate differential methylation between high and low-risk groups, we calculated a delta β = β-valuehigh – β-valuelow. A threshold of delta β>0.1 was required to identify potentially meaningful biologic changes in methylation. Differential methylation between groups was analyzed using Mann-Whitney test and students t-test at level of significance of p-value<0.001. Differential methylation across regions of DNA required ≥5 CpG loci that were significantly different per annotated gene at a p-value<0.05 by Mann-Whitney or T-test. Statistical analysis was performed using the R statistical software package, and MATLAB packages.
RESULTS
Patient Characteristics
We identified 186 patients from RTOG 98-11 randomized to the Mitomycin-C arm with archived tumor tissue available for this study. A total of 121 cases had sufficient DNA for methylation analysis. There were no differences in tumor characteristics and outcomes between patients eligible for the methylation analysis to those in the Mitomycin-C arm that were not included, therefore the sample of 121 anal cancer patients was representative of patients randomized to the Mitomycin-C arm of RTOG 98-11 (data not shown). The median age of the 121 patients was 54 (range 25-79; Table 1). A majority of the patients were women (61%), Caucasian (87%), and highly functional (95% had Karnofsky Performance Status ≥ 80). Sixty-two patients (51%) were classified as low-risk and 59 (49%) as high-risk. There were 88 (73%) and 33 (27%) patients with tumor sizes of ≤ 5cm and >5cm, respectively. Nodal status was determined as node negative (N0) in 85 (70%) patients and node positive (N+) in 36 (30%) patients. There were no statistically significant differences in distribution of gender or age across clinical risk groups (Table 1).
Table 1.
Characteristics of low- and high-risk clinical groups with anal cancer
| Low Riska (n=62) | High Risk (n=59) | Total (n=121) | p-valueb | ||||
|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | ||
| Median Age (min-max) | 54 (37-78) | 53 (25-79) | 54 (25-79) | ||||
| Mean age (SD) | 55.5 (9.7) | 54.4 (10.4) | 55.0 (10.0) | 0.54 | |||
| Gender | |||||||
| Male | 25 | 40% | 22 | 37% | 47 | 39% | 0.73 |
| Female | 37 | 60% | 37 | 63% | 74 | 61% | |
| Race | |||||||
| White | 52 | 84% | 53 | 90% | 105 | 87% | 0.33 |
| African American/Other | 10 | 16% | 6 | 10% | 16 | 13% | |
| KPS | |||||||
| 60-70 | 3 | 5% | 3 | 5% | 6 | 5% | 1.00 |
| 80-100 | 59 | 95% | 56 | 95% | 115 | 95% | |
| Mean tumor size (SD) | 3.5 (0.9) | 5.6 (2.3) | 4.5 (2.0) | --c | |||
| T-Stage | |||||||
| T2 | 60 | 97% | 24 | 41% | 84 | 69% | --c |
| T3/T4 | 2 | 3% | 35 | 59% | 37 | 31% | |
| N-Stage | |||||||
| N0 | 62 | 100% | 23 | 39% | 85 | 70% | --c |
| N1 | 0 | 0% | 11 | 19% | 11 | 9% | |
| N2 | 0 | 0% | 14 | 24% | 14 | 12% | |
| N3 | 0 | 0% | 5 | 8% | 5 | 4% | |
| Nx | 0 | 0% | 6 | 10% | 6 | 5% | |
Abbreviations: min, minimum; max, maximum; SD, standard deviation, KPS, Karnofsky Performance Status
Low-risk = tumors ≤5 cm and N0; High-risk= tumors >5 cm and/or N+
Statistical differences in patient characteristics between the two groups were determined by Chi-square test, Fisher's exact test, or t-test.
Testing not applicable for tumor size and N-stage, as those variables define the risk groups. Similarly for T-stage, which is highly associated with tumor size by AJCC 1997 version definition.
Risk groups associated with patient outcomes
The classification of risk groups as low- and high-risk was confirmed by examining the relationship between clinical risk group and outcome (Figure 1). The 5-year disease-free survival rates were 89% for low-risk and 49% for the high-risk group (p-value<0.0001, Figure 1A). The 5-year overall survival rates were 92% vs. 64% for low- and high-risk groups, respectively (p-value=0.0003, Figure 1B). Similar differences in survival were observed when considering tumor size or nodal status individually (data not shown). These data confirm that within the 121 cases, the risk groups were accurately classified and are representative of the entire Mitomycin-C arm of RTOG 98-11.
Figure 1.
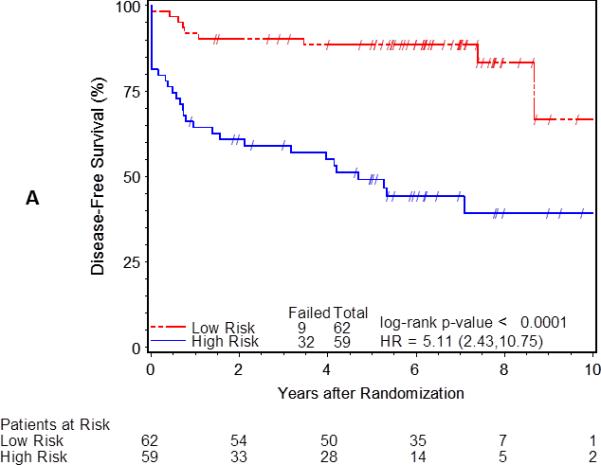
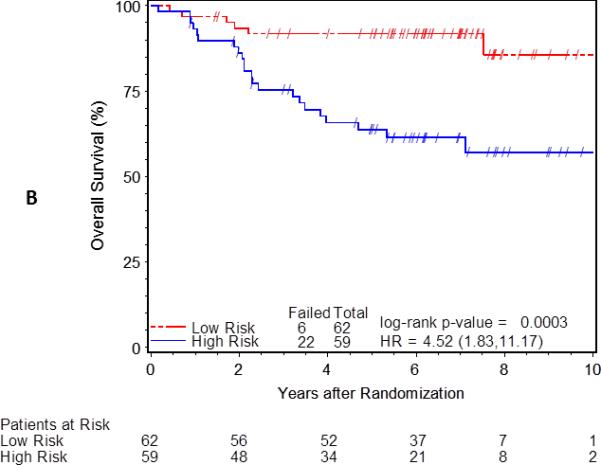
Kaplan-Meier estimates of disease-free (A) and overall (B) survival by clinically low (N=62) and high (N=59) risk group. Differences in probability of disease-free survival (DFS) or overall survival (OS) by low (red) and high-risk (blue) groups tested using the log-rank test and hazard ratio (HR) estimated using cox-proportional hazards models.
Differentially methylated CpG sites by risk group
At a p-value<0.001, 16 CpG loci were differentially methylated between low- and high-risk groups (Table 2). Of these, 14 loci had increased methylation in high-risk tumors. These 16 CpG sites were located in 7 defined genes and one uncharacterized genomic region (See Online Table 1 for gene details). The individual contribution of tumor size and nodal status on methylation differences was also examined. A total of 68 CpG loci from 59 genes were differentially methylated in large tumors (Table 3). There were 61 sites with increased methylation levels within large tumors and 7 sites with decreased methylation. When considering nodal status, 8 CpG loci (5 increased and 3 decreased methylation) from 6 unique genomic regions were differentially methylated in tumors with nodal involvement (data not shown). There was no overlap in individual differentially methylated CpG sites between tumor size and nodal status. Only 2 genes (3.5%) identified in the tumor size analysis were also significantly different by clinical risk group (Table 2 and 3) and 2 genes (33%) from the nodal analysis overlapped with risk group findings (data not shown).
Table 2.
Differentially Methylated CpG Loci in Clinically defined Low and High-risk anal cancers
| Symbol | Product | Association with Disease | CpG Numbera | Median Beta value | p-valuec | |||
|---|---|---|---|---|---|---|---|---|
| Low Risk | High Risk | Difb | MW | T-Test | ||||
| ADAT3d | Adenosine deaminase, tRNA-specific 3 | Not reported as target of methylation or involved in cancer | cg20243900 | 0.64 | 0.75 | 0.11 | 5.48E-04 | 2.01E-04 |
| GSG1L | Germ Cell Associated 1 (GSG1)-Like | Possible susceptibility locus for colon cancer in AOM mouse models; Not reported as target of methylation. | cg09528825 | 0.07 | 0.18 | 0.11 | 3.12E-02 | 8.15E-04 |
| KIAA0319 | KIAA0319 | Dyslexia-associated gene, Not reported as target of methylation or involved in cancer | cg18428180 | 0.24 | 0.09 | −0.15 | 1.29E-03 | 4.87E-04 |
| LOC-728392 | LOC728392 | Not reported as target of methylation or involved in cancer | cg18671949 | 0.11 | 0.40 | 0.29 | 4.96E-05 | 9.84E-06 |
| cg06462347 | 0.24 | 0.53 | 0.28 | 1.43E-04 | 5.17E-05 | |||
| cg27230784 | 0.14 | 0.51 | 0.37 | 4.44E-05 | 4.58E-05 | |||
| cg22298430 | 0.15 | 0.47 | 0.32 | 1.49E-04 | 7.13E-05 | |||
| cg01433610 | 0.38 | 0.52 | 0.15 | 1.66E-03 | 8.04E-04 | |||
| PARD3 | Partitioning defective 3 homolog | Lost in prostate cancer23, involved in cell motility; Associated with WNT/β-catenin pathway24; Not reported as a target of methylation. | cg26795540 | 0.48 | 0.58 | 0.10 | 7.57E-04 | 1.60E-03 |
| SALL3 | SAL-Like 3 | Methylated in HCC25; interacts with DNMT3A to reduce methylation | cg13794993 | 0.22 | 0.38 | 0.17 | 6.89E-04 | 4.15E-04 |
| cg14007067 | 0.14 | 0.31 | 0.17 | 1.27E-03 | 8.36E-04 | |||
| cg06954658 | 0.16 | 0.27 | 0.11 | 1.47E-03 | 4.78E-04 | |||
| cg16433156 | 0.14 | 0.25 | 0.11 | 1.34E-03 | 5.46E-04 | |||
| cg05080154 | 0.11 | 0.29 | 0.18 | 2.36E-03 | 9.22E-04 | |||
| SFRP2 | secreted frizzled-related protein 2 | Methylated in several cancers, including cervical26; head and neck27, and prostate28 | cg22178613 | 0.16 | 0.38 | 0.22 | 1.10E-03 | 8.78E-04 |
| ST6GAL2 | ST6 beta-galactosamide alpha-2,6-sialyltranferase 2 | Upregulated in breast cancer29. Not reported as target of methylation. | cg13168617 | 0.72 | 0.57 | −0.15 | 1.75E-03 | 8.25E-04 |
| SCAMP4d | Secretory Carrier Membrane Protein 4 | Not reported as target of methylation or involved in cancer | cg20243900 | 0.64 | 0.75 | 0.11 | 5.48E-04 | 2.01E-04 |
CpG number is the unique identifier for the loci provided by Illumina; multiple loci within a differentially methylated region listed in descending order according to genomic position.
