Abstract
Detoxification and homeostatic acquisition of metal ions are vital for all living organisms. We have identified PCA1 in yeast Saccharomyces cerevisiae as an overexpression suppressor of copper toxicity. PCA1 possesses signatures of a P1B-type heavy metal-transporting ATPase that is widely distributed from bacteria to humans. Copper resistance conferred by PCA1 is not dependent on catalytic activity, but it appears that a cysteine-rich region located in the N terminus sequesters copper. Unexpectedly, when compared with two independent natural isolates and an industrial S. cerevisiae strain, the PCA1 allele of the common laboratory strains we have examined possesses a missense mutation in a predicted ATP-binding residue conserved in P1B-type ATPases. Consistent with a previous report that identifies an equivalent mutation in a copper-transporting P1B-type ATPase of a Wilson disease patient, the PCA1 allele found in laboratory yeast strains is nonfunctional. Overexpression or deletion of the functional allele in yeast demonstrates that PCA1 is a cadmium efflux pump. Cadmium as well as copper and silver, but not other metals examined, dramatically increase PCA1 protein expression through post-transcriptional regulation and promote subcellular localization to the plasma membrane. Our study has revealed a novel metal detoxification mechanism in yeast mediated by a P1B-type ATPase that is unique in structure, substrate specificity, and mode of regulation.
Excretion and detoxification of nonphysiological metals, homeostatic absorption, and utilization of nutritional yet toxic metals are fundamental biological processes. Metal toxicity and deficiency resulting from defects in metabolism and excess accumulation through environmental contamination are implicated in a number of disorders, including failure in normal growth and development and initiation and progression of degenerative diseases (1–7). For instance, a genetic defect in copper excretion in the liver leads to Wilson disease exhibiting hepatic failure and neuronal dysfunctions (7). Cadmium is a well known industrial and environmental pollutant implicated in cancer, neurological disorders, reproductive defects, and endocrine disruption (8–12). Cadmium exerts its toxic effects by the perturbation of cellular redox balance, inhibition of DNA repair, disruption of metal ion homeostasis, and alterations in signal transduction (8–12). Mechanistic insights into metal metabolism in living organisms would facilitate prevention and treatment of metal-related disorders and develop methods for efficient remediation of toxic metals from the environment.
Organisms have evolved defense mechanisms to combat the toxic effects of heavy metal ions. The P1B-type ATPase family of heavy metal transporters that are distributed from bacteria to humans extrude toxic metal ions such as copper, silver, zinc, cobalt, lead, and/or cadmium from the cell (13–18). Functional characterization of this family of transporters has supported the conclusion that efflux mechanisms play critical roles in metal detoxification in bacterial cells (13–15). Copper-specific P1B-type ATPases have been identified in eukaryotes as well (2, 7, 16, 17). Cloning of genes defective in Menkes and Wilson disease have revealed that two P1B-type ATPases (ATP7a and ATP7b) play essential roles in copper acquisition and excretion in humans (2, 7, 16). Plants express copper-, zinc-, cobalt-, lead-, and/or cadmium-translocating ATPases that appear to be involved in distribution and compartmentalization of these metal ions (17, 18).
Although metal efflux systems have begun to be characterized (16–24), it has been suggested that in eukaryotes metallothionine (MT),2 a Cys-rich low molecular weight protein, and GSH-mediated sequestration appear to be the major mechanism in neutralizing toxic metals (25–29). Ycf1 in Saccharomyces cerevisiae, a vacuolar membrane ATP-binding cassette (ABC) transporter, sequesters glutathione-conjugated cadmium (30). Phytochelatin, a GSH polymer synthesized in plants and yeast Schizosaccharomyces pombe, also detoxifies heavy metals (27). Metal-responsive transcription factor 1 (MTF-1) regulates basal and metal-inducible expression of MTs and other genes in mammals, fruit fly, and fish (31–34). In contrast to mammalian genes encoding MT, copper but not cadmium and zinc induces the Cup1 gene in S. cerevisiae through the ACE1 transcription regulator (35, 36). Consistent with their critical roles in induction of the genes involved in metal detoxification, deletion of MTF1 in mice and ACE1 in S. cerevisiae results in enhanced metal sensitivity (32, 35).
To gain better insights into the mechanisms of heavy metal metabolism, we have selected S. cerevisiae cDNAs that suppress copper sensitivity of the ace1Δ yeast strain. One of the cDNAs identified encodes the putative P1B-type ATPase, PCA1, which has been suggested to be involved in copper and/or iron homeostasis (37, 38). Previously, the cadmium resistance of a mutant yeast strain selected on toxic concentrations of cadmium was mapped to the PCA1 gene that contained multiple mutations (39). However, the role and mechanisms of action of PCA1 in metal metabolism have remained elusive. Here we show that PCA1-mediated copper resistance is dependent on its Cys-rich N-terminal domain. The PCA1 allele in all laboratory yeast strains examined carries a missense mutation in a conserved residue resulting in loss of function. The wild-type PCA1 allele confers cadmium resistance by an efflux mechanism accompanied by a novel mode of metal-dependent post-transcriptional regulation.
EXPERIMENTAL PROCEDURES
Yeast Strains, Media, and Phenotypic Tests
Strain BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and null mutants ace1Δ::KanMX6, pca1Δ::KanMX6 were purchased from Open Biosystems. Strain RM11-1a (40) (MATa leu2Δ0 ura3Δ0 HO::KanMX6) was kindly provided by Leonid Kruglyak (Fred Hutchison Cancer Research Center, Seattle, WA). RM11-1a pca1Δ::URA3 and BY4741 ace1Δ::KanMX6 Δpca1:HIS3 were generated by PCR-based homologous recombination (41). For consistency and simultaneous comparisons of copper and cadmium sensitivity, all experiments unless indicated otherwise were performed using the BY4741 ace1Δ strain. Plasmids were transformed into yeast using the lithium acetate procedure (42). Yeast cells were grown on synthetic complete (SC) media (2% dextrose or galactose, 0.67% yeast nitrogen base, and 0.2% dropout mixture for plasmid selection). For phenotypical analysis, ~5 μl of yeast cells (A600 1.0) were spotted on selective media (1.5% agar) supplemented with the indicated concentrations of cadmium (CdCl2) or copper (CuCl2) (Sigma) and incubated at 30 °C for 2 days.
Selection of cDNAs Conferring Copper Resistance
Strain BY4741 ace1Δ strain was transformed with a yeast cDNA library (43). Transformants (~1 × 106 colonies) growing on SC-Ura media were collected by resuspending in sterilized distilled water, diluted, and plated on galactose-containing SC-Ura media supplemented with copper (0.1 mM CuCl2). Plasmids containing cDNA were isolated from yeast colonies growing on selection media and identified by sequencing. cDNA-dependent copper resistance was confirmed by retrans-formation of the isolated plasmid into ace1Δ yeast cells.
