Sir,
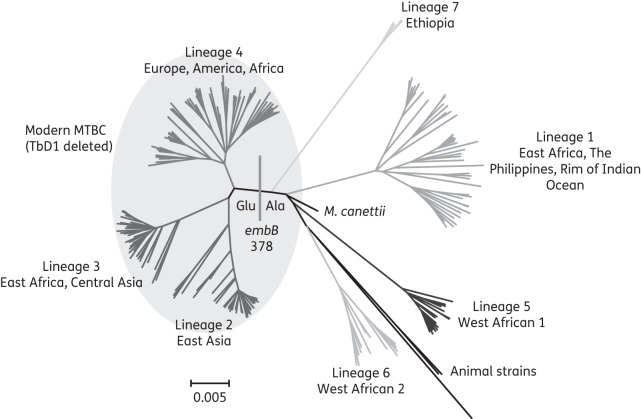
We agree with Moure et al.1 that fast genotypic methods will play an increasingly prominent role in drug susceptibility testing for the Mycobacterium tuberculosis complex (MTBC).2,3 We would, however, like to point out that the embB (Rv3795) Glu378Ala polymorphism, which is detected by probe 3 of their newly developed low-density DNA array, is not a marker for ethambutol resistance.4–7 Instead, Ala represents the ancestral amino acid at this codon (Figure 1), whereas Glu is present in all modern MTBC (lineages 2, 3 and 4).6–9 The MIRU–VNTR data of the 51 ethambutol-resistant isolates from the study by Moure et al.1 are largely congruent with this finding. All 49 phylogenetically modern MTBC isolates had the embB 378 Glu variant. Isolate 5765 was a representative of Mycobacterium bovis, which is consistent with the fact that it harboured the Ala variant and was pyrazinamide resistant. By contrast, it was unclear why isolate 233R, which appeared to be M. bovis based on its MIRU–VNTR signature, had the Glu variant (experimental error or a homoplastic event might account for this discrepancy).
Figure 1.
Whole-genome phylogeny of 219 isolates representative of all major MTBC lineages.9 Glu at codon 378 is a marker for modern MTBC, which all share the TbD1 deletion and include the lineage 4 M. tuberculosis H37Rv laboratory strain that is used as the reference/wild-type sequence for sequence analyses.10
In light of these data, the results of probe 3 would be predicted to lead to systematic false-positive reports, which calls into question the validity of this probe. This underlines that the entire MTBC diversity has to be considered when designing and validating genotypic drug susceptibility testing assays.7,10
Funding
This work was supported by a grant from the Department of Health, Wellcome Trust and the Health Innovation Challenge Fund (HICF-T5-342 and WT098600 to S. J. P.), Public Health England (to S. J. P.), the Medical Research Council (to J. M. B.) and the Wellcome Trust Sanger Institute (WT098051 to J. P. and J. M. B). C. U. K. is a Junior Research Fellow at Wolfson College, Cambridge. I. C. is supported by a Ramón y Cajal fellowship from the Spanish Government (RYC-2012-10627).
Transparency declarations
J. P. has received funding for travel and accommodation from Pacific Biosciences Inc. and Illumina Inc. S. J. P. is a consultant for Pfizer Inc. and has received funding for travel and accommodation from Illumina Inc. All other authors: none to declare.
Disclaimer
This publication presents independent research supported by the Health Innovation Challenge Fund (HICF-T5-342 and WT098600), a parallel funding partnership between the Department of Health and Wellcome Trust. The views expressed in this publication are those of the authors and not necessarily those of the Department of Health or Wellcome Trust.
References
- 1.Moure R, Español M, Tudó G, et al. Characterization of the embB gene in Mycobacterium tuberculosis isolates from Barcelona and rapid detection of main mutations related to ethambutol resistance using a low-density DNA array. J Antimicrob Chemother. 2014;69:947–54. doi: 10.1093/jac/dkt448. [DOI] [PubMed] [Google Scholar]
- 2.Köser CU, Ellington MJ, Cartwright EJ, et al. Routine use of microbial whole genome sequencing in diagnostic and public health microbiology. PLoS Pathog. 2012;8:e1002824. doi: 10.1371/journal.ppat.1002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Köser CU, Bryant JM, Becq J, et al. Whole-genome sequencing for rapid susceptibility testing of M. tuberculosis. N Engl J Med. 2013;369:290–2. doi: 10.1056/NEJMc1215305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sekiguchi J, Miyoshi-Akiyama T, Augustynowicz-Kopeć E, et al. Detection of multidrug resistance in Mycobacterium tuberculosis. J Clin Microbiol. 2007;45:179–92. doi: 10.1128/JCM.00750-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell PJ, Morlock GP, Sikes RD, et al. Molecular detection of mutations associated with first- and second-line drug resistance compared with conventional drug susceptibility testing of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2011;55:2032–41. doi: 10.1128/AAC.01550-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Safi H, Lingaraju S, Amin A, et al. Evolution of high-level ethambutol-resistant tuberculosis through interacting mutations in decaprenylphosphoryl-β-d-arabinose biosynthetic and utilization pathway genes. Nat Genet. 2013;45:1190–7. doi: 10.1038/ng.2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feuerriegel S, Köser CU, Niemann S. Phylogenetic polymorphisms in antibiotic resistance genes of the Mycobacterium tuberculosis complex. J Antimicrob Chemother. 2014;69:1205–10. doi: 10.1093/jac/dkt535. [DOI] [PubMed] [Google Scholar]
- 8.Casali N, Nikolayevskyy V, Balabanova Y, et al. Microevolution of extensively drug-resistant tuberculosis in Russia. Genome Res. 2012;22:735–45. doi: 10.1101/gr.128678.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Comas I, Coscolla M, Luo T, et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat Genet. 2013;45:1176–82. doi: 10.1038/ng.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Köser CU, Feuerriegel S, Summers DK, et al. Importance of the genetic diversity within the Mycobacterium tuberculosis complex for the development of novel antibiotics and diagnostic tests of drug resistance. Antimicrob Agents Chemother. 2012;56:6080–7. doi: 10.1128/AAC.01641-12. [DOI] [PMC free article] [PubMed] [Google Scholar]