Difference in median β-value between high and low risk tumors
Statistical differences in β-values between the two groups were determined by Mann-Whitney or student's t-test
CpG loci located in a genomic sequence that overlaps for ADAT3 and SCAMP4 genes.
Table 3.
Annotation of genes with differentially methylated CpG Loci by diameter of primary tumor (≤ 5 cm vs. >5 cm)
| Symbol | Product | Association with Disease | CpG Numbera | Beta value | p-valuec | |||
|---|---|---|---|---|---|---|---|---|
| ≤5 cm | >5 cm | Dif.b | MW | T-Test | ||||
| ADAT3d | Adenosine deaminase, tRNA-specific 3 | Not reported as target of methylation or involved in cancer | cg01397065 | 0.57 | 0.74 | 0.16 | 2.75E-03 | 2.30E-04 |
| AGFG2 | ArfGAP with FG repeats 2 | Associated with HIV infection; Not reported as target of methylation or involved in cancer | cg03431524 | 0.43 | 0.55 | 0.12 | 6.56E-04 | 1.06E-03 |
| ALDH1L2 | Aldehyde dehydrogenase 1 family, member L2 | Methylated in alcoholism30. Not reported as involved in cancer | cg16527105 | 0.67 | 0.83 | 0.16 | 9.98E-04 | 9.81E-03 |
| ASPSCR1 | Alveolar soft part sarcoma (ASPS) chromosome region, 1 | Translocated in ASPS and renal cell carcinoma; Not reported as target of methylation. | cg11511084 | 0.58 | 0.71 | 0.13 | 4.85E-04 | 5.96E-04 |
| BDNFOS | Brain derived neurotrophic factor (BDNF) antisense RNA | Not reported as target of methylation or involved in cancer | cg23330212 | 0.59 | 0.42 | −0.16 | 8.81E-04 | 4.95E-04 |
| BLVRA | Biliverdin reductase A | Altered expression in HCC31, renal32 and vaginal33 cancer. Upregulated after p53 loss34 and chemotherapy35. Not reported as a target for methylation. | cg14579118 | 0.62 | 0.74 | 0.11 | 1.87E-03 | 6.39E-04 |
| BRE | Brain and reproductive organ-expressed | TNFRSF1A modulator36; Up-regulated in HCC and esophageal cancer; Not reported as target of methylation | cg13861527 | 0.60 | 0.77 | 0.18 | 3.98E-04 | 2.05E-04 |
| C2orf74 | Chromosome 2 open reading frame 74 | Not reported as target of methylation or involved in cancer | cg01648237 | 0.32 | 0.16 | −0.16 | 9.18E-04 | 1.82E-03 |
| C6orf136 | Chromosome 6 open reading frame 136 | Not reported as target of methylation or involved in cancer | cg13016528 | 0.53 | 0.64 | 0.11 | 5.29E-04 | 3.60E-04 |
| CCDC63 | Coiled-coil domain containing 63 | Not reported as target of methylation or involved in cancer | cg02855409 | 0.24 | 0.37 | 0.13 | 4.31E-03 | 8.76E-04 |
| cg10995082 | 0.32 | 0.50 | 0.18 | 8.03E-03 | 4.22E-04 | |||
| cg19006003 | 0.33 | 0.48 | 0.15 | 2.86E-03 | 3.25E-05 | |||
| CDK6 | Cyclin-dependent kinase 6 | Upregulated in lymphoma, leukemia, and melanoma37; inactivation in HPV+ cervical cells critical38; Not reported as a target for methylation. | cg23628117 | 0.71 | 0.83 | 0.12 | 7.60E-04 | 1.51E-03 |
| COX8A | Cytochrome c oxidase subunit VIIIA (ubiquitous) | Related to tumor progression and chemo-resistance in glioma39, breast40 and esophageal41 cancers; loss of COX subunits increases chemo-sensitivity41; HIV inhibits COX activity42; Not reported as a target of methylation. | cg17292384 | 0.60 | 0.72 | 0.12 | 3.56E-04 | 1.09E-04 |
| CYP27C1 | Cytochrome P450, family 27, subfamily C, 1 | Uncharacterized; Not reported as target of methylation or involved in cancer. | cg08022717 | 0.63 | 0.76 | 0.13 | 1.38E-03 | 5.10E-04 |
| DMRT3 | Doublesex and MAB-3 related transcription factor 3 | Silenced or lost in lung SCC43. Not reported as target of methylation. | cg14176274 | 0.27 | 0.39 | 0.12 | 2.60E-04 | 4.45E-04 |
| EBF3 | Early B-cell factor 3 | Methylated in pancreatic44, gastric45; and HPV+ head and neck SCC46 | cg04804618 | 0.16 | 0.34 | 0.18 | 8.81E-04 | 1.59E-03 |
| cg14737286 | 0.28 | 0.43 | 0.14 | 1.41E-03 | 8.06E-04 | |||
| FAM150A | Family With Sequence Similarity 150, Member A | Increased methylation by Infinium in more aggressive renal cell cancers47 | cg07076175 | 0.45 | 0.60 | 0.15 | 1.53E-04 | 1.14E-04 |
| cg10094616 | 0.17 | 0.42 | 0.24 | 2.91E-04 | 1.43E-04 | |||
| cg22862746 | 0.18 | 0.36 | 0.18 | 6.56E-04 | 9.57E-04 | |||
| cg09442654 | 0.36 | 0.61 | 0.25 | 6.70E-04 | 1.62E-04 | |||
| FOS | FBJ murine osteosarcoma viral oncogene homolog | Oncogene; AP-1 activates transcription and methylation of TSG and HPV48; Induces EMT | cg23404711 | 0.52 | 0.63 | 0.11 | 5.40E-04 | 5.38E-04 |
| FZD10 | Frizzled family receptor 10 | Member of WNT/β-catenin signaling pathway; Not reported as target of methylation | cg13859208 | 0.49 | 0.61 | 0.12 | 4.92E-05 | 2.07E-06 |
| GET4 | Golgi to ER traffic protein 4 homolog (S. cerevisiae) | Associated with ubiquitination and ER; Not reported as target of methylation or involved in cancer | cg24580076 | 0.46 | 0.66 | 0.20 | 1.46E-04 | 9.10E-05 |
| GSG1L | Germ Cell Associated 1 (GSG1)-Like | Possible susceptibility locus for colon cancer in AOM mouse models; Not reported as a target of methylation. | cg03921753 | 0.15 | 0.30 | 0.15 | 5.76E-04 | 8.10E-04 |
| cg09528825 | 0.07 | 0.37 | 0.29 | 1.38E-03 | 3.35E-04 | |||
| cg03394150 | 0.08 | 0.29 | 0.21 | 2.31E-03 | 8.97E-04 | |||
| GULP1 | Engulfment adaptor PTB domain 1 | Possible TSG; inhibits TGF-B induced growth | cg19202813 | 0.72 | 0.57 | −0.15 | 5.52E-04 | 1.02E-03 |
| HDAC2 | histone deacetylase 2 | Overexpressed in several cancers, including prostate49 and ovarian50 cancer; regulator of DNA methylation. Not reported as a target of methylation. | cg15069235 | 0.55 | 0.69 | 0.15 | 6.28E-04 | 5.73E-04 |
| HORMAD 2 | HORMA domain containing 2 | Novel cancer/testis gene; expressed in lung cancer (Chinese)51. Not reported as a target of methylation. | cg24211826 | 0.59 | 0.71 | 0.12 | 5.40E-04 | 7.70E-04 |
| HOXA6 | homeobox A6 | Methylated in meningioma52 and lymphoid malignancies53. | cg23129930 | 0.45 | 0.60 | 0.15 | 5.89E-04 | 9.02E-04 |
| HS6ST1 | heparan sulfate 6-O-sulfotransferase 1 | HPV binds heparan sulfate protoglycans for cell entry54; activity reported in ovarian cells55; Not reported as target for methylation. | cg08472795 | 0.44 | 0.58 | 0.14 | 5.64E-04 | 5.79E-04 |
| KIR3DX1 | killer cell immunoglobulin-like receptor, three domains, X1 | Regulated by DNA methylation56. KIR3DX1 not reported to be a target for methylation or involved in cancer. | cg10731960 | 0.64 | 0.77 | 0.13 | 1.02E-03 | 4.58E-04 |
| KLHL29 | Kelch-like family member 29 | Not reported as target of methylation or involved in cancer | cg00537210 | 0.44 | 0.58 | 0.14 | 7.45E-04 | 1.27E-03 |
| MECOM | MDS1 and EVI1 complex locus | Amplification early event in several cancers, including ovary, head and neck, and cervical cancers. Not reported as a target of methylation | cg20528780 | 0.51 | 0.40 | −0.12 | 2.85E-04 | 7.94E-04 |