Plasmid Construction
The PCA1 open reading frame (ORF) was PCR-amplified from yeast genomic DNA and cloned into BamHI and XhoI restriction sites of a single-copy yeast expression vector for GPD (p413 GPD) (44) or GAL1 (p413 GAL1) (45) gene promoter-mediated expression. The coding sequences of the PCA1 N-terminal domains (amino acids 1–392 and 1–452) were PCR-amplified with a primer set containing a start codon and an artificially generated stop codon and were subcloned into BamHI and EcoRI sites of p413-GPD. The PCA1 truncation mutant (amino acids 393–1216) was generated by PCR amplification of the sequences, including start and stop codons, and was subcloned into p413 GPD at BamHI and XhoI restriction sites. PCA1-GFP and PCA1-FLAG were constructed by generation of a NotI restriction site after the start codon for insertion of NotI-flanked green fluorescent protein (GFP) without start and stop codons or two tandem FLAG epitopes, respectively. For natural promoter-driven expression, the PCA1 ORF, including 810 bp upstream of the start position, was cloned into SacI and XhoI restriction sites of plasmid pRS413 (46). Site-directed mutagenesis was conducted by the primer overlap extension method (47). All constructs were confirmed by sequencing.
Fluorescence Microscopy
Yeast cells were transformed with PCA1-GFP or PCA1-WT-GFP expression plasmids and cultured in SC-His media at 30 °C with agitation until mid-log phase. Metals were added to the culture media for 15 min to 2 h prior to imaging. Cells were collected, washed in phosphate-buffered saline (PBS), and imaged on a confocal microscope (Olympus FV500).
Metal Measurements
Yeast cells were cultured until mid-log phase, and metal ions were added to the culture media for 6 h. Cells were collected in 2-ml aliquots and washed two times in PBS containing 10 mM EDTA. Cell pellets were dissolved in 70% nitric acid at 60 °C for 30 min and diluted to 10% nitric acid. To measure cadmium excretion, cells were cultured with 5 mM CdCl2 for 30 min, washed two times in PBS containing 10 mM EDTA, and resuspended in fresh media prior to sample collection at the indicated time points. Total cellular metal levels were measured by inductively coupled plasma mass spectrometry (ICPMS) at the Department of Geological Sciences, University of Michigan.
Immunoblotting
Total protein extracts were prepared from yeast cells using glass beads and Triton X-100 (1%) as described previously (48). Protein concentrations were measured by the BCA method (Pierce) according to the manufacturer’s specifications. Protein samples were resolved by reducing SDS-PAGE and transferred to a nitrocellulose membrane. PCA1-GFP was detected by chemiluminescence using a primary rabbit anti-GFP polyclonal antibody (1:2000) (Santa Cruz Biotechnology) and secondary goat anti-rabbit horseradish peroxidase-conjugated antibody (1:5000) (Santa Cruz Biotechnology). PCA1-FLAG was probed with primary mouse anti-FLAG M2 monoclonal antibody (1:1000) (Sigma) and secondary sheep anti-mouse IgG horseradish peroxidase-conjugated antibody (Amersham Biosciences) (1:5000). Loading control, phosphoglycerate kinase (PGK), was detected by chemiluminescence using mouse monoclonal anti-PGK antibodies (1:4000) (Molecular Probes) and secondary sheep anti-mouse IgG horseradish peroxidase-conjugated antibody (1:5000) (Amersham Biosciences).
Northern Blotting
Yeast cells were cultured until mid-log phase and supplemented with either 50 μM CdCl2 or 50 μM CuCl2 for 15 and 60 min. Total RNA was extracted from cells, and 25 μg was separated on an RNA gel (0.75% agarose and 2% formaldehyde) and transferred to a nitrocellulose membrane (Protran). Gene-specific DNA probes labeled with [α-32P]dCTP (Amersham Biosciences) were generated using the random primer labeling system (Invitrogen). Hybridization of radiolabeled probes with RNA transcripts was performed by methods described previously (49). Relative mRNA levels were detected by autoradiography.
RESULTS
PCA1 N-terminal Domain Confers Copper Resistance in Yeast
To identify new factors involved in heavy metal defense, we carried out a selection of S. cerevisiae cDNAs that suppress lethality of the ace1Δ yeast strain on toxic copper media. A cDNA encoding PCA1, one of two P1B-type ATPases in the S. cerevisiae genome (37, 50), was identified. Yeast GPD gene promoter-driven constitutive expression of PCA1 confers resistance in the ace1Δ strain to copper (0.1 mM CuCl2) compared with empty vector transformed control cells (Fig. 1A). Deletion of PCA1 in the ace1Δ strain resulted in a slight increase in sensitivity to copper toxicity (Fig. 1A). However, in a wild-type strain, copper resistance was not easily discernible upon either overexpression or deletion of PCA1 (data not shown).
FIGURE 1. Copper resistance is mediated by the Cys-rich N-terminal domain of PCA1.
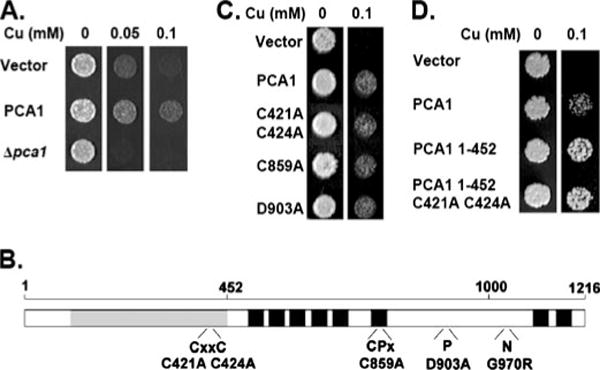
A, PCA1 overexpression confers copper resistance. BY4741 ace1Δ transformed with empty vector (p413-GPD) or PCA1 expression vector and BY4741 ace1Δ pca1Δ yeast cells were cultured in SC (SC-His) media and spotted on solid media supplemented with copper (CuCl2) at the indicated concentrations. B, schematic depiction of PCA1 sequence features. PCA1 contains the signatures of a P1B-type ATPase, eight predicted transmembrane domains (black boxes), intra-membranous CPX motif within the sixth transmembrane domain, a CXXC metal binding domain, a phosphorylation domain (P), and a nucleotide binding domain (N). The N-terminal Cys-rich domain is shaded gray. Conserved residues that have been mutated for characterization are indicated below each domain. C, copper resistance conferred by PCA1 is not dependent on metal translocation. BY4741 ace1Δ cells transformed with empty vector, a PCA1 expression vector, or PCA1 with mutations in the CXXC motif (C421A and C424A), the CPX motif (C859A), or the aspartyl phosphorylation residue (D903A) were spotted on SC-His media with or without copper supplementation. D, copper resistance is conferred by the PCA1 N terminus. Cells expressing empty vector, PCA1, PCA1 residues 1–452, or PCA1 residues 1–452 with site-directed mutations of the predicted metal-binding cysteines (C421A, C424A) were spotted on SC-His media with or without copper supplementation.