| MFI2 | Melanotransferrin | Expressed in melanoma57 and other cancers; Not reported as target of methylation | cg00477017 | 0.38 | 0.48 | 0.10 | 4.54E-04 | 8.00E-04 |
| MIR200B | MicroRNA 200b | Regulated by DNA methylation; epigenetic silencing promotes EMT58; Altered in oral59 and head and neck60. | cg14161399 | 0.77 | 0.91 | 0.14 | 1.87E-03 | 1.19E-05 |
| MIR200Ad | MicroRNA 200a | cg02825344 | 0.60 | 0.72 | 0.12 | 2.75E-03 | 7.00E-04 | |
| MMP9 | Matrix metallopeptidase 9 | Tumor-associated tissue remodeling and metastasis. Not reported as target of methylation. | cg17664577 | 0.26 | 0.46 | 0.19 | 7.77E-04 | 8.27E-04 |
| MRPS22 | Mitochondrial ribosomal protein S22 | Overexpressed in breast cancer61; Not reported as a target of methylation. | cg11277156 | 0.66 | 0.76 | 0.10 | 2.36E-05 | 1.09E-06 |
| MSI2 | Musashi RNA-binding protein 2 | Overexpressed in leukemia62 and medulloblastoma63; downstream regulator of WNT-related modulation of p2164. Not reported as target of methylation | cg04486382 | 0.64 | 0.84 | 0.20 | 6.99E-04 | 1.54E-03 |
| cg19810715 | 0.64 | 0.82 | 0.18 | 4.44E-04 | 3.26E-03 | |||
| NPTX1 | Neuronal pentraxin I | Methylated in cancer, including colon65 and cervical cancer66,67. | cg20853771 | 0.68 | 0.79 | 0.11 | 2.18E-03 | 2.43E-04 |
| OR2C3 | Olfactory receptor, family 2, subfamily C, 3 | Not reported as target of methylation or involved in cancer | cg20320823 | 0.56 | 0.69 | 0.12 | 6.99E-04 | 4.33E-04 |
| PAK1 | p21 protein-activated kinase 1 | Upregulated in several cancers68,69; involved in WNT/β-catenin and RAS pathways; Increased cell motility; interaction with HPV70 | cg24536703 | 0.62 | 0.72 | 0.11 | 3.98E-04 | 6.91E-05 |
| PDE3Bd | Phosphodiesterase 3B, | Pro-apoptotic in CML71; associated with cisplatin resistance in head and neck SCC72 | cg21901307 | 0.24 | 0.35 | 0.11 | 2.72E-04 | 1.36E-03 |
| PLA2G2A | Phospholipase A2, group IIA | Regulated by WNT/β-catenin pathway; expression associated with improved survival in gastric73 and esophageal74 cancers | cg13211559 | 0.72 | 0.83 | 0.12 | 3.98E-04 | 1.88E-03 |
| PLEKHG1 | Pleckstrin homology domain containing, family G member 1 | Oncogene; activation leads to aberrant Rho GTPase targets; Not reported as a target for methylation. | cg26852242 | 0.66 | 0.80 | 0.14 | 8.27E-04 | 4.47E-04 |
| PLEKHG3 | Pleckstrin homology domain containing, family G member 3 | Oncogene; activation leads to aberrant Rho GTPase targets; Not reported as a target for methylation. | cg11802553 | 0.66 | 0.77 | 0.11 | 2.43E-04 | 3.02E-04 |
| PON3 | Paraoxonase 3 | Imprinted in mouse genome; Over-expressed in several cancers75; anti-apoptotic | cg04080282 | 0.48 | 0.37 | −0.10 | 4.39E-03 | 7.39E-04 |
| cg08898155 | 0.41 | 0.24 | −0.16 | 3.20E-03 | 5.54E-04 | |||
| cg11435506 | 0.33 | 0.16 | −0.17 | 2.91E-03 | 2.10E-04 | |||
| PPP1R9A | Protein phosphatase 1, regulatory subunit 9 | Imprinted gene; Not reported to be involved in cancer. | cg09724492 | 0.38 | 0.52 | 0.14 | 4.47E-03 | 5.00E-04 |
| RERE | Arginine-glutamic acid dipeptide (RE) repeats | Expression reduced in neuroblastoma and leukemia76. Not reported to be a target of methylation. | cg19679865 | 0.62 | 0.75 | 0.13 | 3.64E-04 | 2.97E-04 |
| PSMA1d | Proteasome subunit, alpha type, 1 | Mediates bortezomib resistance in AML77; may promote tumor growth; loss radio-sensitized NSCLCs. Not reported as a target for methylation. | cg21901307 | 0.24 | 0.35 | 0.11 | 2.72E-04 | 1.36E-03 |
| RNASEN | Ribonuclease type III, nuclear | Regulates cell proliferation; increased copy number in cervical cancer78 | cg04590036 | 0.68 | 0.82 | 0.14 | 4.44E-04 | 5.74E-04 |
| RREB1 | Ras responsive element binding protein 1 | Tumor marker in melanoma; downregulated in pancreatic cancer79; regulated by miRNA80 | cg03137792 | 0.62 | 0.83 | 0.21 | 1.28E-03 | 7.00E-04 |
| SCAMP4d | Secretory Carrier Membrane Protein 4 | Not reported as target of methylation or involved in cancer | cg01397065 | 0.57 | 0.74 | 0.16 | 2.75E-03 | 2.30E-04 |
| SEPP1 | Selenoprotein P 1 | SNPs in prostate81 and esophageal risk82; mercury induced hypomethylation83. | cg08626131 | 0.51 | 0.66 | 0.15 | 2.79E-04 | 2.32E-04 |
| SLC9A3 | Solute carrier family 9, subfamily A, member 3 | Downregulated in Ulcerative Colitis84; Not a reported target for methylation. | cg06058576 | 0.68 | 0.91 | 0.23 | 1.98E-04 | 3.64E-04 |
| SNTB1 | Syntrophin, beta 1, basic component 1 | Expression associated with lung cancer survival85; Interacts with the HTLV-1 TAX protein86; Not reported as a target of methylation. | cg04318006 | 0.12 | 0.23 | 0.10 | 8.10E-04 | 1.68E-03 |
| STMN2 | Stathmin-like 2 | Involved in WNT/β-catenin pathway; novel β-catenin transcriptional target in HCC87; age-related methylation in germ cells88. | cg00398130 | 0.45 | 0.57 | 0.12 | 3.08E-03 | 7.09E-04 |
| SURF6 | Surfeit 6 | Not reported as target of methylation or involved in cancer. | cg01832218 | 0.57 | 0.72 | 0.14 | 1.93E-04 | 5.13E-05 |
| TMEM196 | Transmembrane protein 196 | Not reported as target of methylation or involved in cancer. | cg18505401 | 0.33 | 0.49 | 0.16 | 8.45E-04 | 5.94E-04 |
| TRA2B | Transformer 2 beta homolog | Oncogenic; amplified in lung, ovary, cervix and head/neck89; downregulation-improved survival. | cg12825509 | 0.52 | 0.62 | 0.10 | 5.40E-04 | 1.33E-04 |
| TTC9 | Tetratricopeptide Repeat Domain-Containing 9 | Upregulation leads to increased cell motility in breast cancer; Not reported as target of methylation. | cg01634544 | 0.35 | 0.52 | 0.17 | 3.19E-04 | 4.04E-04 |
| VWDE | von Willebrand factor D and EGF domains | Not reported as target of methylation or involved in cancer. | cg02935154 | 0.21 | 0.42 | 0.21 | 7.60E-04 | 6.10E-04 |
| cg16278512 | 0.15 | 0.37 | 0.21 | 2.72E-04 | 8.68E-04 | |||
| WNT9A | wingless-type MMTV integration site family, member 9A | Bi-directional promoter for WNT9A/CD558500 silenced by methylation90; mutated in colon cancer91 | cg26452081 | 0.42 | 0.53 | 0.12 | 1.89E-04 | 1.97E-04 |
| ZHX2 | ZHX2 zinc fingers and homeoboxes 2 | TSG; methylated in HCC92 | cg01801603 | 0.29 | 0.41 | 0.12 | 7.45E-04 | 1.29E-03 |
CpG number is the unique identifier for the loci provided by Illumina; multiple loci within a differentially methylated region listed in descending order according to genomic position.
Difference in median β-value between large and small tumors
Statistical differences in β-values between the two groups were determined by Mann-Whitney or student's t-test
CpG loci located in a genomic sequence that overlaps for ADAT3/SCAMP4, MIR200A/MIR200B; and PDE3B/PSMA1 genes.
Differential methylation within genomic regions
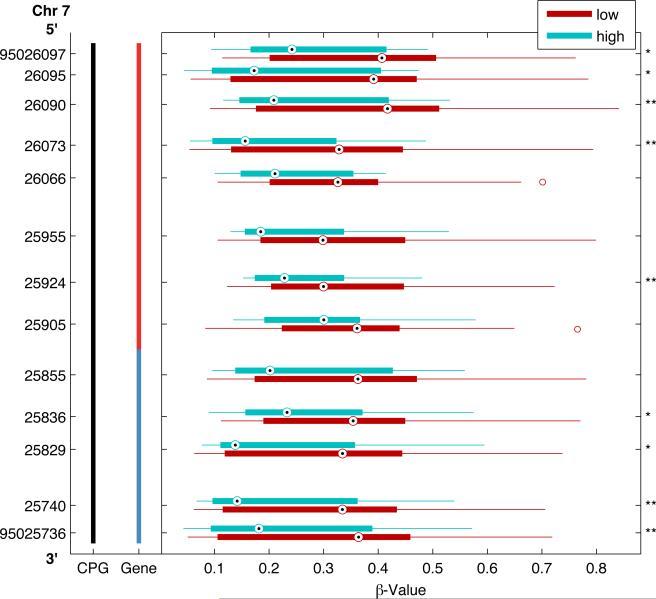
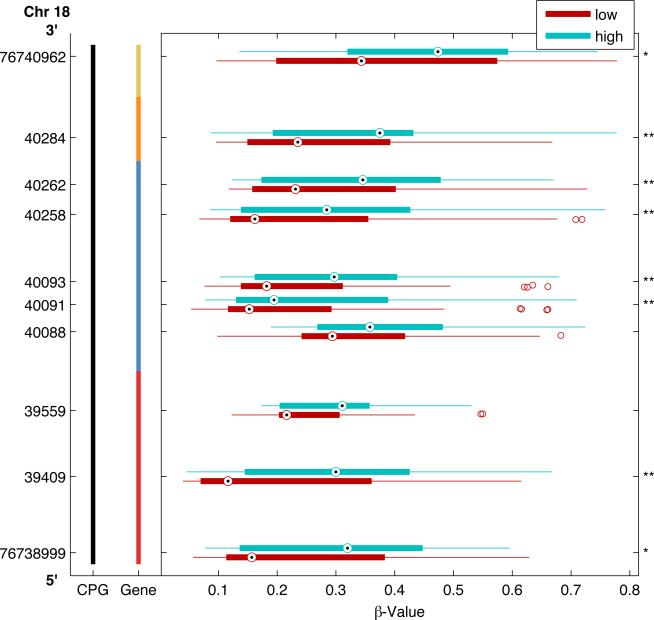
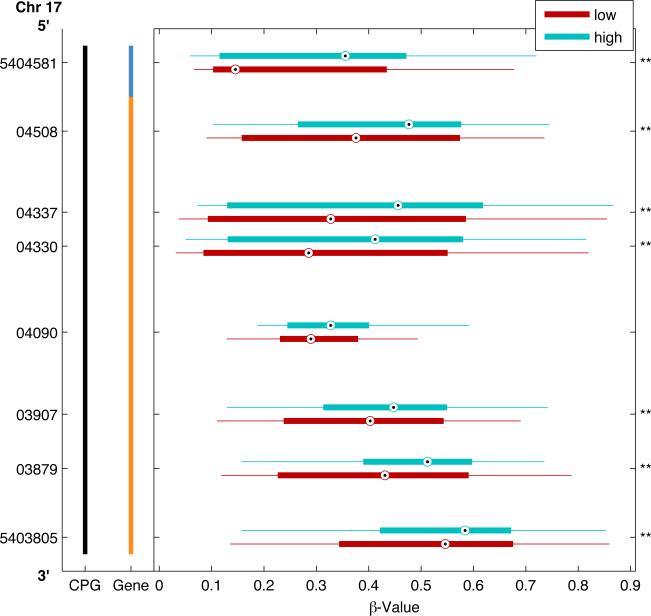
An individual methylated CpG site itself can serve as a detectable biomarker regardless of downstream impact on gene expression. However, prognostic biomarkers with functional biological relevance are of great interest. Given that functionally significant methylation is often associated with methylation across clusters of CpG sites, we next examined whether there were methylation differences in clusters of CpG sites within genomic regions between the high and low risk groups. At a p-value<0.05, we identified 6 genes with clusters of CpG sites differentially methylated in high-risk tumors (Table 4). Figure 2 plots a 361 bp region of the genome encoding the Paraoxonase 3 (PON3) gene that includes 13 CpG sites examined within the CpG island. Box plots represent the median and interquartile range (25th and 75th percentiles) of methylation levels in high (aqua blue boxes) and low-risk (maroon red boxes) tumors. The median methylation for each of the 9 CpG sites (32% of CpG sites examined in the PON3 gene) was significantly lower in high-risk tumors than low-risk tumors. Figure 3 plots a ~2kb region of the SAL-Like 3 gene (SALL3) that includes 10 CpG sites within the CpG island. Overall, 8 out of 27 total sites examined had significantly higher methylation in the high-risk tumors. The genomic region with the most differentially methylated sites (LOC728392) does not have a defined function, but does have predicted gene coding regions and an identified CpG island. Figure 4 plots 8 out of 27 CpG sites examined across 776 bps of a CpG island that all have higher methylation in high-risk tumors. When examining clusters of differentially methylated sites by tumor size, we identified 20 regions (Table 4), of which 14 genes had exclusively higher methylation in large tumors and 3 had exclusively lower methylation. Only three genomic regions had clusters of significantly different CpG methylation by nodal status, all of which were increased in node positive cases (Table 4).
Table 4.
Differentially methylated CpG loci across genomic regionsa by risk group, tumor size and nodal status
| Gene | Number of Methylated Loci / Total CpG sites (%) | Loci with decreased methylationb | Loci with Increased methylation |
|---|---|---|---|
| Risk Group | |||
| LOC728392 | 7 / 15 (46.7) | 0 | 7 |
| PON3 | 9 / 28 (32.1) | 9 | 0 |
| VWC2 | 8 / 26 (30.8) | 0 | 8 |
| PREX2 | 6 / 20 (30.0) | 0 | 6 |
| SALL3 | 8 / 27 (29.6) | 0 | 8 |
| SLITRK3 | 7 / 25 (28.0) | 0 | 7 |
| Tumor Size | |||
| SVIP | 10 / 12 (83.3) | 0 | 10 |
| C2orf43 | 8 / 14 (57.1) | 8 | 0 |
| ENPP5 | 7 / 13 (53.8) | 0 | 7 |
| PON3 | 14 / 28 (50.0) | 14 | 0 |
| C2orf74 | 7 / 16 (43.8) | 7 | 0 |
| DPY19L2P4 | 7 / 16 (43.8) | 0 | 7 |
| FOXI2 | 9 / 22 (40.9) | 0 | 9 |
| TMEM178 | 6 / 15 (40.0) | 0 | 6 |
| STK32A | 7 / 18 (38.9) | 1 | 6 |
| VWC2 | 10 / 26 (38.5) | 0 | 10 |
| STK33 | 8 / 22 (36.4) | 0 | 8 |
| PREX2 | 7 / 20 (35.0) | 0 | 7 |
| RGS17 | 6 / 20 (30.0) | 0 | 6 |
| NHSL1 | 8 / 44 (18.2) | 0 | 8 |
| NKAIN3 | 6 / 38 (15.8) | 0 | 6 |
| GRID2 | 6 / 47 (12.8) | 1 | 5 |
| SGK1 | 6 / 51 (11.8) | 0 | 6 |
| HOXB3 | 6 / 54 (11.1) | 0 | 6 |
| CDK6 | 7 / 66 (10.6) | 0 | 7 |
| HOXC4 | 8 / 126 (6.3) | 4 | 4 |
| Nodal Status | |||
| LOC728392 | 7 / 15 (46.7) | 0 | 7 |
| SALL3 | 6 / 27 (22.2) | 0 | 6 |
| SLITRK3 | 6 / 25 (24.0) | 1 | 5 |
Gene regions defined by Illumina.
Number of CpG Loci that had significantly decreased methylation (lower β-value in high-risk, >5cm, or N+ groups) and increased methylation (higher β-value in high-risk, >5cm, or N+ groups)
Figure 2.
Differentially methylated CpG Loci by clinical risk group across genomic regions of the Paraoxonase 3 (PON3) gene. A representative region of the genome from chromosome 7 (Chr 7) encoding the Paraoxonase 3 (PON3) gene is presented spanning 361 base pairs (bp) from 5’ (top) to 3’ (bottom) with genomic coordinates on vertical axis. This region includes 13 CpG loci within a CpG island located 1500 bp (blue) and 200 bp (red) from the gene transcriptional start site (vertical bar). For each CpG loci, boxplots illustrate the median (dot) and interquartile range [25th (low boundary of box) and 75th (upper boundary of box) percentiles] of β-values in high (aqua blue boxes) and low-risk (maroon red boxes) tumors. Significantly different median methylation at each CpG loci is noted: * p<0.05; ** p<0.01.
Figure 3.
Differentially methylated CpG Loci by clinical risk group across genomic regions of the SAL-Like 3 (SALL3) gene. A representative region of the genome from chromosome 18 (Chr 18) encoding the SALL3 gene is presented spanning 2Kb from 3’ (top) to 5’ (bottom) with genomic coordinates on vertical axis. This region includes 10 CpG loci within a CpG island (black bar) located 200 bp (red) and 1500 bp (blue vertical bar) from the gene transcriptional start site or within the first exon (orange) or body (mustard) of the gene. For each CpG loci, boxplots illustrate the median (dot) and interquartile range [25th (low boundary of box) and 75th (upper boundary of box) percentiles] of β-values in high (aqua blue boxes) and low-risk (maroon red boxes) tumors. Significantly different median methylation at each CpG loci is noted: * p<0.05; ** p<0.01.
Figure 4.