PCA1 contains all of the conserved features of the P1B-type ATPase family, which is specific for heavy metal transport (16 – 18, 51) (Fig. 1B). These features include eight predicted transmembrane domains, where an intramembranous metal-transporting CPX motif is located in the sixth transmembrane domain (Fig. 1B). The catalytic domains reside within cytosolic loops that include the nucleotide binding domain (N-domain) and the phosphorylation domain (P-domain) (Fig. 1B). PCA1 contains a single CXXC heavy metal-binding motif located within the N-terminal region. An intriguing feature of PCA1 compared with other P1B-type ATPases is a Cys-rich N-terminal extension ~550 amino acids before the first transmembrane domain (Fig. 1B). To ascertain whether the observed copper resistance by PCA1 is dependent on ATPase function, we carried out site-directed mutagenesis of conserved residues of this family of proteins and assayed for copper sensitivity. However, copper resistance remained unchanged in all mutants (Fig. 1C), suggesting that copper resistance is not dependent on metal translocation. Given that the N terminus contains several cysteine residues (33 Cys), which could serve as copper ligands, we tested whether this domain could independently confer resistance. A peptide corresponding to amino acid residues 1–452, which includes the CXXC motif (Fig. 1B), was expressed in the ace1Δ strain. Expression of this peptide resulted in copper resistance above that of full-length PCA1, presumably by chelating copper ions in a manner analogous to MT (Fig. 1D). Copper resistance is maintained even after mutation of the conserved CXXC motif (C421A, C423A) (Fig. 1D) implying the existence of other copper coordination site(s) in addition to this well characterized metal-binding motif.
Common Laboratory Yeast Strains Possess a G970R Mutation in PCA1
S288c is a commonly used S. cerevisiae haploid strain in which its entire genome has been sequenced (52). Comparisons of the PCA1 sequence of S288c with natural isolates, RM11-1a (40) and YJM789 (53), using BLAST (NCBI, www.ncbi.nlm.nih.gov) revealed that the S288c strain carries a single nucleotide change in the PCA1 gene, which results in a G970R substitution. (Fig. 2). Our sequencing results of PCA1 cloned from the RM11-1a strain as well as an industrial bakers’ yeast strain (Fleischmann’s) confirmed the G970R substitution. To determine whether this mutation was unique to the S288c strain, we sequenced the PCA1 ORF of several laboratory yeast strains, BY4741 (Open Biosystems), DTY1 (54), EG103 (55), SKY9 (56), and YPH98 (46). All contained the G970R mutation (data not shown), which suggests a common lineage among theses strains. Furthermore, sequence alignments of prokaryotic, yeast, plant, and human P1B-type ATPases show that this Gly residue is invariantly conserved among this family of proteins (Fig. 2). Recently, structural studies of the nucleotide binding domain (N-domain) of two P1B-type ATPases (CopA and ATP7b) places this Gly residue within an ATP-binding pocket (57, 58). The functional importance of this residue is further underscored by an equivalent G1101R substitution in ATP7b previously reported in patients with Wilson disease (59). Together, our data along with recent reports strongly support the conclusion that many laboratory S. cerevisiae strains contain a mutation of a critical residue in PCA1 that may play essential roles in function, expression, and/or regulation.
FIGURE 2. Yeast laboratory strains contain a G970R mutation in PCA1.

Aregion of the predicted nucleotide binding domain (N-domain) of PCA1 from strains S288c, RM11-1a, and YJM789 were aligned with the corresponding region of other P1B-type ATPases, including human copper-transporting ATP7a and ATP7b, S. cerevisiae CCC2, A. thaliana HMA4, S. aureus CadA, and Archaeoglobus fulgidus CopA. Boxes highlight conserved residues. Asterisk marks G970R substitution identified in the S288C and other laboratory S. cerevisiae strains. Sequences were obtained from data bases at The National Center for Biotechnological Information (NCBI), and alignments were performed with ClustalX (1.81).
PCA1 Natural Allele Confers Hyper-resistance to Cadmium
To characterize the function of PCA1, we expressed the natural allele (PCA1-WT) and the G970R mutant allele (PCA1) in a laboratory yeast strain and examined metal tolerance. Indeed, expression of PCA1-WT allows cells to grow on media containing a cadmium concentration over 10-fold higher compared with vector and PCA1 carrying the G970R mutation (Fig. 3A). However, growth on media supplemented with copper was indistinguishable, which is consistent for copper resistance being dependent on metal binding rather than ATPase activity (Fig. 3A). No growth advantages in cells expressing PCA1-WT were observed when toxic concentrations of zinc, manganese, cobalt, or iron were added to growth media (data not shown), suggesting specificity in metal binding and translocation by PCA1-WT.
FIGURE 3. PCA1 natural allele (PCA1-WT) plays a critical role in cadmium defense.
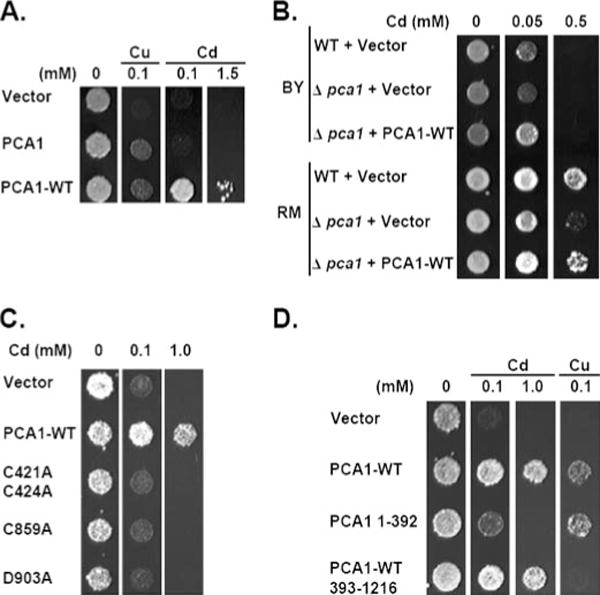
A, PCA1-WT confers resistance to cadmium toxicity. BY4741 ace1Δ cells expressing PCA1 cloned from a laboratory strain (PCA1) or natural isolate (PCA1-WT) under control of a constitutive promoter of the GPD gene were spotted on SC-His media supplemented with CuCl2 or CdCl2 at the indicated concentrations. B, PCA1-WT plays a physiological role in cadmium defense. Cadmium resistance of laboratory strain BY4741 (BY) was compared with natural isolate RM11-1a (RM). Wild-type BY4741 and RM11-1a strains or PCA1 null mutants were transformed with empty vector or PCA1-WT under control of its natural promoter. Cells were spotted on SC-Leu media supplemented with CdCl2 at indicated concentrations. C, cadmium resistance conferred by PCA1-WT is dependent on P1B-type ATPase catalytic activity. Amino acid substitutions in PCA1-WT were made in the N-terminal CXXC motif (C421A and C424A), the CPX motif (C859A), or the aspartyl phosphorylation residue (D903A). Cells expressing PCA1-WT or mutants were spotted on SC-His media supplemented with indicated concentrations of CdCl2. D, the Cys-rich N terminus of PCA1-WT is not essential for cadmium resistance. BY4741 ace1Δ cells expressing empty vector, PCA1-WT, PCA1 (amino acids 1–392), and truncated PCA1-WT (amino acids 393–1216) were spotted on SC-His solid media supplemented with the indicated concentrations of CuCl2 or CdCl2.
Because the RM11-1a strain contains a functional PCA1 allele, it would be predicted that this strain is more resistant to cadmium compared with laboratory strains. Cadmium sensitivity was compared between the RM11-1a strain and our BY4741 laboratory strain. Indeed, the BY4741 strain is much more sensitive to cadmium (Fig. 3B). Deletion of PCA1-WT in the RM11-1a strain reduces cadmium resistance compared with control cells (Fig. 3B) thus underscoring its significance in cadmium defense. Cadmium tolerance returns to wild-type levels in the RM11-1a pca1Δ strain transformed with a natural promoter-driven PCA1-WT plasmid confirming that cadmium sensitivity was specific for deletion of PCA1-WT (Fig. 3B). As expected, deletion of the nonfunctional PCA1 in the BY4741 strain did not significantly affect cadmium resistance. However, introduction of PCA1-WT under control of its natural promoter rescued growth on cadmium media (Fig. 3B). Interestingly, the RM11-1a strain displayed a cadmium growth advantage over the BY4741 strain when PCA1-WT is null-mutated, implying that there may be other factor(s) contributing to cadmium resistance (Fig. 3B).