Differentially methylated CpG Loci by clinical risk group across the LOC728392 genomic region. A representative region of the genome from chromosome 17 (Chr 17) encoding the uncharacterized transcript LOC728392 spanning from 5’ (top) to 3’ (bottom) with genomic coordinates on vertical axis. This region includes 8 CpG loci within a CpG island located 1500 bp (blue vertical bar) from the gene transcriptional start site or within the first exon (orange vertical bar). For each CpG loci, boxplots illustrate the median (dot) and interquartile range [25th (low boundary of box) and 75th (upper boundary of box) percentiles] of β-values in high (aqua blue boxes) and low-risk (maroon red boxes) tumors. Significantly different median methylation at each CpG loci is noted: * p<0.05; ** p<0.01.
DISCUSSION
This study investigated whether DNA methylation differed between locally advanced (high-risk) or locoregionally-confined (low-risk) anal cancers. To address this question, we conducted the first genome-wide investigation of DNA methylation in anal cancer using a well annotated set of tumors archived within the RTOG 98-11 trial. We identified individual CpG sites and clusters of CpG sites that were differentially methylated in locally advanced tumors. Furthermore, we identified a large number of CpG sites with differential methylation in larger (>5cm) tumors, regardless of nodal status. These differentially methylated regions provide clues for future studies that can examine whether this dense DNA methylation leads to transcriptional silencing of genes within large or advanced tumors. While the biological impact of methylation at an individual CpG site is unknown, the differentially methylated or unmethylated CpG sites identified in advanced or large tumors may be developed as clinically-applicable biomarkers
The findings of this study are in line with the growing appreciation that aberrant epigenetic events are critical in the process of cancer growth and progression. For example, differential methylation patterns have been reported in the progression of several malignancies, including prostate93,94 bladder,95,96 renal cell,47 esophageal,97 HPV-positive and HPV-negative head and neck,98,99 and cervical cancers.100,101 In general, these papers reported that increased DNA methylation within CpG islands was associated with increasing tumor aggressiveness94,101 and some epigenetic events appear to be early markers of progression.100 In cervical cancer, methylation of RASSF2 was associated with increased tumor vascular invasion and shorter survival time, independent of tumor stage.100 Such differences in methylation provide biological insight into the mechanisms of carcinogenesis.
DNA methylation differences were observed in locally advanced anal cancers compared to early, less advanced tumors. Tumor suppressor genes are often targets of DNA methylation-mediated inactivation which in turn, contributes to cancer progression. A loss of methylation can also be associated with re-expression of suppressed oncogenic elements which can then also drive neoplastic growth. We identified 2 tumor suppressor genes with differentially methylated individual CpG sites or clusters of methylated sites in locally advanced anal cancers. SAL-like 3 (SALL3) has been reported to be methylated in hepatocellular cancer.25 Secreted frizzled-related protein 2 (SFRP2) has been reported to be frequently methylated in several cancers, including cervical34, HPV-positive and negative head and neck27, and prostate28 (Online-Table 1). Paraoxonase 3 (PON 3) has been reported as an imprinted gene and observed here to have reduction methylation in advanced anal cancer. Furthermore, there were a large number of differentially methylated CpG loci in large tumors, suggesting an accumulation of methylation events with progressive growth of a tumor. Of these, 7 genes are tumor suppressor genes previously reported as methylated in other cancers (Online-Table 2). Specifically, Early B-cell factor 3 (EBF3)37 and Neuronal pentraxin I (NPTX1)66,67 have been reported as epigenetically altered in HPV-associated oropharyngeal and cervical cancers, respectively. Increased methylation of EBF3 was highly correlated with HPV-16 infection in head and neck SCC.46 In addition, several genes found to be methylated in locally advanced or large anal tumors encode for proteins that interact with HPV oncogenes. For example, cyclin-dependent kinase 6 (CDK6) regulates the activity of tumor suppressor pRb, which is a target of HPV oncoprotein E7. In HPV-positive cervical cancer cell lines, the inactivation of CDK6 was critical for HPV-associated carcinogenesis.38 Notably, a large number of differentially methylated loci identified in this study occurred within genes that are not well characterized and may represent novel methylation targets. Overall, this genome-wide methylation analysis identified several biologically-relevant methylated genes that have been consistently found to be methylated in other cancers (including those associated with HPV). This epigenetic silencing that may promote anal tumor growth and progression to advanced disease.
A striking number of epigenetic alterations in CpG loci were identified within components of the WNT/β-catenin pathway102 in anal tumors. The clustering of epigenetic alterations in the WNT/β-catenin pathway among larger or high-risk anal cancers is similar to what has been reported in HPV-associated cervical cancer.17,26,103-105 The WNT signaling pathway appears to be a target of HPV, with both epigenetic106 and gene expression-related alterations.107 It is well established that WNT/β-catenin signaling is a critical component of cancer progression and epigenetic alterations of both activators and inhibitors may promote aberrant cellular proliferation and carcinogenesis. These epigenetic changes within the WNT pathway warrant further exploration.
There were relatively few significant differentially methylated regions identified by nodal status or clinical risk groups. There are several possible explanations for the relatively few methylation differences by nodal status. First, epigenetic alterations may not play a role in nodal invasion and thus our limited number of aberrantly methylated loci represents the only underlying changes. Second, due to clinical lymph node staging (e.g. physical exam and/or radiological imaging) in anal cancer, there is a possibility that clinically node-negative patients had occult, undetected micrometastatic disease. This would not only dilute the molecular comparison but result in minimizing differences in outcomes between node-positive and node-negative patients. The observation of significantly better outcomes among node-negative patients provides evidence that misclassification of nodal status does not entirely explain these findings. Finally, the cross-sectional design of this study limited our ability to identify the sequence of epigenetic alterations in anal cancer progression and to distinguish alterations which may be drivers or bystanders of neoplastic progression. Future studies that determine the extent of epigenetic differences by nodal involvement and identify epigenomic signatures of patient outcomes are warranted.
This study represents the largest comprehensive genome-wide molecular analysis (in this case, DNA methylation) of anal cancers; a rare malignancy with low tissue availability. The RTOG 98-11 specimen archive is one of the largest fully annotated pre-treatment anal SCC tissue repositories available. Cases in this trial may not represent the general population with anal SCC, especially high-risk populations such as HIV or immunocompromised patients. HIV status of cancer patients was unknown. This study is limited to patients in the mitomycin-C arm with available tumor tissue; however, this sample is representative of the mitomycin-C arm when comparing the distribution of demographic, pathological factors and outcomes (Figure 2). Due to the limited availability of anal cancer tissues (especially fresh frozen), this study was unable to determine whether differences in methylation resulted in decreased mRNA transcription. However, differential methylation events at specific loci may still represent potential biomarkers of HPV-associated carcinogenesis. Furthermore, the characterization of clusters of differentially methylated CpG loci within CpG islands allows for the identification of biologically relevant targets for which expression is likely modulated by methylation and can be further investigated using in-vitro and in-vivo laboratory models. The molecular biology of anal cancers is not well characterized and based on this study alone; we cannot determine the most important epigenetic alterations among those identified. However, this study provides important candidate targets for future validation in other patient populations and using different methods, such as identifying loss of mRNA or protein expression.
Using a genome-wide methylation analysis, this study has demonstrated that significant epigenetic alterations occur in the progression from early to later stage locally advanced anal cancer. The overall differences in methylation may lend clues to understanding the molecular alterations that occur with the malignant progression of anal cancer. Effective methylation-related biomarkers may ultimately guide modification of treatment for high risk patients (~50% of RTOG 98-11 patients), including radiation dose intensification, closer monitoring of dose completions and/or gaps in treatment and even possibly the development of novel targeted, radiosensitizing agents. An emerging option for dose modification for anal cancer patients is intensity modulated radiation therapy, which has been associated with less acute toxicity as reported in the RTOG-0529 trial108 and theoretically less potential for accelerated tumor repopulation due to treatment breaks. Furthermore, these findings are also concordant with observations that HPV infection may be associated with extensive epigenetic modifications in the host genome that may impact on tumor development and behavior. Similar to other malignancies, these data suggest that the WNT pathway may play an important role in the progression of anal cancer. Further exploration of the potential roles of methylation-related biomarkers including the development of refined and validated epigenetic signatures of prognosis will be useful in optimizing outcomes in patients with anal cancer.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Sean Yoder and Chaomei Zhang in the Moffitt Molecular Genomics Core for running the Infinium HumanMethylation 450K arrays.
Funding: This was funded by a grant from American Society of Colon and Rectal Surgeons Norman Nigro LPG-09 grant (Shibata/Siegel PI); Miles for Moffitt Milestone Awards (Siegel and Shibata), and LeSportsac Cancer Research Fund (Shibata/Hoffe). Radiation Therapy Oncology Group (RTOG) conducted the RTOG98-11 trial that was supported by RTOG grant U10 CA21661 and CCOP grant U10 CA37422 from the National Cancer Institute (NCI). This manuscript's contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute.
Footnotes
Author Contribution:
A: Contributions to conception and design (EMS, SE, KW, AB, JA, JM, DS), Acquisition of data (KW, BR, AA, JS, JM, JA, AM, AE), Data Analysis and interpretation of data (EMS, SE, KW, AB, SH, DS)
B. Drafting the article (EMS, SE, KW, BR, AB, DS) or revising it critically for important intellectual content (EMS, ES, KW, BR, AB, AA, JS, JM, JA, AM, AE, SH, DS).
C. Final approval of the version to be published (EMS, ES, KW, BR, AB, AA, JS, JM, JA, AM, AE, SH, DS).
Manuscript Category: Colorectal/Anal neoplasia
Conference Proceedings: Presented in part at the 2013 Meeting of the American Society of Colon and Rectal Surgeons, April 29th-May 1st, in Phoenix, AZ; Podium S27 and International Anal Neoplasia Society, November 22-24, 2013 in San Francisco, Abstract # 42
The authors have no financial disclosures.