We then asked whether cadmium resistance is coupled with active metal transport of a P1B-type ATPase. Site-directed mutagenesis of conserved amino acids among P1B-type ATPases, including the CXXC motif (C421, C424), the CPX motif (C859), and the phosphorylation site (Asp-903), completely abolished cadmium resistance (Fig. 3C). These results suggest that cadmium resistance is dependent on an active metal transport mechanism.
The Cys-rich N-terminal Domain of PCA1 Is Not Essential for Cadmium Resistance
PCA1 carries a Cys-rich N-terminal domain that does not have significant sequence homology with any characterized metal-binding protein. We examined the role of this domain in cadmium resistance by expressing the N-terminal 392 amino acid domain and a truncation PCA1-WT allele lacking this domain. The truncated PCA1-WT allele (amino acids 393–1216) includes the CXXC motif because it is critical for cadmium resistance (Fig. 3C). Cells expressing the N-terminal 392-amino acid peptide are more resistant to both cadmium and copper compared with empty vector-transformed cells (Fig. 3D). Truncation of this N-terminal domain did not significantly affect cadmium resistance as compared with full-length PCA1-WT (Fig. 3D). These data suggest that the Cys-rich N-terminal domain itself reduces copper and cadmium toxicity, but it is not critical for cadmium transport. In contrast, copper resistance conferred by PCA1-WT was abolished in the truncated mutant (Fig. 3D), which further supports the conclusion that copper resistance conferred by PCA1 is not dependent on ATPase activity but rather metal sequestration by this domain.
PCA1-mediated Cadmium Export
We addressed three different possible mechanisms governing PCA1-WT-mediated cadmium resistance. First, PCA1-WT could be involved in cadmium sequestration and/or compartmentalization to intracellular organelles. To test this possibility, total cellular cadmium levels were measured by inductively coupled plasma mass spectrometry (ICPMS) of cells expressing empty vector, PCA1, or PCA1-WT when cultured in cadmium-supplemented media. Cells expressing PCA1-WT as compared with empty vector and PCA1 displayed a marked decrease in total cellular cadmium accumulation (Fig. 4A). These results indicate that PCA1-WT either reduces cadmium uptake or enhances efflux. Second, to address the possibility that PCA1-WT may indirectly reduce cellular cadmium uptake, yeast cells expressing empty vector or GAL1 promoter-mediated expression of PCA1-WT were pretreated with toxic amounts of cadmium under glucose repression. Cells were then washed to remove extracellular cadmium and spotted on solid SC media containing either glucose or galactose. Empty vector-transformed cells and PCA1-WT displayed similar growth under glucose repression (Fig. 4B). However, cells expressing PCA1-WT displayed an obvious growth advantage on galactose media in which PCA1-WT is expressed (Fig. 4B). Because PCA1-WT expression was repressed during cadmium exposure, its role in cadmium defense must occur after cadmium has already entered the cell. Third, to conclusively demonstrate an active efflux role, yeast cells were pretreated with cadmium, washed to remove extracellular cadmium, resuspended in fresh media, and collected at 30-min intervals to determine cellular cadmium content. Cadmium levels in cells expressing empty vector remained relatively constant for 60 min after cadmium removal (Fig. 4C and inset). In contrast, cadmium levels rapidly declined in cells expressing PCA1-WT (Fig. 4C and inset). Altogether, these data strongly support the conclusion that PCA1-WT functions as a cadmium efflux pump.
FIGURE 4. PCA1-WT cadmium resistance is dependent on an efflux mechanism.
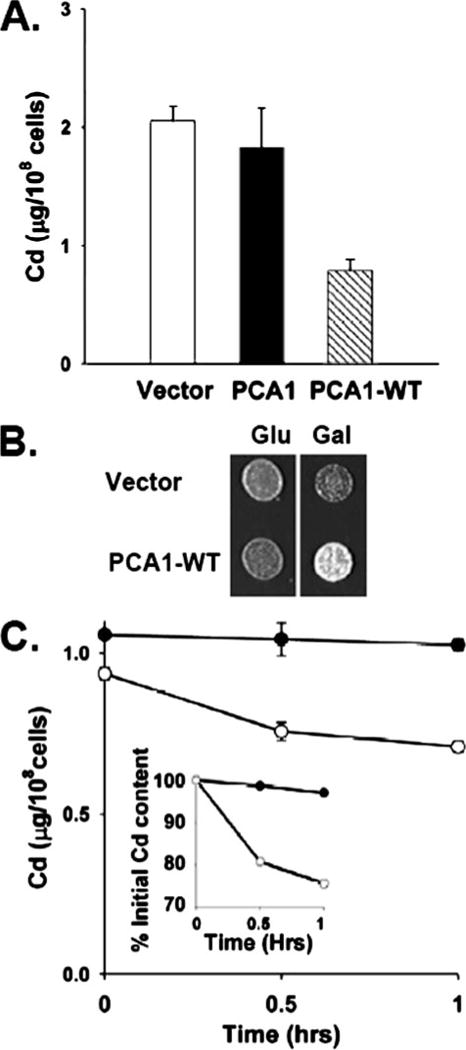
A, overexpression of PCA1-WT reduces total cellular cadmium content. BY4741 ace1Δ cells transformed with empty vector (p413-GPD) or PCA1 or PCA1-WT expression vectors were cultured in SC-His media supplemented with cadmium (50 μM CdCl2) for 6 h. Cells were collected and washed, and total cellular cadmium levels were measured by ICPMS. Bar graphs represent the average ± S.D. of four independent measurements. B, BY4741 ace1Δ cells were transformed with an expression plasmid of PCA1-WT under control of the GAL1 gene promoter or control plasmid. Cells were cultured in SC-His media supplemented with 1 mM CdCl2 for 1 h under glucose repression, washed, and spotted on SC-His media containing either glucose (Glu) or galactose (Gal). C, PCA1-WT enhances cadmium efflux. BY4741 ace1Δ cells transformed with empty vector (closed circles) or a GPD promoter-mediated PCA1-WT expression vector (open circles). Cells were cultured in SC-His media supplemented with 0.5 mM CdCl2 for 30 min, washed, and resuspended in fresh media. Aliquots were collected at indicated time points, and cadmium levels were measured. Each data point represents the average ± S.D. of four independent samples. Inset indicates percent of initial cadmium content.