References
- 1.Eng C. Anal cancer: current and future methodology. Cancer Invest. 2006;24:535–544. doi: 10.1080/07357900600815208. [DOI] [PubMed] [Google Scholar]
- 2.Joseph DA, Miller JW, Wu X, et al. Understanding the burden of human papillomavirus-associated anal cancers in the US. Cancer. 2008;113:2892–2900. doi: 10.1002/cncr.23744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson LG, Madeleine MM, Newcomer LM, Schwartz SM, Daling JR. Anal cancer incidence and survival: the surveillance, epidemiology, and end results experience, 1973-2000. Cancer. 2004;101:281–288. doi: 10.1002/cncr.20364. [DOI] [PubMed] [Google Scholar]
- 4.Nelson RA, Levine AM, Bernstein L, Smith DD, Lai LL. Changing patterns of anal canal carcinoma in the United States. J Clin Oncol. 2013;31:1569–1575. doi: 10.1200/JCO.2012.45.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.SEER Cancer Statistics Factsheets: Anal Cancer. 2013 Available at: http://seer.cancer.gov/statfacts/html/anus.html2014.
- 6.Ajani JA, Winter KA, Gunderson LL, et al. Fluorouracil, mitomycin, and radiotherapy vs fluorouracil, cisplatin, and radiotherapy for carcinoma of the anal canal: a randomized controlled trial. Jama. 2008;299:1914–1921. doi: 10.1001/jama.299.16.1914. [DOI] [PubMed] [Google Scholar]
- 7.Gunderson LL, Winter KA, Ajani JA, et al. Long-term update of US GI intergroup RTOG 98-11 phase III trial for anal carcinoma: survival, relapse, and colostomy failure with concurrent chemoradiation involving fluorouracil/mitomycin versus fluorouracil/cisplatin. J Clin Oncol. 2012;30:4344–4351. doi: 10.1200/JCO.2012.43.8085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.James RD, Glynne-Jones R, Meadows HM, et al. Mitomycin or cisplatin chemoradiation with or without maintenance chemotherapy for treatment of squamous-cell carcinoma of the anus (ACT II): a randomised, phase 3, open-label, 2 × 2 factorial trial. Lancet Oncol. 2013;14:516–524. doi: 10.1016/S1470-2045(13)70086-X. [DOI] [PubMed] [Google Scholar]
- 9.Peiffert D, Tournier-Rangeard L, Gerard JP, et al. Induction chemotherapy and dose intensification of the radiation boost in locally advanced anal canal carcinoma: final analysis of the randomized UNICANCER ACCORD 03 trial. J Clin Oncol. 2012;30:1941–1948. doi: 10.1200/JCO.2011.35.4837. [DOI] [PubMed] [Google Scholar]
- 10.Wietfeldt ED, Thiele J. Malignancies of the anal margin and perianal skin. Clin Colon Rectal Surg. 2009;22:127–135. doi: 10.1055/s-0029-1223845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bird A. The essentials of DNA methylation. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- 13.Sahasrabuddhe VV, Luhn P, Wentzensen N. Human papillomavirus and cervical cancer: biomarkers for improved prevention efforts. Future Microbiol. 2011;6:1083–1098. doi: 10.2217/fmb.11.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wentzensen N, Sherman ME, Schiffman M, Wang SS. Utility of methylation markers in cervical cancer early detection: appraisal of the state-of-the-science. Gynecologic Oncol. 2009;112:293–299. doi: 10.1016/j.ygyno.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shivapurkar N, Sherman ME, Stastny V, et al. Evaluation of candidate methylation markers to detect cervical neoplasia. Gynecologic Oncol. 2007;107:549–553. doi: 10.1016/j.ygyno.2007.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang N, Nijhuis ER, Volders HH, et al. Gene promoter methylation patterns throughout the process of cervical carcinogenesis. Cell Oncol. 2010;32:131–143. doi: 10.3233/CLO-2009-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delmas AL, Riggs BM, Pardo CE, et al. WIF1 is a frequent target for epigenetic silencing in squamous cell carcinoma of the cervix. Carcinogenesis. 2011;32:1625–1633. doi: 10.1093/carcin/bgr193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J, Martins CR, Fansler ZB, et al. DNA methylation in anal intraepithelial lesions and anal squamous cell carcinoma. Clin Cancer Res. 2005;11:6544–6549. doi: 10.1158/1078-0432.CCR-05-0374. [DOI] [PubMed] [Google Scholar]
- 19.Hernandez JM, Siegel EM, Riggs B, et al. DNA methylation profiling across the spectrum of HPV-associated anal squamous neoplasia. PloS One. 2012;7:e50533. doi: 10.1371/journal.pone.0050533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bibikova M, Lin Z, Zhou L, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16:383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sandoval J, Heyn H, Moran S, et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6:692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 22.Pidsley R, Wong CC, Volta M, Lunnon K, Mill J, Schalkwyk LC. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC genomics. 2013;14:293. doi: 10.1186/1471-2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kunnev D, Ivanov I, Ionov Y. Par-3 partitioning defective 3 homolog (C. elegans) and androgen-induced prostate proliferative shutoff associated protein genes are mutationally inactivated in prostate cancer cells. BMC Cancer. 2009;9:318. doi: 10.1186/1471-2407-9-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang L, Muthuswamy SK. Polarity protein alterations in carcinoma: a focus on emerging roles for polarity regulators. Curr Opin Genet Dev. 2010;20:41–50. doi: 10.1016/j.gde.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang XX, Sun JZ, Li FX, et al. Aberrant methylation and downregulation of sall3 in human hepatocellular carcinoma. World J Gastroenterol Cancer. 2012;18:2719–2726. doi: 10.3748/wjg.v18.i21.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chung MT, Sytwu HK, Yan MD, et al. Promoter methylation of SFRPs gene family in cervical cancer. Gynecologic Oncol. 2009;112:301–306. doi: 10.1016/j.ygyno.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 27.Marsit CJ, McClean MD, Furniss CS, Kelsey KT. Epigenetic inactivation of the SFRP genes is associated with drinking, smoking and HPV in head and neck squamous cell carcinoma. Int J Cancer. 2006;119:1761–1766. doi: 10.1002/ijc.22051. [DOI] [PubMed] [Google Scholar]
- 28.Perry AS, O'Hurley G, Raheem OA, et al. Gene expression and epigenetic discovery screen reveal methylation of SFRP2 in prostate cancer. Int J Cancer. 2013;132:1771–1780. doi: 10.1002/ijc.27798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castellana B, Escuin D, Peiro G, et al. ASPN and GJB2 Are Implicated in the Mechanisms of Invasion of Ductal Breast Carcinomas. J Cancer. 2012;3:175–183. doi: 10.7150/jca.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao R, Zhang R, Li W, et al. Genome-wide DNA methylation patterns in discordant sib pairs with alcohol dependence. Asia-Pacific Psychiatry. 2013;5:39–50. doi: 10.1111/appy.12010. [DOI] [PubMed] [Google Scholar]
- 31.Melle C, Ernst G, Scheibner O, et al. Identification of specific protein markers in microdissected hepatocellular carcinoma. J Proteome Res. 2007;6:306–315. doi: 10.1021/pr060439b. [DOI] [PubMed] [Google Scholar]
- 32.Maines MD, Mayer RD, Erturk E, Huang TJ, Disantagnese A. The oxidoreductase, biliverdin reductase, is induced in human renal carcinoma--pH and cofactor-specific increase in activity. J Urol. 1999;162:1467–1472. [PubMed] [Google Scholar]
- 33.Hellman K, Alaiya AA, Becker S, et al. Differential tissue-specific protein markers of vaginal carcinoma. Br J Cancer. 2009;100:1303–1314. doi: 10.1038/sj.bjc.6604975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barone E, Cenini G, Sultana R, et al. Lack of p53 decreases basal oxidative stress levels in the brain through upregulation of thioredoxin-1, biliverdin reductase-A, manganese superoxide dismutase, and nuclear factor kappa-B. Antioxid Redox Signal. 2012;16:1407–1420. doi: 10.1089/ars.2011.4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Florczyk U, Golda S, Zieba A, Cisowski J, Jozkowicz A, Dulak J. Overexpression of biliverdin reductase enhances resistance to chemotherapeutics. Cancer Lett. 2011;300:40–47. doi: 10.1016/j.canlet.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 36.Chan JY, Li L, Miao J, Cai DQ, Lee KK, Chui YL. Differential expression of a novel gene BRE (TNFRSF1A modulator/BRCC45) in response to stress and biological signals. Mol Biol Rep. 2010;37:363–368. doi: 10.1007/s11033-009-9796-8. [DOI] [PubMed] [Google Scholar]
- 37.Kollmann K, Heller G, Schneckenleithner C, et al. A Kinase-Independent Function of CDK6 Links the Cell Cycle to Tumor Angiogenesis. Cancer Cell. 2013;24:167–181. doi: 10.1016/j.ccr.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McLaughlin-Drubin ME, Park D, Munger K. Tumor suppressor p16INK4A is necessary for survival of cervical carcinoma cell lines. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1310432110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Griguer CE, Cantor AB, Fathallah-Shaykh HM, et al. Prognostic relevance of cytochrome C oxidase in primary glioblastoma multiforme. PloS One. 2013;8:e61035. doi: 10.1371/journal.pone.0061035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sen S, Kawahara B, Chaudhuri G. Mitochondrial-associated nitric oxide synthase activity inhibits cytochrome c oxidase: implications for breast cancer. Free Radic Biol Med. 2013;57:210–220. doi: 10.1016/j.freeradbiomed.2012.10.545. [DOI] [PubMed] [Google Scholar]
- 41.Aichler M, Elsner M, Ludyga N, et al. Clinical response to chemotherapy in oesophageal adenocarcinoma patients is linked to defects in mitochondria. J Pathol. 2013;230:410–419. doi: 10.1002/path.4199. [DOI] [PubMed] [Google Scholar]
- 42.Lecoeur H, Borgne-Sanchez A, Chaloin O, et al. HIV-1 Tat protein directly induces mitochondrial membrane permeabilization and inactivates cytochrome c oxidase. Cell Death Dis. 2012;3:e282. doi: 10.1038/cddis.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kang JU, Koo SH, Kwon KC, Park JW. Frequent silence of chromosome 9p, homozygous DOCK8, DMRT1 and DMRT3 deletion at 9p24.3 in squamous cell carcinoma of the lung. Int J Oncol. 2010;37:327–335. [PubMed] [Google Scholar]
- 44.Hong SM, Omura N, Vincent A, et al. Genome-wide CpG island profiling of intraductal papillary mucinous neoplasms of the pancreas. Clin Cancer Res. 2012;18:700–712. doi: 10.1158/1078-0432.CCR-11-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim J, Min SY, Lee HE, Kim WH. Aberrant DNA methylation and tumor suppressive activity of the EBF3 gene in gastric carcinoma. Int J Cancer. 2012;130:817–826. doi: 10.1002/ijc.26038. [DOI] [PubMed] [Google Scholar]
- 46.Bennett KL, Lee W, Lamarre E, et al. HPV status-independent association of alcohol and tobacco exposure or prior radiation therapy with promoter methylation of FUSSEL18, EBF3, IRX1, and SEPT9, but not SLC5A8, in head and neck squamous cell carcinomas. Genes Chromosomes Cancer. 2010;49:319–326. doi: 10.1002/gcc.20742. [DOI] [PubMed] [Google Scholar]
- 47.Arai E, Chiku S, Mori T, et al. Single-CpG-resolution methylome analysis identifies clinicopathologically aggressive CpG island methylator phenotype clear cell renal cell carcinomas. Carcinogenesis. 2012;33:1487–1493. doi: 10.1093/carcin/bgs177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ordway JM, Williams K, Curran T. Transcription repression in oncogenic transformation: common targets of epigenetic repression in cells transformed by Fos, Ras or Dnmt1. Oncogene. 2004;23:3737–3748. doi: 10.1038/sj.onc.1207483. [DOI] [PubMed] [Google Scholar]
- 49.Weichert W, Roske A, Gekeler V, et al. Histone deacetylases 1, 2 and 3 are highly expressed in prostate cancer and HDAC2 expression is associated with shorter PSA relapse time after radical prostatectomy. Br J Cancer. 2008;98:604–610. doi: 10.1038/sj.bjc.6604199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gu Y, Yang P, Shao Q, et al. Investigation of the expression patterns and correlation of DNA methyltransferases and class I histone deacetylases in ovarian cancer tissues. Oncol Lett. 2013;5:452–458. doi: 10.3892/ol.2012.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu M, Chen J, Hu L, et al. HORMAD2/CT46.2, a novel cancer/testis gene, is ectopically expressed in lung cancer tissues. Mol Hum Reprod. 2012;18:599–604. doi: 10.1093/molehr/gas033. [DOI] [PubMed] [Google Scholar]
- 52.Gao F, Shi L, Russin J, et al. DNA methylation in the malignant transformation of meningiomas. PloS One. 2013;8:e54114. doi: 10.1371/journal.pone.0054114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strathdee G, Holyoake TL, Sim A, et al. Inactivation of HOXA genes by hypermethylation in myeloid and lymphoid malignancy is frequent and associated with poor prognosis. Clin Cancer Res. 2007;13:5048–5055. doi: 10.1158/1078-0432.CCR-07-0919. [DOI] [PubMed] [Google Scholar]
- 54.Raff AB, Woodham AW, Raff LM, et al. The evolving field of human papillomavirus receptor research: a review of binding and entry. J Virol. 2013;87:6062–6072. doi: 10.1128/JVI.00330-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Backen AC, Cole CL, Lau SC, et al. Heparan sulphate synthetic and editing enzymes in ovarian cancer. Br J Cancer. 2007;96:1544–1548. doi: 10.1038/sj.bjc.6603747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chan HW, Miller JS, Moore MB, Lutz CT. Epigenetic control of highly homologous killer Ig-like receptor gene alleles. J Immunol. 2005;175:5966–5974. doi: 10.4049/jimmunol.175.9.5966. [DOI] [PubMed] [Google Scholar]
- 57.Suryo Rahmanto Y, Dunn LL, Richardson DR. The melanoma tumor antigen, melanotransferrin (p97): a 25-year hallmark--from iron metabolism to tumorigenesis. Oncogene. 2007;26:6113–6124. doi: 10.1038/sj.onc.1210442. [DOI] [PubMed] [Google Scholar]
- 58.Davalos V, Moutinho C, Villanueva A, et al. Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene. 2012;31:2062–2074. doi: 10.1038/onc.2011.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wiklund ED, Gao S, Hulf T, et al. MicroRNA alterations and associated aberrant DNA methylation patterns across multiple sample types in oral squamous cell carcinoma. PloS One. 2011;6:e27840. doi: 10.1371/journal.pone.0027840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zidar N, Bostjancic E, Gale N, et al. Down-regulation of microRNAs of the miR-200 family and miR-205, and an altered expression of classic and desmosomal cadherins in spindle cell carcinoma of the head and neck--hallmark of epithelial-mesenchymal transition. Hum Pathol. 2011;42:482–488. doi: 10.1016/j.humpath.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 61.Sotgia F, Whitaker-Menezes D, Martinez-Outschoorn UE, et al. Mitochondria “fuel” breast cancer metabolism: fifteen markers of mitochondrial biogenesis label epithelial cancer cells, but are excluded from adjacent stromal cells. Cell Cycle (Georgetown, Tex) 2012;11:4390–4401. doi: 10.4161/cc.22777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ito T, Kwon HY, Zimdahl B, et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 2010;466:765–768. doi: 10.1038/nature09171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cox JL, Wilder PJ, Gilmore JM, Wuebben EL, Washburn MP, Rizzino A. The SOX2-interactome in brain cancer cells identifies the requirement of MSI2 and USP9X for the growth of brain tumor cells. PloS One. 2013;8:e62857. doi: 10.1371/journal.pone.0062857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hoverter NP, Ting JH, Sundaresh S, Baldi P, Waterman ML. A WNT/p21 circuit directed by the C-clamp, a sequence-specific DNA binding domain in TCFs. Mol Cell Biol. 2012;32:3648–3662. doi: 10.1128/MCB.06769-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mori Y, Olaru AV, Cheng Y, et al. Novel candidate colorectal cancer biomarkers identified by methylation microarray-based scanning. Endocr Relat Cancer. 2011;18:465–478. doi: 10.1530/ERC-11-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ongenaert M, Wisman GB, Volders HH, et al. Discovery of DNA methylation markers in cervical cancer using relaxation ranking. BMC medical genomics. 2008;1:57. doi: 10.1186/1755-8794-1-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang N, Eijsink JJ, Lendvai A, et al. Methylation markers for CCNA1 and C13ORF18 are strongly associated with high-grade cervical intraepithelial neoplasia and cervical cancer in cervical scrapings. Cancer Epidemiol Biomarkers Prev. 2009;18:3000–3007. doi: 10.1158/1055-9965.EPI-09-0405. [DOI] [PubMed] [Google Scholar]
- 68.Hu X, Guo J, Zheng L, et al. The heterochronic microRNA let-7 inhibits cell motility by regulating the genes in the actin cytoskeleton pathway in breast cancer. Mol Cancer Res. 2013;11:240–250. doi: 10.1158/1541-7786.MCR-12-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Licciulli S, Maksimoska J, Zhou C, et al. FRAX597, a small molecule inhibitor of the p21-activated kinases, inhibits tumorigenesis of neurofibromatosis type 2 (NF2)-associated Schwannomas. J Biol Chem. 2013;288:29105–29114. doi: 10.1074/jbc.M113.510933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schelhaas M, Shah B, Holzer M, et al. Entry of human papillomavirus type 16 by actin-dependent, clathrin- and lipid raft-independent endocytosis. PLoS Pathog. 2012;8:e1002657. doi: 10.1371/journal.ppat.1002657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moon E, Lee R, Near R, Weintraub L, Wolda S, Lerner A. Inhibition of PDE3B augments PDE4 inhibitor-induced apoptosis in a subset of patients with chronic lymphocytic leukemia. Clin Cancer Res. 2002;8:589–595. [PubMed] [Google Scholar]
- 72.Yamano Y, Uzawa K, Saito K, et al. Identification of cisplatin-resistance related genes in head and neck squamous cell carcinoma. Int J Cancer. 2010;126:437–449. doi: 10.1002/ijc.24704. [DOI] [PubMed] [Google Scholar]
- 73.Wang X, Huang CJ, Yu GZ, et al. Expression of group IIA phospholipase A2 is an independent predictor of favorable outcome for patients with gastric cancer. Hum Pathol. 2013 doi: 10.1016/j.humpath.2013.01.027. [DOI] [PubMed] [Google Scholar]