Metal-dependent Regulation of PCA1 Expression
Because cadmium concentration in the environment likely fluctuates, it is reasonable to predict that cadmium regulates PCA1 expression. However, Northern blot analysis did not indicate any induction of the PCA1 gene after the addition of either cadmium or copper to culture media (Fig. 5A). Both cadmium and copper induced GSH1 encoding an enzyme required for GSH synthesis under the same experimental conditions (Fig. 5A). We next examined the possibility of post-transcriptional regulation. Total protein extracts from cells expressing N-terminal FLAG epitope-tagged PCA1-WT were subjected to Western blot analysis. PCA1-WT-FLAG and nontagged PCA1-WT exhibit indistinguishable cadmium resistance indicating that epitope tagging does not perturb function (data not shown). Although PCA1-WT expression was mediated by a relatively strong GPD gene promoter, PCA1-WT-FLAG was hardly detectable. However, cadmium supplementation to culture media dramatically enhanced PCA1-WT-FLAG protein levels in a time- and dose-dependent manner (Fig. 5B). Not only cadmium but also copper and silver specifically increased PCA1-WT-FLAG protein levels (Fig. 5C). Because the coding sequences of PCA1-WT were inserted into an expression vector for constitutive expression by a GPD gene promoter and CYC1 terminator, it is likely that cadmium, copper, and silver stabilize PCA1 rather than regulate translation. CCC2, a copper-transporting P1B-type ATPase, inserted into the same vector, was not regulated by metal ions supporting the conclusion that the observed metal regulation of PCA1 is independent of the GPD promoter and/or CYC1 terminator (data not shown).
FIGURE 5. Metal-dependent regulation of PCA1 expression.
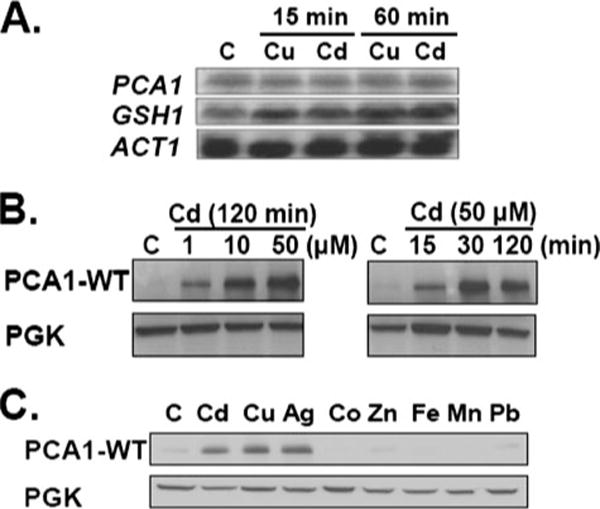
A, cadmium and copper do not induce transcription of PCA1. BY4741 ace1Δ cells were cultured in SC media supplemented with cadmium (50 μM CdCl2) or copper (50 μM CuCl2) for 15 or 60 min prior to total RNA extraction. Transcripts of PCA1, GSH1 (a positive control), and ACT1 (a loading control) were detected by Northern blot analysis using gene-specific 32P-labeled probes. B, post-transcriptional regulation of PCA1-WT. BY4741 ace1Δ cells transformed with PCA1-WT-FLAG were cultured in SC-His media supplemented with cadmium (1, 10, or 50 μM, CdCl2) for 120 min (left panel) or 50 μM CdCl2 (15, 30, or 120 min) (right panel). PCA1-WT-FLAG expression was detected by Western blot analysis. PGK was probed as a loading control. C, metal-specific post-transcriptional regulation of PCA1-WT. BY4741 ace1Δ cells transformed with a PCA1-WT-FLAG expression construct were cultured for 120 min in SC-His media supplemented with cadmium (50 μM CdCl2), copper (50 μM CuCl2), silver (25 μM AgNO3), cobalt (4 mM CoSO4), zinc (15 mM ZnCl2), iron (2 mM iron citrate HCl), manganese (15 mM MnCl2), and lead (0.2 mM Pb(NO3)2 HCl). PCA1-WT-FLAG expression was analyzed by Western blot using anti-FLAG antibodies. PGK was probed as a loading control. C, control.
Subcellular Localization of PCA1-WT
The function of PCA1-WT as a cadmium efflux pump suggests that it is localized at the plasma membrane. To test this prediction, both PCA1-WT and PCA1 fused with GFP at the N terminus were localized by confocal fluorescence microscopy. The cadmium resistance conferred by tagged PCA1-WT-GFP and untagged PCA1-WT was indistinguishable (data not shown). Under normalconditions both PCA1 and PCA1-WT display faint fluorescent signals distributed throughout the cytoplasm (Fig. 6A). In accordance with the enhanced protein expression determined by Western blotting (Fig. 5, B and C), the addition of cadmium (Fig. 6A) as well as copper and silver (data not shown) to culture media dramatically enhanced the fluorescent signal in cells. Furthermore, consistent with a predicted role in cadmium efflux, PCA1-WT-GFP is primarily localized at the plasma membrane in the presence of cadmium (Fig. 6A) as well as copper and silver (data not shown). Addition of zinc, iron, cobalt, lead, and manganese to the culture media had no significant effect on fluorescent signals for PCA1-WT-GFP or PCA1-GFP (data not shown). Expression of the nonfunctional PCA1-GFP was enhanced by cadmium; however, it did not properly localize to the plasma membrane (Fig. 6, A and B), suggesting that the G970R mutation perturbs trafficking. Western blot analysis of PCA1-GFP and PCA1-WT-GFP detected a product of the predicted size (Fig. 6B), indicating that the GFP signal reflects full-length PCA1 protein. These data demonstrate that metal ions regulate PCA1-WT protein levels and stimulate plasma membrane localization.
FIGURE 6. Metal-and functionality-dependent changes in subcellular distribution of PCA1.
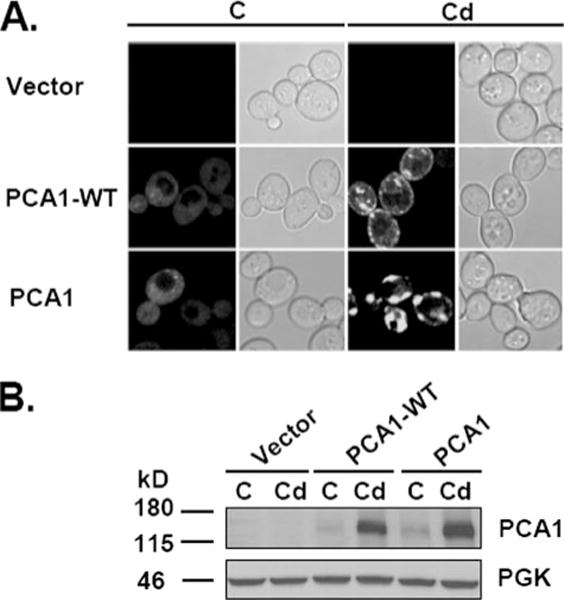
A, subcellular localization of wild-type and nonfunctional PCA1. BY4741 ace1Δ cells transformed with empty vector, PCA1-WT-GFP, or PCA1-GFP expression constructs were cultured to mid-log phase with (Cd)or without (C) cadmium (50 μM CdCl2) supplementation for 120 min. Cells were washed in PBS, and fluorescence was visualized by confocal microscopy (left panels) and differential interference contrast (right panels). B, Western analysis of PCA1-GFP fusion proteins. Total protein extract was prepared from cells with or without cadmium treatment, and PCA1-WT-GFP or PCA1-GFP was detected by Western blot analysis using anti-GFP antibodies. PGK was probed as a loading control.
DISCUSSION
The P-type ATPase family of integral membrane proteins translocates various ions and phospholipids across membranes using the chemical energy of ATP hydrolysis (15–18, 51, 60–62). Virtually all organisms rely on these transporters for maintaining a transmembrane gradient of various ions, which are vital for nutrient uptake, neurotransmission, signaling, and excretion of toxic metals (15–18, 51, 60–62). The P1B-type ATPase specifically transports soft metals, such as copper, cadmium, zinc, and lead (13–18). Our data presented here support the conclusion that PCA1, one of the two P1B-type ATPases in the S. cerevisiae genome, is a cadmium efflux pump that is nonfunctional in all five laboratory yeast strains that we have examined. Metal-induced stabilization and distribution of PCA1 at the plasma membrane are likely to be important mechanisms of rapid response to excess cadmium. This is a novel mode of regulation identified in this family of proteins. Function, structural features, and metal-dependent regulation patterns of PCA1 revealed important new insights into the classification, substrate specificities, mechanisms of action, and regulation of P1B-type ATPases. Moreover, this study also uncovered that the S. cerevisiae laboratory strains that have been used widely as an outstanding model system in biomedical sciences do not mirror, at least in cellular cadmium defense, the strains living in natural habitats.
PCA1 has been proposed and classified as a copper-transporting P1B-type ATPase (37). However our data do not support a copper-transporting function. Instead the Cys-rich N-terminal domain appears to be the contributing factor involved in copper resistance. The abundant Cys residues may serve as ligands for copper binding. Expression of this domain itself confers copper tolerance in yeast, which supports the hypothesis that these abundant Cys residues participate in metal coordination. However, detoxification of copper-generated reactive oxygen species (4) by this domain cannot be excluded as superoxide dismutase activity has been reported for the similar Cys-rich MT (63). It is interesting to note that AtHMA2 and AtHMA4, two P1B-type ATPases of Arabidopsis thaliana, possess a similar Cys/His-rich domain at the C terminus (19, 21, 22). However, these domains do not show significant sequence identity with each other or with any other known metal-binding proteins. It appears that this region is not essential for AtHMA2/4 function (64, 65). Consistently, truncation of the Cys-rich N-terminal domain, excluding the CXXC motif, does not alter PCA1-WT mediated cadmium resistance. This region may play regulatory roles through interaction with metals and/or with other proteins. Several Ca2+-transporting P-type ATPases were shown to carry autoinhibitory domains (51, 60). Binding of regulators and phosphorylation of C-terminal residues of H+-transporting ATPases in plants has also been reported (51, 61, 66). We are currently testing the regulatory roles of the Cys-rich N-terminal domain of PCA1.
A conserved signature of P1B-type ATPase family is the one to six CXXC motifs located within the N-terminal region (67). For instance, human ATP7a and ATP7b contain six such motifs. The two Cys residues bind a single copper (Cu+) ion in a linear bicoordinate manner (68). Although copper binding to these motifs is firmly established by several methods, their roles in the function and regulation of ATP7a and ATP7b remain unclear. These motifs have been suggested to influence copper translocation and/or subcellular trafficking of ATP7a and ATP7b (67). Site-directed mutagenesis of the CXXC motif in PCA1 showed that the two Cys residues are necessary for cadmium resistance, which suggests cadmium binding to this domain is critical for PCA1 function. Cadmium coordination unlikely occurs through two Cys residues but rather with multiple ligands. Future metal binding studies and structural characterization of the Cys-rich domain would reveal how PCA1 coordinates cadmium or other metal ions. Distinct structural features for metal coordination of this domain may explain substrate specificity of P1B-type ATPases. Given that the cytosolic copper carrier, Atox1, directly interacts with the metal binding domain of ATP7a and ATP7b, PCA1 may acquire cadmium from a carrier molecule. Because cadmium binds thiol groups (25–29), glutathione and/or other cytosolic cadmium carriers may specifically interact with the PCA1 metal binding domain to transfer cadmium. For instance, YCF1, a vacuolar ABC transporter utilizes bis(glutathionato)cadmium as its substrate (30).
P1B-type ATPases are categorized into several subfamilies depending on substrates and structural features (69), although the molecular determinant(s) for the substrate specificities is not known. Given that PCA1 deletion or overexpression specifically alters cellular accumulation and resistance of cadmium, PCA1 appears to be the first cadmium-specific P1B-type ATPase. It is possible that PCA1 may efflux other heavy metal ions, but because they do not enhance protein expression, substrate specificity cannot be deciphered by metal resistance. We have addressed this issue by the generation of a functional PCA1 mutant that is constitutively expressed even in the absence of cadmium.3 Additionally, in vitro biochemical ATPase assays would further define PCA1 substrate specificity.
Sequence analysis of the PCA1 ORF of two independent natural strains and an industrial strain clearly identified a G970R mutation to exist in all examined laboratory strains. Although a few other polymorphisms exist, all carried the G970R substitution. It is likely that the ancestral S. cerevisiae laboratory strain carrying the original G970R mutation was propagated to research laboratories across the world. Interestingly, a PCA1 allele containing several (27 amino acids) missense mutations was identified in a yeast strain that was selected on media containing lethal concentrations of cadmium (39). Among these mutations, an R970G mutation was implicated in cadmium resistance conferred by the mutant PCA1 (39). Thus, in this study it appears that a nonfunctional PCA1 mutant was reverted to the functional allele under cadmium stress. The nonfunctional PCA1 allele, at least in part, explains cadmium sensitivity of a laboratory strain (BY4741) compared with a natural isolate (RM11-1a). However, the RM11-1a strain was still hyper-resistant compared with BY4741 even when PCA1 was deleted in these strains, implicating the existence of other contributing factors involved in cadmium defense. Elucidation of the underlying reason of cadmium hyper-resistance in the RM11-1a strain would lead to a better understanding of the mechanisms involved in cadmium detoxification.
Regulation of subcellular distribution and transcription of P1B-type ATPases have been reported previously, suggesting the significance of active regulation of this family of transporters. ATP7a and ATP7b localize to the endoplasmic reticulum-Golgi membrane where they transport copper across the membrane for incorporation into proteins in the secretory pathway (7, 16, 50, 67). Excess cellular accumulation of copper induces trafficking of ATP7a to the plasma membrane and ATP7b to cytoplasmic vesicles (67). In addition to post-translational regulation, several examples of metal-dependent transcriptional regulation of genes encoding P1B-type ATPases have been reported in plant, yeast, and bacteria (19, 21, 70–73). PCA1 mRNA levels are not regulated by cadmium or copper. However, cadmium dramatically regulates PCA1 protein levels and promotes localization to the plasma membrane, which is a unique mode of regulation for this family of transporters. Interestingly, copper and silver also regulate PCA1 protein expression, although PCA1 does not transport these metals. Because the N terminus of PCA1 possesses a Cys-rich region that likely binds copper, it is plausible that metal binding to this domain controls stability of PCA1.
Given that the Gly-970 residue in PCA1 is conserved among this family of proteins and has been positioned in an ATP-binding pocket of two independent structures of P1B-type ATPases (58, 74), it is likely that this residue is involved in ATP binding. Interestingly, the G970R mutant mislocalizes to a vesicle-like compartment. An equivalent mutation identified in ATP7b of Wilson disease patients might also lead to defects in subcellular trafficking. A specific conformation resulting from ATP binding and metal translocation may determine proper trafficking of PCA1. This may occur through PCA1 conformation-dependent attraction of secretory and/or endocytosis machinery. We are currently exploring these possibilities.