- 74.Ren P, Zhang JG, Xiu L, Yu ZT. Clinical significance of phospholipase A2 group IIA (PLA2G2A) expression in primary resected esophageal squamous cell carcinoma. Eur Rev Med Pharmacol Sci. 2013;17:752–757. [PubMed] [Google Scholar]
- 75.Schweikert EM, Devarajan A, Witte I, et al. PON3 is upregulated in cancer tissues and protects against mitochondrial superoxide-mediated cell death. Cell Death Differ. 2012;19:1549–1560. doi: 10.1038/cdd.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Waerner T, Gardellin P, Pfizenmaier K, Weith A, Kraut N. Human RERE is localized to nuclear promyelocytic leukemia oncogenic domains and enhances apoptosis. Cell Growth Differ. 2001;12:201–210. [PubMed] [Google Scholar]
- 77.Fang J, Rhyasen G, Bolanos L, et al. Cytotoxic effects of bortezomib in myelodysplastic syndrome/acute myeloid leukemia depend on autophagy-mediated lysosomal degradation of TRAF6 and repression of PSMA1. Blood. 2012;120:858–867. doi: 10.1182/blood-2012-02-407999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Narayan G, Murty VV. Integrative genomic approaches in cervical cancer: implications for molecular pathogenesis. Future oncology. 2010;6:1643–1652. doi: 10.2217/fon.10.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Costello LC, Zou J, Desouki MM, Franklin RB. Evidence for changes in RREB-1, ZIP3, and Zinc in the early development of pancreatic adenocarcinoma. Journal of Gastrointest Cancer. 2012;43:570–578. doi: 10.1007/s12029-012-9378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kent OA, Fox-Talbot K, Halushka MK. RREB1 repressed miR-143/145 modulates KRAS signaling through downregulation of multiple targets. Oncogene. 2013;32:2576–2585. doi: 10.1038/onc.2012.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Steinbrecher A, Meplan C, Hesketh J, et al. Effects of selenium status and polymorphisms in selenoprotein genes on prostate cancer risk in a prospective study of European men. Cancer Epidemiol Biomarkers Prev. 2010;19:2958–2968. doi: 10.1158/1055-9965.EPI-10-0364. [DOI] [PubMed] [Google Scholar]
- 82.Takata Y, Kristal AR, Santella RM, et al. Selenium, selenoenzymes, oxidative stress and risk of neoplastic progression from Barrett's esophagus: results from biomarkers and genetic variants. PloS One. 2012;7:e38612. doi: 10.1371/journal.pone.0038612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goodrich JM, Basu N, Franzblau A, Dolinoy DC. Mercury biomarkers and DNA methylation among Michigan dental professionals. Environmental and Mol Mutagen. 2013;54:195–203. doi: 10.1002/em.21763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fonseca-Camarillo G, Yamamoto-Furusho JK. Gene expression of solute carrier family 9 (sodium/hydrogen exchanger) 3, (SLC9A3) is downregulated in patients with ulcerative colitis. Inflamm Bowel Dis. 2012;18:1197–1198. doi: 10.1002/ibd.22968. [DOI] [PubMed] [Google Scholar]
- 85.Galvan A, Frullanti E, Anderlini M, et al. Gene expression signature of non-involved lung tissue associated with survival in lung adenocarcinoma patients. Carcinogenesis. 2013 doi: 10.1093/carcin/bgt294. [DOI] [PubMed] [Google Scholar]
- 86.Rousset R, Fabre S, Desbois C, Bantignies F, Jalinot P. The C-terminus of the HTLV-1 Tax oncoprotein mediates interaction with the PDZ domain of cellular proteins. Oncogene. 1998;16:643–654. doi: 10.1038/sj.onc.1201567. [DOI] [PubMed] [Google Scholar]
- 87.Lee HS, Lee DC, Park MH, et al. STMN2 is a novel target of beta-catenin/TCF-mediated transcription in human hepatoma cells. Biochem Biophys Res Commun. 2006;345:1059–1067. doi: 10.1016/j.bbrc.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 88.Flanagan JM, Popendikyte V, Pozdniakovaite N, et al. Intra- and interindividual epigenetic variation in human germ cells. Am J Hum Genet. 2006;79:67–84. doi: 10.1086/504729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Best A, Dagliesh C, Ehrmann I, Kheirollahi-Kouhestani M, Tyson-Capper A, Elliott DJ. Expression of Tra2 beta in Cancer Cells as a Potential Contributory Factor to Neoplasia and Metastasis. Int J Cell Biol. 2013;2013:843781. doi: 10.1155/2013/843781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shu J, Jelinek J, Chang H, et al. Silencing of bidirectional promoters by DNA methylation in tumorigenesis. Cancer Res. 2006;66:5077–5084. doi: 10.1158/0008-5472.CAN-05-2629. [DOI] [PubMed] [Google Scholar]
- 91.An CH, Kim SS, Kang MR, et al. Frameshift mutations of ATBF1, WNT9A, CYLD and PARK2 in gastric and colorectal carcinomas with high microsatellite instability. Pathology. 2010;42:583–585. doi: 10.3109/00313025.2010.508735. [DOI] [PubMed] [Google Scholar]
- 92.Lv Z, Zhang M, Bi J, Xu F, Hu S, Wen J. Promoter hypermethylation of a novel gene, ZHX2, in hepatocellular carcinoma. Am J Clin Pathol. 2006;125:740–746. doi: 10.1309/09B4-52V7-R76K-7D6K. [DOI] [PubMed] [Google Scholar]
- 93.Lin PC, Giannopoulou EG, Park K, et al. Epigenomic alterations in localized and advanced prostate cancer. Neoplasia. 2013;15:373–383. doi: 10.1593/neo.122146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Delgado-Cruzata L, Hruby GW, Gonzalez K, et al. DNA methylation changes correlate with Gleason score and tumor stage in prostate cancer. DNA Cell Biol. 2012;31:187–192. doi: 10.1089/dna.2011.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Marsit CJ, Houseman EA, Christensen BC, et al. Identification of methylated genes associated with aggressive bladder cancer. PloS One. 2010;5:e12334. doi: 10.1371/journal.pone.0012334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim YJ, Yoon HY, Kim JS, et al. HOXA9, ISL1 and ALDH1A3 methylation patterns as prognostic markers for nonmuscle invasive bladder cancer: Array-based DNA methylation and expression profiling. Int J Cancer. 2013;133:1135–1142. doi: 10.1002/ijc.28121. [DOI] [PubMed] [Google Scholar]
- 97.Guo W, Zhu T, Dong Z, Cui L, Zhang M, Kuang G. Decreased expression and aberrant methylation of Gadd45G is associated with tumor progression and poor prognosis in esophageal squamous cell carcinoma. Clin Exp Metastasis. 2013 doi: 10.1007/s10585-013-9597-2. [DOI] [PubMed] [Google Scholar]
- 98.Colacino JA, Dolinoy DC, Duffy SA, et al. Comprehensive analysis of DNA methylation in head and neck squamous cell carcinoma indicates differences by survival and clinicopathologic characteristics. PloS One. 2013;8:e54742. doi: 10.1371/journal.pone.0054742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Poage GM, Butler RA, Houseman EA, et al. Identification of an epigenetic profile classifier that is associated with survival in head and neck cancer. Cancer Res. 2012;72:2728–2737. doi: 10.1158/0008-5472.CAN-11-4121-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Guerrero-Setas D, Perez-Janices N, Blanco-Fernandez L, et al. RASSF2 hypermethylation is present and related to shorter survival in squamous cervical cancer. Mod Pathol. 2013;26:1111–1122. doi: 10.1038/modpathol.2013.32. [DOI] [PubMed] [Google Scholar]
- 101.Lendvai A, Johannes F, Grimm C, et al. Genome-wide methylation profiling identifies hypermethylated biomarkers in high-grade cervical intraepithelial neoplasia. Epigenetics. 2012;7:1268–1278. doi: 10.4161/epi.22301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Niehrs C. The complex world of WNT receptor signalling. Nat Rev Mol Cell Biol. 2012;13:767–779. doi: 10.1038/nrm3470. [DOI] [PubMed] [Google Scholar]
- 103.Lee J, Yoon YS, Chung JH. Epigenetic silencing of the WNT antagonist DICKKOPF-1 in cervical cancer cell lines. Gynecologic Oncol. 2008;109:270–274. doi: 10.1016/j.ygyno.2008.01.034. [DOI] [PubMed] [Google Scholar]
- 104.Lee EJ, Jo M, Rho SB, et al. Dkk3, downregulated in cervical cancer, functions as a negative regulator of beta-catenin. Int J Cancer. 2009;124:287–297. doi: 10.1002/ijc.23913. [DOI] [PubMed] [Google Scholar]
- 105.van der Meide WF, Snellenberg S, Meijer CJ, et al. Promoter methylation analysis of WNT/beta-catenin signaling pathway regulators to detect adenocarcinoma or its precursor lesion of the cervix. Gynecologic Oncol. 2011;123:116–122. doi: 10.1016/j.ygyno.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 106.Leonard SM, Wei W, Collins SI, et al. Oncogenic human papillomavirus imposes an instructive pattern of DNA methylation changes which parallel the natural history of cervical HPV infection in young women. Carcinogenesis. 2012;33:1286–1293. doi: 10.1093/carcin/bgs157. [DOI] [PubMed] [Google Scholar]
- 107.Fragoso-Ontiveros V, Maria Alvarez-Garcia R, Contreras-Paredes A, et al. Gene expression profiles induced by E6 from non-European HPV18 variants reveals a differential activation on cellular processes driving to carcinogenesis. Virology. 2012;432:81–90. doi: 10.1016/j.virol.2012.05.029. [DOI] [PubMed] [Google Scholar]
- 108.Call JA, Haddock MG, Quevedo JF, Larson DW, Miller RC. Intensity-modulated radiotherapy for squamous cell carcinoma of the anal canal: efficacy of a low daily dose to clinically negative regions. Radiat Oncol. 2011;6:134. doi: 10.1186/1748-717X-6-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.