Cadmium is one of the most toxic environmental pollutants. It is removed slowly from the body, which may be due to high affinity binding to cellular ligands, such as thiol groups in proteins, glutathione, methallothionein, and cysteine. Cadmium transporters in hepatocytes and renal epithelial cells, such as ATP-binding cassette, multidrug resistance protein, GSH-cadmium complex transporter, have been proposed to efflux GSH or Cys-conjugated cadmium (23, 24). However, no report has identified other P1B-type ATPases in the human genome in addition to ATP7a and ATP7b. Further characterization of cadmium efflux systems in higher eukaryotes would advance our ability to combat cadmium-related disorders and develop methods for remediation of toxic heavy metals.
Acknowledgments
We are grateful to the members of Lee laboratory, Angela Adle, Patrick Kennedy, Jesse Cox, and Yoshio Toda, for helpful suggestions and technical assistance. We thank Dr. Kruglyak for kindly providing the RM11-1a yeast strain.
Footnotes
This work was supported by Project 3 (to J. L.), National Institutes of Health Grant P20 RR-17675, and by funds provided through Hatch Act in the University of Nebraska Agricultural Research Division.
The abbreviations used are: MT, metallothionine; GFP, green fluorescent protein; PGK, phosphoglycerate kinase; ORF, open reading frame; ICPMS, inductively coupled plasma mass spectrometry; PBS, phosphate-buffered saline; GPD, glyceraldehyde-3-phosphate dehydrogenase.
References
- 1.Nelson N. EMBO J. 1999;18:4361–4371. doi: 10.1093/emboj/18.16.4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pena MM, Lee J, Thiele DJ. J Nutr. 1999;129:1251–1260. doi: 10.1093/jn/129.7.1251. [DOI] [PubMed] [Google Scholar]
- 3.Hentze MW, Muckenthaler MU, Andrews NC. Cell. 2004;117:285–297. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- 4.Halliwell B, Gutteridge JM. Methods Enzymol. 1990;186:1–85. doi: 10.1016/0076-6879(90)86093-b. [DOI] [PubMed] [Google Scholar]
- 5.Vallee BL, Ulmer DD. Annu Rev Biochem. 1972;41:91–128. doi: 10.1146/annurev.bi.41.070172.000515. [DOI] [PubMed] [Google Scholar]
- 6.Bush AI. Curr Opin Chem Biol. 2000;4:184–191. doi: 10.1016/s1367-5931(99)00073-3. [DOI] [PubMed] [Google Scholar]
- 7.Tao TY, Gitlin JD. Hepatology. 2003;37:1241–1247. doi: 10.1053/jhep.2003.50281. [DOI] [PubMed] [Google Scholar]
- 8.Ercal N, Gurer-Orhan H, Aykin-Burns N. Curr Top Med Chem. 2001;1:529–539. doi: 10.2174/1568026013394831. [DOI] [PubMed] [Google Scholar]
- 9.Henson MC, Chedrese PJ. Exp Biol Med (Maywood) 2004;229:383–392. doi: 10.1177/153537020422900506. [DOI] [PubMed] [Google Scholar]
- 10.McMurray CT, Tainer JA. Nat Genet. 2003;34:239–241. doi: 10.1038/ng0703-239. [DOI] [PubMed] [Google Scholar]
- 11.Darbre PD. J Appl Toxicol. 2006;26:191–197. doi: 10.1002/jat.1135. [DOI] [PubMed] [Google Scholar]
- 12.Stoica A, Katzenellenbogen BS, Martin MB. Mol Endocrinol. 2000;14:545–553. doi: 10.1210/mend.14.4.0441. [DOI] [PubMed] [Google Scholar]
- 13.Silver S, Phung LT. Annu Rev Microbiol. 1996;50:753–789. doi: 10.1146/annurev.micro.50.1.753. [DOI] [PubMed] [Google Scholar]
- 14.Nies DH. Appl Microbiol Biotechnol. 1999;51:730–750. doi: 10.1007/s002530051457. [DOI] [PubMed] [Google Scholar]
- 15.Gatti D, Mitra B, Rosen BP. J Biol Chem. 2000;275:34009–34012. doi: 10.1074/jbc.R000012200. [DOI] [PubMed] [Google Scholar]
- 16.Solioz M, Vulpe C. Trends Biochem Sci. 1996;21:237–241. [PubMed] [Google Scholar]
- 17.Axelsen KB, Palmgren MG. Plant Physiol. 2001;126:696–706. doi: 10.1104/pp.126.2.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams LE, Mills RF. Trends Plant Sci. 2005;10:491–502. doi: 10.1016/j.tplants.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 19.Papoyan A, Kochian LV. Plant Physiol. 2004;136:3814–3823. doi: 10.1104/pp.104.044503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hussain D, Haydon MJ, Wang Y, Wong E, Sherson SM, Young J, Camakaris J, Harper JF, Cobbett CS. Plant Cell. 2004;16:1327–1339. doi: 10.1105/tpc.020487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eren E, Argüello JM. Plant Physiol. 2004;136:3712–3723. doi: 10.1104/pp.104.046292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verret F, Gravot A, Auroy P, Leonhardt N, David P, Nussaume L, Vavasseur A, Richaud P. FEBS Lett. 2004;576:306–312. doi: 10.1016/j.febslet.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 23.Ballatori N. Environ Health Perspect. 2002;110(Suppl. 5):689–694. doi: 10.1289/ehp.02110s5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zalups RK, Ahmad S. Toxicol Appl Pharmacol. 2003;186:163–188. doi: 10.1016/s0041-008x(02)00021-2. [DOI] [PubMed] [Google Scholar]
- 25.Hamer DH. Annu Rev Biochem. 1986;55:913–951. doi: 10.1146/annurev.bi.55.070186.004405. [DOI] [PubMed] [Google Scholar]
- 26.Klaassen CD, Liu J, Choudhuri S. Annu Rev Pharmacol Toxicol. 1999;39:267–294. doi: 10.1146/annurev.pharmtox.39.1.267. [DOI] [PubMed] [Google Scholar]
- 27.Cobbett C, Goldsbrough P. Annu Rev Plant Biol. 2002;53:159–182. doi: 10.1146/annurev.arplant.53.100301.135154. [DOI] [PubMed] [Google Scholar]
- 28.Singhal RK, Anderson ME, Meister A. FASEB J. 1987;1:220–223. doi: 10.1096/fasebj.1.3.2887478. [DOI] [PubMed] [Google Scholar]
- 29.Wimmer U, Wang Y, Georgiev O, Schaffner W. Nucleic Acids Res. 2005;33:5715–5727. doi: 10.1093/nar/gki881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li ZS, Lu YP, Zhen RG, Szczypka M, Thiele DJ, Rea PA. Proc Natl Acad Sci U S A. 1997;94:42–47. doi: 10.1073/pnas.94.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lichtlen P, Schaffner W. BioEssays. 2001;23:1010–1017. doi: 10.1002/bies.1146. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, Wimmer U, Lichtlen P, Inderbitzin D, Stieger B, Meier PJ, Hunziker L, Stallmach T, Forrer R, Rulicke T, Georgiev O, Schaffner W. FASEB J. 2004;18:1071–1079. doi: 10.1096/fj.03-1282com. [DOI] [PubMed] [Google Scholar]
- 33.Heuchel R, Radtke F, Georgiev O, Stark G, Aguet M, Schaffner W. EMBO J. 1994;13:2870–2875. doi: 10.1002/j.1460-2075.1994.tb06581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palmiter RD. Proc Natl Acad Sci U S A. 1994;91:1219–1223. doi: 10.1073/pnas.91.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thiele DJ. Mol Cell Biol. 1988;8:2745–2752. doi: 10.1128/mcb.8.7.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou P, Thiele DJ. Biofactors. 1993;4:105–115. [PubMed] [Google Scholar]
- 37.Rad MR, Kirchrath L, Hollenberg CP. Yeast. 1994;9:1217–1225. doi: 10.1002/yea.320100910. [DOI] [PubMed] [Google Scholar]
- 38.De Freitas JM, Kim JH, Poynton H, Su T, Wintz H, Fox T, Holman P, Loguinov A, Keles S, van der Laan M, Vulpe C. J Biol Chem. 2004;279:4450–4458. doi: 10.1074/jbc.M212308200. [DOI] [PubMed] [Google Scholar]
- 39.Shiraishi E, Inouhe M, Joho M, Tohoyama H. Curr Genet. 2000;37:79–86. doi: 10.1007/s002940050013. [DOI] [PubMed] [Google Scholar]
- 40.Brem RB, Yvert R, Clinton R, Kruglyak L. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 41.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Phillippsen P, Pringle JR. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 42.Geitz DR, Schiestl RH, Willems AR, Woods RA. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 43.Liu H, Krizek J, Bretscher A. Genetics. 1992;132:665–673. doi: 10.1093/genetics/132.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mumberg D, Müller R, Funk M. Gene (Amst) 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 45.Johnston M, Davis RW. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sikorski RS, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Gene (Amst) 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 48.Pena MM, Puig S, Thiele DJ. J Biol Chem. 2000;275:33244–33251. doi: 10.1074/jbc.M005392200. [DOI] [PubMed] [Google Scholar]
- 49.Lee J, Petris MJ, Thiele DJ. J Biol Chem. 2002;277:40253–40259. doi: 10.1074/jbc.M208002200. [DOI] [PubMed] [Google Scholar]
- 50.Yuan DS, Stearman R, Dancis A, Dunn T, Beeler T, Klausner RD. Proc Natl Acad Sci U S A. 1995;92:2632–2636. doi: 10.1073/pnas.92.7.2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuhlbrandt W. Nat Rev Mol Cell Biol. 2004;5:282–295. doi: 10.1038/nrm1354. [DOI] [PubMed] [Google Scholar]
- 52.Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnson M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin, Oliver SG. Science. 1996;274:563–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 53.Tawfik OW, Papasian CJ, Dixon AY, Potter LM. J Clin Microbiol. 1989;27:1689–1691. doi: 10.1128/jcm.27.7.1689-1691.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rymond BC, Zitomer RS, Schumperli D, Rosenberg M. Gene (Amst) 1983;25:249–262. doi: 10.1016/0378-1119(83)90229-9. [DOI] [PubMed] [Google Scholar]
- 55.Gralla EB, Valentine JS. J Bacteriol. 1991;173:5918–5920. doi: 10.1128/jb.173.18.5918-5920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Knight SA, Labbé S, Kwon LF, Kosman DJ, Thiele DJ. Genes Dev. 1996;10:1917–1929. doi: 10.1101/gad.10.15.1917. [DOI] [PubMed] [Google Scholar]
- 57.Sazinsky MH, Mandal AK, Argüello JM, Rosenwieg AC. J Biol Chem. 2006;281:11161–11166. doi: 10.1074/jbc.M510708200. [DOI] [PubMed] [Google Scholar]
- 58.Dmitriev O, Tsivkovskii R, Abildgaard F, Morgan CT, Markley JL, Lutsenko S. Proc Nat Acad Sci U S A. 2006;103:5302–5307. doi: 10.1073/pnas.0507416103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thomas GR, Forbes JR, Roberts EA, Walshe JM, Cox DW. Nat Genet. 1995;9:210–217. doi: 10.1038/ng0295-210. [DOI] [PubMed] [Google Scholar]
- 60.Toyoshima C, Inesi G. Annu Rev Biochem. 2004;73:269–292. doi: 10.1146/annurev.biochem.73.011303.073700. [DOI] [PubMed] [Google Scholar]
- 61.Palmgren MG. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:817–845. doi: 10.1146/annurev.arplant.52.1.817. [DOI] [PubMed] [Google Scholar]
- 62.Kaplan JH. Annu Rev Biochem. 2002;71:511–535. doi: 10.1146/annurev.biochem.71.102201.141218. [DOI] [PubMed] [Google Scholar]
- 63.Tamai KT, Gralla EB, Ellerby LM, Valentine JS, Thiele DJ. Proc Natl Acad Sci U S A. 1993;90:8013–8017. doi: 10.1073/pnas.90.17.8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mills RF, Francini A, Ferreira da Rocha PS, Baccarini PJ, Aylett M, Krijger GC, Williams LE. FEBS Lett. 2005;579:783–791. doi: 10.1016/j.febslet.2004.12.040. [DOI] [PubMed] [Google Scholar]
- 65.Eren E, Kennedy DC, Maroney MJ, Argüello JM. J Biol Chem. 2006;281:33881–33891. doi: 10.1074/jbc.M605218200. [DOI] [PubMed] [Google Scholar]
- 66.Portillo F. Biochim Biophys Acta. 2000;1469:31–42. doi: 10.1016/s0304-4157(99)00011-8. [DOI] [PubMed] [Google Scholar]
- 67.Lutsenko S, Petris MJ. J Membr Biol. 2003;191:1–12. doi: 10.1007/s00232-002-1040-6. [DOI] [PubMed] [Google Scholar]
- 68.Gitschier J, Moffat B, Reilly D, Wood WI, Fairbrother WJ. Nat Struct Biol. 1998;5:47–54. doi: 10.1038/nsb0198-47. [DOI] [PubMed] [Google Scholar]
- 69.Argüello JM. J Membr Biol. 2003;195:93–108. doi: 10.1007/s00232-003-2048-2. [DOI] [PubMed] [Google Scholar]
- 70.Mills RF, Krijger GC, Baccarini PJ, Hall JL, Williams LE. Plant J. 2003;35:164–176. doi: 10.1046/j.1365-313x.2003.01790.x. [DOI] [PubMed] [Google Scholar]
- 71.Wunderli-Ye H, Solioz M. Adv Exp Med Biol. 1999;448:255–264. doi: 10.1007/978-1-4615-4859-1_23. [DOI] [PubMed] [Google Scholar]
- 72.Rensing C, Fan B, Sharma R, Mitra B, Rosen BP. Proc Natl Acad Sci U S A. 2000;97:652–656. doi: 10.1073/pnas.97.2.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Andres-Colas N, Sancenon V, Rodriguez-Navarro S, Mayo S, Thiele DJ, Ecker JR, Puig S, Penarrubia L. Plant J. 2006;45:225–236. doi: 10.1111/j.1365-313X.2005.02601.x. [DOI] [PubMed] [Google Scholar]
- 74.Sazinsky MH, Mandal AK, Argüello JM, Rosenzweig AC. J Biol Chem. 2006;281:11161–11166. doi: 10.1074/jbc.M510708200. [DOI] [PubMed] [Google Scholar]


